DXR DE Analysis
Emma M Pfortmiller
2025-05-14
Last updated: 2025-07-11
Checks: 6 1
Knit directory: Recovery_5FU/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20250217) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Using absolute paths to the files within your workflowr project makes it difficult for you and others to run your code on a different machine. Change the absolute path(s) below to the suggested relative path(s) to make your code more reproducible.
| absolute | relative |
|---|---|
| C:/Users/emmap/RDirectory/Recovery_RNAseq/Recovery_5FU/data/new/counts_raw_matrix_EMP_250514.csv | data/new/counts_raw_matrix_EMP_250514.csv |
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 1998e95. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/Anat HM/
Ignored: data/CDKN1A_geneplot_Dox.RDS
Ignored: data/Cormotif_dox.RDS
Ignored: data/Cormotif_prob_gene_list.RDS
Ignored: data/Cormotif_prob_gene_list_doxonly.RDS
Ignored: data/DMSO_TNN13_plot.RDS
Ignored: data/DOX_TNN13_plot.RDS
Ignored: data/DOXgeneplots.RDS
Ignored: data/ExpressionMatrix_EMP.csv
Ignored: data/Ind1_DOX_Spearman_plot.RDS
Ignored: data/Ind6REP_Spearman_list.csv
Ignored: data/Ind6REP_Spearman_plot.RDS
Ignored: data/Ind6REP_Spearman_set.csv
Ignored: data/Ind6_Spearman_plot.RDS
Ignored: data/MDM2_geneplot_Dox.RDS
Ignored: data/Metadata.csv
Ignored: data/SIRT1_geneplot_Dox.RDS
Ignored: data/Sample_annotated.csv
Ignored: data/SpearmanHeatmapMatrix_EMP
Ignored: data/VennDiagram_DEGs.csv.2025-05-19_13-04-29.204911.log
Ignored: data/VolcanoPlot_D144R_original_EMP_250623.png
Ignored: data/VolcanoPlot_D24R_original_EMP_250623.png
Ignored: data/VolcanoPlot_D24T_original_EMP_250623.png
Ignored: data/annot_dox.RDS
Ignored: data/annot_list_hm.csv
Ignored: data/cormotifARclust_pp.RDS
Ignored: data/cormotif_dxr_1.RDS
Ignored: data/cormotif_dxr_2.RDS
Ignored: data/counts/
Ignored: data/counts_DE_df_dox.RDS
Ignored: data/counts_DE_raw_data.RDS
Ignored: data/counts_raw_filt.RDS
Ignored: data/counts_raw_matrix.RDS
Ignored: data/counts_raw_matrix_EMP_250514.csv
Ignored: data/d24_Spearman_plot.RDS
Ignored: data/dge_calc_dxr.RDS
Ignored: data/dge_calc_matrix.RDS
Ignored: data/ensembl_backup_dox.RDS
Ignored: data/fC_AllCounts.RDS
Ignored: data/fC_DOXCounts.RDS
Ignored: data/featureCounts_Concat_Matrix_DOXSamples_EMP_250430.csv
Ignored: data/filcpm_colnames_matrix.csv
Ignored: data/filcpm_matrix.csv
Ignored: data/filt_gene_list_dox.RDS
Ignored: data/filter_gene_list_final.RDS
Ignored: data/final_data/
Ignored: data/gene_clustlike_motif.RDS
Ignored: data/gene_postprob_motif.RDS
Ignored: data/genedf_dxr.RDS
Ignored: data/genematrix_dox.RDS
Ignored: data/genematrix_dxr.RDS
Ignored: data/heartgenes.csv
Ignored: data/heartgenes_dox.csv
Ignored: data/heatmap_group_Anat.RDS
Ignored: data/ind_num_dox.RDS
Ignored: data/ind_num_dxr.RDS
Ignored: data/initial_cormotif.RDS
Ignored: data/initial_cormotif_dox.RDS
Ignored: data/new/
Ignored: data/new_cormotif_dox.RDS
Ignored: data/plot_leg_d.RDS
Ignored: data/plot_leg_d_horizontal.RDS
Ignored: data/plot_leg_d_vertical.RDS
Ignored: data/process_gene_data_funct.RDS
Ignored: data/ruv/
Ignored: data/tableED_GOBP.RDS
Ignored: data/tableESR_GOBP_postprob.RDS
Ignored: data/tableLD_GOBP.RDS
Ignored: data/tableLR_GOBP_postprob.RDS
Ignored: data/tableNR_GOBP.RDS
Ignored: data/tableNR_GOBP_postprob.RDS
Ignored: data/table_motif1_GOBP_d.RDS
Ignored: data/table_motif1_genes_GOBP.RDS
Ignored: data/table_motif2_GOBP_d.RDS
Ignored: data/table_motif3_genes_GOBP.RDS
Ignored: data/top.table_V.D144r_dox.RDS
Ignored: data/top.table_V.D24_dox.RDS
Ignored: data/top.table_V.D24r_dox.RDS
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/DXR_Project_Analysis.Rmd)
and HTML (docs/DXR_Project_Analysis.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 1998e95 | emmapfort | 2025-07-11 | Updated Analysis 07/11/25 |
| html | 0ae67cb | emmapfort | 2025-07-10 | Build site. |
| Rmd | 73c8641 | emmapfort | 2025-07-10 | Updated Analysis 07/10/25 EMP |
| html | 54b2620 | emmapfort | 2025-07-08 | Build site. |
| Rmd | 425d3aa | emmapfort | 2025-07-08 | Updated Analysis 07/08/25 |
| Rmd | a43fb2c | emmapfort | 2025-07-07 | Updated Analysis 07/07/25 EMP Proportion Barplots Cormotif p53, DDR, DIC gene sets |
| html | 1b8f045 | emmapfort | 2025-07-03 | Build site. |
| Rmd | 10b885f | emmapfort | 2025-07-03 | Updated Website 07/03/25 EMP |
| html | 14cdab3 | emmapfort | 2025-07-02 | Build site. |
| Rmd | abdf4fe | emmapfort | 2025-07-02 | Updated Website with Streamlined Analysis 07/02/25 EMP |
| html | 0b1ffad | emmapfort | 2025-07-02 | Build site. |
| Rmd | 35bbd4f | emmapfort | 2025-07-02 | Updated Analysis all in one place EMP - 07/02/25 |
| Rmd | 81e180f | emmapfort | 2025-06-27 | RUVs Cormotif + updated analysis 06/27/25 |
| Rmd | 82c6704 | emmapfort | 2025-06-23 | Updated Analysis 06/23/25 |
| Rmd | 98a546f | emmapfort | 2025-06-16 | 06/16/25 |
| Rmd | f530175 | emmapfort | 2025-06-16 | Updating Analysis 06/16/25 |
| Rmd | 9f609f6 | emmapfort | 2025-06-13 | Update 06/13/25 |
| html | 1d4d1de | emmapfort | 2025-05-16 | Build site. |
| html | 29a5c35 | emmapfort | 2025-05-16 | Build site. |
| Rmd | 5db2858 | emmapfort | 2025-05-16 | Removing copied pages + including final analysis |
| Rmd | b6f4e9c | emmapfort | 2025-05-16 | Updated Analysis 05/15/25 |
| Rmd | 32de50d | emmapfort | 2025-05-14 | Analysis Overhaul 05/14/25 |
editor_options: chunk_output_type: inline
I did this part with Sayan according to his analysis to ensure that my matrix was consistent with his - allowing me to work on downstream analysis #Read in Libraries
#Read in Counts Files from FeatureCounts
#load in libraries needed
#these counts files are from featureCounts, all saved as RDS objects
# ####Individual 1 - 84-1####
# Counts_84_DOX_24 <- readRDS("data/counts/Counts_84_DOX_24.RDS")
# Counts_84_DMSO_24 <- readRDS("data/counts/Counts_84_DMSO_24.RDS")
# Counts_84_DOX_24rec <- readRDS("data/counts/Counts_84_DOX_24rec.RDS")
# Counts_84_DMSO_24rec <- readRDS("data/counts/Counts_84_DMSO_24rec.RDS")
# Counts_84_DOX_144rec <- readRDS("data/counts/Counts_84_DOX_144rec.RDS")
# Counts_84_DMSO_144rec <- readRDS("data/counts/Counts_84_DMSO_144rec.RDS")
#
# ####Individual 2 - 87-1####
# Counts_87_DOX_24 <- readRDS("data/counts/Counts_87_DOX_24.RDS")
# Counts_87_DMSO_24 <- readRDS("data/counts/Counts_87_DMSO_24.RDS")
# Counts_87_DOX_24rec <- readRDS("data/counts/Counts_87_DOX_24rec.RDS")
# Counts_87_DMSO_24rec <- readRDS("data/counts/Counts_87_DMSO_24rec.RDS")
# Counts_87_DOX_144rec <- readRDS("data/counts/Counts_87_DOX_144rec.RDS")
# Counts_87_DMSO_144rec <- readRDS("data/counts/Counts_87_DMSO_144rec.RDS")
#
# ####Individual 3 - 78-1####
# Counts_78_DOX_24 <- readRDS("data/counts/Counts_78_DOX_24.RDS")
# Counts_78_DMSO_24 <- readRDS("data/counts/Counts_78_DMSO_24.RDS")
# Counts_78_DOX_24rec <- readRDS("data/counts/Counts_78_DOX_24rec.RDS")
# Counts_78_DMSO_24rec <- readRDS("data/counts/Counts_78_DMSO_24rec.RDS")
# Counts_78_DOX_144rec <- readRDS("data/counts/Counts_78_DOX_144rec.RDS")
# Counts_78_DMSO_144rec <- readRDS("data/counts/Counts_78_DMSO_144rec.RDS")
#
# ####Individual 4 - 75-1####
# Counts_75_DOX_24 <- readRDS("data/counts/Counts_75_DOX_24.RDS")
# Counts_75_DMSO_24 <- readRDS("data/counts/Counts_75_DMSO_24.RDS")
# Counts_75_DOX_24rec <- readRDS("data/counts/Counts_75_DOX_24rec.RDS")
# Counts_75_DMSO_24rec <- readRDS("data/counts/Counts_75_DMSO_24rec.RDS")
# Counts_75_DOX_144rec <- readRDS("data/counts/Counts_75_DOX_144rec.RDS")
# Counts_75_DMSO_144rec <- readRDS("data/counts/Counts_75_DMSO_144rec.RDS")
#
# ####Individual 5 - 17-3####
# Counts_17_DOX_24 <- readRDS("data/counts/Counts_17_DOX_24.RDS")
# Counts_17_DMSO_24 <- readRDS("data/counts/Counts_17_DMSO_24.RDS")
# Counts_17_DOX_24rec <- readRDS("data/counts/Counts_17_DOX_24rec.RDS")
# Counts_17_DMSO_24rec <- readRDS("data/counts/Counts_17_DMSO_24rec.RDS")
# Counts_17_DOX_144rec <- readRDS("data/counts/Counts_17_DOX_144rec.RDS")
# Counts_17_DMSO_144rec <- readRDS("data/counts/Counts_17_DMSO_144rec.RDS")
#
# ####Individual 6 - 90-1####
# Counts_90_DOX_24 <- readRDS("data/counts/Counts_90_DOX_24.RDS")
# Counts_90_DMSO_24 <- readRDS("data/counts/Counts_90_DMSO_24.RDS")
# Counts_90_DOX_24rec <- readRDS("data/counts/Counts_90_DOX_24rec.RDS")
# Counts_90_DMSO_24rec <- readRDS("data/counts/Counts_90_DMSO_24rec.RDS")
# Counts_90_DOX_144rec <- readRDS("data/counts/Counts_90_DOX_144rec.RDS")
# Counts_90_DMSO_144rec <- readRDS("data/counts/Counts_90_DMSO_144rec.RDS")
#
# ####Individual 7 - 90-1REP####
# Counts_90REP_DOX_24 <- readRDS("data/counts/Counts_90REP_DOX_24.RDS")
# Counts_90REP_DMSO_24 <- readRDS("data/counts/Counts_90REP_DMSO_24.RDS")
# Counts_90REP_DOX_24rec <- readRDS("data/counts/Counts_90REP_DOX_24rec.RDS")
# Counts_90REP_DMSO_24rec <- readRDS("data/counts/Counts_90REP_DMSO_24rec.RDS")
# Counts_90REP_DOX_144rec <- readRDS("data/counts/Counts_90REP_DOX_144rec.RDS")
# Counts_90REP_DMSO_144rec <- readRDS("data/counts/Counts_90REP_DMSO_144rec.RDS")##Create my Dataframe from Counts Files
# counts_raw_df <-
# data.frame(
# Counts_84_DOX_24,
# Counts_84_DMSO_24$MCW_EMP_JT_R29_R1.bam,
# Counts_84_DOX_24rec$MCW_EMP_JT_R30_R1.bam,
# Counts_84_DMSO_24rec$MCW_EMP_JT_R32_R1.bam,
# Counts_84_DOX_144rec$MCW_EMP_JT_R33_R1.bam,
# Counts_84_DMSO_144rec$MCW_EMP_JT_R35_R1.bam,
# Counts_87_DOX_24$MCW_EMP_JT_R36_R1.bam,
# Counts_87_DMSO_24$MCW_EMP_JT_R38_R1.bam,
# Counts_87_DOX_24rec$MCW_EMP_JT_R39_R1.bam,
# Counts_87_DMSO_24rec$MCW_EMP_JT_R41_R1.bam,
# Counts_87_DOX_144rec$MCW_EMP_JT_R42_R1.bam,
# Counts_87_DMSO_144rec$MCW_EMP_JT_R44_R1.bam,
# Counts_78_DOX_24$MCW_EMP_JT_R45_R1.bam,
# Counts_78_DMSO_24$MCW_EMP_JT_R47_R1.bam,
# Counts_78_DOX_24rec$MCW_EMP_JT_R48_R1.bam,
# Counts_78_DMSO_24rec$MCW_EMP_JT_R50_R1.bam,
# Counts_78_DOX_144rec$MCW_EMP_JT_R51_R1.bam,
# Counts_78_DMSO_144rec$MCW_EMP_JT_R53_R1.bam,
# Counts_75_DOX_24$MCW_EMP_JT_R54_R1.bam,
# Counts_75_DMSO_24$MCW_EMP_JT_R56_R1.bam,
# Counts_75_DOX_24rec$MCW_EMP_JT_R57_R1.bam,
# Counts_75_DMSO_24rec$MCW_EMP_JT_R59_R1.bam,
# Counts_75_DOX_144rec$MCW_EMP_JT_R60_R1.bam,
# Counts_75_DMSO_144rec$MCW_EMP_JT_R62_R1.bam,
# Counts_17_DOX_24$MCW_EMP_JT_R63_R1.bam,
# Counts_17_DMSO_24$MCW_EMP_JT_R65_R1.bam,
# Counts_17_DOX_24rec$MCW_EMP_JT_R66_R1.bam,
# Counts_17_DMSO_24rec$MCW_EMP_JT_R68_R1.bam,
# Counts_17_DOX_144rec$MCW_EMP_JT_R69_R1.bam,
# Counts_17_DMSO_144rec$MCW_EMP_JT_R71_R1.bam,
# Counts_90_DOX_24$MCW_EMP_JT_R72_R1.bam,
# Counts_90_DMSO_24$MCW_EMP_JT_R74_R1.bam,
# Counts_90_DOX_24rec$MCW_EMP_JT_R75_R1.bam,
# Counts_90_DMSO_24rec$MCW_EMP_JT_R77_R1.bam,
# Counts_90_DOX_144rec$MCW_EMP_JT_R78_R1.bam,
# Counts_90_DMSO_144rec$MCW_EMP_JT_R80_R1.bam,
# Counts_90REP_DOX_24$MCW_EMP_JT_R81_R1.bam,
# Counts_90REP_DMSO_24$MCW_EMP_JT_R83_R1.bam,
# Counts_90REP_DOX_24rec$MCW_EMP_JT_R84_R1.bam,
# Counts_90REP_DMSO_24rec$MCW_EMP_JT_R86_R1.bam,
# Counts_90REP_DOX_144rec$MCW_EMP_JT_R87_R1.bam,
# Counts_90REP_DMSO_144rec$MCW_EMP_JT_R89_R1.bam
# )
#now save this as a matrix
#counts_raw_matrix <- counts_raw_df %>% column_to_rownames(var = "X") %>% as.matrix()
counts_raw_matrix <- readRDS("data/new/counts_raw_matrix.RDS")
dim(counts_raw_matrix)[1] 28395 42#28395 is my initial amount of genes prior to filtering
#write this to a csv so I can save it for later
#write.csv(counts_raw_matrix, "C:/Users/emmap/RDirectory/Recovery_RNAseq/Recovery_5FU/data/new/counts_raw_matrix_EMP_250514.csv")
#I also want to save this as an R object so I don't have to run the counts every time
#saveRDS(counts_raw_matrix, "data/new/counts_raw_matrix.RDS")###Include colors and factors for analysis
#I want to include the color schemes I have for my treatment, individuals, and timepoints
####Colors####
tx_col <- c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")
col_tx_large <- rep(c("#499FBD" , "#BBBBBC"), 21)
col_tx_large_2 <- c(rep("#499FBD" , 3), rep("#BBBBBC", 3), 21)
ind_col <- c("#003F5C", "#45AE91", "#58209D", "#8B3E9B", "#FF6361", "#BC4169", "#FF2362")
ind_col_norep <- c("#003F5C", "#45AE91", "#58209D", "#8B3E9B", "#FF6361", "#BC4169")
time_col <- c("#238B45", "#74C476", "#C7E9C0")
cond_col <- c("#003F5C", "#45AE91", "#58209D", "#8B3E9B", "#FF6361", "#BC4169")#QC Mapping Plots
#this dataframe contains my alignment percentages from featureCounts
##already filtered to only include DOX + DMSO samples
fC_DOXCounts <- readRDS("data/fC_DOXCounts.RDS")
#Now I want to plot these values out
####Reads by Sample####
reads_by_sample <- c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")
fC_DOXCounts %>%
ggplot(., aes (x = Conditions, y = Total_Align, fill = Treatment, group_by = Line))+
geom_col()+
geom_hline(aes(yintercept=20000000))+
scale_fill_manual(values=reads_by_sample)+
ggtitle(expression("Total number of reads by sample"))+
xlab("")+
ylab(expression("RNA-sequencing reads"))+
theme_bw()+
theme(plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.text.y = element_text(size =10, color = "black", angle = 0, hjust = 0.8, vjust = 0.5),
axis.text.x = element_text(size =10, color = "black", angle = 90, hjust = 1, vjust = 0.2),
#strip.text.x = element_text(size = 15, color = "black", face = "bold"),
strip.text.y = element_text(color = "white"))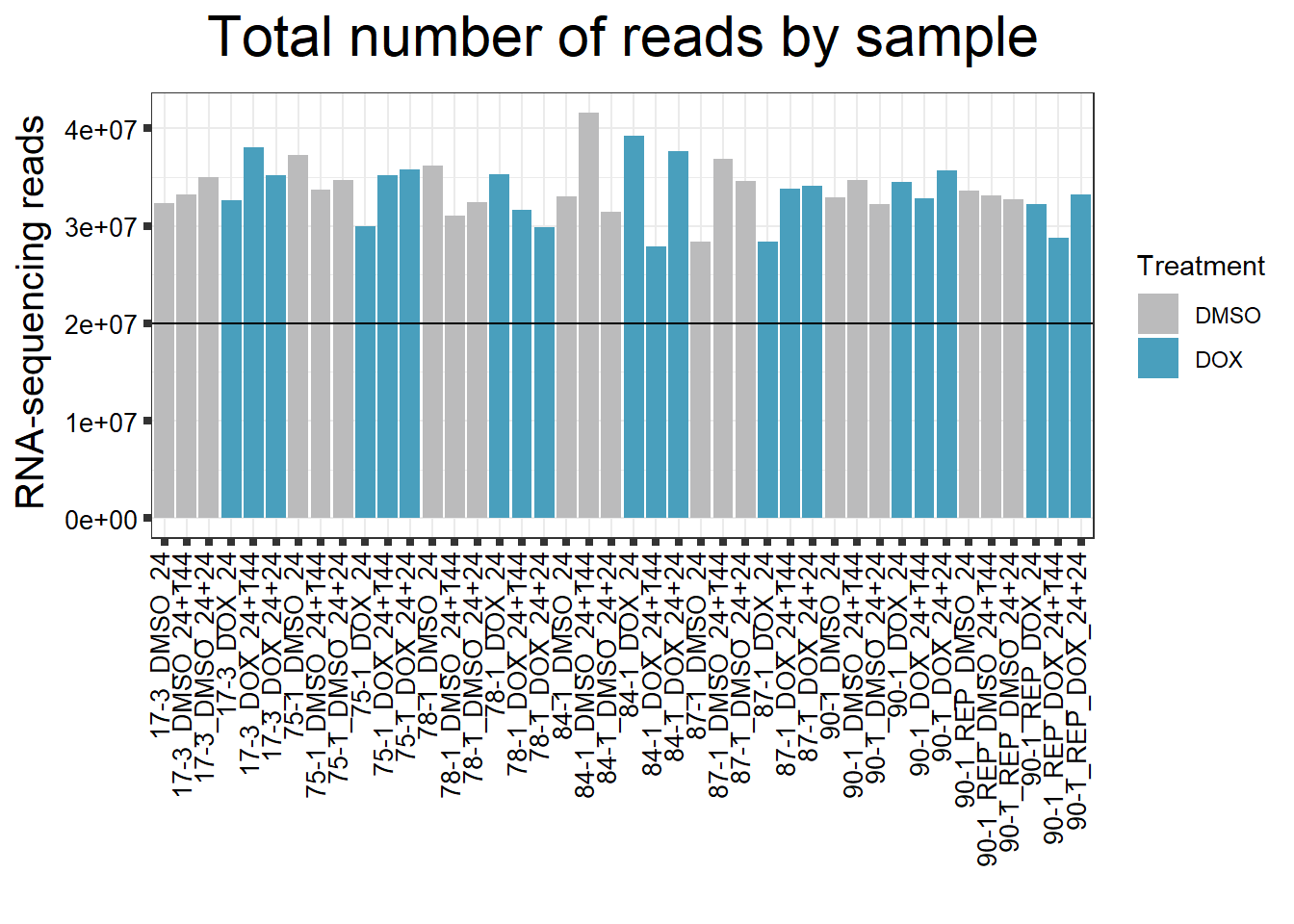
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
####Read Counts by Treatment####
fC_DOXCounts %>%
ggplot(., aes (x =Treatment, y= Total_Align, fill = Treatment))+
geom_boxplot()+
scale_fill_manual(values=reads_by_sample)+
ggtitle(expression("Total number of reads by treatment"))+
xlab("")+
ylab(expression("RNA-sequencing reads"))+
theme_bw()+
theme(plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.text.y = element_text(size =10, color = "black", angle = 0, hjust = 0.8, vjust = 0.5),
axis.text.x = element_text(size =10, color = "black", angle = 90, hjust = 1, vjust = 0.2),
#strip.text.x = element_text(size = 15, color = "black", face = "bold"),
strip.text.y = element_text(color = "white"))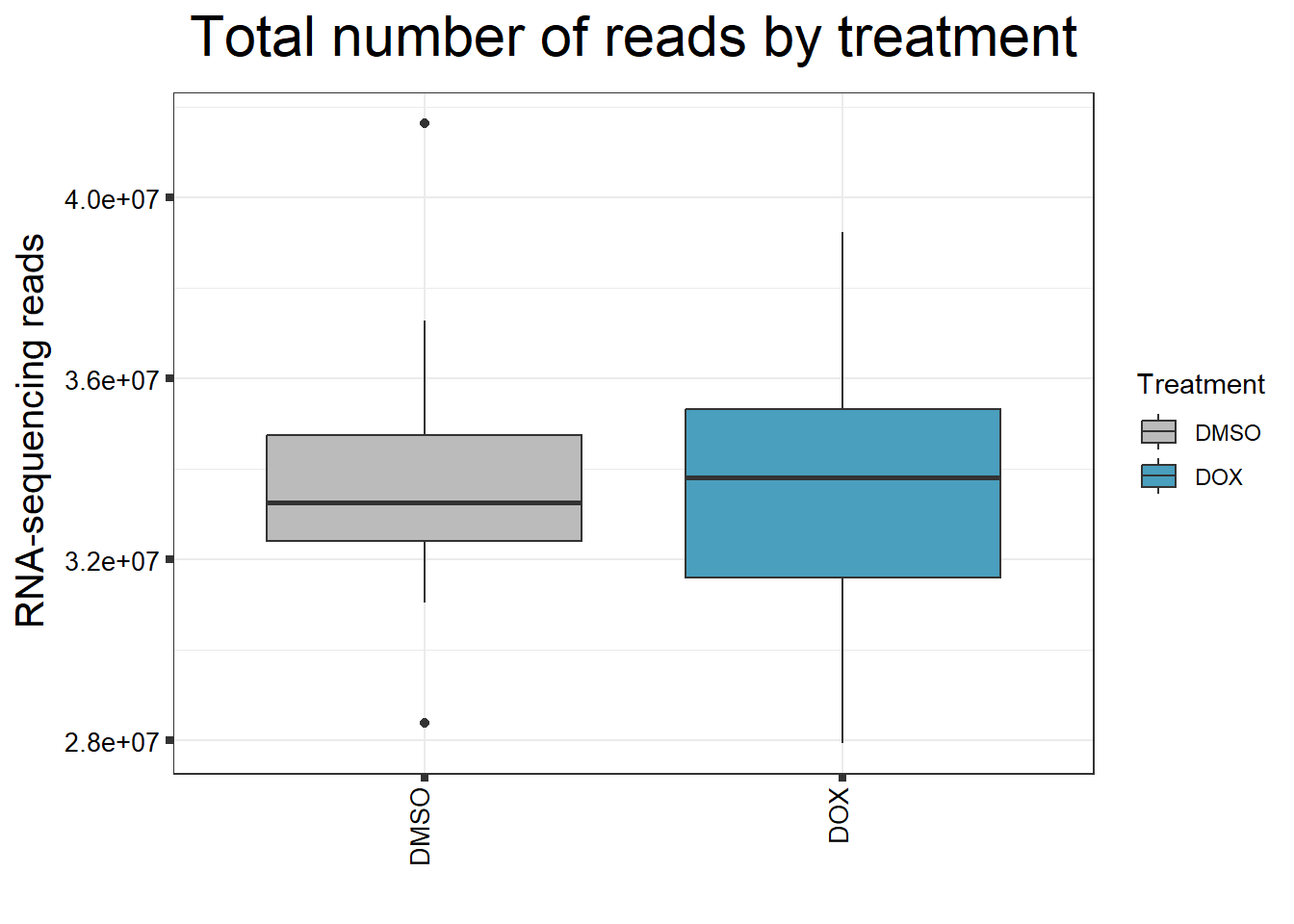
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
####Total Reads Per Individual####
fC_DOXCounts %>%
ggplot(., aes (x =as.factor(Line), y=Total_Align))+
geom_boxplot(aes(fill=as.factor(Line)))+
scale_fill_brewer(palette = "Dark2", name = "Individual")+
ggtitle(expression("Total number of reads by individual"))+
xlab("")+
ylab(expression("RNA-sequencing reads"))+
theme_bw()+
theme(plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.text.y = element_text(size =10, color = "black", angle = 0, hjust = 0.8, vjust = 0.5),
axis.text.x = element_text(size =10, color = "black", angle = 0, hjust = 1),
#strip.text.x = element_text(size = 15, color = "black", face = "bold"),
strip.text.y = element_text(color = "white"))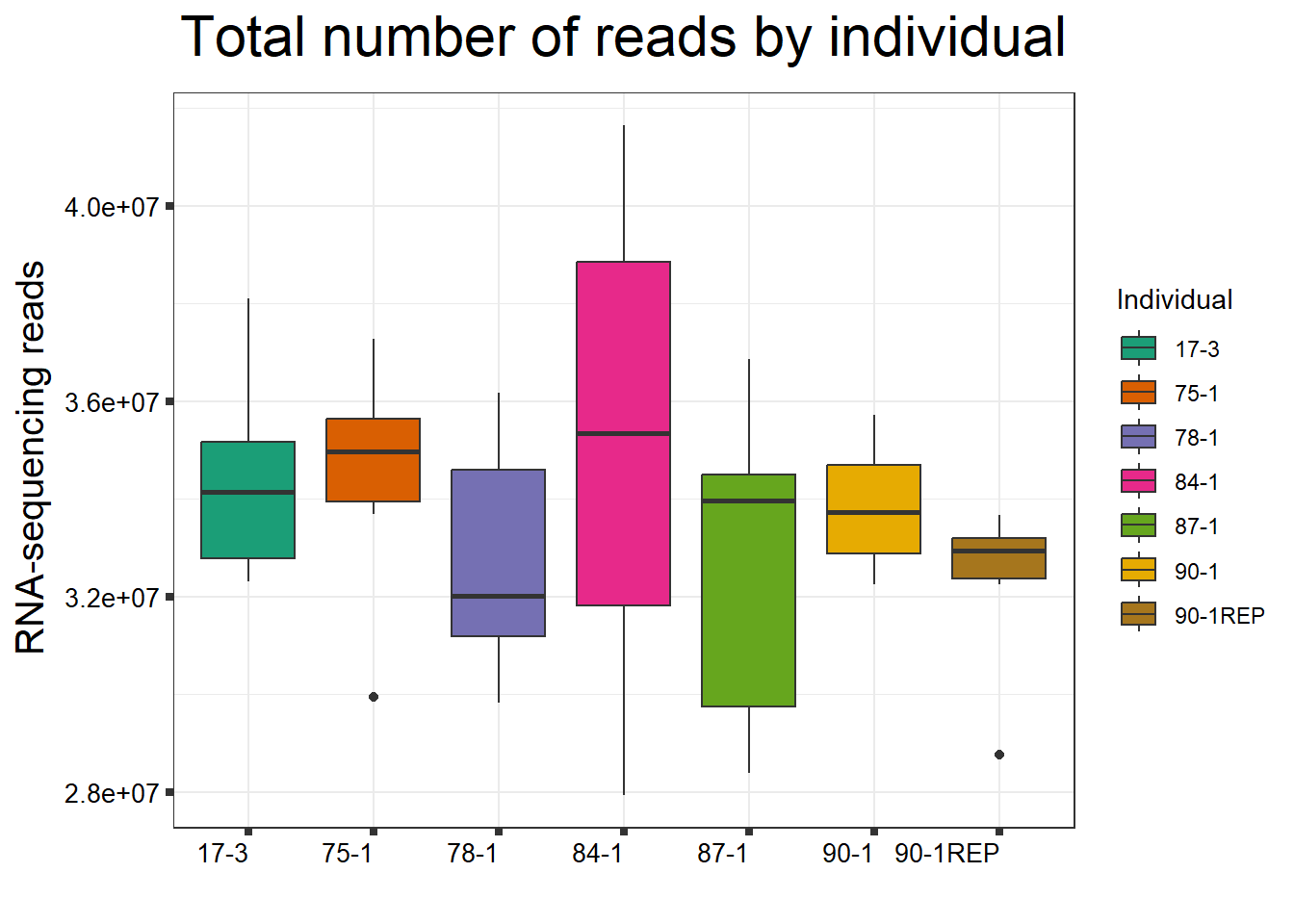
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
####Total Mapped Reads Per Drug####
reads_by_sample <- c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")
fC_DOXCounts %>%
ggplot(., aes (x = Conditions, y = Assigned_Align, fill = Treatment, group_by = Line))+
geom_col()+
geom_hline(aes(yintercept=20000000))+
scale_fill_manual(values=reads_by_sample)+
ggtitle(expression("Total number of mapped reads by sample"))+
xlab("")+
ylab(expression("RNA-sequencing reads"))+
theme_bw()+
theme(plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.text.y = element_text(size =10, color = "black", angle = 0, hjust = 0.8, vjust = 0.5),
axis.text.x = element_text(size =10, color = "black", angle = 90, hjust = 1, vjust = 0.2),
#strip.text.x = element_text(size = 15, color = "black", face = "bold"),
strip.text.y = element_text(color = "white"))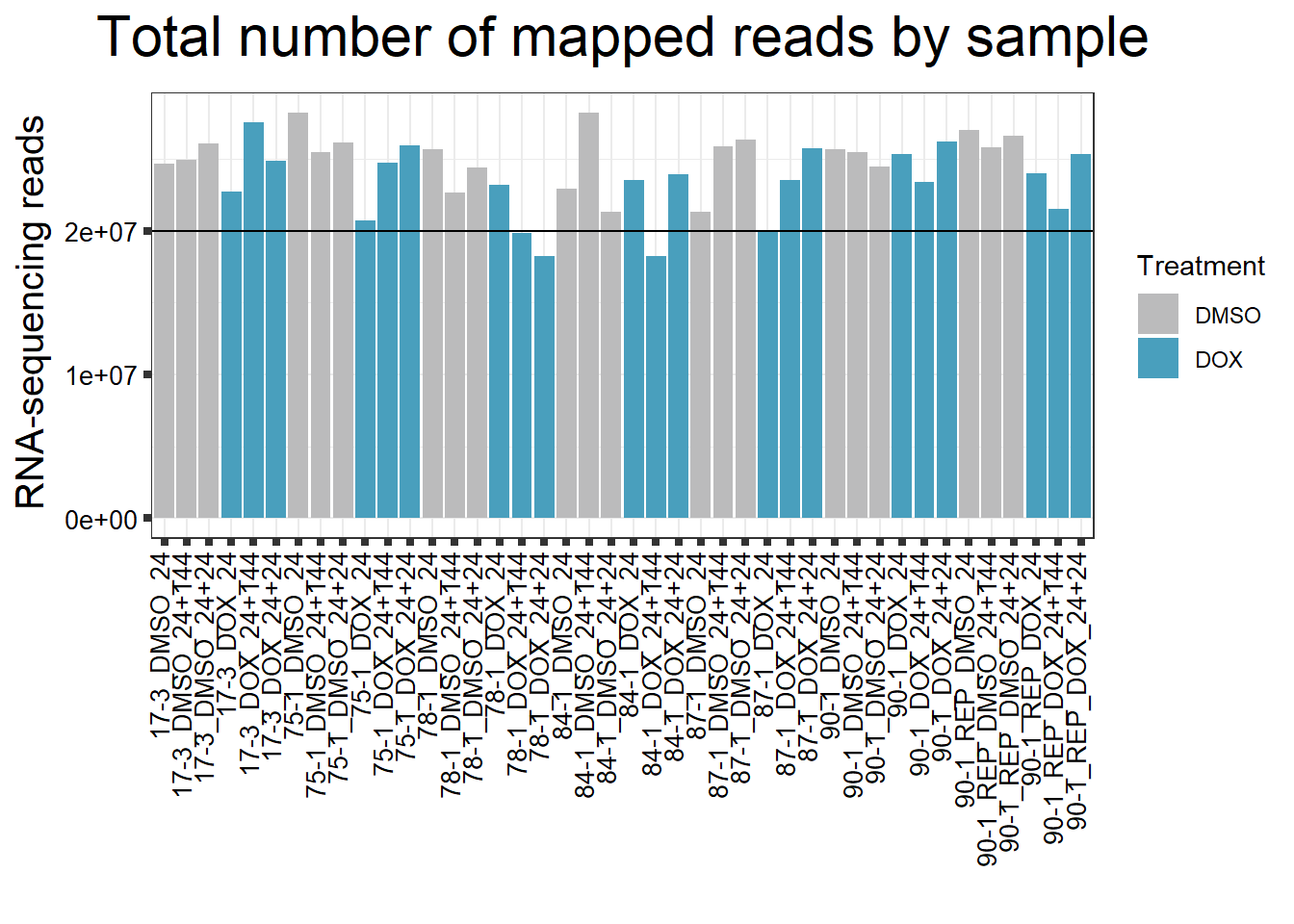
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
####Read Counts by Treatment####
fC_DOXCounts %>%
ggplot(., aes (x =Treatment, y= Assigned_Align, fill = Treatment))+
geom_boxplot()+
scale_fill_manual(values=reads_by_sample)+
ggtitle(expression("Total number of mapped reads by treatment"))+
xlab("")+
ylab(expression("RNA-sequencing reads"))+
theme_bw()+
theme(plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.text.y = element_text(size =10, color = "black", angle = 0, hjust = 0.8, vjust = 0.5),
axis.text.x = element_text(size =10, color = "black", angle = 90, hjust = 1, vjust = 0.2),
#strip.text.x = element_text(size = 15, color = "black", face = "bold"),
strip.text.y = element_text(color = "white"))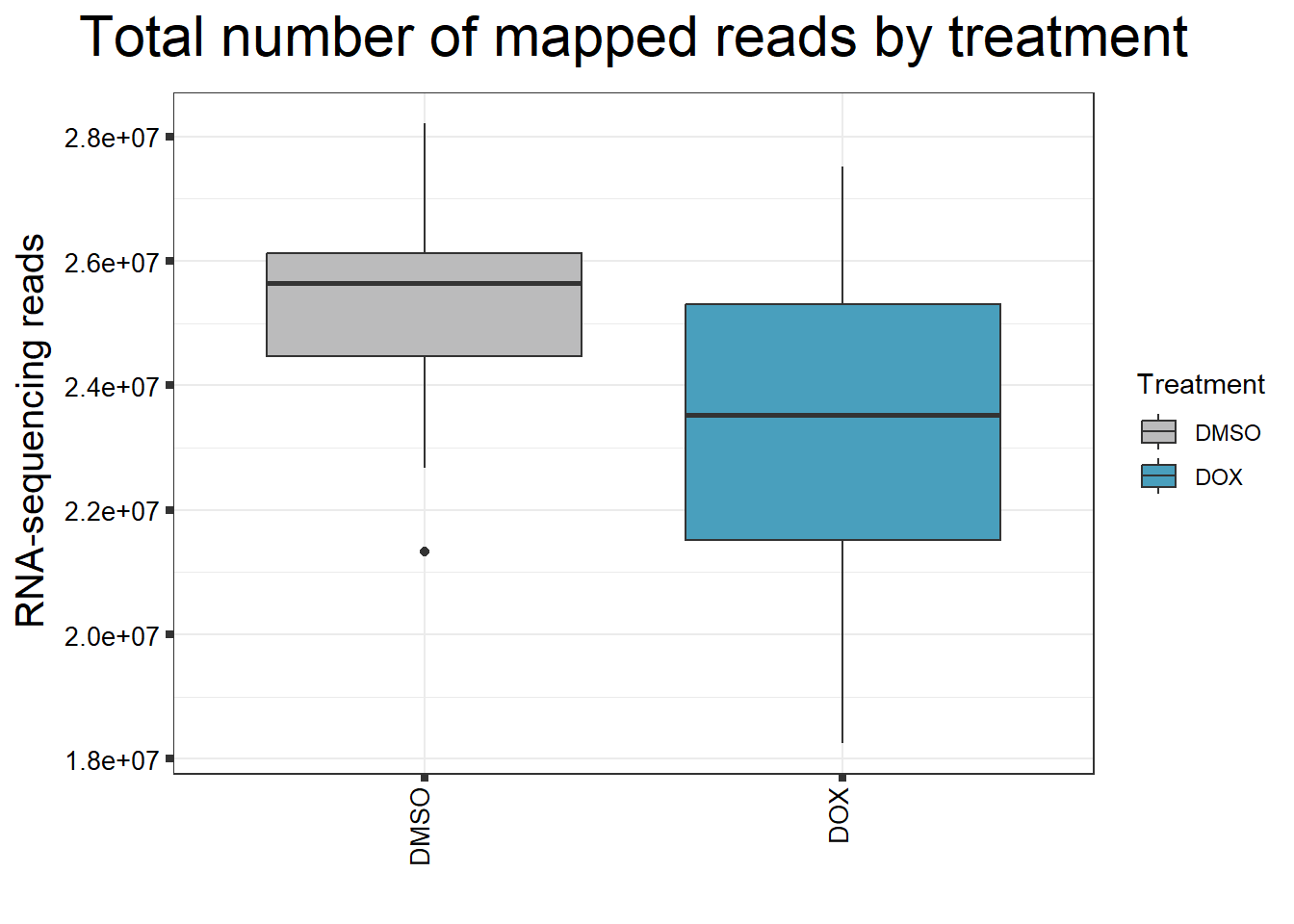
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
####Total Reads Per Individual####
fC_DOXCounts %>%
ggplot(., aes (x =as.factor(Line), y=Assigned_Align))+
geom_boxplot(aes(fill=as.factor(Line)))+
scale_fill_brewer(palette = "Dark2", name = "Individual")+
ggtitle(expression("Total number of mapped reads by individual"))+
xlab("")+
ylab(expression("RNA-sequencing reads"))+
theme_bw()+
theme(plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.text.y = element_text(size =10, color = "black", angle = 0, hjust = 0.8, vjust = 0.5),
axis.text.x = element_text(size =10, color = "black", angle = 0, hjust = 1),
#strip.text.x = element_text(size = 15, color = "black", face = "bold"),
strip.text.y = element_text(color = "white"))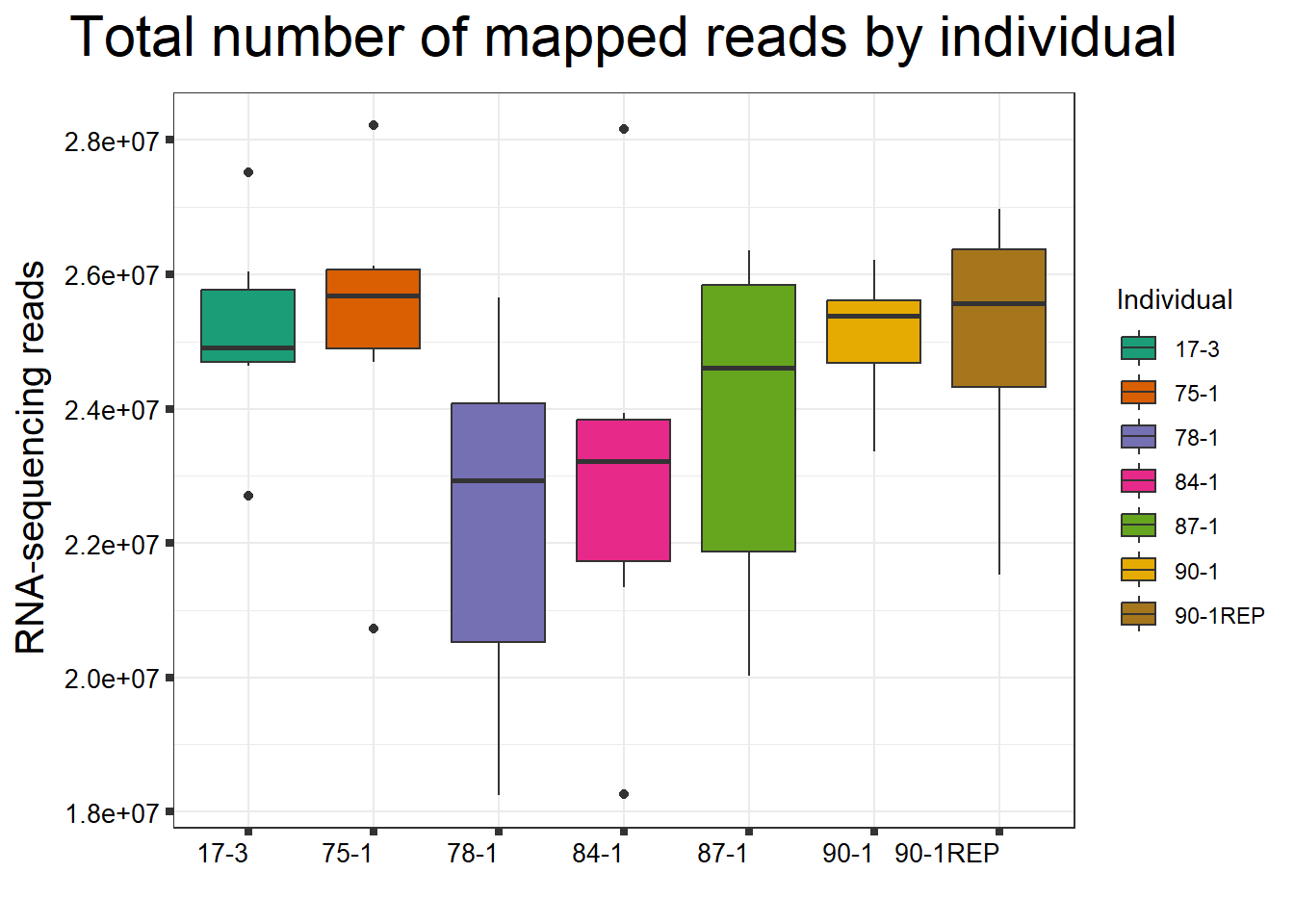
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
Now, I want to filter my dataframe Before I can filter by rowMeans, I must convert to log2cpm #Filter my Dataframe and Convert to log2cpm
#transform counts to cpm as a first step
counts_cpm_unfilt <- cpm(counts_raw_matrix, log = TRUE)
dim(counts_cpm_unfilt)[1] 28395 42#I should have 28395 genes here since this is unfiltered
hist(counts_cpm_unfilt,
main = "Histogram of Unfiltered Counts",
xlab = expression("Log"[2]*" counts-per-million"),
col = 4)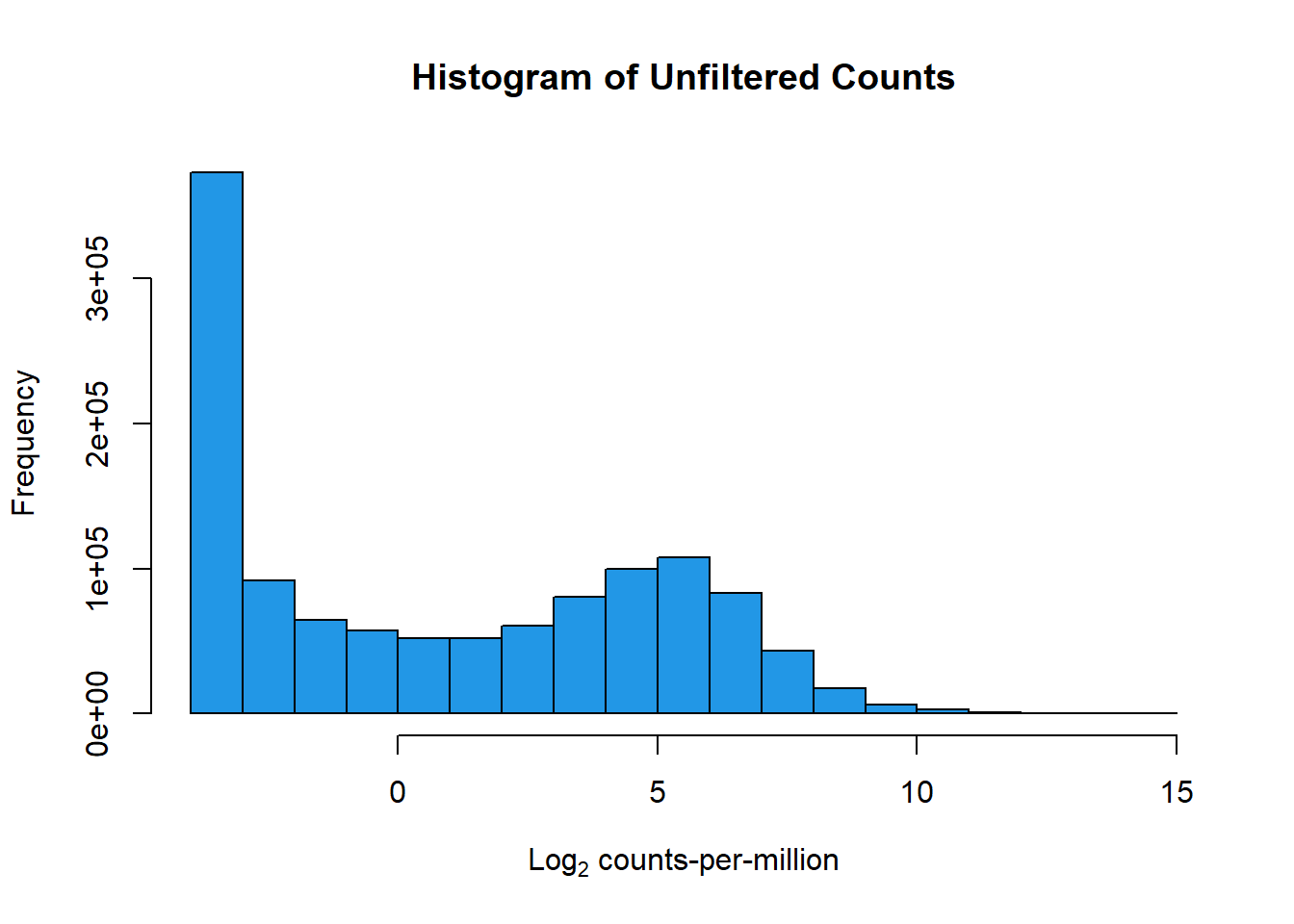
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
###filter my data by rowMeans > 0 to exclude lowly expressed genes
filcpm_matrix <- subset(counts_cpm_unfilt, (rowMeans(counts_cpm_unfilt) > 0))
dim(filcpm_matrix)[1] 14319 42#I should have 14319 genes here
#now let's make a histogram of this to check the difference
hist(filcpm_matrix,
main = "Histogram of Filtered Counts by rowMeans > 0",
xlab = expression("Log"[2]*" counts-per-million"),
col = 2)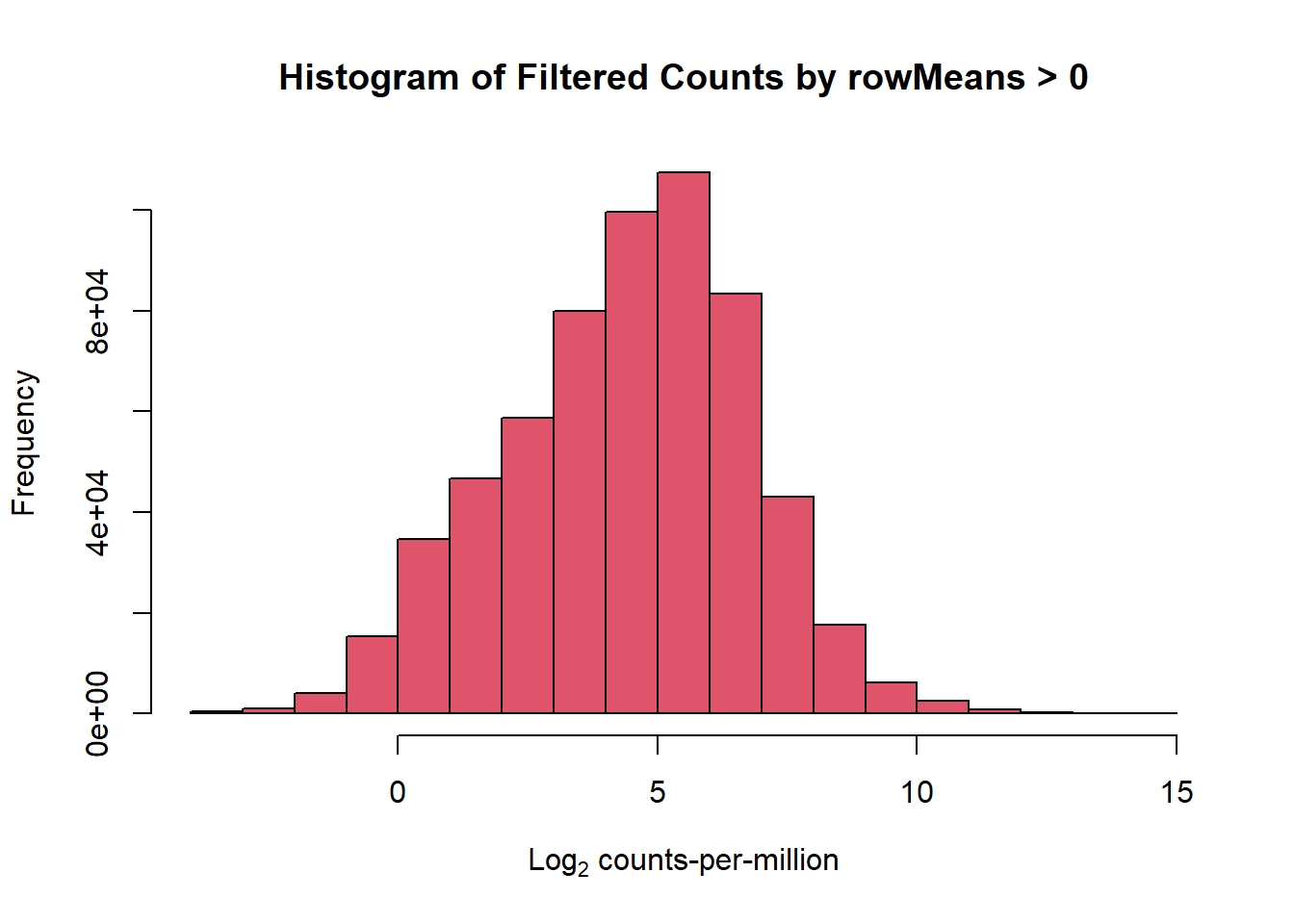
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
#change the column names to match my samples - make sure that they are in the right order
#Individual 1 = 84-1 (M)
#Individual 2 = 87-1 (F)
#Individual 3 = 78-1 (F)
#Individual 4 = 75-1 (F)
#Individual 5 = 17-3 (M)
#Individual 6 = 90-1 (M)
#Individual 6REP = 90-1REP (M)
#Treatment/time should follow this order:
#DOX24tx
#DMSO24tx
#DOX24rec
#DMSO24rec
#DOX144rec
#DMSO144rec
colnames(filcpm_matrix) <- c("DOX_24T_Ind1",
"DMSO_24T_Ind1",
"DOX_24R_Ind1",
"DMSO_24R_Ind1",
"DOX_144R_Ind1",
"DMSO_144R_Ind1",
"DOX_24T_Ind2",
"DMSO_24T_Ind2",
"DOX_24R_Ind2",
"DMSO_24R_Ind2",
"DOX_144R_Ind2",
"DMSO_144R_Ind2",
"DOX_24T_Ind3",
"DMSO_24T_Ind3",
"DOX_24R_Ind3",
"DMSO_24R_Ind3",
"DOX_144R_Ind3",
"DMSO_144R_Ind3",
"DOX_24T_Ind4",
"DMSO_24T_Ind4",
"DOX_24R_Ind4",
"DMSO_24R_Ind4",
"DOX_144R_Ind4",
"DMSO_144R_Ind4",
"DOX_24T_Ind5",
"DMSO_24T_Ind5",
"DOX_24R_Ind5",
"DMSO_24R_Ind5",
"DOX_144R_Ind5",
"DMSO_144R_Ind5",
"DOX_24T_Ind6",
"DMSO_24T_Ind6",
"DOX_24R_Ind6",
"DMSO_24R_Ind6",
"DOX_144R_Ind6",
"DMSO_144R_Ind6",
"DOX_24T_Ind6REP",
"DMSO_24T_Ind6REP",
"DOX_24R_Ind6REP",
"DMSO_24R_Ind6REP",
"DOX_144R_Ind6REP",
"DMSO_144R_Ind6REP")
#export this as a csv
#write.csv(filcpm_matrix, "data/new/filcpm_final_matrix.csv")##QC Boxplots of Filtered vs Unfiltered Data
#make boxplots of all counts vs log2cpm filtered counts
#set the margins so the x axis isn't cut off
##I don't mind if this one is partially cut off since all you need is the library number and not the whole name
par(mar = c(8,4,2,2))
#boxplot of unfiltered cpm matrix
boxplot(counts_cpm_unfilt,
main = "Boxplots of Unfiltered log2cpm",
names = colnames(counts_cpm_unfilt),
adj=1, las = 2, cex.axis = 0.7)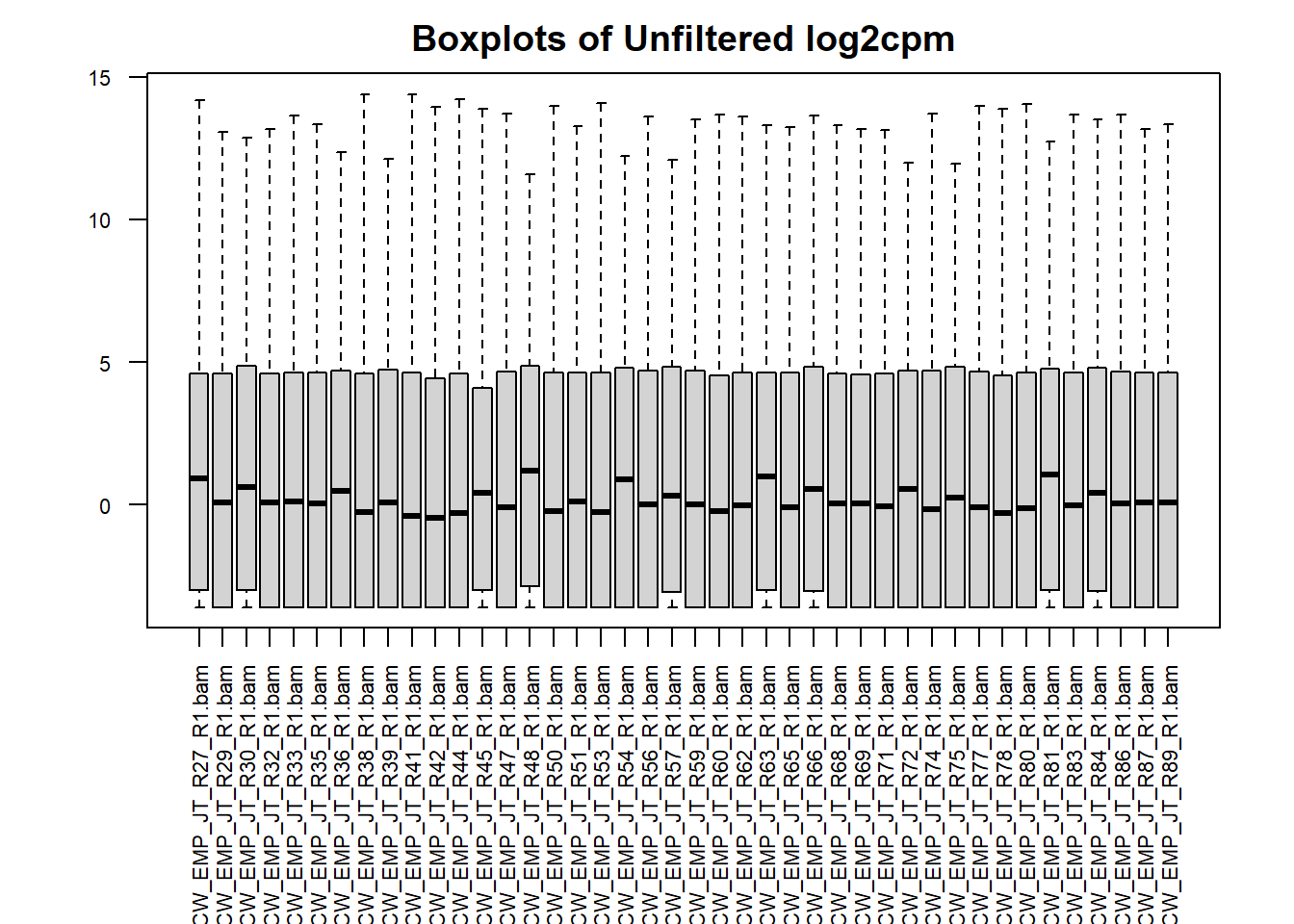
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
#set the margins so the x axis isn't cut off
par(mar = c(8,4,2,2))
#boxplot of filtered cpm matrix
boxplot(filcpm_matrix,
main = "Boxplots of Filtered log2cpm (rowMeans > 0)",
names = colnames(filcpm_matrix),
adj=1, las = 2, cex.axis = 0.7)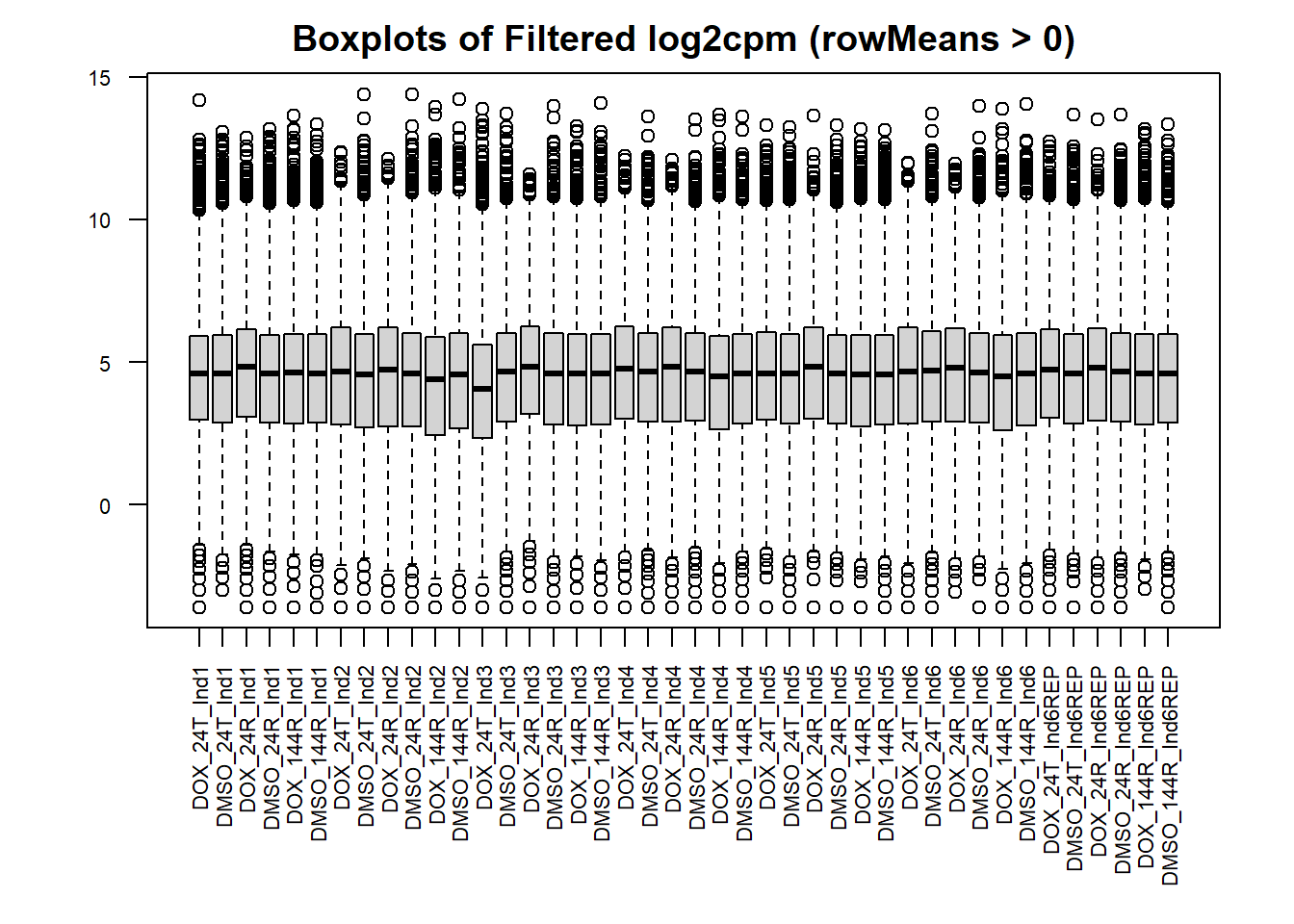
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
After making my final matrix, I pulled the gene symbols from the entrez IDs I had as my rownames I ran this initially and then moved the column into my final matrix My final matrix is called filcpm_final_matrix.csv saved under data #Put together my Data for Filtered Gene List
##I did this earlier so don't run again, I put the list into the filcpm_final_matrix.csv
# # ----------------- Load Required Libraries -----------------
# library(dplyr)
# library(readr)
# library(org.Hs.eg.db)
# library(AnnotationDbi)
# # ----------------- Load Data -----------------
# sample_data <- read_csv("data/filcpm_final_matrix.csv", show_col_types = FALSE)
# # ----------------- Ensure Entrez_ID is Present and in Character Format -----------------
# # Check column names
# print(colnames(sample_data))
# # Rename if needed (adjust if the column name is not exactly 'Entrez_ID')
# # sample_data <- sample_data %>% rename(Entrez_ID = `actual_column_name`)
# # Convert Entrez_ID to character
# sample_data <- sample_data %>%
# mutate(Entrez_ID = as.character(Entrez_ID))
# # ----------------- Map Entrez_ID to Gene Symbol -----------------
# gene_symbols <- AnnotationDbi::select(
# org.Hs.eg.db,
# keys = sample_data$Entrez_ID,
# columns = c("SYMBOL"),
# keytype = "ENTREZID"
# )
# # ----------------- Join Back to Main Data -----------------
# sample_annotated <- left_join(sample_data, gene_symbols, by = c("Entrez_ID" = "ENTREZID"))
# # ----------------- Save Annotated Output -----------------
# #write_csv(sample_annotated, "data/Sample_annotated.csv")
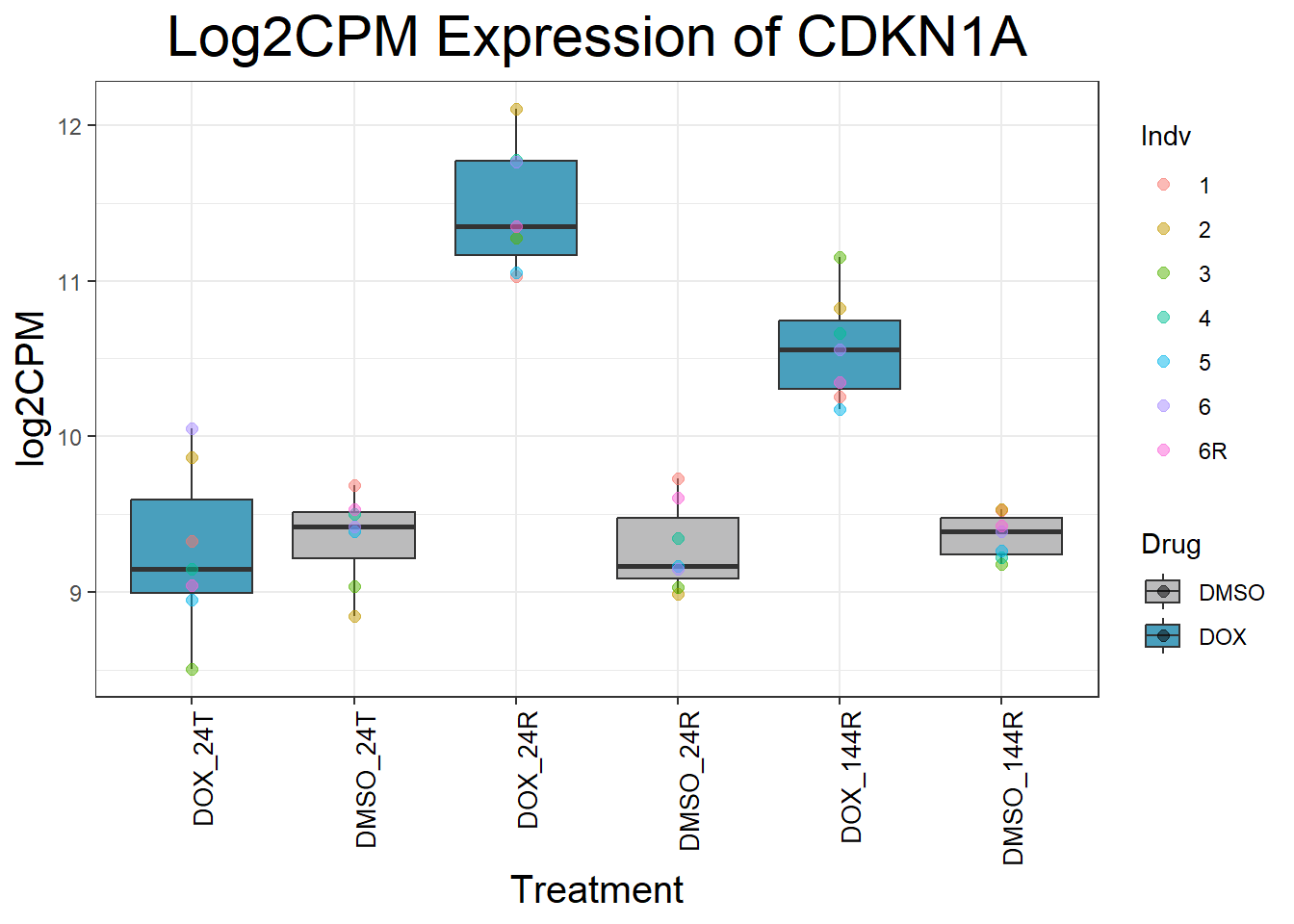
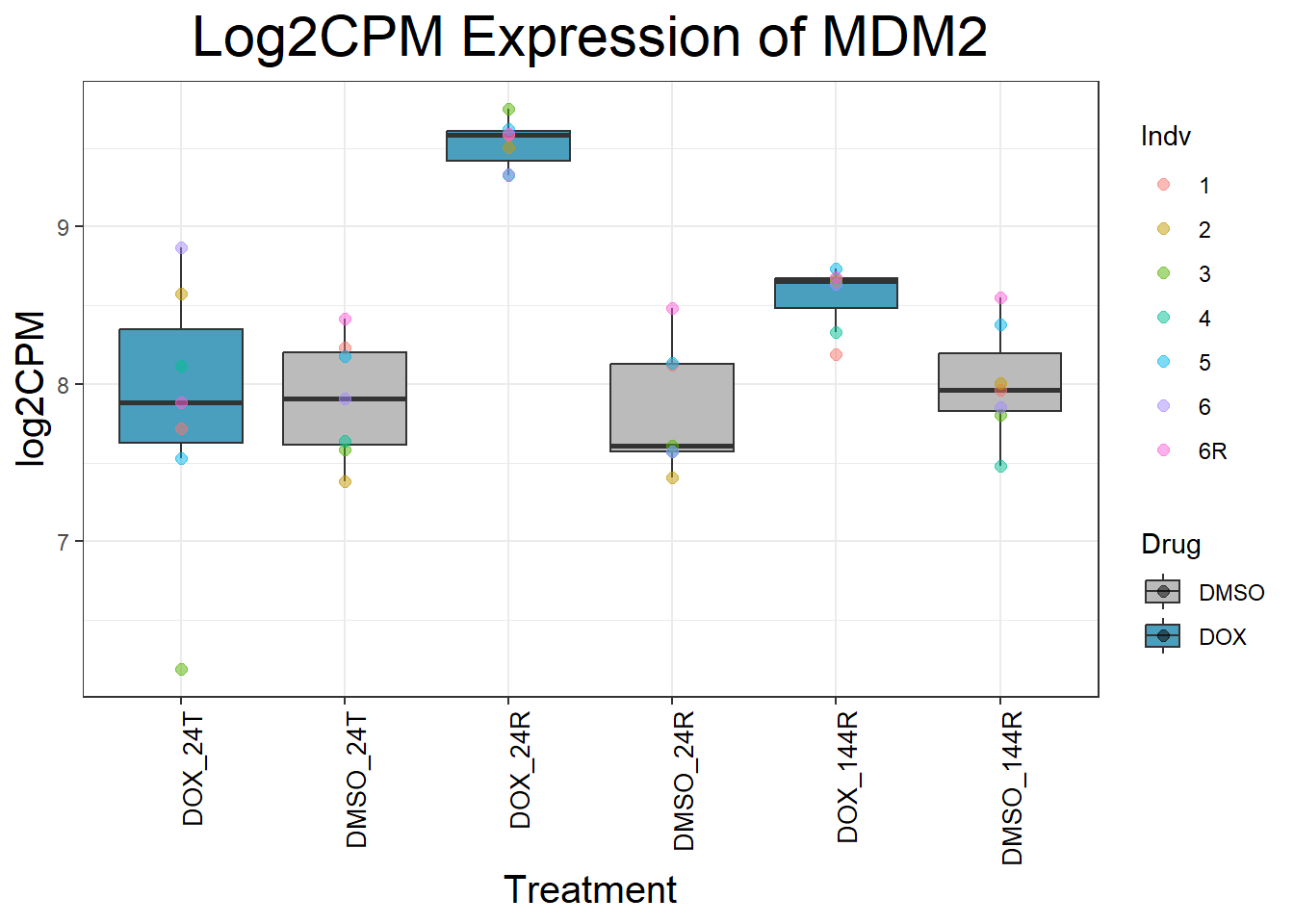
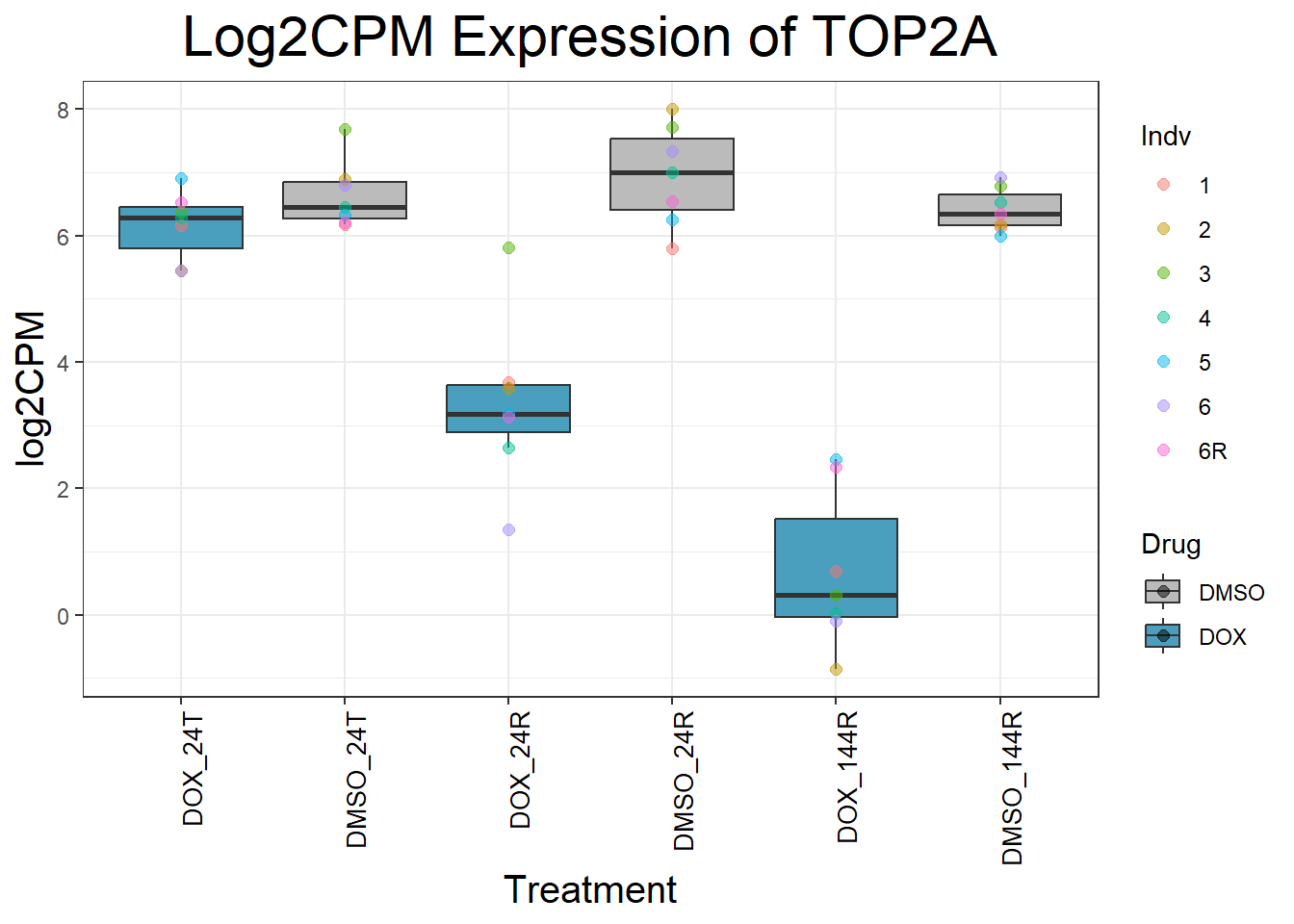
#Since I ran this before, Sample_annotated.csv columns of EntrezID and Symbol have been copied into my final matrix - so disregard this file except for record-keepingNow that I have my final matrix, I would like to check some key genes I want to make sure that these genes are responding as we expect We have triple checked this dataset to ensure that columns are in order #Check my Response Genes in log2cpm format
#Load in my count matrix
boxplot1 <- read.csv("data/new/filcpm_final_matrix.csv") %>%
as.data.frame()
#save boxplot1 as an object filcpm_matrix_genes
#saveRDS(boxplot1, "data/new/filcpm_matrix_genes.RDS")
#Define gene list(s)
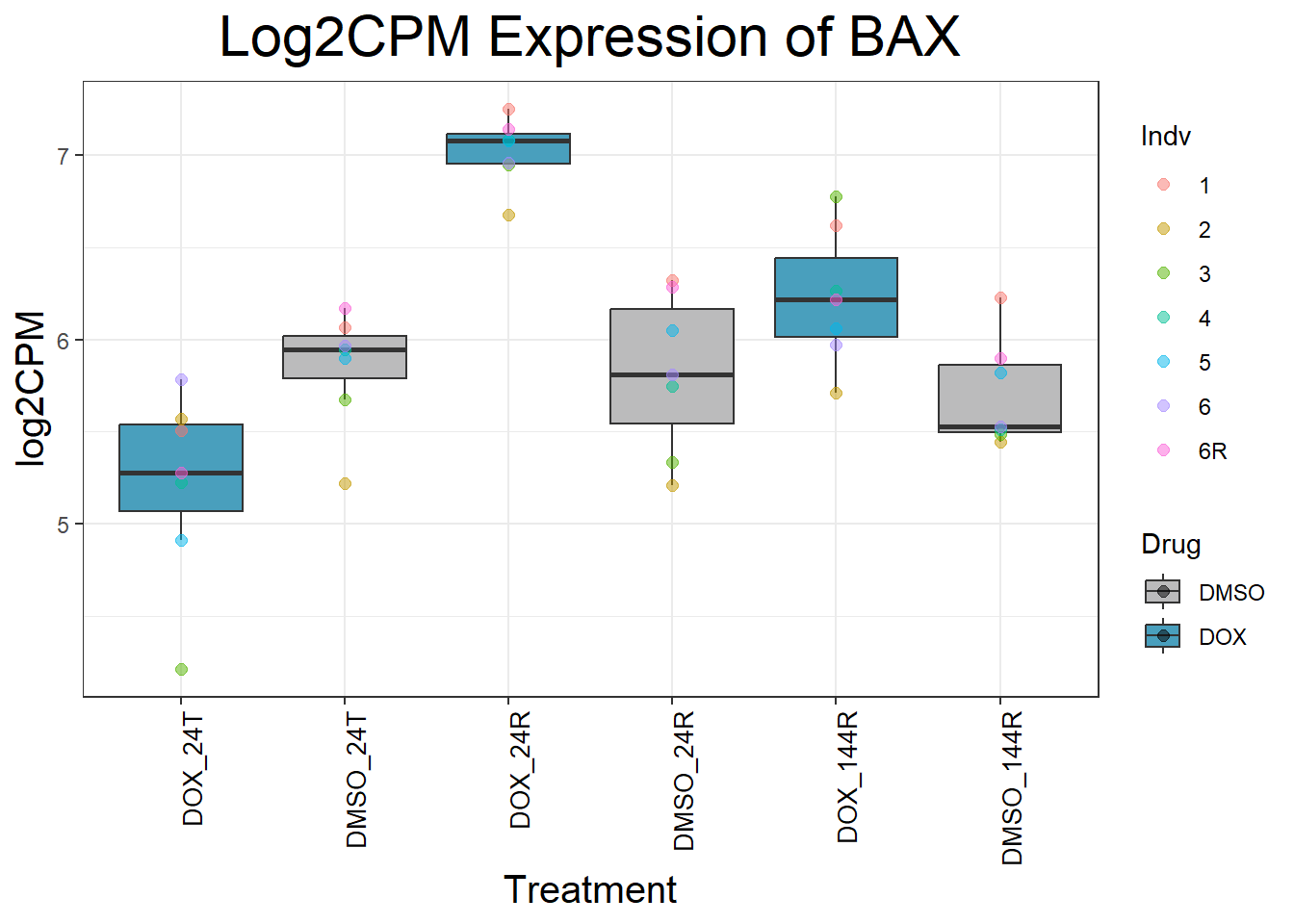
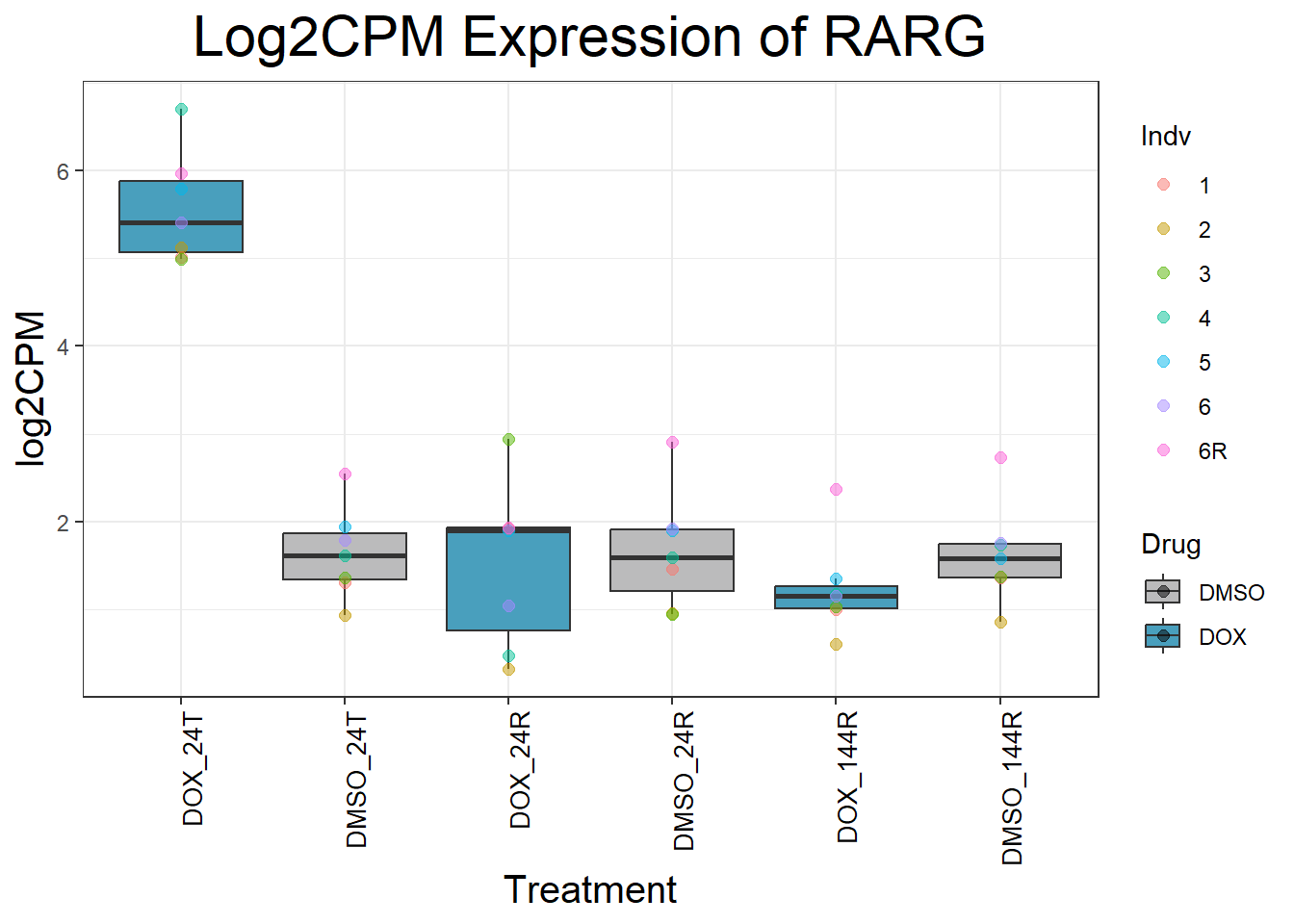
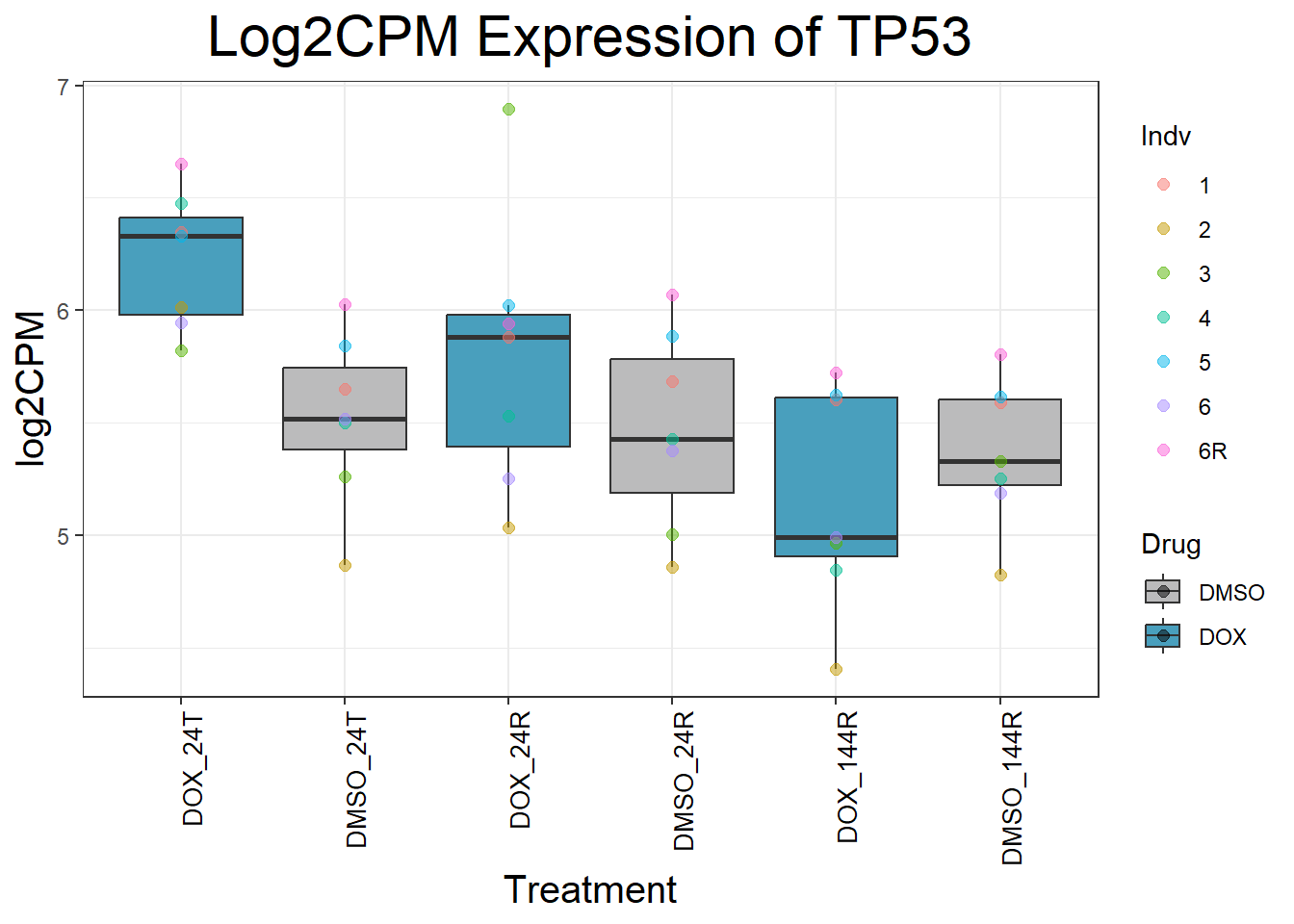
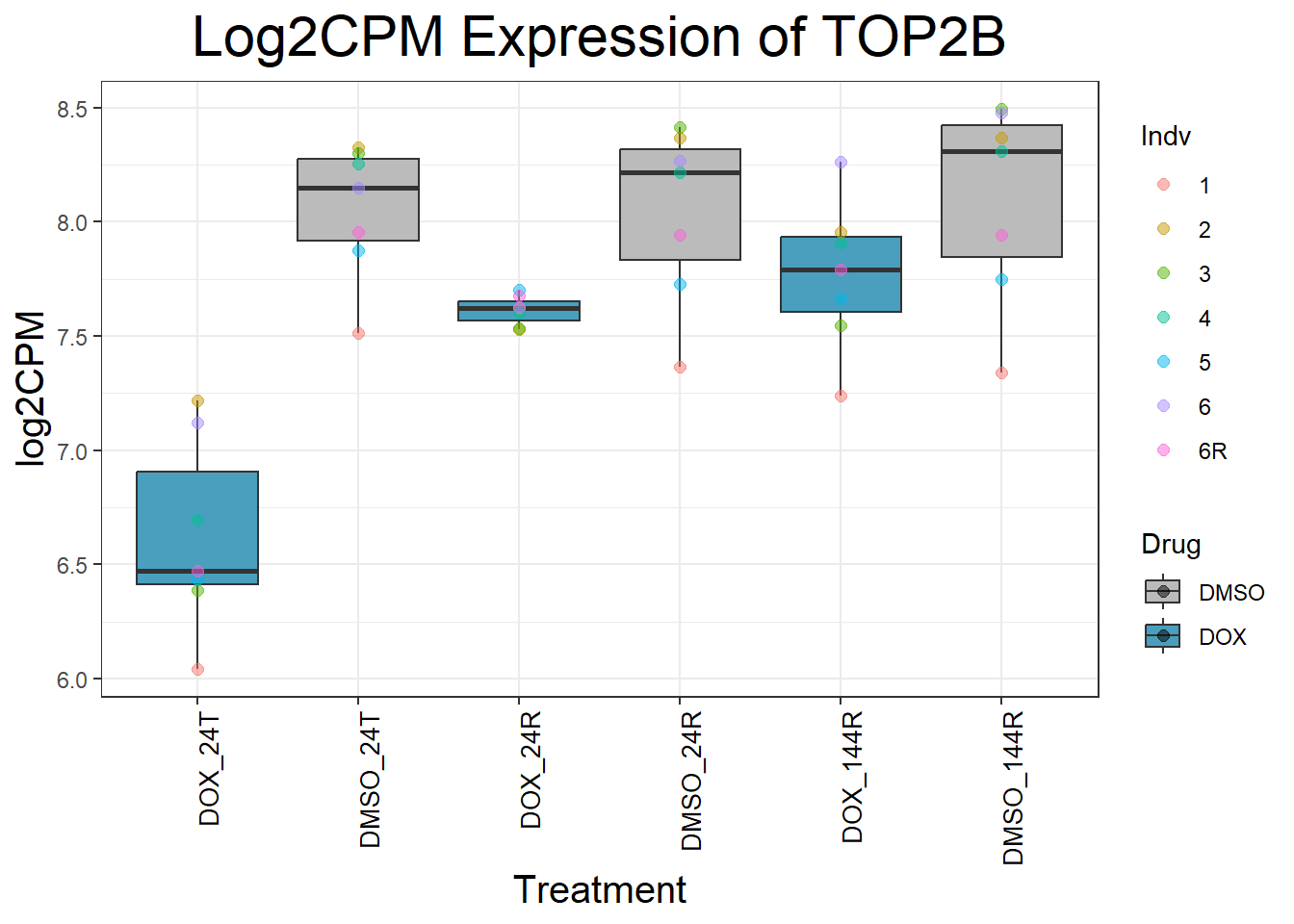
initial_test_genes <- c("CDKN1A", "MDM2", "BAX", "RARG", "TP53", "TOP2B", "TOP2A")
#Add more gene symbols as needed or add more categories
#Now put in the function I want to use to generate boxplots of genes
process_gene_data <- function(gene) {
gene_data <- boxplot1 %>% filter(SYMBOL == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2CPM") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6REP$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#this function is saved under process_gene_data so I will save as an R object
#saveRDS(process_gene_data, "data/new/process_gene_data_funct.RDS")
#Generate Boxplots from the above function using our gene list above
for (gene in initial_test_genes) {
gene_data <- process_gene_data(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2CPM, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2CPM Expression of", gene)) +
labs(x = "Treatment", y = "log2CPM") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
Now I’ve confirmed with some boxplots that my genes are present and (mostly) behaving as they should - Sayan’s CDKN1A and MDM2 are initially high at 24hr in DOX 0.5 - My CDKN1A and MDM2 are similar to DMSO at 24hr DOX 0.5 - These genes increase at DOX24R - These genes are also high at DOX144R but not as high as 24R However, TP53 and BAX are acting similarly across our data #PCA Analysis
#Now I want to check if my data is as expected on a PCA plot
#perform PCA calculations
prcomp_res_unfilt <- prcomp(t(counts_cpm_unfilt %>% as.matrix()), center = TRUE)
prcomp_res_filt <- prcomp(t(filcpm_matrix %>% as.matrix()), center = TRUE)
#read in my metadata annotations
Metadata <- read.csv("data/new/Metadata.csv")
#add in labels for individual numbers
ind_num <- c("1", "1", "1", "1", "1", "1",
"2", "2", "2", "2", "2", "2",
"3", "3", "3", "3", "3", "3",
"4", "4", "4", "4", "4", "4",
"5", "5", "5", "5", "5", "5",
"6", "6", "6", "6", "6", "6",
"6R", "6R", "6R", "6R", "6R", "6R")
# saveRDS(ind_num, "data/new/ind_num.RDS")
#now plot my PCA for unfiltered log2cpm
####PC1/PC2####
ggplot2::autoplot(prcomp_res_unfilt, data = Metadata, colour = "Condition", shape = "Time", size =4, x=1, y=2) +
ggrepel::geom_text_repel(label=ind_num, vjust = -.5, max.overlaps = 30) +
scale_color_manual(values=cond_col) +
ggtitle(expression("PCA of Unfiltered log"[2]*"cpm")) +
theme_bw()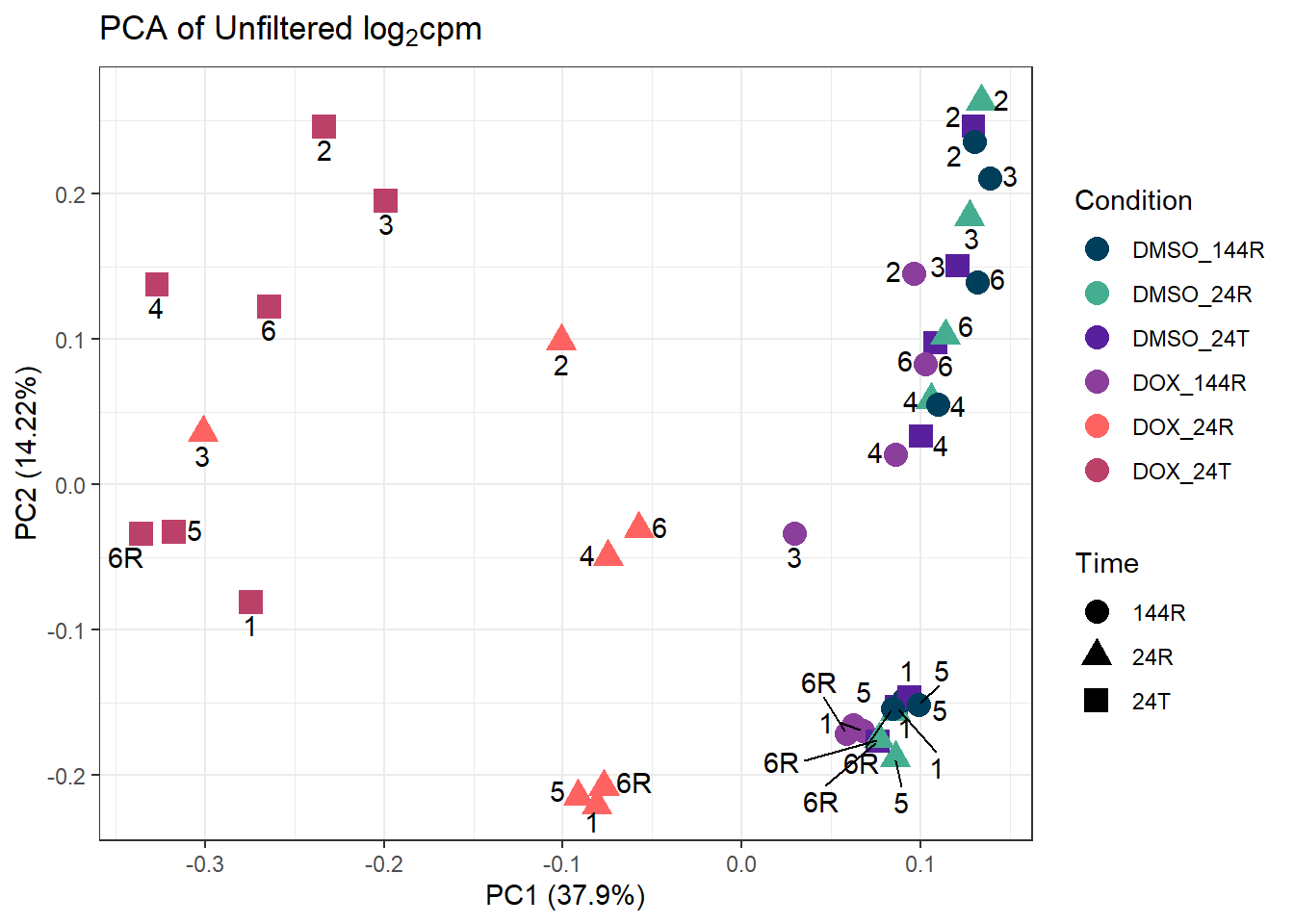
####PC2/PC3####
ggplot2::autoplot(prcomp_res_unfilt, data = Metadata, colour = "Condition", shape = "Time", size =4, x=2, y=3) +
ggrepel::geom_text_repel(label=ind_num, vjust = -.5, max.overlaps = 30) +
scale_color_manual(values=cond_col) +
ggtitle(expression("PCA of Unfiltered log"[2]*"cpm")) +
theme_bw()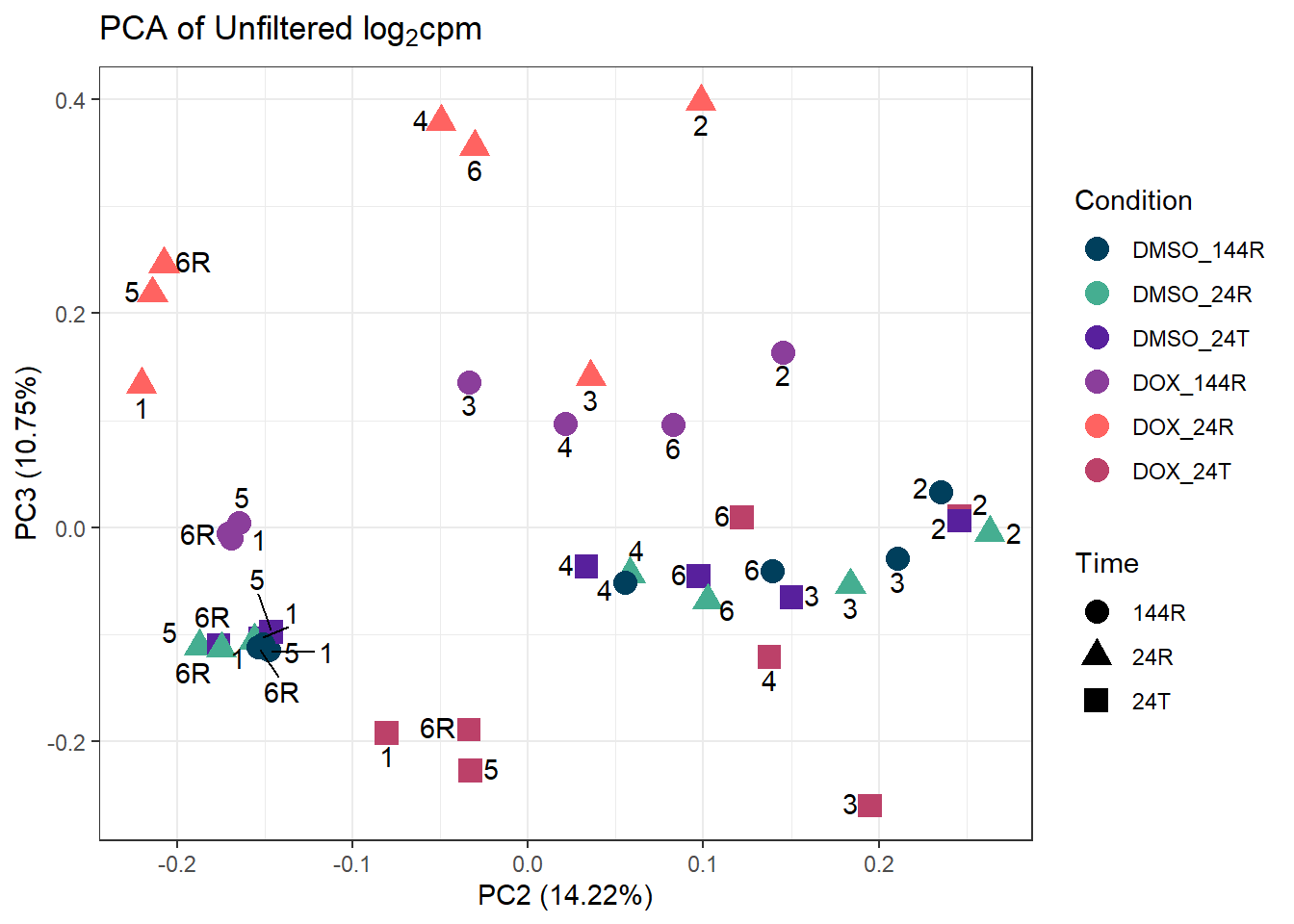
####PC3/PC4####
ggplot2::autoplot(prcomp_res_unfilt, data = Metadata, colour = "Condition", shape = "Time", size =4, x=3, y=4) +
ggrepel::geom_text_repel(label=ind_num, vjust = -.5, max.overlaps = 30) +
scale_color_manual(values=cond_col) +
ggtitle(expression("PCA of Unfiltered log"[2]*"cpm")) +
theme_bw()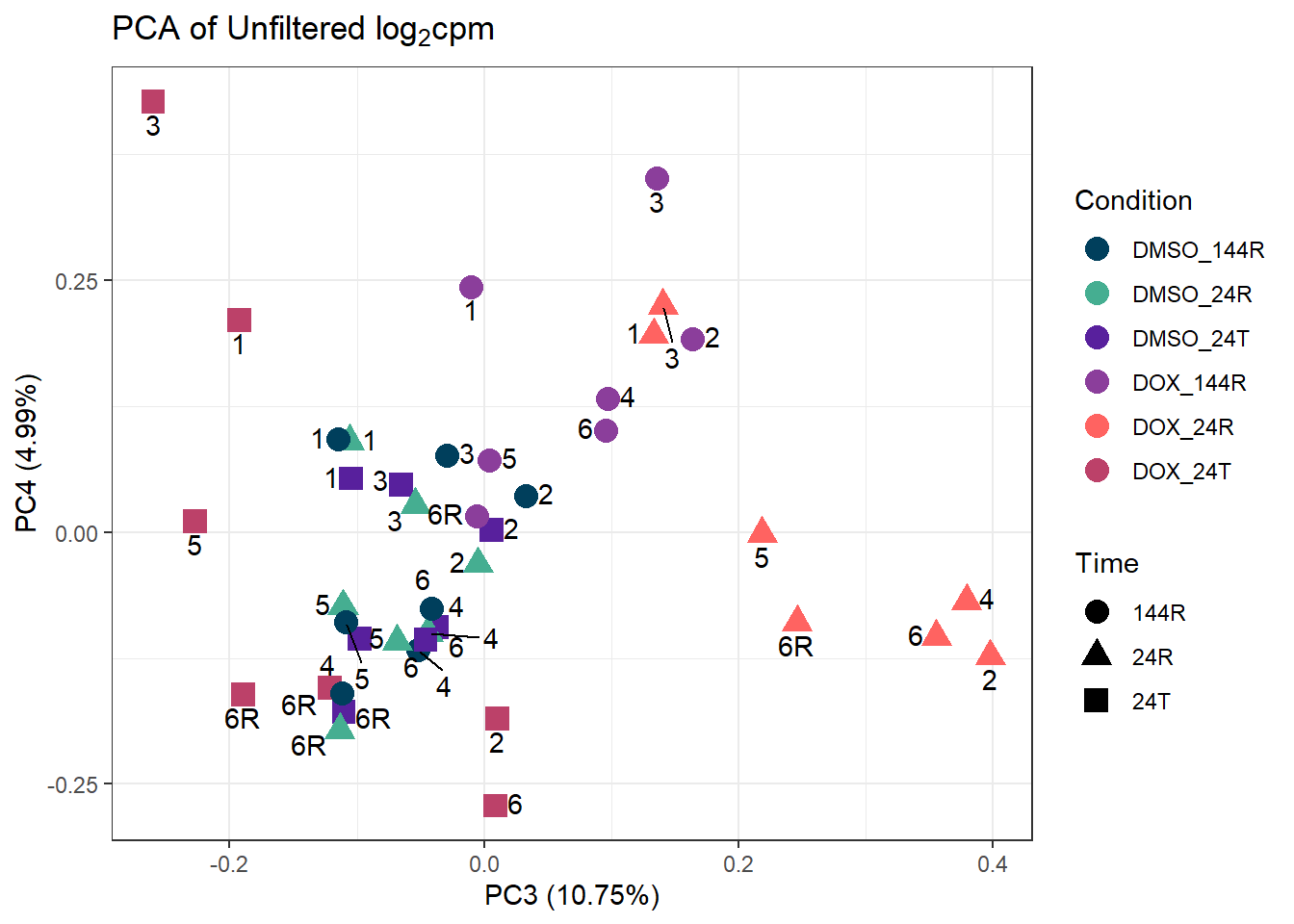
#Now plot my PCA for filtered log2cpm
####PC1/PC2####
ggplot2::autoplot(prcomp_res_filt, data = Metadata, colour = "Condition", shape = "Time", size =4, x=1, y=2) +
ggrepel::geom_text_repel(label=ind_num, vjust = -.5, max.overlaps = 30) +
scale_color_manual(values=cond_col) +
ggtitle(expression("PCA of Filtered log"[2]*"cpm")) +
theme_bw()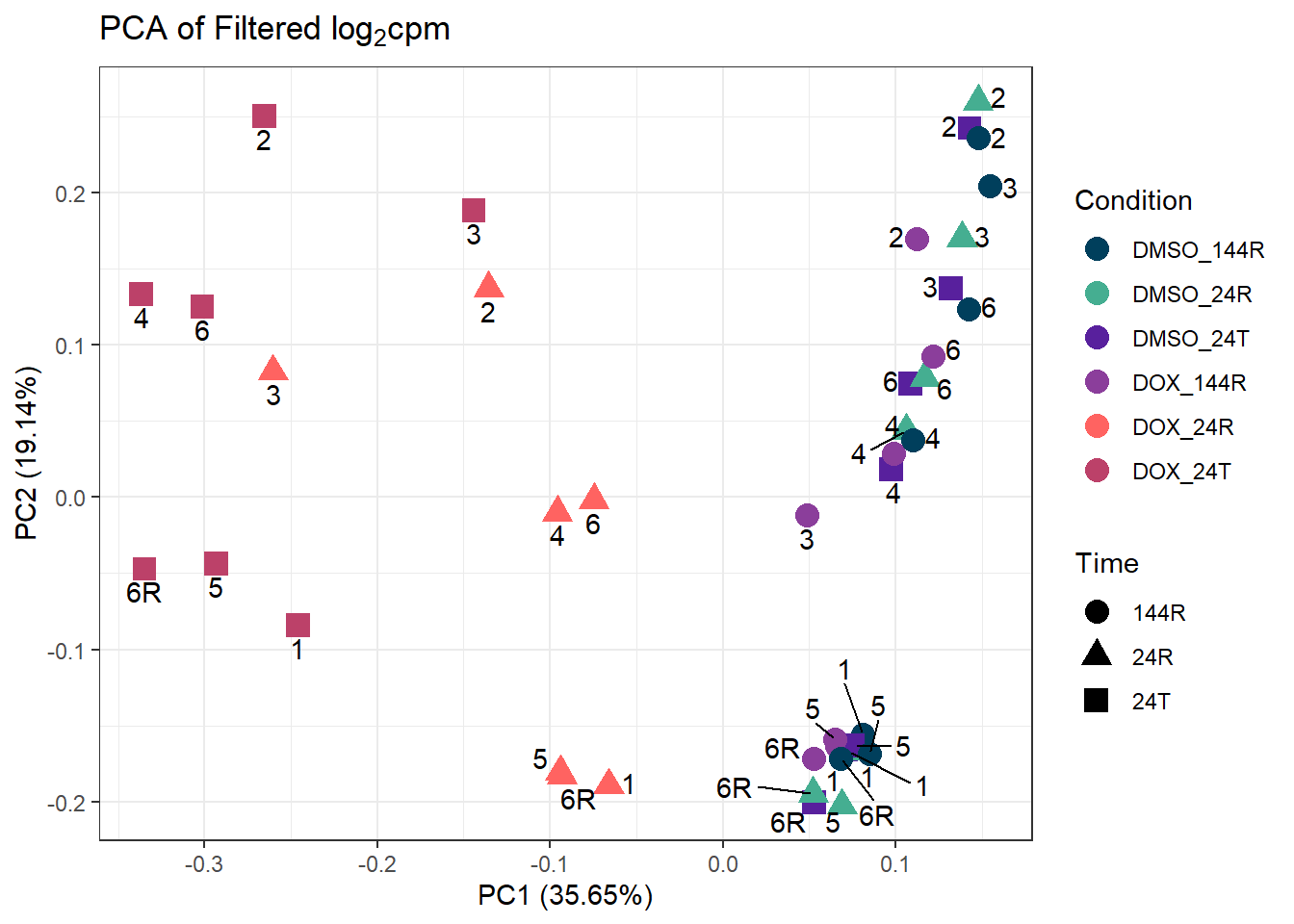
ggplot2::autoplot(prcomp_res_filt, data = Metadata, colour = "Condition", shape = "Time", size =4, x=1, y=2) +
ggrepel::geom_text_repel(label=ind_num, vjust = -.5, max.overlaps = 30) +
scale_color_manual(values=cond_col) +
ggtitle(expression("PCA of Filtered log"[2]*"cpm")) +
theme_bw()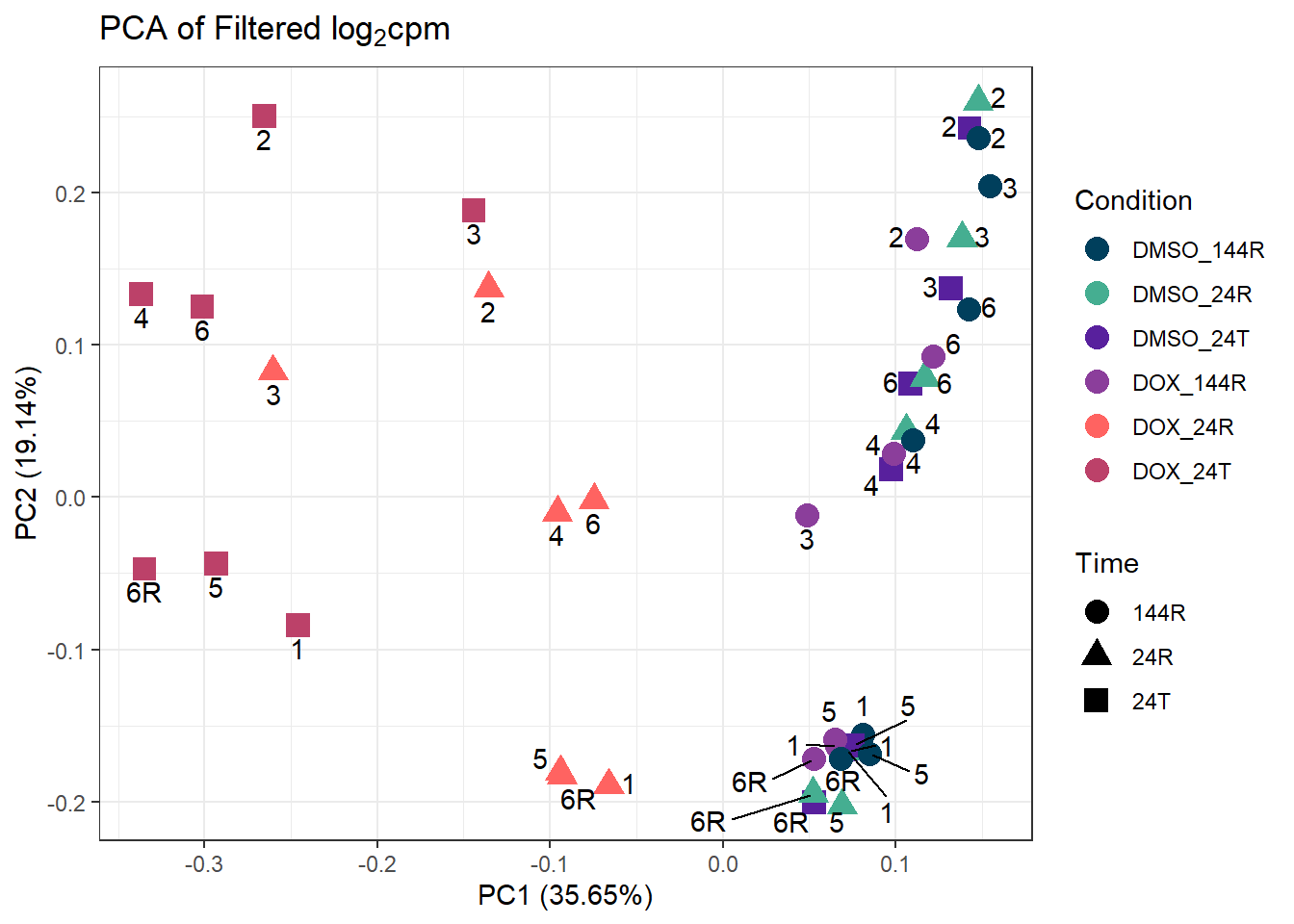
####PC2/PC3####
ggplot2::autoplot(prcomp_res_filt, data = Metadata, colour = "Condition", shape = "Time", size =4, x=2, y=3) +
ggrepel::geom_text_repel(label=ind_num, vjust = -.5, max.overlaps = 30) +
scale_color_manual(values=cond_col) +
ggtitle(expression("PCA of Filtered log"[2]*"cpm")) +
theme_bw()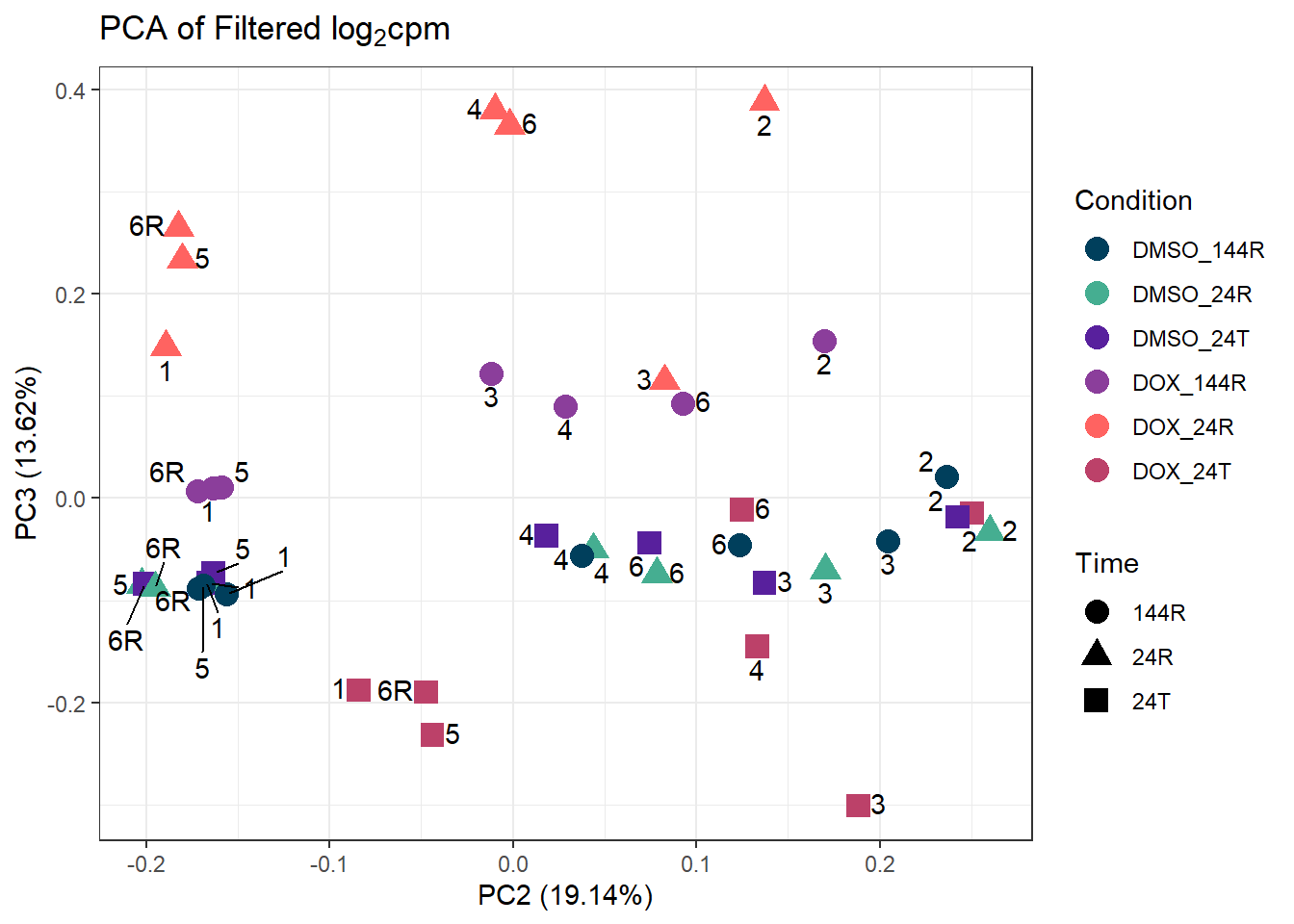
####PC3/PC4####
ggplot2::autoplot(prcomp_res_filt, data = Metadata, colour = "Condition", shape = "Time", size =4, x=3, y=4) +
ggrepel::geom_text_repel(label=ind_num, vjust = -.5, max.overlaps = 30) +
scale_color_manual(values=cond_col) +
ggtitle(expression("PCA of Filtered log"[2]*"cpm")) +
theme_bw()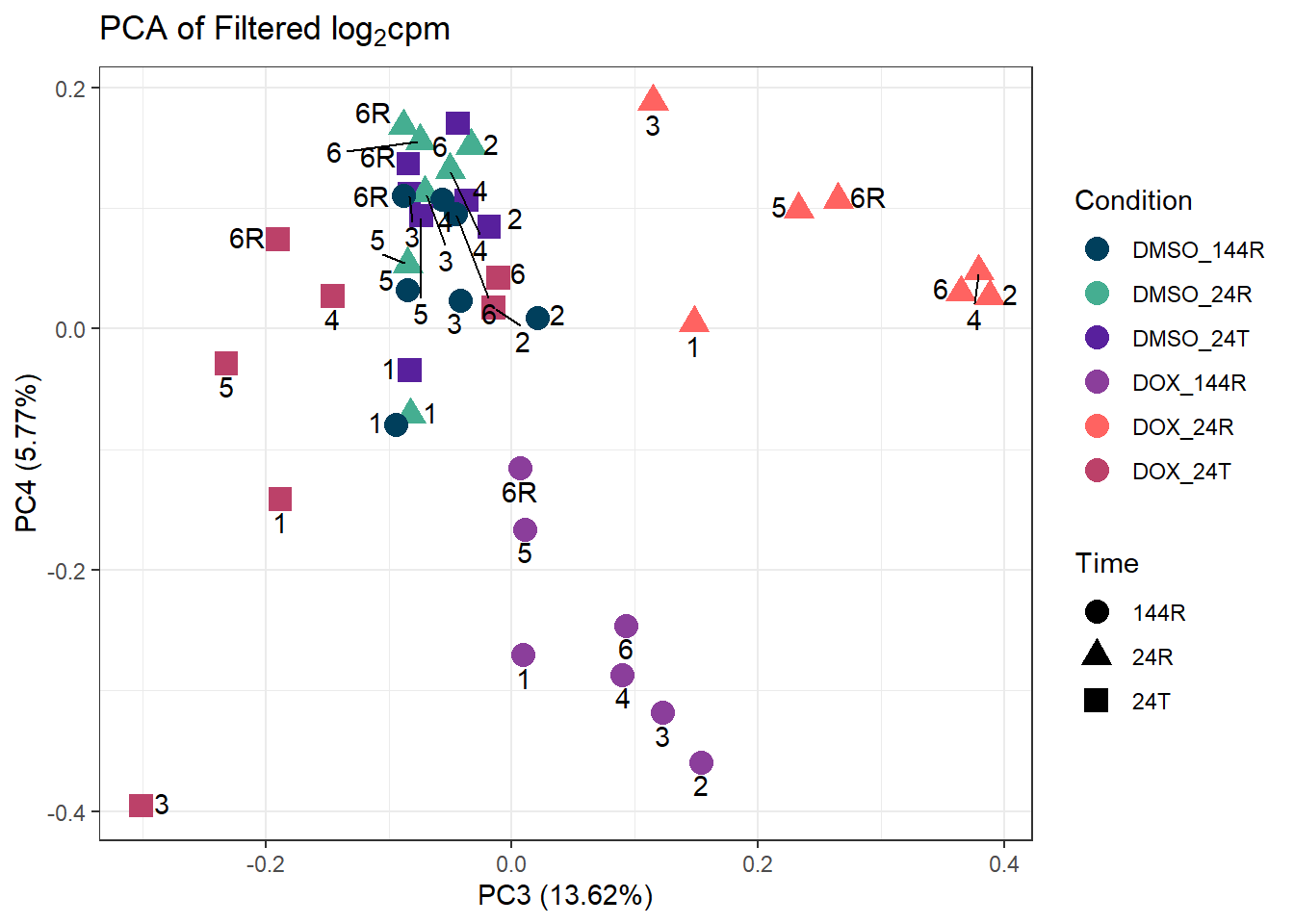
#Correlation Heatmaps
#check to make sure that the column names are correct
lcpm_2 <- filcpm_matrix
colnames(lcpm_2) <- Metadata$Final_sample_name
#compute the correlation matrices, one pearson and one spearman
cor_matrix_pearson <- cor(lcpm_2,
y = NULL,
use = "everything",
method = "pearson")
cor_matrix_spearman <- cor(lcpm_2,
y = NULL,
use = "everything",
method = "spearman")
# Extract metadata columns
Individual <- as.character(Metadata$Ind)
Time <- as.character(Metadata$Time)
Treatment <- as.character(Metadata$Drug)
# Define color palettes for annotations
annot_col_cor = list(drugs = c("DOX" = "#499FBD",
"DMSO" = "#BBBBBC"),
individuals = c("1" = "#003F5C",
"2" = "#45AE91",
"3" = "#58209D",
"4" = "#8B3E9B",
"5" = "#FF6361",
"6" = "#BC4169",
"6R" = "#FF2362"),
timepoints = c("24T" = "#238B45",
"24R" = "#74C476",
"144R" = "#C7E9C0"))
drug_colors <- c("DOX" = "#499FBD",
"DMSO" = "#BBBBBC")
ind_colors <- c("1" = "red",
"2" = "orange",
"3" = "yellow",
"4" = "green",
"5" = "blue",
"6" = "violet",
"6R" = "purple")
time_colors <- c("24T" = "#238B45",
"24R" = "#74C476",
"144R" = "#C7E9C0")
# Create annotations
top_annotation <- HeatmapAnnotation(
Individual = Individual,
Time = Time,
Treatment = Treatment,
col = list(
Individual = ind_colors,
Time = time_colors,
Treatment = drug_colors
)
)
####ANNOTATED HEATMAPS####
# pheatmap(cor_matrix_pearson, border_color = "black", legend = TRUE, angle_col = 90, display_numbers = FALSE, number_color = "black", fontsize = 10, fontsize_number = 5, annotation_col = top_annotation, annotation_colors = annot_col)
####Pearson Heatmap####
heatmap_pearson <- Heatmap(cor_matrix_pearson,
name = "Pearson",
top_annotation = top_annotation,
show_row_names = TRUE,
show_column_names = TRUE,
cluster_rows = TRUE,
cluster_columns = TRUE,
border = TRUE)
# Draw the heatmap
draw(heatmap_pearson)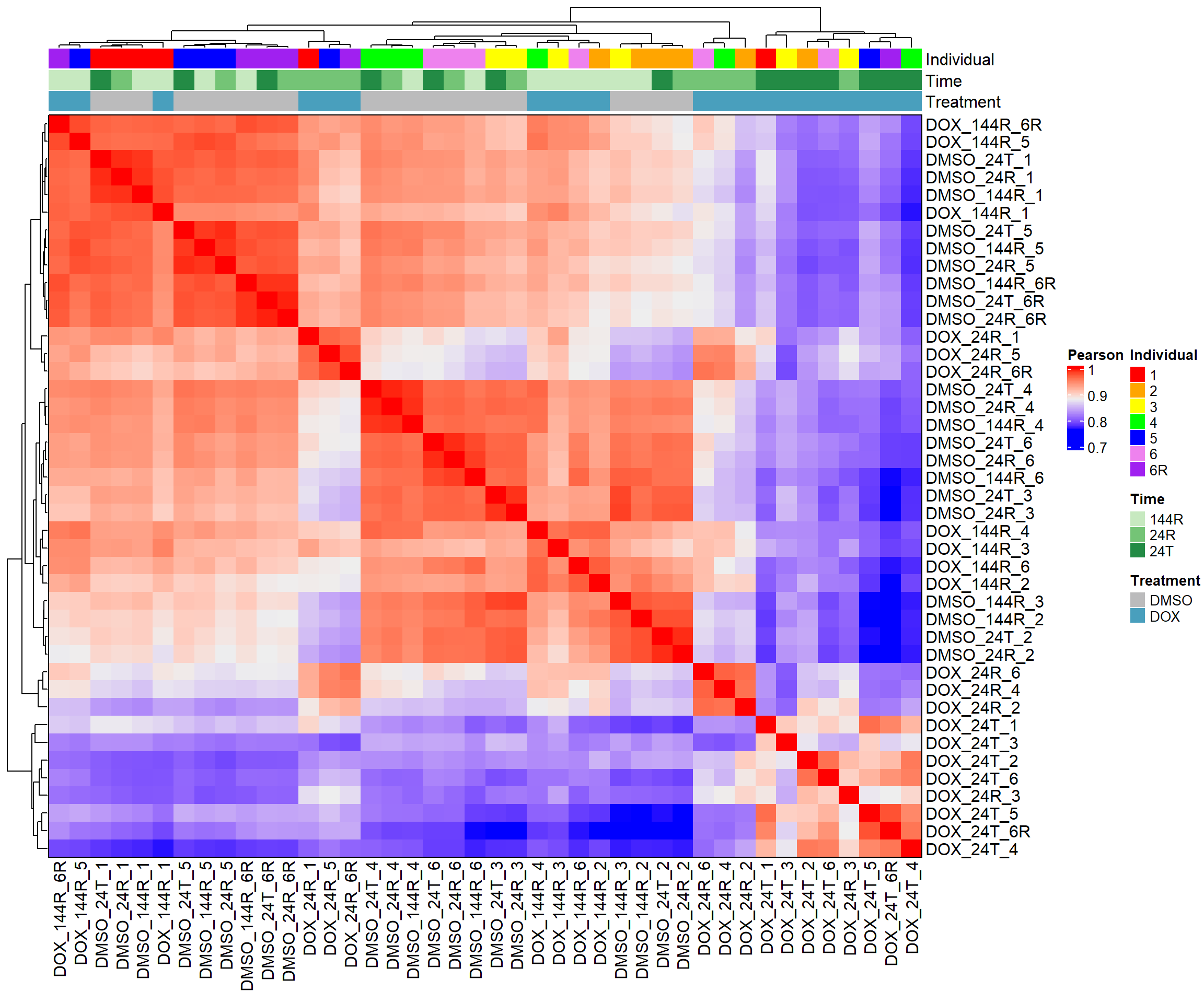
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
####Spearman Heatmap####
heatmap_spearman <- Heatmap(cor_matrix_spearman,
name = "Spearman",
top_annotation = top_annotation,
show_row_names = TRUE,
show_column_names = TRUE,
cluster_rows = TRUE,
cluster_columns = TRUE,
border = TRUE)
# Draw the heatmap
draw(heatmap_spearman)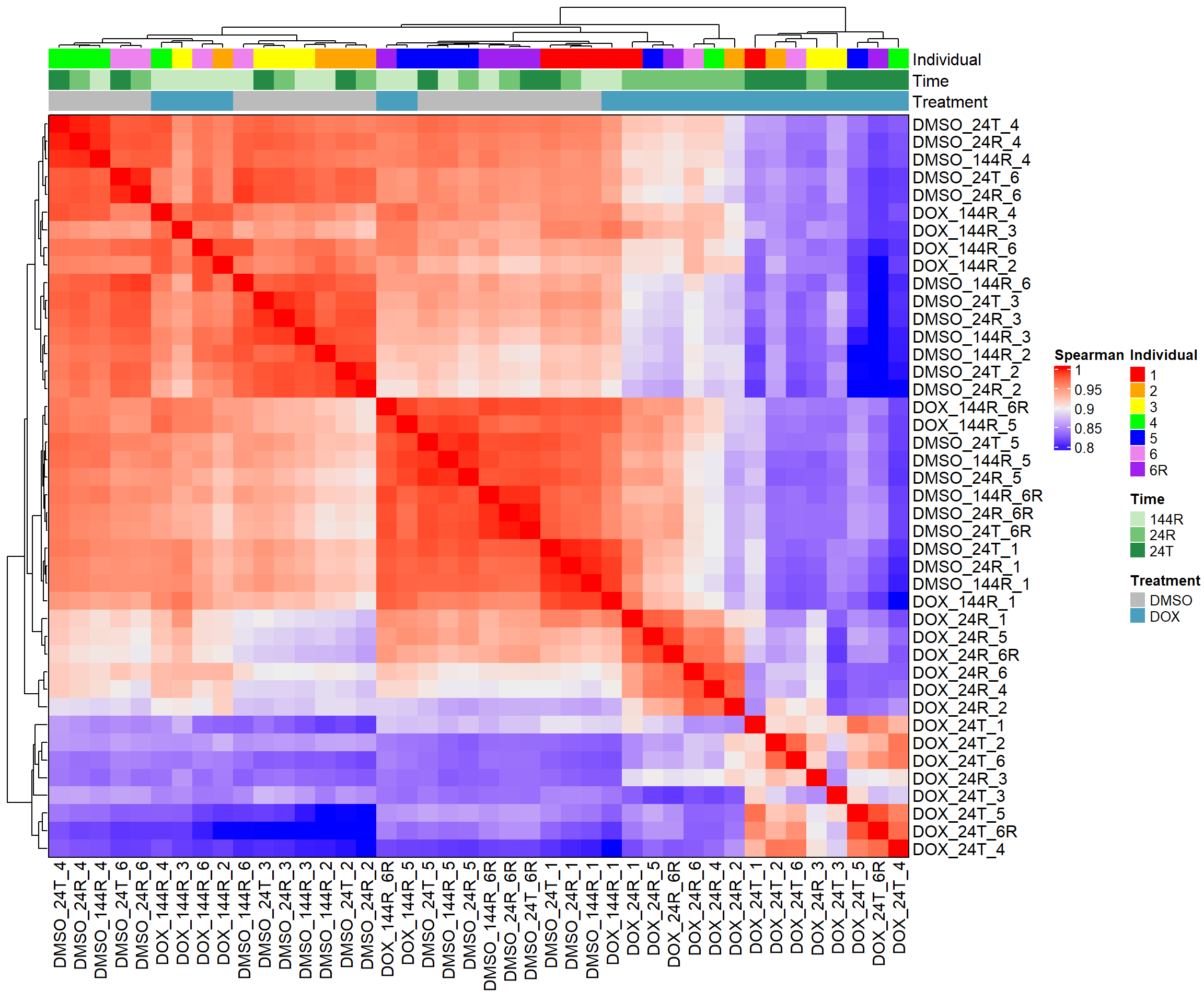
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
#Make my Filtered Gene List
#Now I want to make a filtered gene list (my rownames)
##I will use this to filter my counts for limma + Cormotif
filt_gene_list <- rownames(filcpm_matrix)
#save this filtered gene list as I'll use it to filter my counts
#saveRDS(filt_gene_list, "data/new/filt_gene_list.RDS")##Filter my Counts For DE Analysis
counts_raw_matrix <- readRDS("data/new/counts_raw_matrix.RDS")
#change column names to match samples for my raw counts matrix
colnames(counts_raw_matrix) <- c("DOX_24T_Ind1",
"DMSO_24T_Ind1",
"DOX_24R_Ind1",
"DMSO_24R_Ind1",
"DOX_144R_Ind1",
"DMSO_144R_Ind1",
"DOX_24T_Ind2",
"DMSO_24T_Ind2",
"DOX_24R_Ind2",
"DMSO_24R_Ind2",
"DOX_144R_Ind2",
"DMSO_144R_Ind2",
"DOX_24T_Ind3",
"DMSO_24T_Ind3",
"DOX_24R_Ind3",
"DMSO_24R_Ind3",
"DOX_144R_Ind3",
"DMSO_144R_Ind3",
"DOX_24T_Ind4",
"DMSO_24T_Ind4",
"DOX_24R_Ind4",
"DMSO_24R_Ind4",
"DOX_144R_Ind4",
"DMSO_144R_Ind4",
"DOX_24T_Ind5",
"DMSO_24T_Ind5",
"DOX_24R_Ind5",
"DMSO_24R_Ind5",
"DOX_144R_Ind5",
"DMSO_144R_Ind5",
"DOX_24T_Ind6",
"DMSO_24T_Ind6",
"DOX_24R_Ind6",
"DMSO_24R_Ind6",
"DOX_144R_Ind6",
"DMSO_144R_Ind6",
"DOX_24T_Ind6REP",
"DMSO_24T_Ind6REP",
"DOX_24R_Ind6REP",
"DMSO_24R_Ind6REP",
"DOX_144R_Ind6REP",
"DMSO_144R_Ind6REP")
#subset my count matrix based on filtered CPM matrix
x <- counts_raw_matrix[row.names(filcpm_matrix),]
dim(x)[1] 14319 42#14319 genes as expected!
#this is still in counts form
#remove my replicate individual at this time
x_norep <- x[,1:36]
#modify my metadata to match
Metadata_2 <- Metadata[1:36,]
rownames(Metadata_2) <- Metadata_2$Sample_bam
colnames(x_norep) <- Metadata_2$Sample_ID
rownames(Metadata_2) <- Metadata_2$Sample_ID
Metadata_2$Condition <- make.names(Metadata_2$Condition)
Metadata_2$Ind <- as.character(Metadata_2$Ind)
#saveRDS(Metadata_2, "data/new/Metadata_2_norep.RDS")#Perform Differential Expression Analysis
#create DGEList object
dge <- DGEList(counts = x_norep)
dge$samples$group <- factor(Metadata_2$Condition)
dge <- calcNormFactors(dge, method = "TMM")
#saveRDS(dge, "data/new/dge_matrix.RDS")
#check normalization factors from TMM normalization of LIBRARIES
dge$samples group lib.size norm.factors
84-1_DOX_24 DOX_24T 23393931 0.9745263
84-1_DMSO_24 DMSO_24T 22853195 0.9565797
84-1_DOX_24+24 DOX_24R 23846995 1.1659432
84-1_DMSO_24+24 DMSO_24R 21299355 0.9649641
84-1_DOX_24+144 DOX_144R 18222568 0.9913625
84-1_DMSO_24+144 DMSO_144R 28115884 0.9653464
87-1_DOX_24 DOX_24T 19935097 1.0526605
87-1_DMSO_24 DMSO_24T 21302879 0.9773889
87-1_DOX_24+24 DOX_24R 25636959 1.0751043
87-1_DMSO_24+24 DMSO_24R 26319662 0.9940323
87-1_DOX_24+144 DOX_144R 23463426 0.9003102
87-1_DMSO_24+144 DMSO_144R 25840938 0.9888449
78-1_DOX_24 DOX_24T 23085807 0.7676077
78-1_DMSO_24 DMSO_24T 25610495 1.0077383
78-1_DOX_24+24 DOX_24R 18083930 1.1682704
78-1_DMSO_24+24 DMSO_24R 24331177 0.9906872
78-1_DOX_24+144 DOX_144R 19754391 0.9941834
78-1_DMSO_24+144 DMSO_144R 22641509 1.0010734
75-1_DOX_24 DOX_24T 20583626 1.0676786
75-1_DMSO_24 DMSO_24T 28166198 1.0031906
75-1_DOX_24+24 DOX_24R 25831427 1.1530208
75-1_DMSO_24+24 DMSO_24R 26081158 1.0058953
75-1_DOX_24+144 DOX_144R 24659898 0.9261599
75-1_DMSO_24+144 DMSO_144R 25412931 0.9703454
17-3_DOX_24 DOX_24T 22518848 0.9766893
17-3_DMSO_24 DMSO_24T 24589534 0.9612345
17-3_DOX_24+24 DOX_24R 24797547 1.1703079
17-3_DMSO_24+24 DMSO_24R 25977536 0.9509690
17-3_DOX_24+144 DOX_144R 27447106 0.9422729
17-3_DMSO_24+144 DMSO_144R 24893583 0.9356377
90-1_DOX_24 DOX_24T 25187428 1.0311957
90-1_DMSO_24 DMSO_24T 25630519 1.0283437
90-1_DOX_24+24 DOX_24R 26138399 1.1183471
90-1_DMSO_24+24 DMSO_24R 24430396 0.9988688
90-1_DOX_24+144 DOX_144R 23323463 0.9496884
90-1_DMSO_24+144 DMSO_144R 25424152 0.9872926#create my design matrix for DE
design <- model.matrix(~ 0 + Metadata_2$Condition)
colnames(design) <- gsub("Metadata_2\\$Condition", "", colnames(design))
#take care that the matrix automatically sorts cols alphabetically
##currently DMSO144R, DMSO24R, DMSO24T, DOX144R, DOX24R, DOX24T
#run duplicate correlation for individual effect
corfit <- duplicateCorrelation(object = dge$counts, design = design, block = Metadata_2$Ind)
#voom transformation and plot
v <- voom(dge, design, block = Metadata_2$Ind, correlation = corfit$consensus.correlation, plot = TRUE)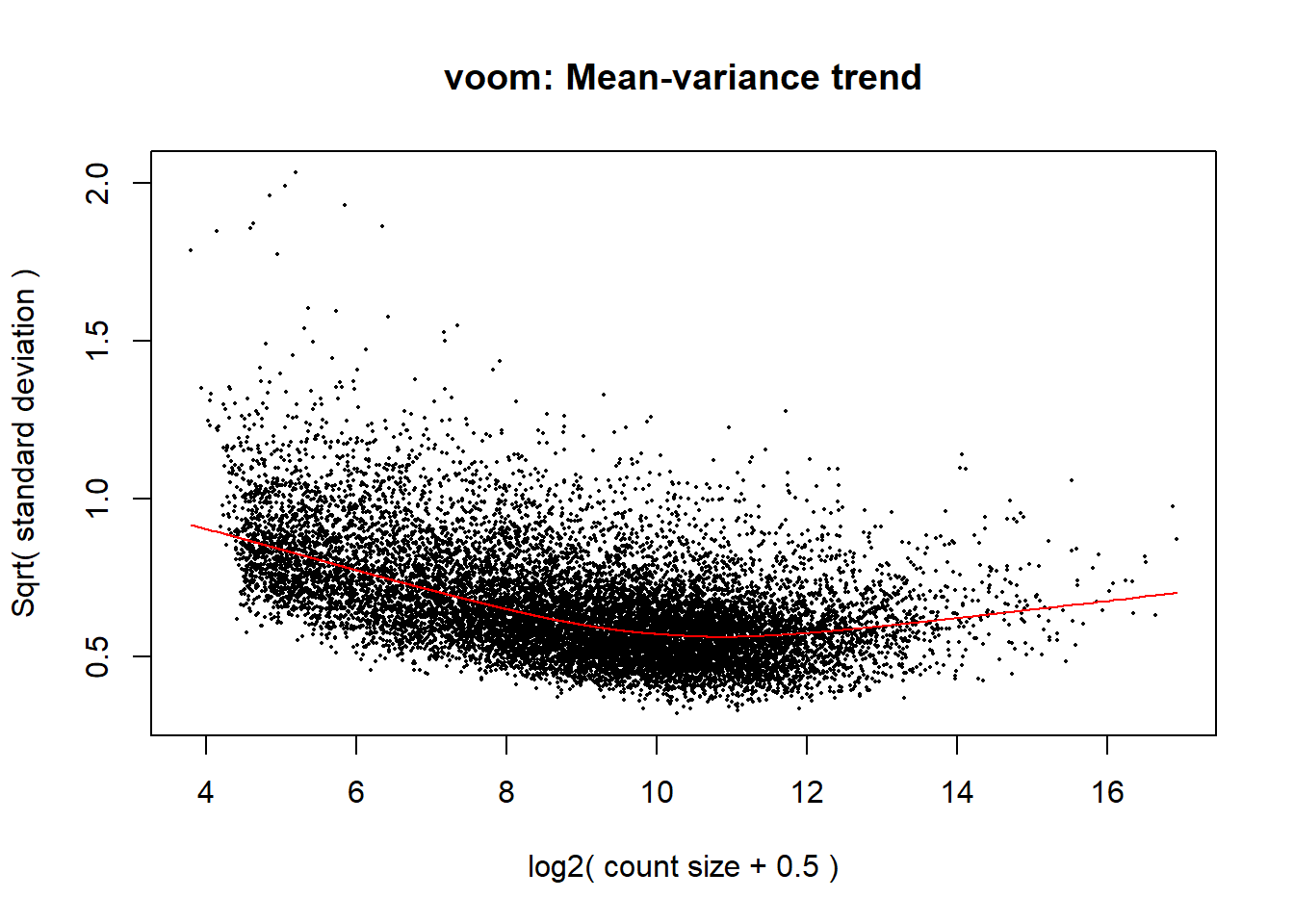
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
#fit my linear model
fit <- lmFit(v, design, block = Metadata_2$Ind, correlation = corfit$consensus.correlation)
#make my contrast matrix to compare across tx and veh
contrast_matrix <- makeContrasts(
V.D24T = DOX_24T - DMSO_24T,
V.D24R = DOX_24R - DMSO_24R,
V.D144R = DOX_144R - DMSO_144R,
levels = design
)
#apply these contrasts to compare DOX to DMSO VEH
fit2 <- contrasts.fit(fit, contrast_matrix)
fit2 <- eBayes(fit2)
#plot the mean-variance trend
plotSA(fit2, main = "Final model: Mean-Variance trend")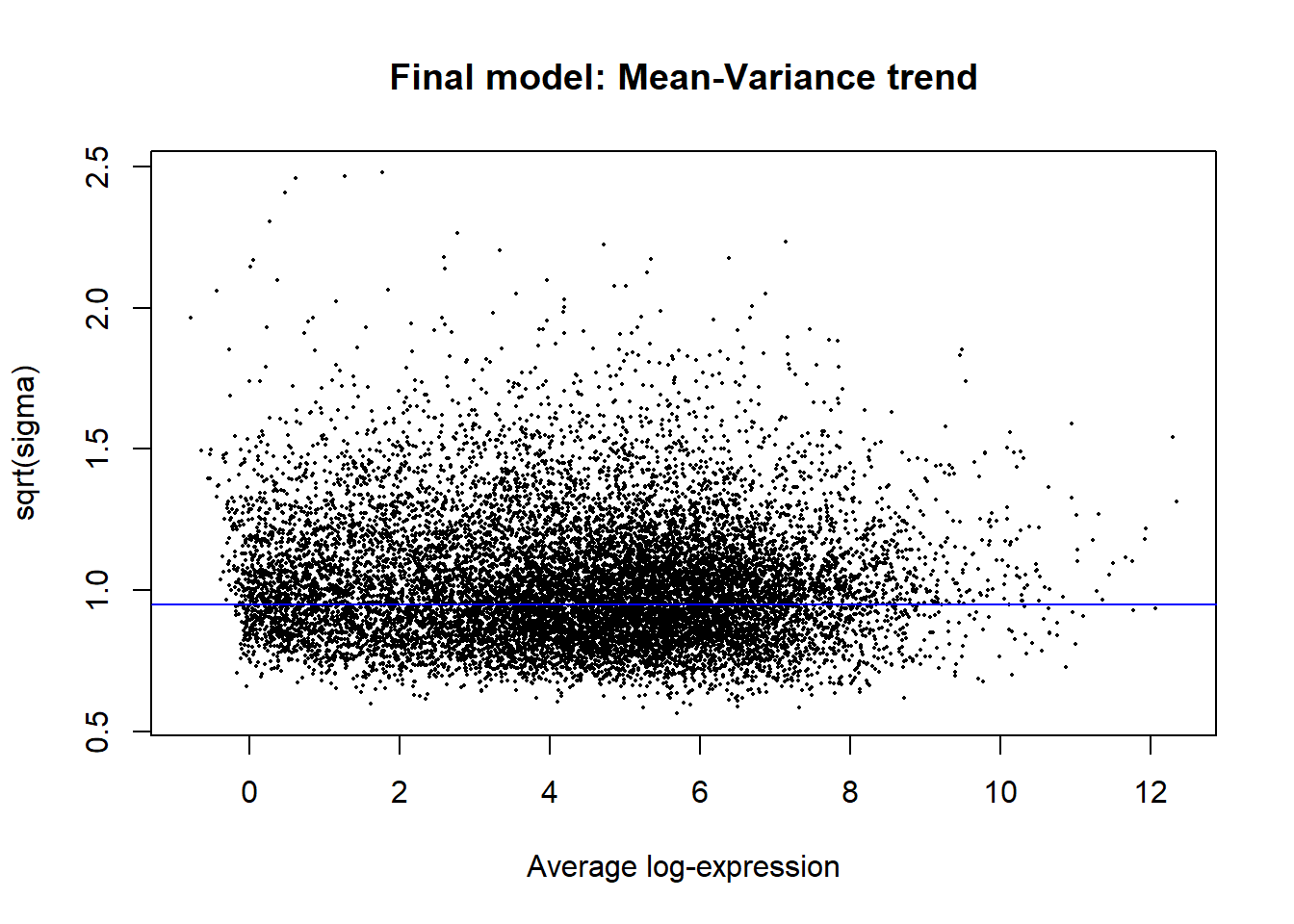
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
#look at the summary of your results
##this tells you the number of DEGs in each condition
results_summary <- decideTests(fit2, adjust.method = "BH", p.value = 0.05)
summary(results_summary) V.D24T V.D24R V.D144R
Down 4723 3593 359
NotSig 5076 7151 13810
Up 4520 3575 150# V.D24 V.D24r V.D144r
# Down 4723 3593 359
# NotSig 5076 7151 13810
# Up 4520 3575 150
vennDiagram(object = results_summary, include = c("up", "down"))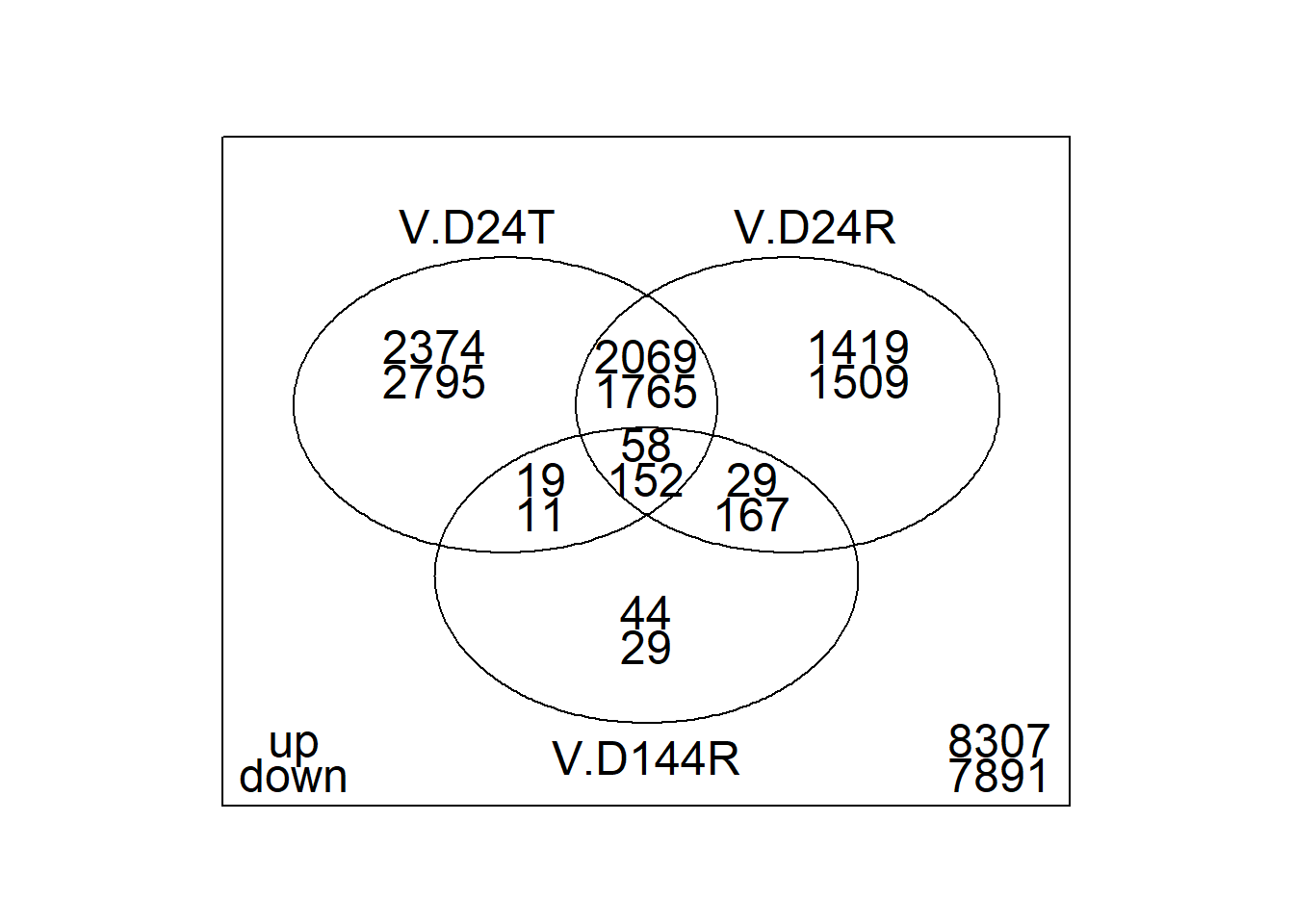
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
##Create Toptables of my DEGs
# Generate Top Table for Specific Comparisons
Toptable_V.D24T <- topTable(fit = fit2, coef = "V.D24T", number = nrow(x), adjust.method = "BH", p.value = 1, sort.by = "none")
#write.csv(Toptable_V.D24T, "data/new/DEGs/Toptable_V.D24T.csv")
Toptable_V.D24R <- topTable(fit = fit2, coef = "V.D24R", number = nrow(x), adjust.method = "BH", p.value = 1, sort.by = "none")
#write.csv(Toptable_V.D24R, "data/new/DEGs/Toptable_V.D24R.csv")
Toptable_V.D144R <- topTable(fit = fit2, coef = "V.D144R", number = nrow(x), adjust.method = "BH", p.value = 1, sort.by = "none")
#write.csv(Toptable_V.D144R, "data/new/DEGs/Toptable_V.D144R.csv")
#save all of these toptables as R objects
# saveRDS(list(
# V.D24T = Toptable_V.D24T,
# V.D24R = Toptable_V.D24R,
# V.D144R = Toptable_V.D144R
# ), file = "data/new/Toptable_list.RDS")
Toptable_list <- readRDS("data/new/Toptable_list.RDS")##Top 5 DEGs after DE Analysis per Condition
#use your three toptables so I can pull out top 5 genes from each based on adj. p val
top5_D24T <- Toptable_V.D24T[order(Toptable_V.D24T$adj.P.Val), ][1:5,] %>%
rownames_to_column(., var = "Entrez_ID")
top5_D24R <- Toptable_V.D24R[order(Toptable_V.D24R$adj.P.Val), ][1:5,] %>%
rownames_to_column(., var = "Entrez_ID")
top5_D144R <- Toptable_V.D144R[order(Toptable_V.D144R$adj.P.Val), ][1:5,] %>%
rownames_to_column(., var = "Entrez_ID")
#now that I've pulled the top 5 DEGs from each, make a list to pull them from my log2cpm data
boxplot1 <- read.csv("data/new/filcpm_final_matrix.csv") %>%
as.data.frame()
#Define gene list
#these are the top 5 genes pulled from my toptables
top5_D24T_geneslist <- c(top5_D24T$Entrez_ID)
top5_D24R_geneslist <- c(top5_D24R$Entrez_ID)
top5_D144R_geneslist <- c(top5_D144R$Entrez_ID)
#Add more gene symbols as needed or add more categories
#now pull these from my log2cpm matrix
top5_D24T_genes <- boxplot1[boxplot1$Entrez_ID %in% top5_D24T_geneslist,]
dim(top5_D24T_genes)[1] 5 44#5 genes in 44 cols
top5_D24R_genes <- boxplot1[boxplot1$Entrez_ID %in% top5_D24R_geneslist,]
dim(top5_D24R_genes)[1] 5 44#5 genes in 44 cols
top5_D144R_genes <- boxplot1[boxplot1$Entrez_ID %in% top5_D144R_geneslist,]
dim(top5_D144R_genes)[1] 5 44#5 genes in 44 cols
#Now put in the function I want to use to generate boxplots of genes
#####D24T#####
process_top5_D24T <- function(gene) {
gene_data <- top5_D24T_genes %>% filter(Entrez_ID == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2cpm") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6R$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#Generate Boxplots from the above function using our gene list above
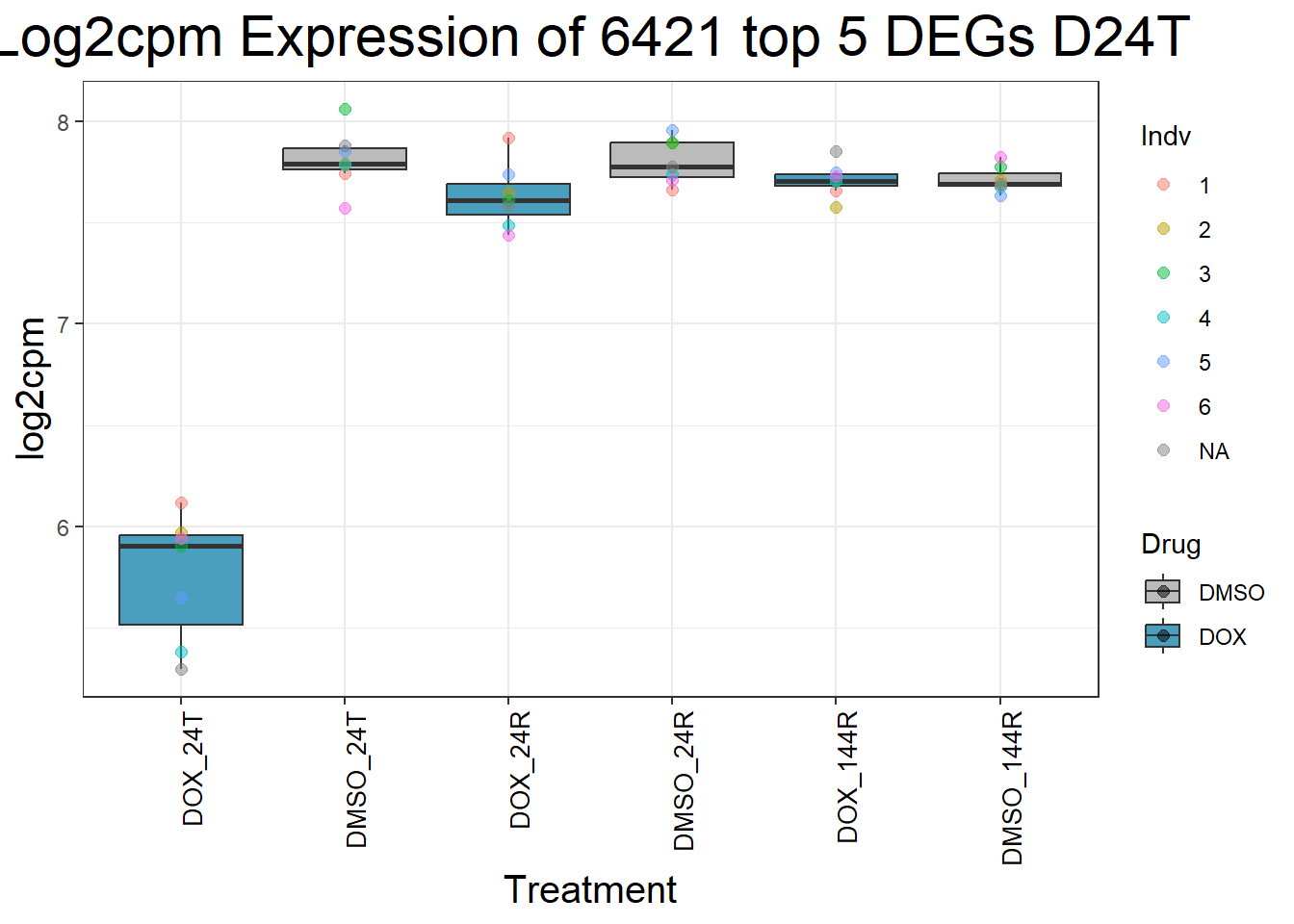
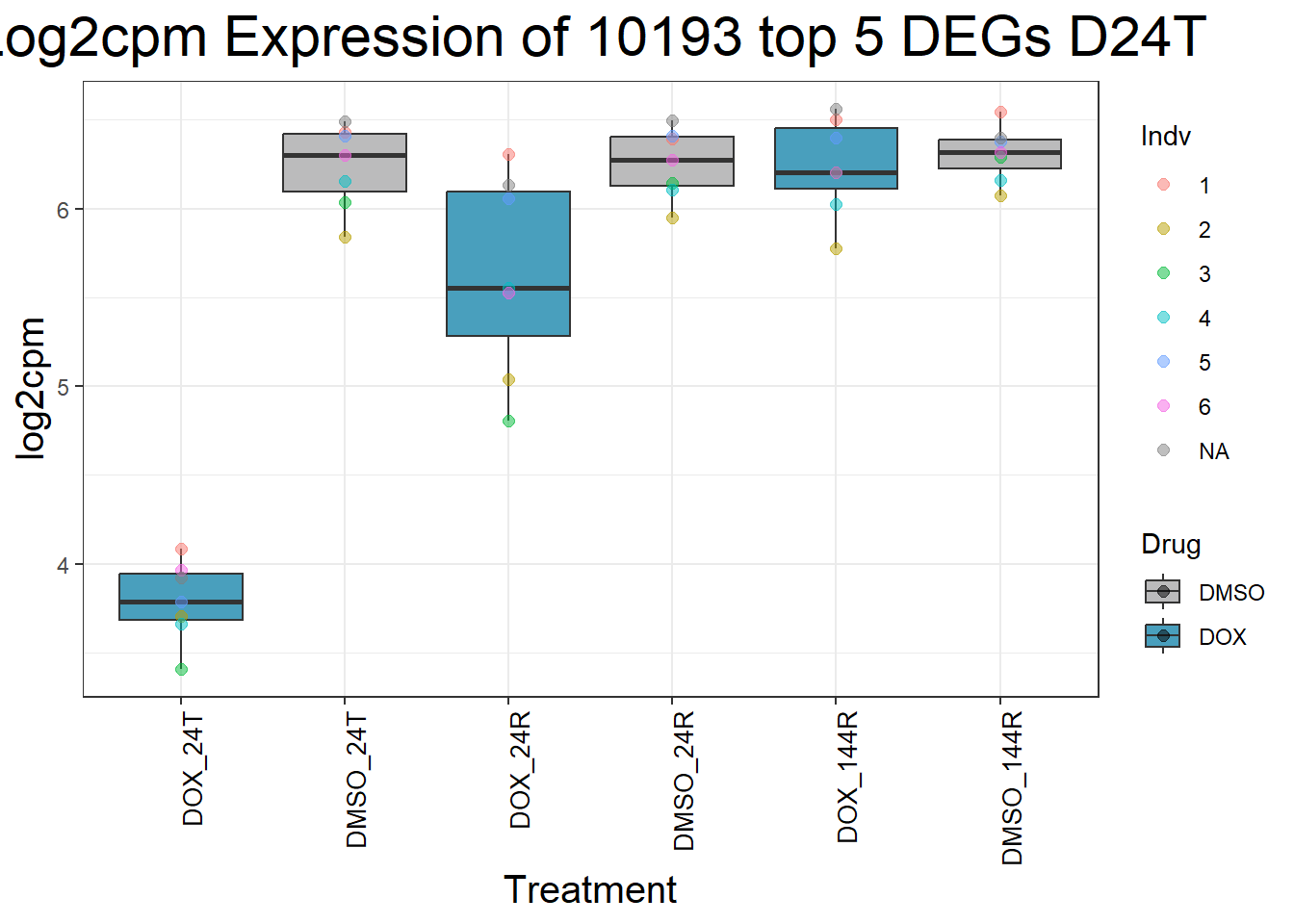
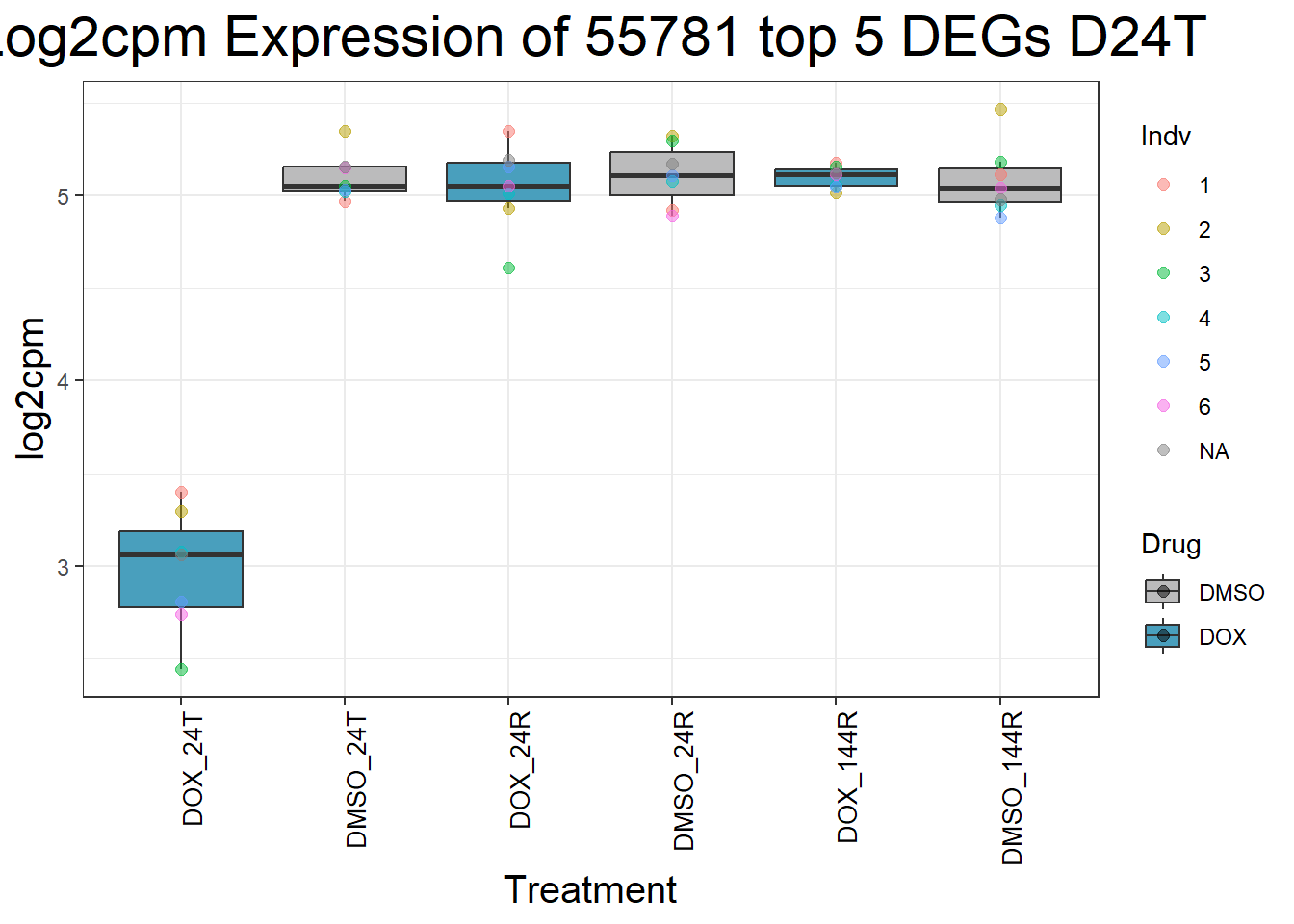
for (gene in top5_D24T_geneslist) {
gene_data <- process_top5_D24T(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2cpm, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2cpm Expression of", gene, "top 5 DEGs D24T")) +
labs(x = "Treatment", y = "log2cpm") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
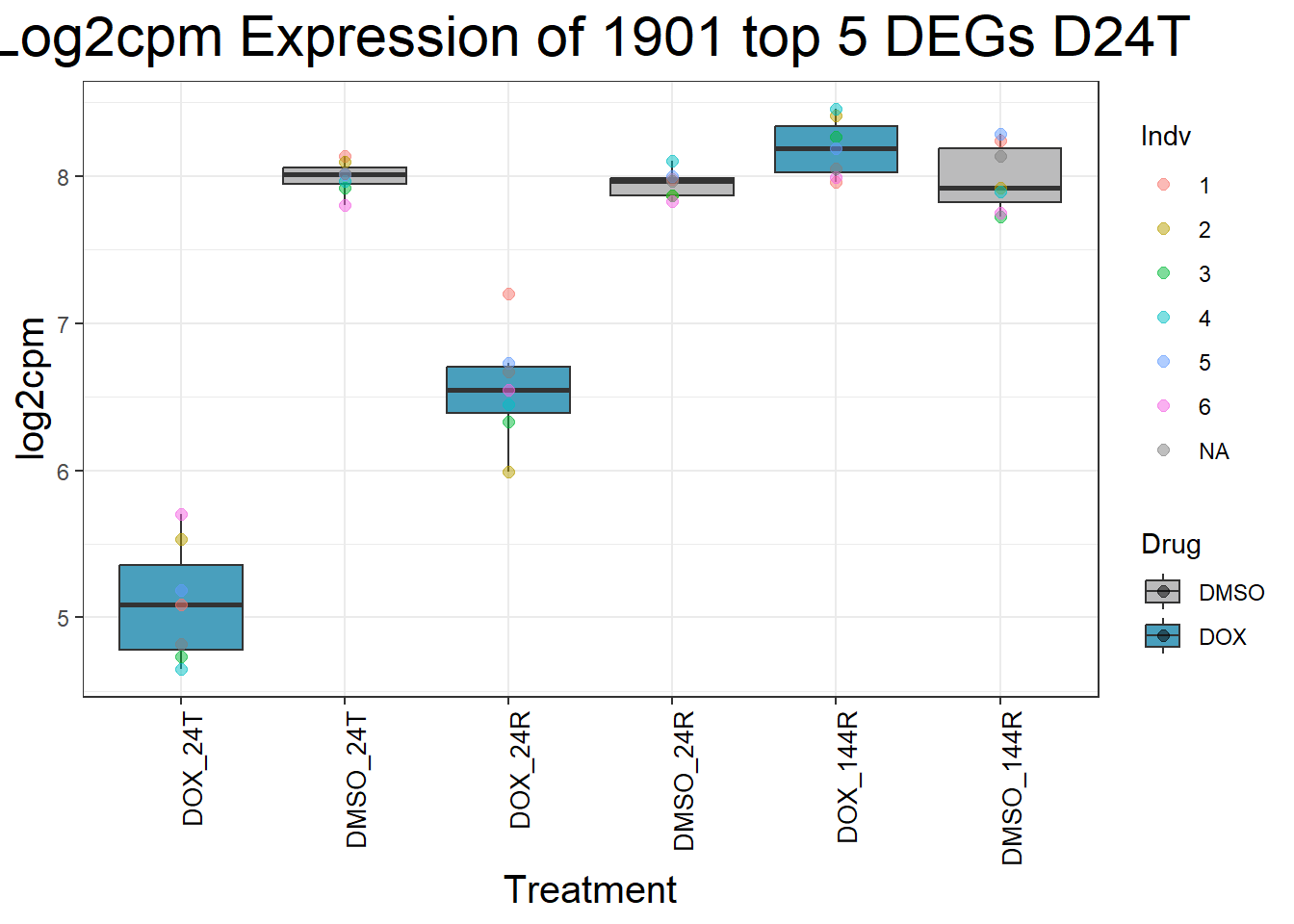
#####D24R#####
process_top5_D24R <- function(gene) {
gene_data <- top5_D24R_genes %>% filter(Entrez_ID == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2cpm") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6R$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#Generate Boxplots from the above function using our gene list above
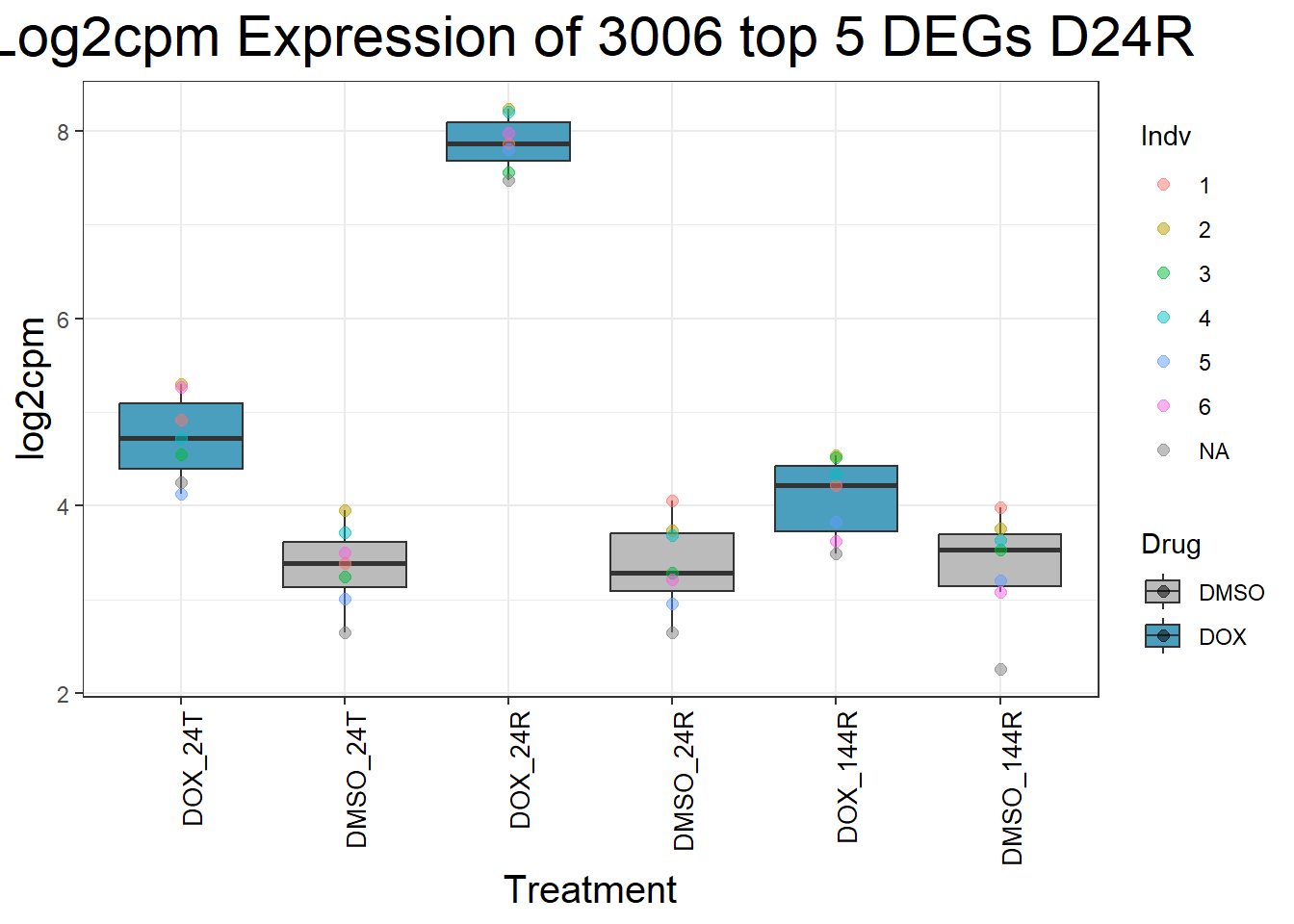
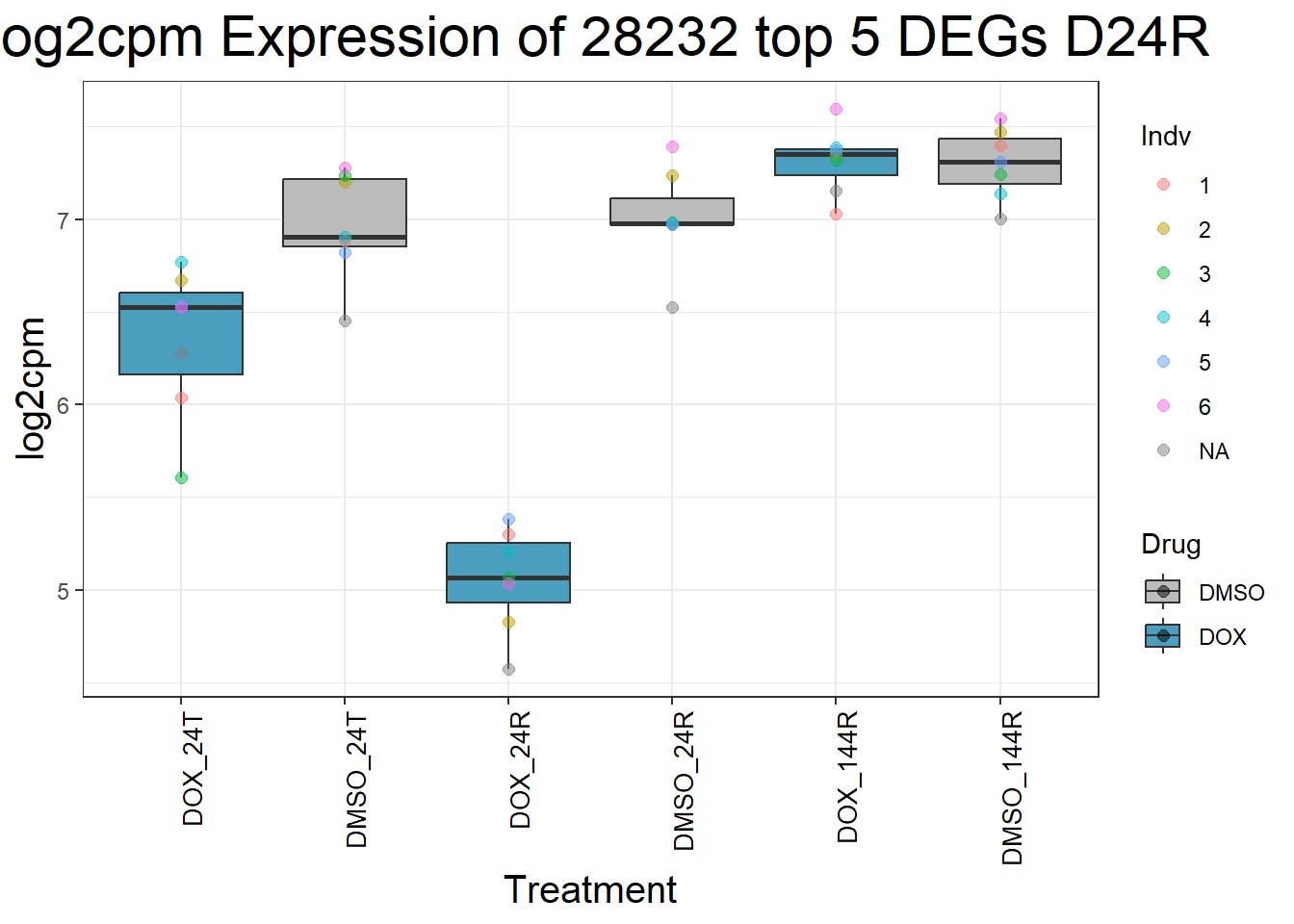
for (gene in top5_D24R_geneslist) {
gene_data <- process_top5_D24R(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2cpm, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2cpm Expression of", gene, "top 5 DEGs D24R")) +
labs(x = "Treatment", y = "log2cpm") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
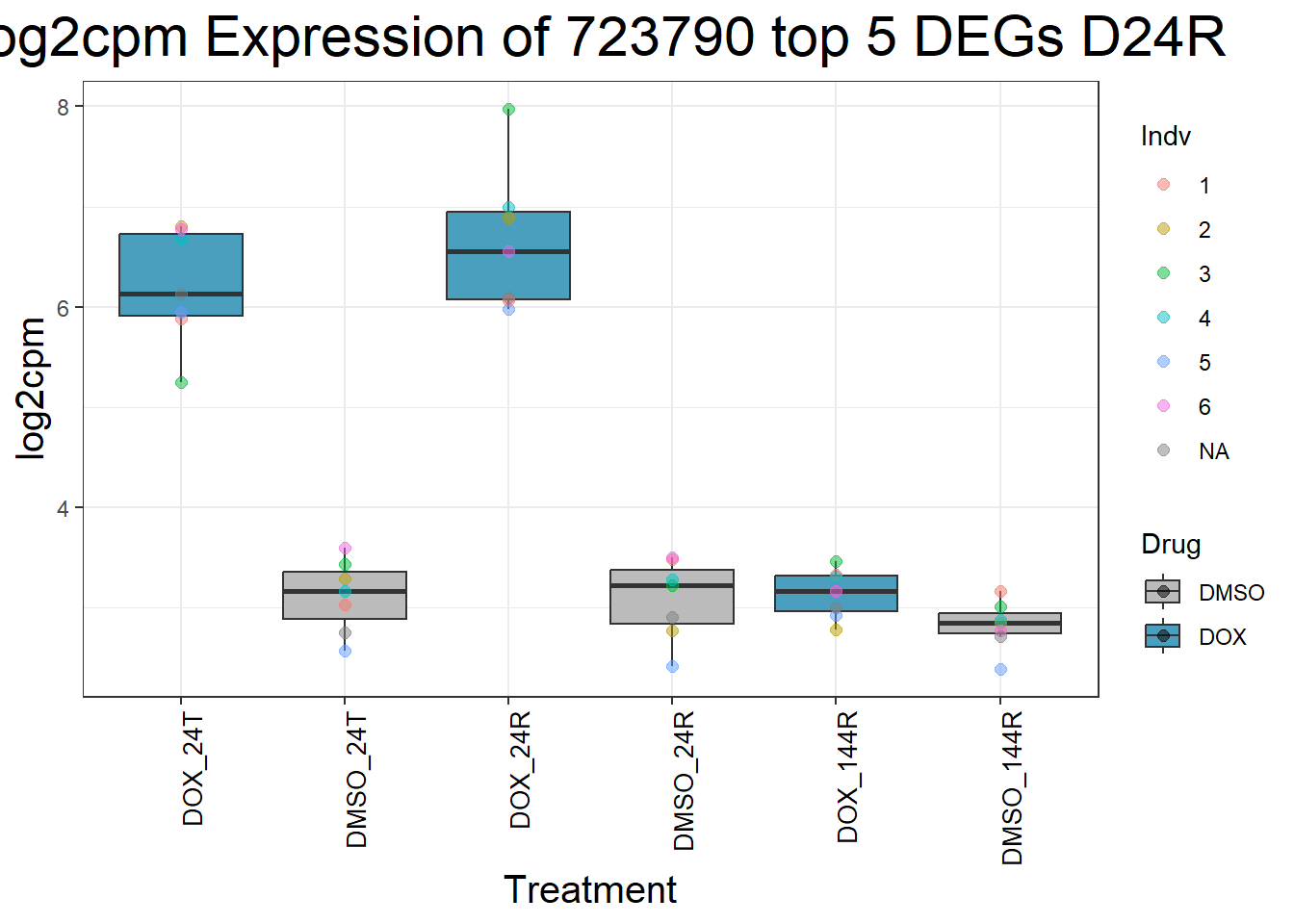
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
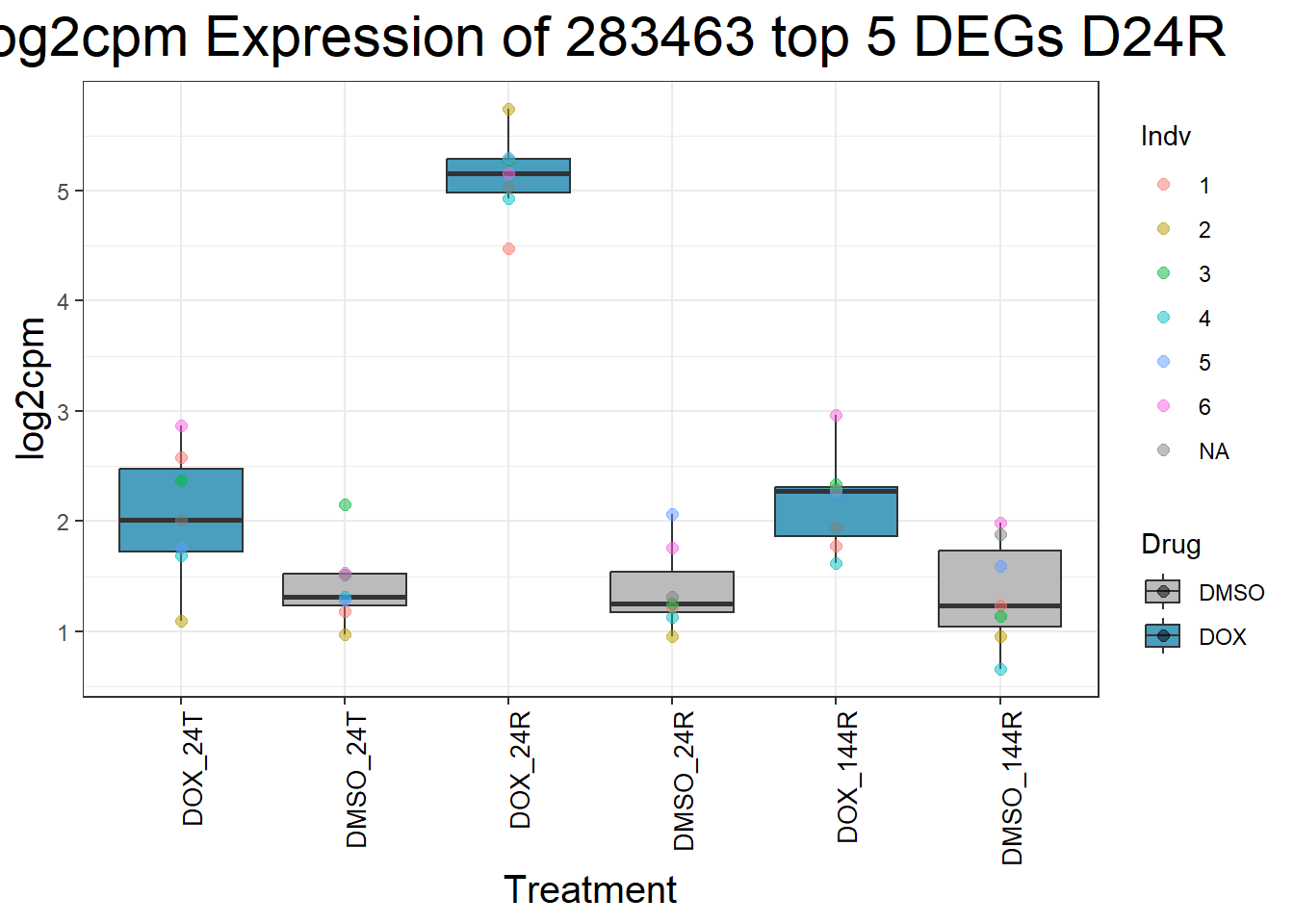
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
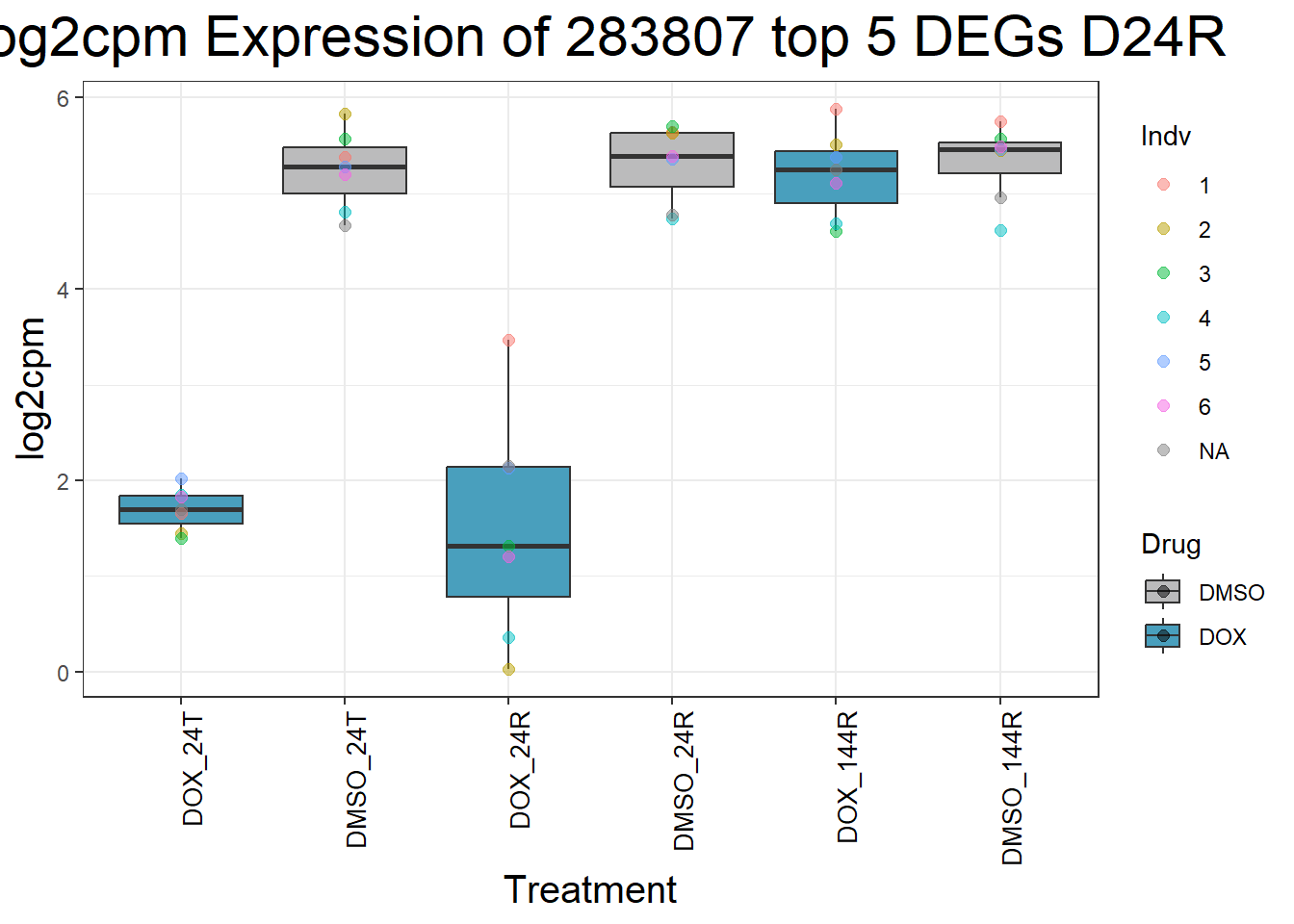
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
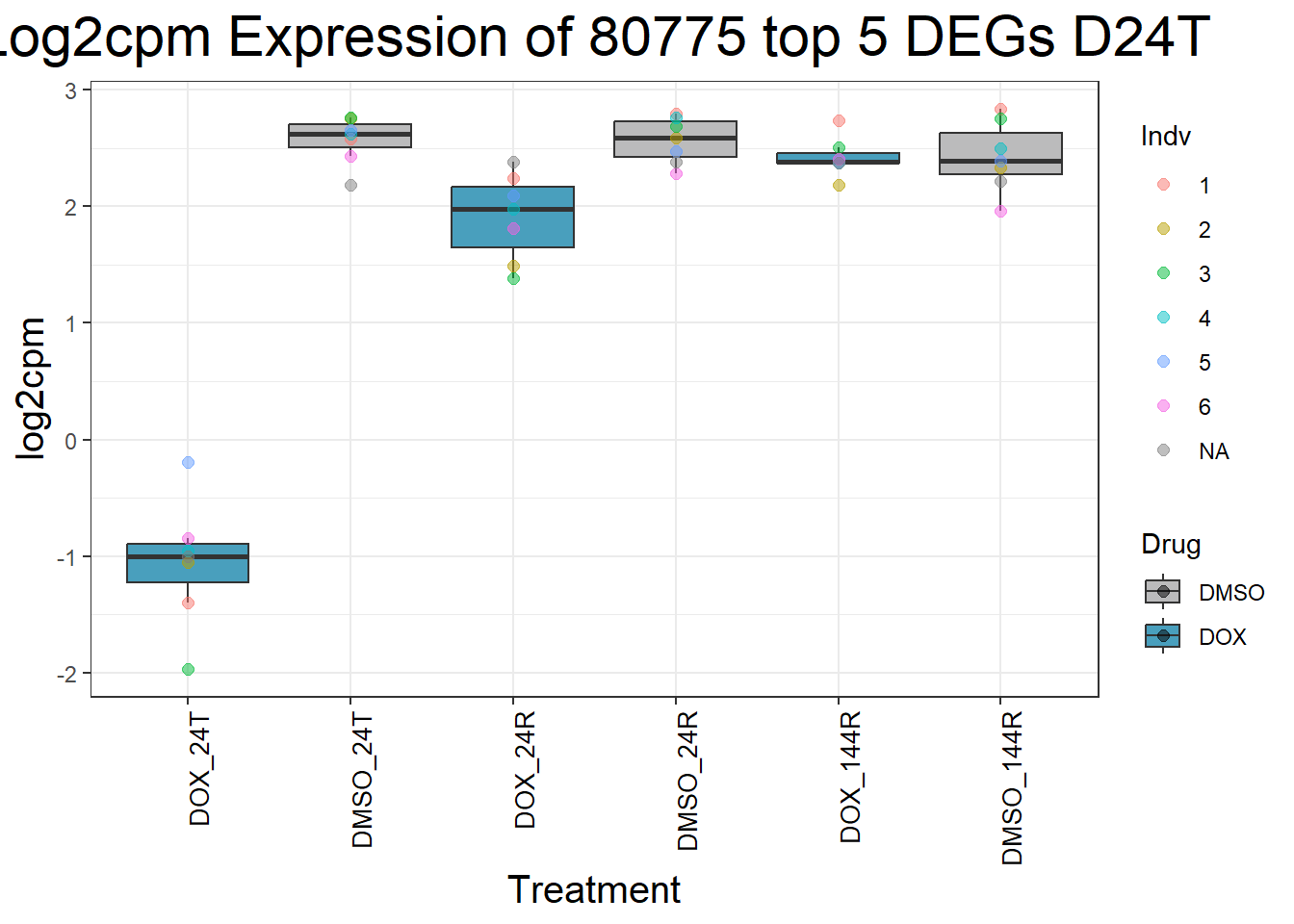
#####D144R#####
process_top5_D144R <- function(gene) {
gene_data <- top5_D144R_genes %>% filter(Entrez_ID == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2cpm") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6R$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#Generate Boxplots from the above function using our gene list above
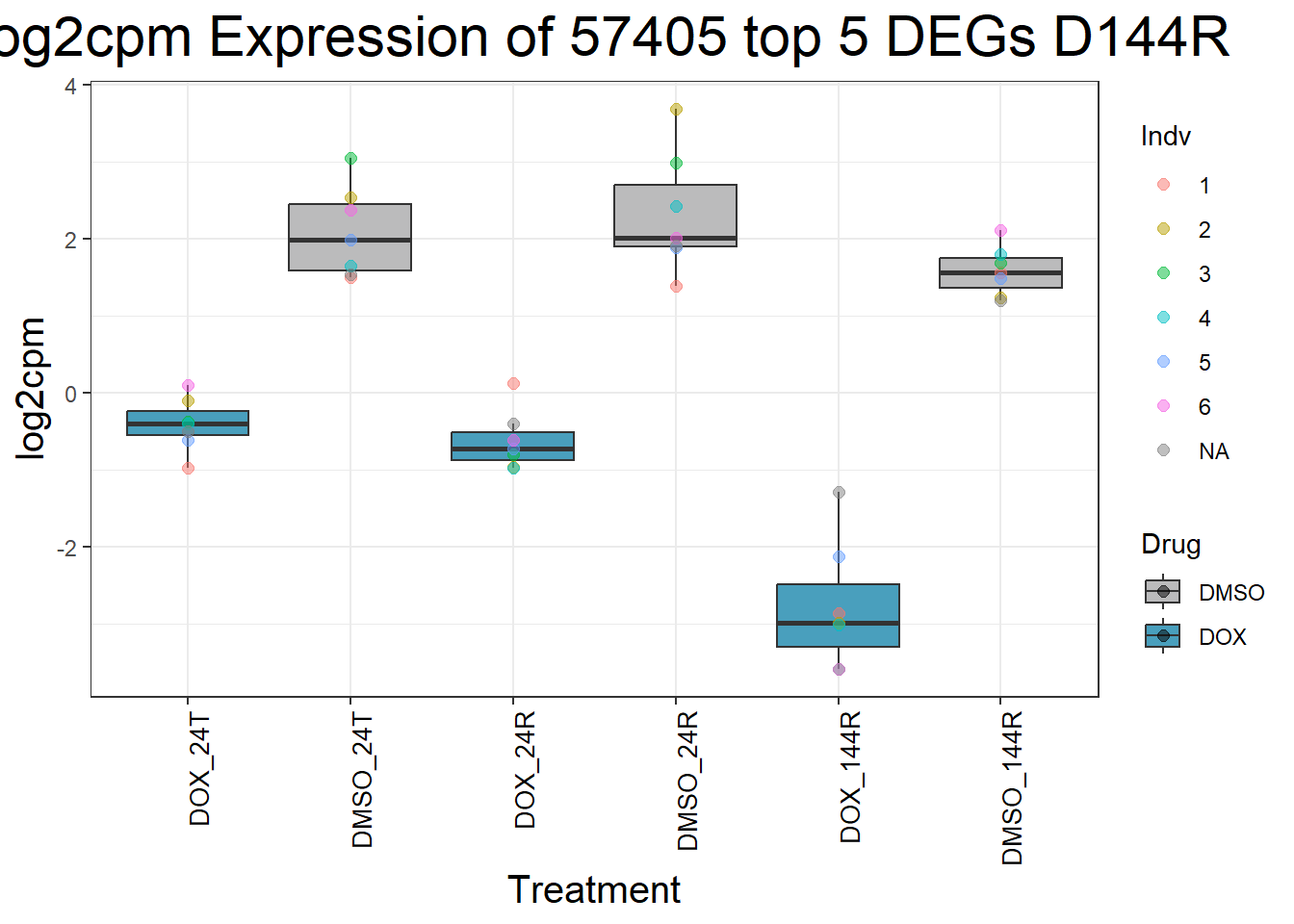
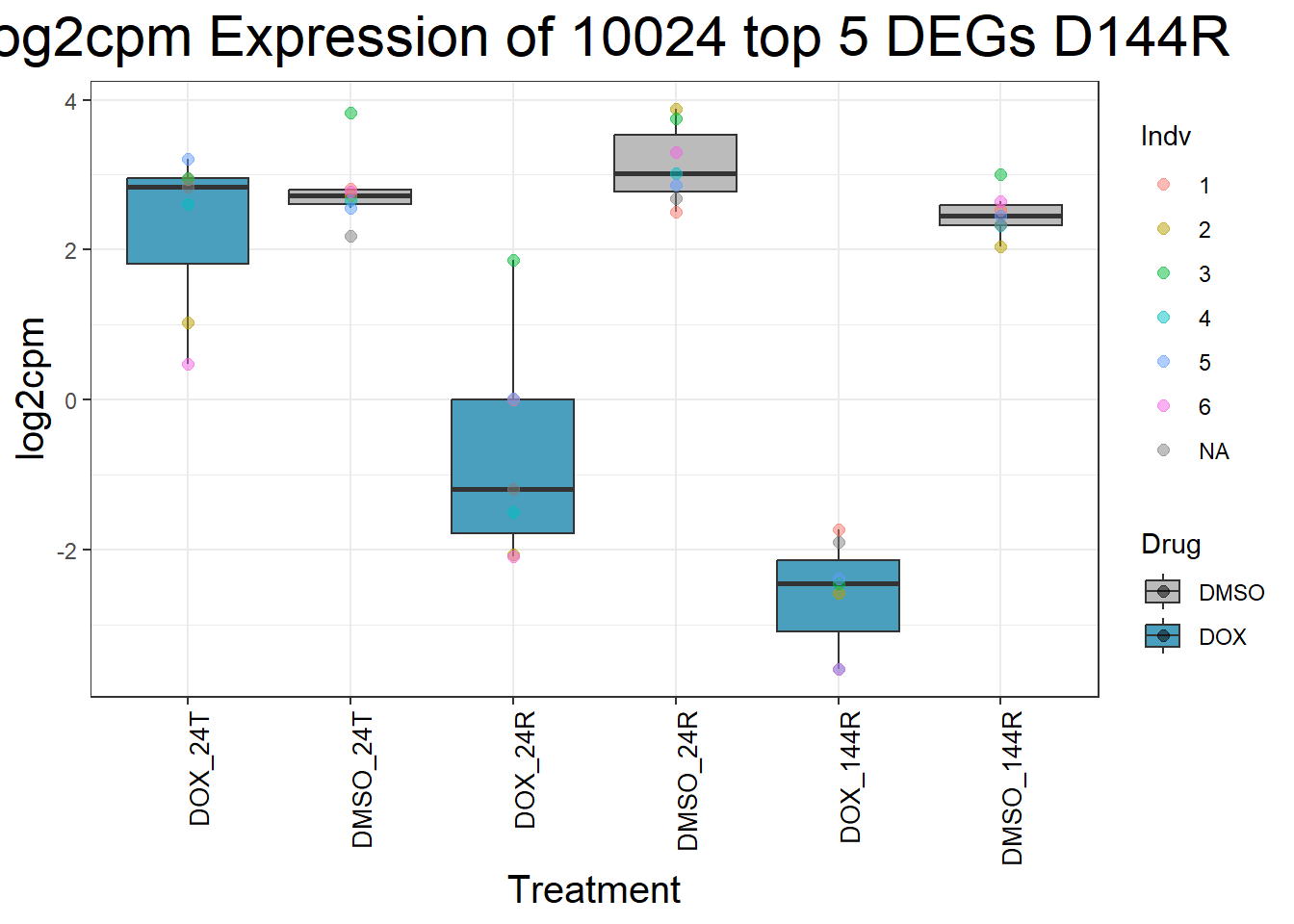
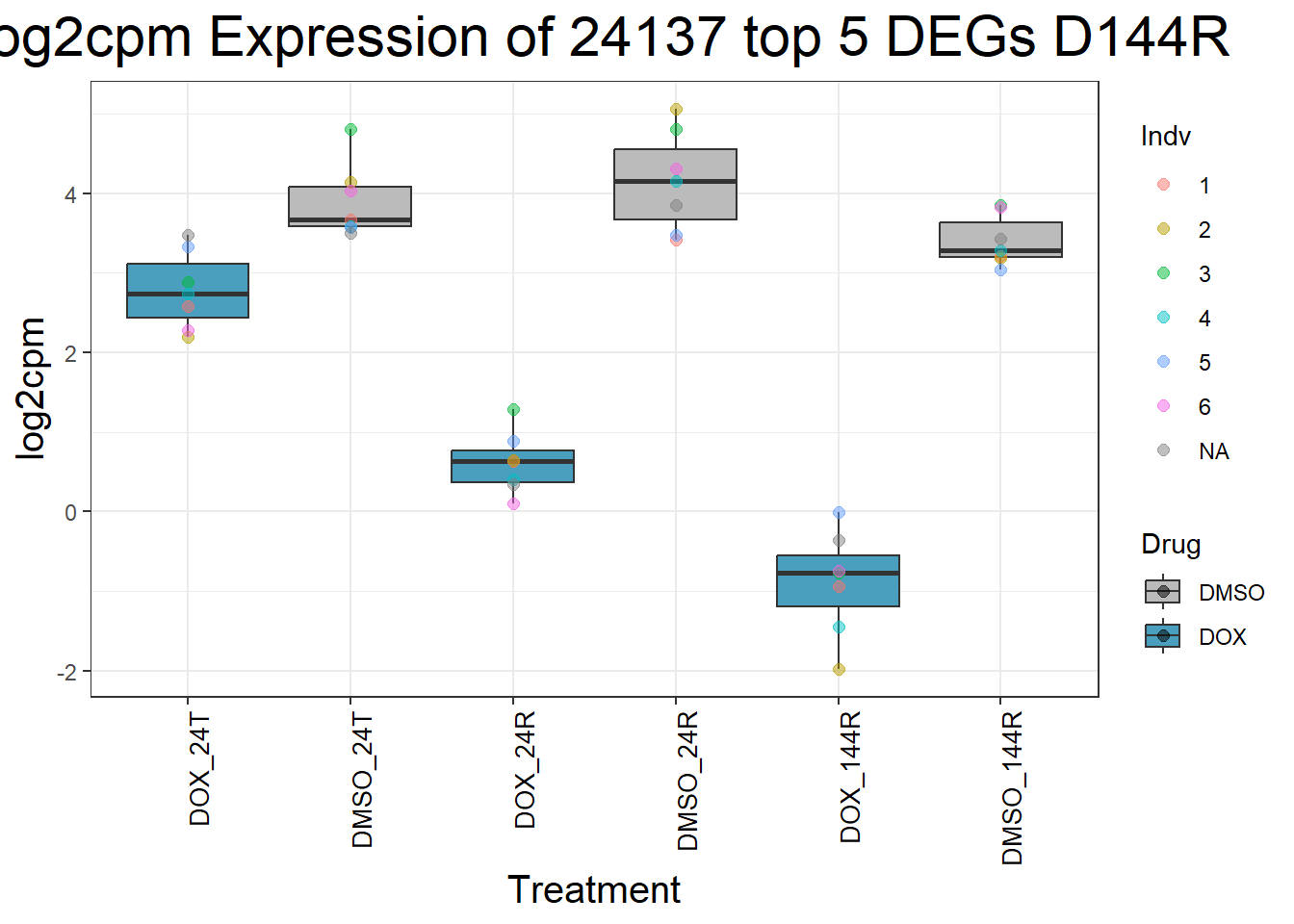
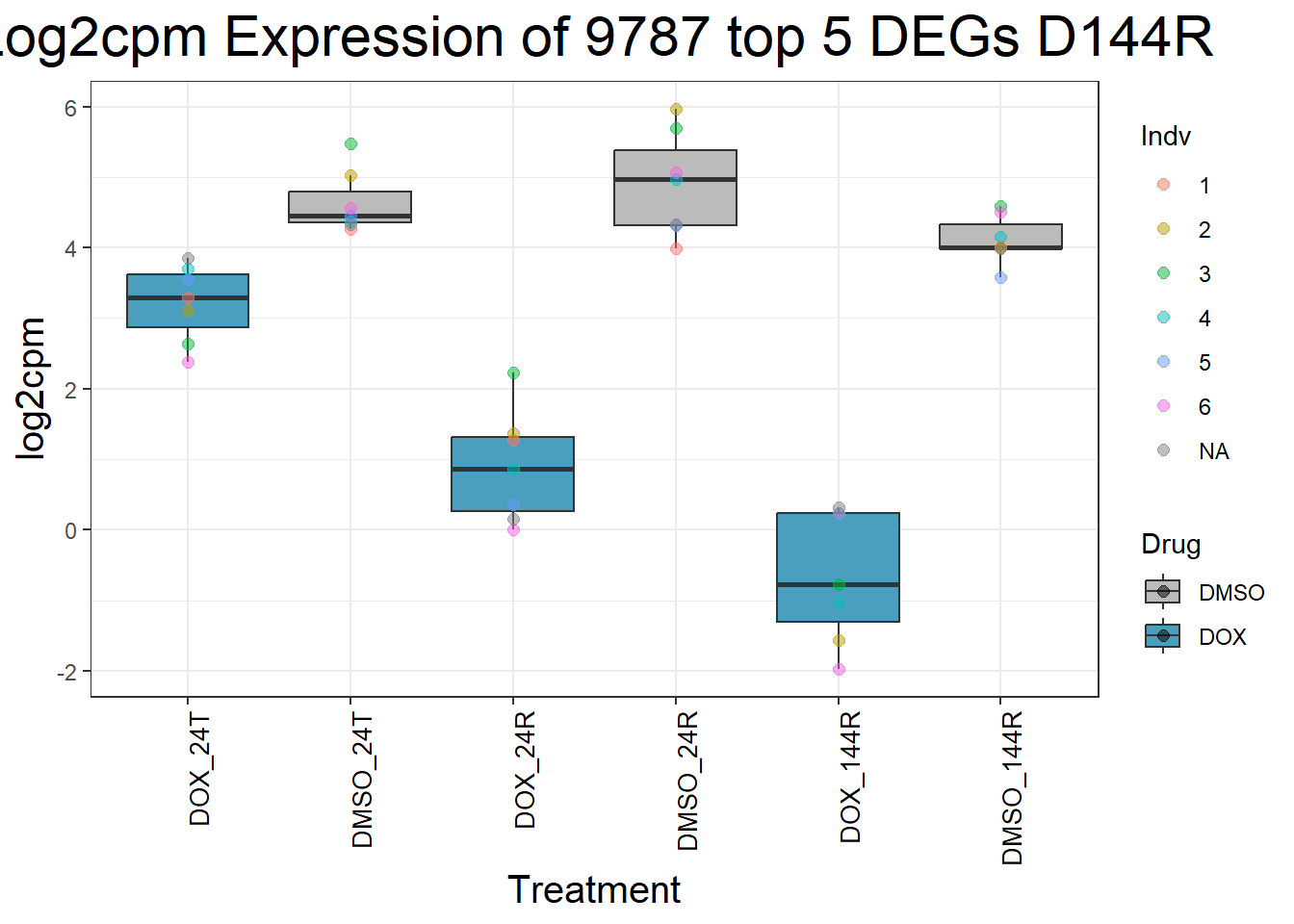
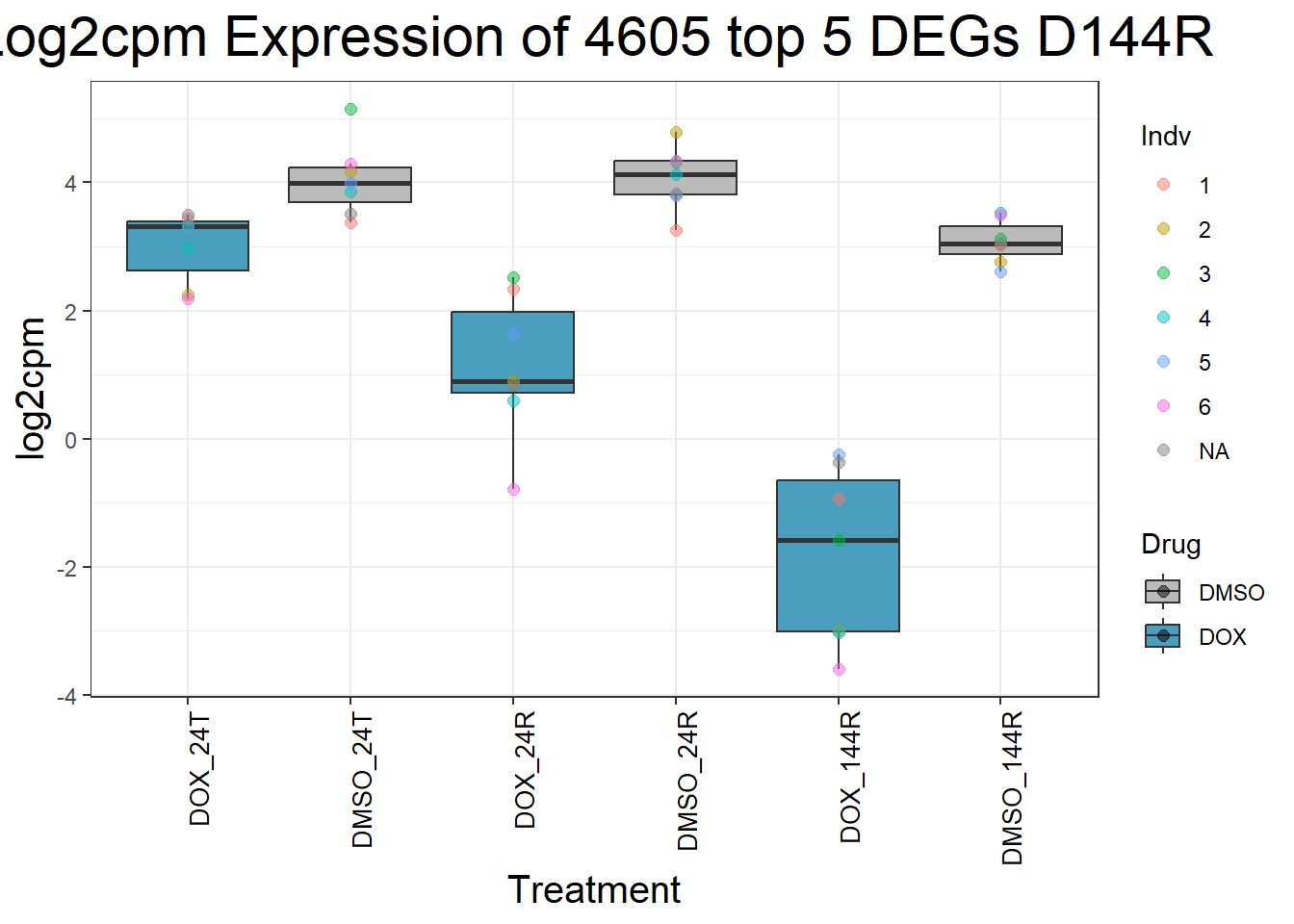
for (gene in top5_D144R_geneslist) {
gene_data <- process_top5_D144R(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2cpm, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2cpm Expression of", gene, "top 5 DEGs D144R")) +
labs(x = "Treatment", y = "log2cpm") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
##LogFC Boxplots for All Conditions
# Toptable_V.D24T
# Load DEGs Data
DOX_24T <- read.csv("data/new/DEGs/Toptable_V.D24T.csv")
DOX_24R <- read.csv("data/new/DEGs/Toptable_V.D24R.csv")
DOX_144R <- read.csv("data/new/DEGs/Toptable_V.D144R.csv")
#make a list of all of the genes in this set so I can plot the logFC in other sets
D24T_DEGs <- DOX_24T$Entrez_ID[DOX_24T$adj.P.Val < 0.05]
length(D24T_DEGs)[1] 9243#9243 genes in length after adj. p value cutoff
#if I did p value only - 9658 genes
#now that I have a list of my DEGs from D24T - pull these genes out from the other DEG lists
# D24R_DEGs <- Toptable_V.D24R[Toptable_V.D24R$adj.P.Val < 0.05,]
# D24R_DEGs_D24T <- D24R_DEGs %>%
# rownames_to_column(., var = "Entrez_ID")
# D24R_DEGs_D24T <- D24R_DEGs_D24T[D24R_DEGs_D24T$Entrez_ID %in% D24T_DEGs,]
# dim(D24R_DEGs_D24T)
#4831 genes in common here after adj p value cutoff
# D144R_DEGs <- Toptable_V.D144R[Toptable_V.D144R$adj.P.Val < 0.05,]
#
# D144R_DEGs_D24T <- D144R_DEGs %>%
# rownames_to_column(., var = "Entrez_ID")
#
# D144R_DEGs_D24T <- D144R_DEGs_D24T[D144R_DEGs_D24T$Entrez_ID %in% D24T_DEGs,]
# dim(D144R_DEGs_D24T)
#322 genes in common after adj p value cutoff
#now I want to plot the logFC of these
#ignore the above for now, just plot those full gene sets in logFC
D24T_DEGs <- DOX_24T$Entrez_ID[DOX_24T$adj.P.Val < 0.05]
length(D24T_DEGs)[1] 9243D24R_DEGs <- DOX_24R$Entrez_ID[DOX_24R$adj.P.Val < 0.05]
length(D24R_DEGs)[1] 7168D144R_DEGs <- DOX_144R$Entrez_ID[DOX_144R$adj.P.Val < 0.05]
length(D144R_DEGs)[1] 509#now that I have the full list of genes, I want to plot the logFC across conditions
#to do this - make a combined toptable
# Toptable_list
#plot the set of genes across conditions
#
# logFC_long_allsets <- imap_dfr(Toptable_list, function(tbl, condition) {
# tbl %>%
# rownames_to_column(var = "Entrez_ID") %>%
# dplyr::select(Entrez_ID, logFC) %>%
# mutate(Condition = condition)
# })
#
#
# ggplot(logFC_long_allsets, aes(x = Condition, y = logFC, group = Entrez_ID, color = Entrez_ID)) +
# geom_boxplot() +
# geom_point(size = 2) +
# labs(
# title = "logFC of Genes Across Conditions",
# x = "Condition",
# y = "logFC"
# ) +
# theme_minimal()
####Try this####
#Combine the toptables I have from pairwise analysis into a single dataframe
d24_toptable_dxr <- Toptable_V.D24T %>%
rownames_to_column(var = "Entrez_ID") %>%
mutate(Time = "24")
d24r_toptable_dxr <- Toptable_V.D24R %>%
rownames_to_column(var = "Entrez_ID") %>%
mutate(Time = "24R")
d144r_toptable_dxr <- Toptable_V.D144R %>%
rownames_to_column(var = "Entrez_ID") %>%
mutate(Time = "144R")
combined_toptables_dxr <- bind_rows(
d24_toptable_dxr,
d24r_toptable_dxr,
d144r_toptable_dxr)
#Filter the data based on each motif
filt_toptable_dxr <- combined_toptables_dxr %>%
dplyr::filter(Entrez_ID %in% D24T_DEGs) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC for D24T DEGs Across Conditions")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#now do the same with the other conditions
filt_toptable_dxr_24r <- combined_toptables_dxr %>%
dplyr::filter(Entrez_ID %in% D24R_DEGs) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC for D24R DEGs Across Conditions")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#D144R
filt_toptable_dxr_144r <- combined_toptables_dxr %>%
dplyr::filter(Entrez_ID %in% D144R_DEGs) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC for D144R DEGs Across Conditions")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#now put the names of these graphs to print them
filt_toptable_dxr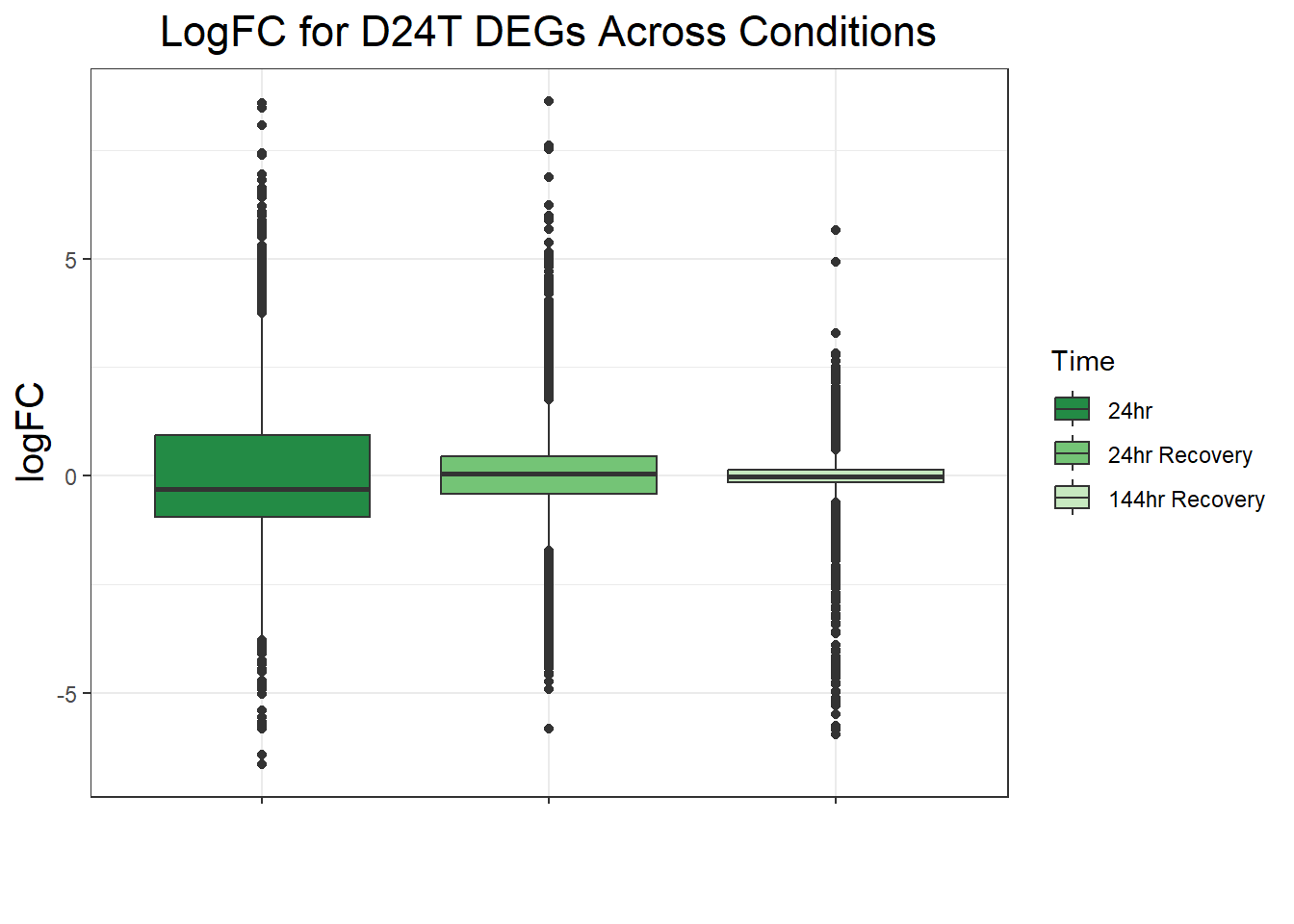
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
filt_toptable_dxr_24r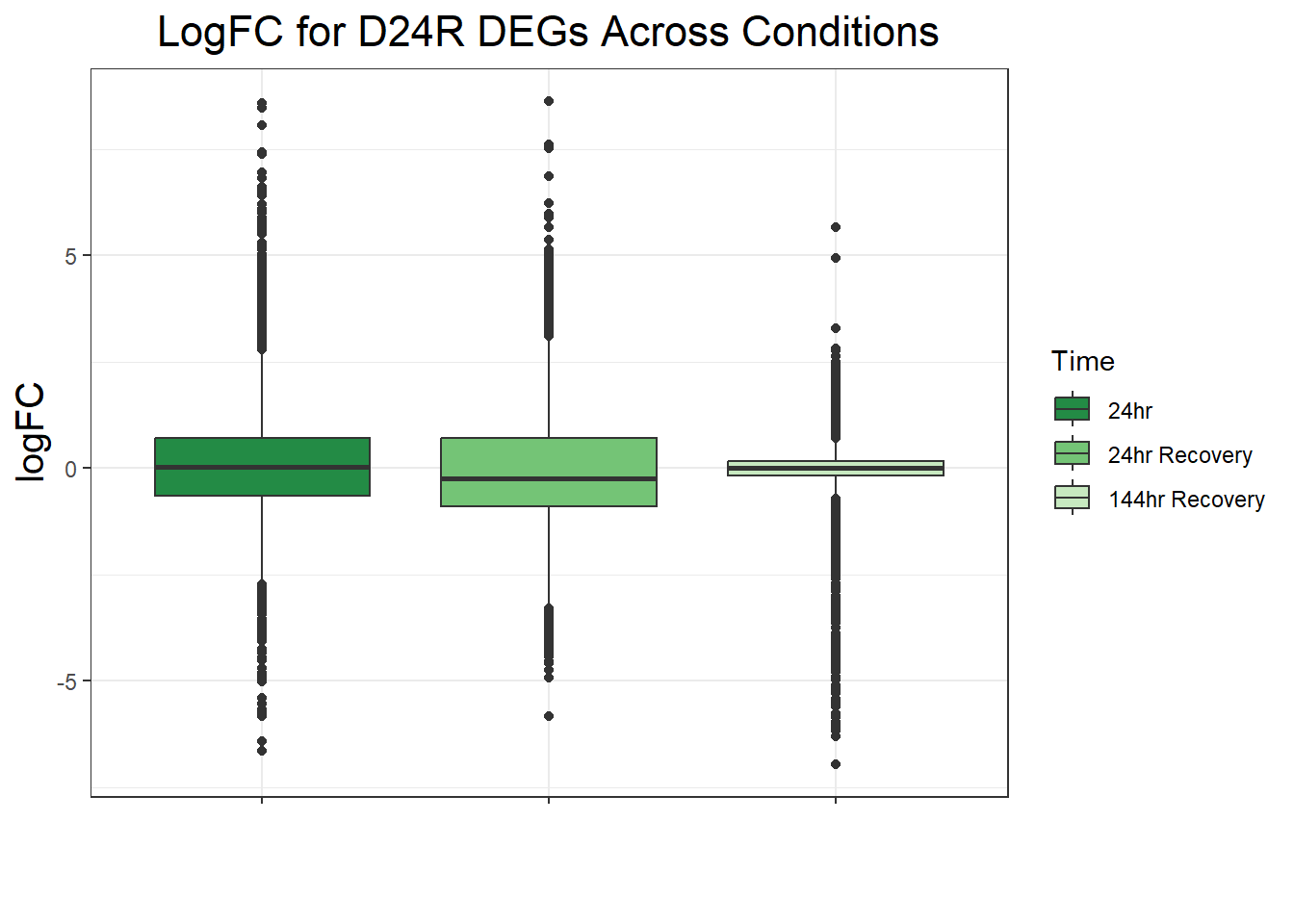
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
filt_toptable_dxr_144r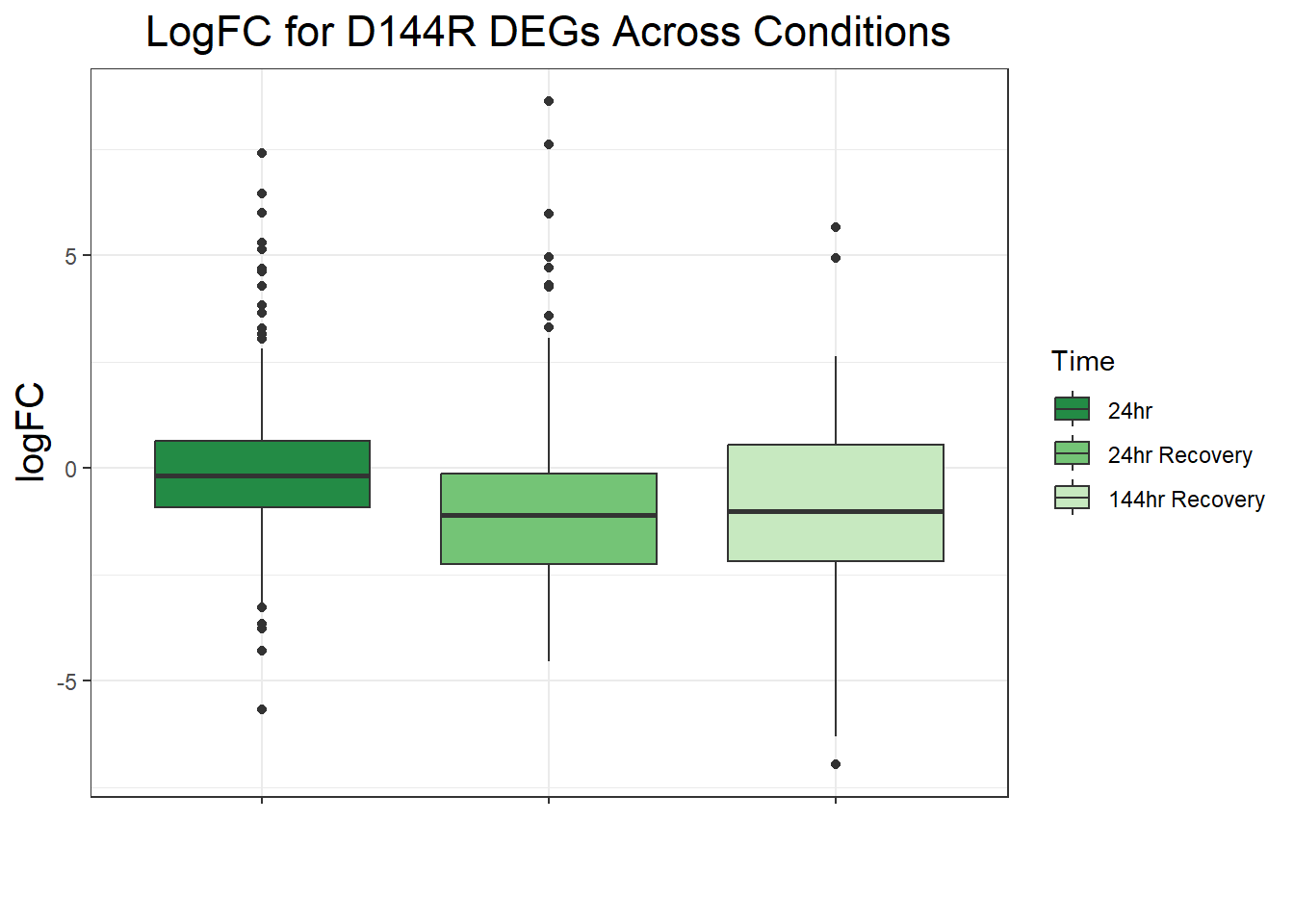
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#Perform RUVs Correction ##Set Up Data and Perform PCAs on Un-normalized Data
filt_gene_list <- rownames(filcpm_matrix)
#14319 genes as usual
#in order to make this match with annot later down the line, change the col names for counts_raw_matrix to match final_sample_names in annot
#i'll also want to make sure I keep the replicate for this set
colnames(counts_raw_matrix) <- Metadata$Final_sample_name
RUV_filt_counts <- counts_raw_matrix %>%
as.data.frame() %>%
dplyr::filter(., row.names(.)%in% filt_gene_list)
#add in the annotation files
ind_num <- readRDS("data/new/ind_num.RDS")
annot <- read.csv("data/new/Metadata.csv")
# counts need to be integer values and in a numeric matrix
# note: the log transformation needs to be accounted for in the isLog argument in RUVs function.
counts <- as.matrix(RUV_filt_counts)
#saveRDS(counts, "data/new/RUV/filt_counts_matrix.RDS")
# Create a DataFrame for the phenoData
phenoData <- DataFrame(annot)
# Now create the RangedSummarizedExperiment necessary for RUVs input
# looks like it did need both the phenodata and the counts.
set <- SummarizedExperiment(assays = counts, metadata = phenoData)
# Generate a background matrix
# The column "Cond" holds the comparisons that you actually want to make. DOX_24, DMSO_24,5FU_24, DOX_3,etc.
scIdx <-RUVSeq::makeGroups(phenoData$Condition)
scIdx [,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 6 12 18 24 30 36 42
[2,] 4 10 16 22 28 34 40
[3,] 2 8 14 20 26 32 38
[4,] 5 11 17 23 29 35 41
[5,] 3 9 15 21 27 33 39
[6,] 1 7 13 19 25 31 37#now I've made all of the data I need for this - they are located in each section for k values
#DO NOT USE THESE COUNTS FOR LINEAR MODELING
#colors for all of the plots
fill_col_ind <- c("#66C2A5", "#FC8D62", "#1F78B4", "#E78AC3", "#A6D854", "#FFD92A", "#8B3E9B")
fill_col_ind_dark <- c("#003F5C", "#45AE91", "#58508D", "#BC4099", "#8B3E9B", "#FF6361", "#FF2362")
fill_col_tx <- c("#63666D", "#499FBD", "#DCACED")
fill_col_txtime <- c("#003F5C", "#45AE91", "#58508D", "#BC4099", "#8B3E9B", "#FF6361", "#FF2362", "#A6D854", "#FC8D62")
# before ruv (counts PCA)
prcomp_res_counts <- prcomp(t(counts), scale. = FALSE, center = TRUE)
annot_prcomp_res <- prcomp_res_counts$x %>% cbind(., annot)
group_2 <- annot$Condition
#now plot my PCA for filtered counts
####PC1/PC2####
ggplot2::autoplot(prcomp_res_counts,
data = annot,
colour = "Condition",
shape = "Time",
size =4,
x=1,
y=2) +
ggrepel::geom_text_repel(label=ind_num,
vjust = -0.5,
max.overlaps = 30) +
scale_color_manual(values=cond_col) +
ggtitle(expression("PCA of Filtered Counts")) +
theme_bw()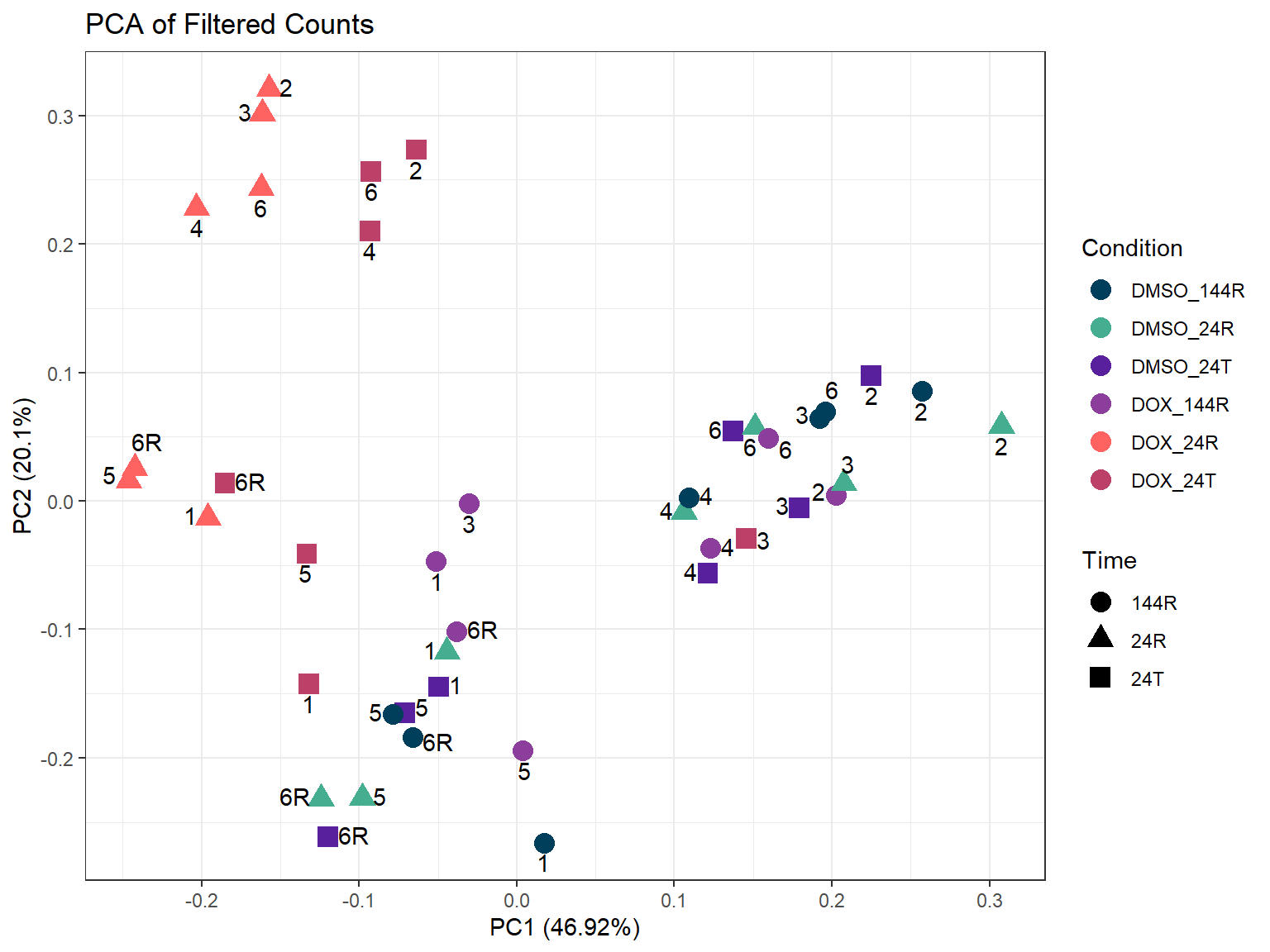
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
####PC2/PC3####
ggplot2::autoplot(prcomp_res_counts,
data = annot,
colour = "Condition",
shape = "Time",
size =4,
x=2,
y=3) +
ggrepel::geom_text_repel(label=ind_num,
vjust = -0.5,
max.overlaps = 30) +
scale_color_manual(values=cond_col) +
ggtitle(expression("PCA of Filtered Counts")) +
theme_bw()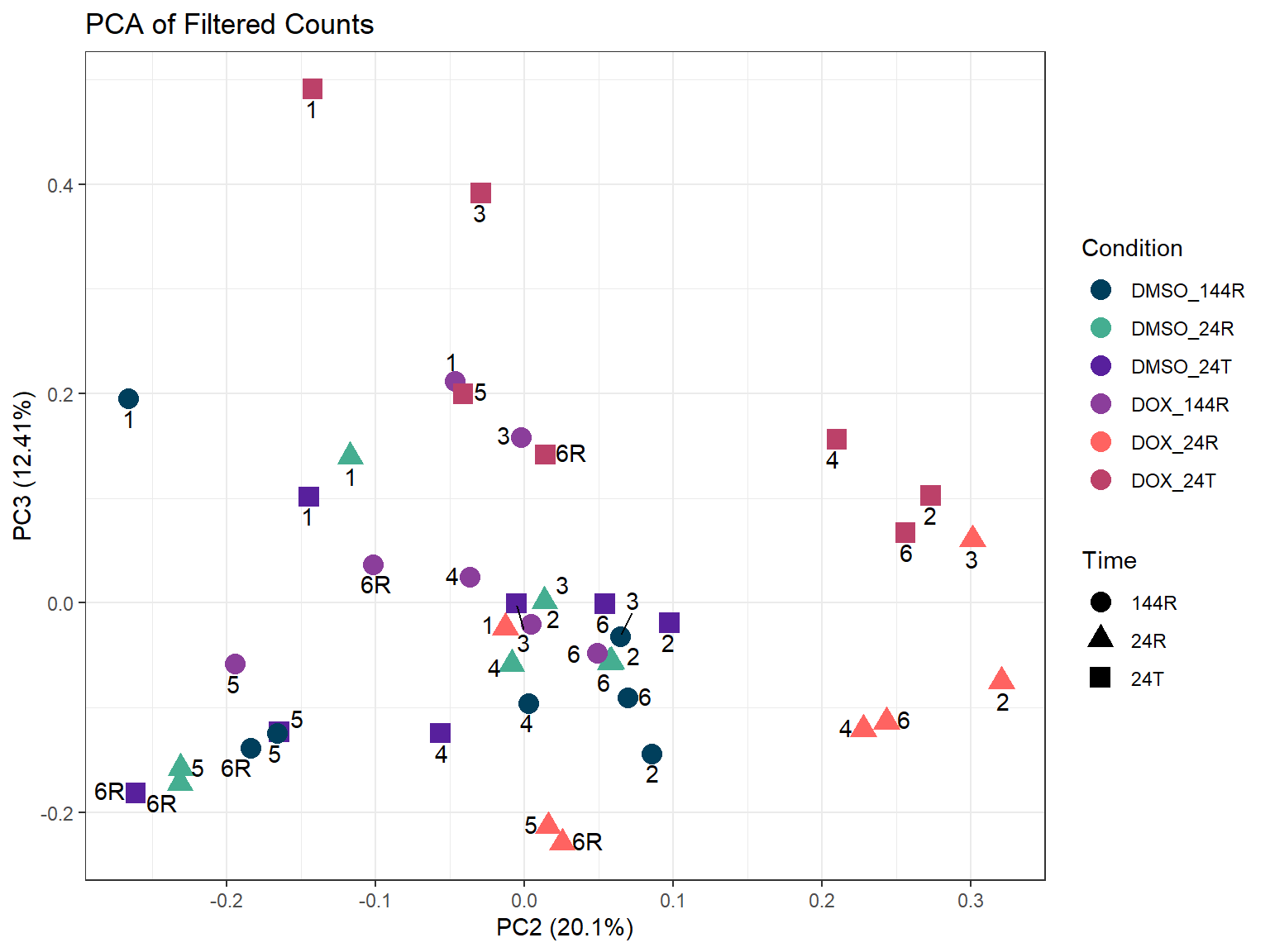
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#go ahead and plot PCA of log2cpm to compare (somewhat) later since the norm counts output isn't possible with these data since they don't undergo correction
prcomp_res_cpm <- prcomp(t(filcpm_matrix %>% as.matrix()), center = TRUE)
####PC1/PC2####
ggplot2::autoplot(prcomp_res_cpm,
data = annot,
colour = "Condition",
shape = "Time",
size =4,
x=1,
y=2) +
ggrepel::geom_text_repel(label=ind_num,
vjust = -0.5,
max.overlaps = 30) +
scale_color_manual(values=cond_col) +
ggtitle(expression("PCA of log2cpm no RUVs")) +
theme_bw()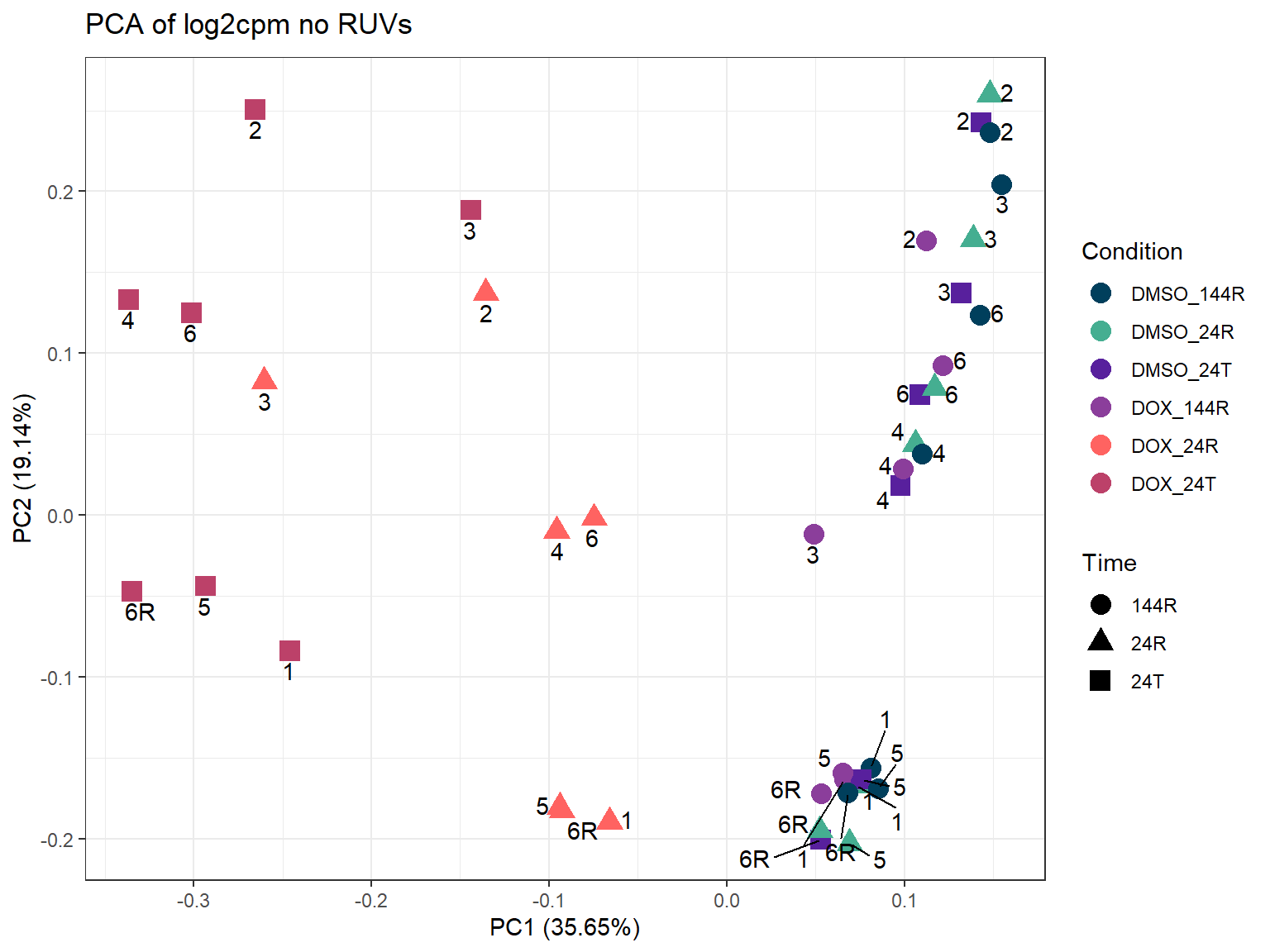
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
####PC2/PC3####
ggplot2::autoplot(prcomp_res_cpm,
data = annot,
colour = "Condition",
shape = "Time",
size =4,
x=2,
y=3) +
ggrepel::geom_text_repel(label=ind_num,
vjust = -0.5,
max.overlaps = 30) +
scale_color_manual(values=cond_col) +
ggtitle(expression("PCA of log2cpm no RUVs")) +
theme_bw()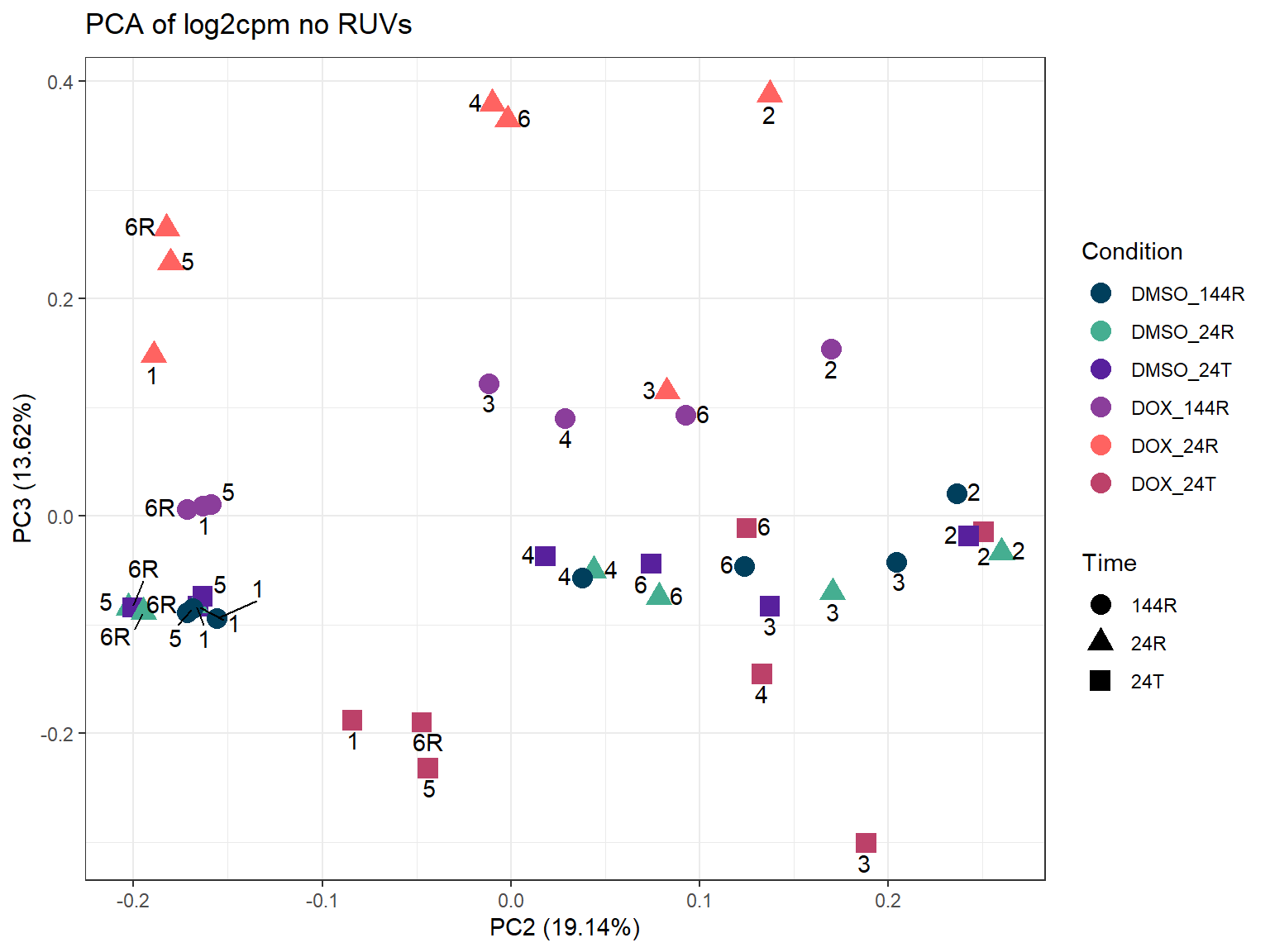
####new PCA plots no correction####
#PCA plots for each value of k attached in each section
#####Now start performing RUV 1-3#####RUVs with k = 1
#Apply RUVs function from RUVSeq
#"k" will be iteratively adjusted over time depending on your PCA.
set1 <- RUVSeq::RUVs(x = counts, k =1, scIdx = scIdx, isLog = FALSE)
#get the ruv weights to put into the linear model. n weights = k.
#k=1
RUV_df1 <- set1$W %>% as.data.frame()
RUV_df1$Names <- rownames(RUV_df1)
#Check that the names match
#k=1
RUV_df_rm1 <- RUV_df1[RUV_df1$Names %in% annot$Final_sample_name, ]
RUV_1 <- RUV_df_rm1$W_1
#saveRDS(RUV_df_rm1, "data/new/RUV_df_rm1.RDS")
#saveRDS(RUV_1, "data/new/RUV_1.RDS")
#PCA checks
#k=1
prcomp_res_1 <- prcomp(t(set1$normalizedCounts), scale. = FALSE, center = TRUE)
annot_prcomp_res_1 <- prcomp_res_1$x %>% cbind(., annot)
ggplot2::autoplot(prcomp_res_1, data = annot, colour = "Condition", shape = "Time", size =4, x = 1, y = 2)+
theme_bw()+
scale_color_manual(values=cond_col)+
ggrepel::geom_text_repel(label= ind_num, vjust = -0.5, max.overlaps = 30)+
ggtitle("RUVs Correction k=1 NormCounts")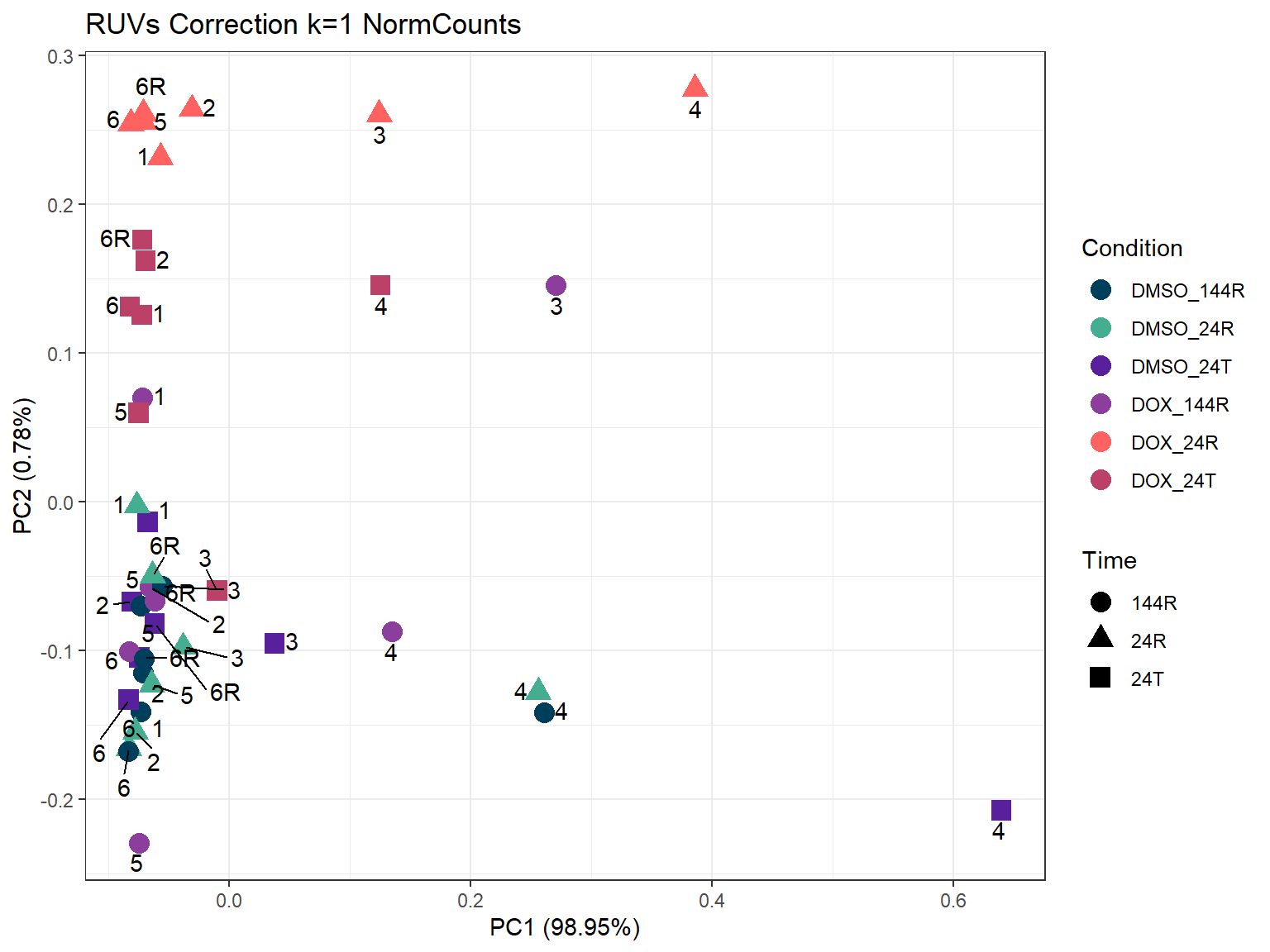
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
ggplot2::autoplot(prcomp_res_1, data = annot, colour = "Condition", shape = "Time", size =4, x=2, y=3)+
theme_bw()+
scale_color_manual(values=cond_col)+
ggrepel::geom_text_repel(label= ind_num, vjust = -0.5, max.overlaps = 30)+
ggtitle("RUVs Correction k=1 NormCounts")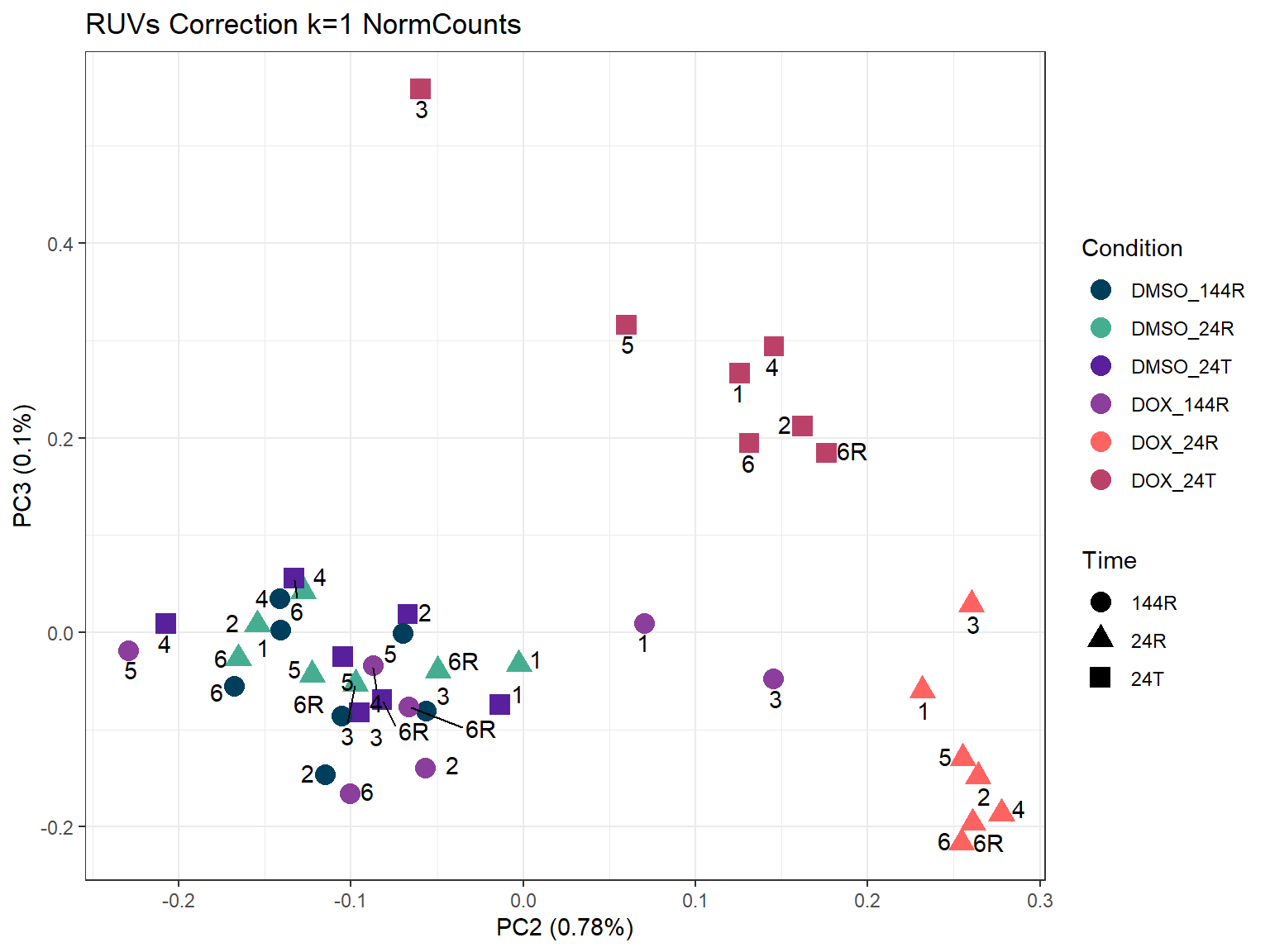
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#also try by converting these values to log2cpm
RUV_df_rm1_cpm <- cpm(set1$normalizedCounts, log = TRUE)
prcomp_res_1_cpm <- prcomp(t(RUV_df_rm1_cpm), scale. = FALSE, center = TRUE)
annot_prcomp_res_1_cpm <- prcomp_res_1_cpm$x %>% cbind(., annot)
##PC1/2
ggplot2::autoplot(prcomp_res_1_cpm,
data = annot,
colour = "Condition",
shape = "Time",
size = 4,
x=1,
y=2)+
theme_bw()+
scale_color_manual(values=cond_col)+
ggrepel::geom_text_repel(label= ind_num,
vjust = -0.5,
max.overlaps = 30)+
ggtitle("RUVs Correction k=1 log2cpm")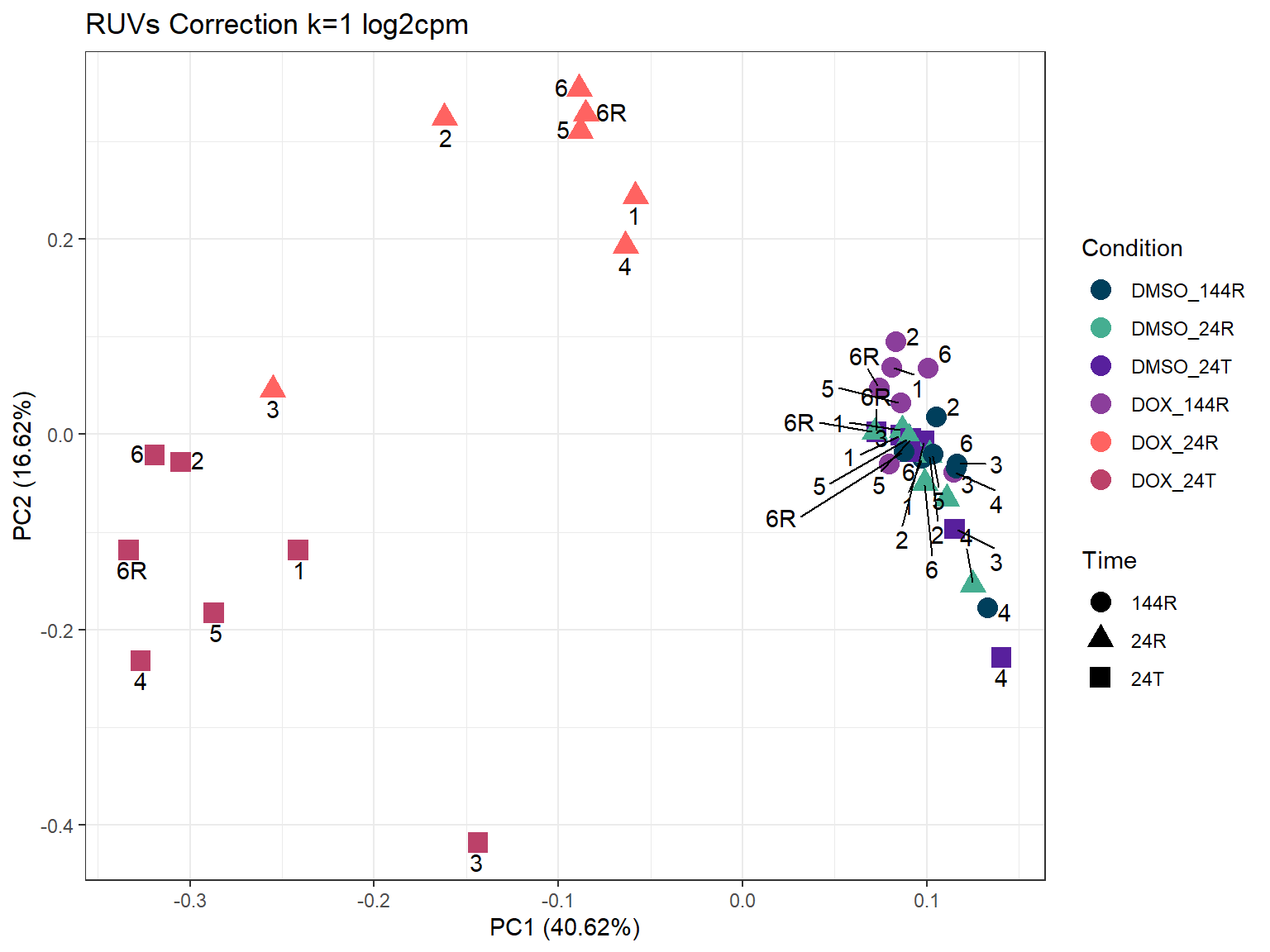
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
###PC2/3
ggplot2::autoplot(prcomp_res_1_cpm,
data = annot,
colour = "Condition",
shape = "Time",
size = 4,
x=2,
y=3)+
theme_bw()+
scale_color_manual(values=cond_col)+
ggrepel::geom_text_repel(label= ind_num,
vjust = -0.5,
max.overlaps = 30)+
ggtitle("RUVs Correction k=1 log2cpm")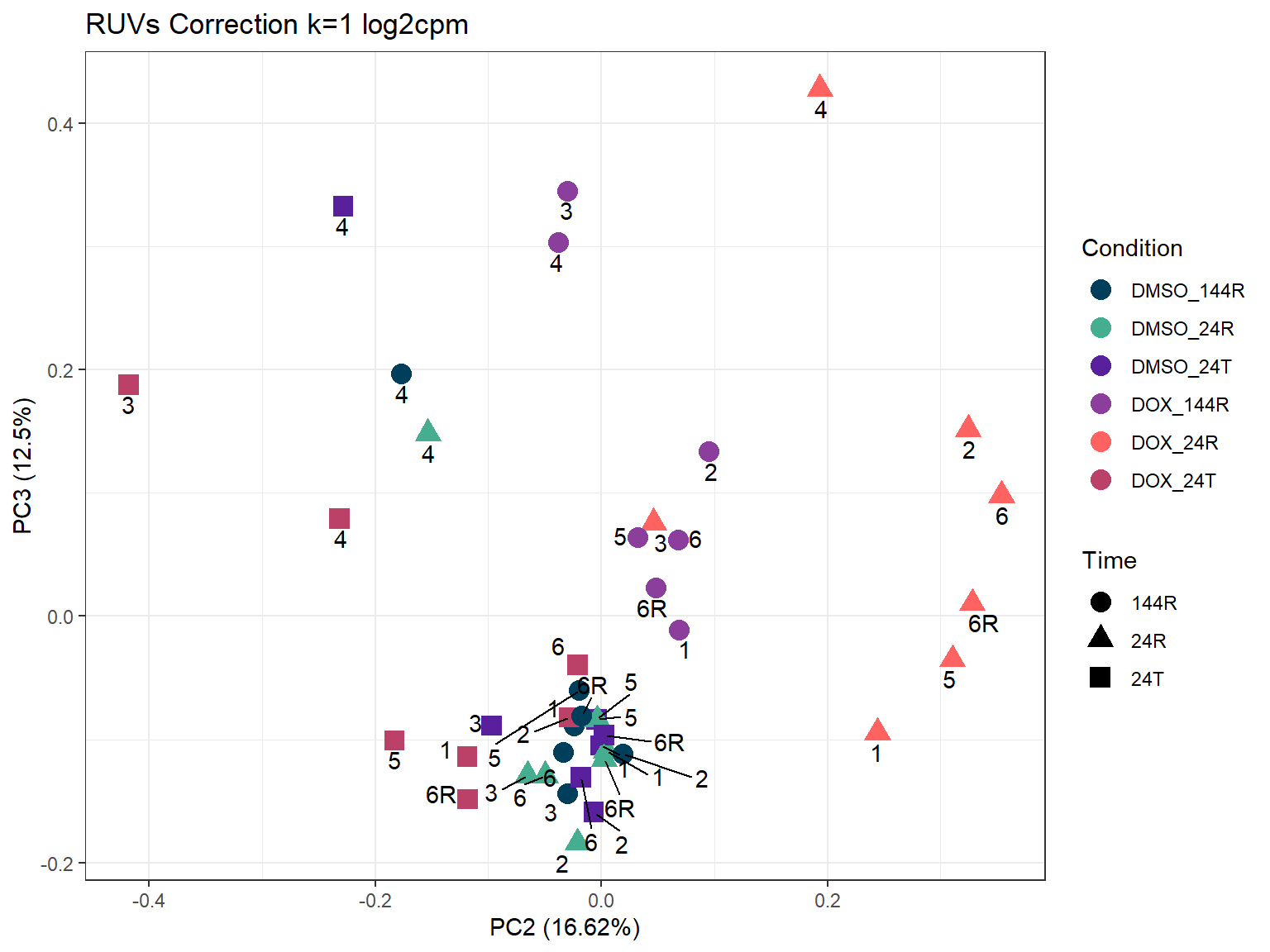
##RUVs with k = 2
#Apply RUVs function from RUVSeq
#"k" will be iteratively adjusted over time depending on your PCA.
set2 <- RUVSeq::RUVs(x = counts, k =2, scIdx = scIdx, isLog = FALSE)
#get the ruv weights to put into the linear model. n weights = k.
#k=2
RUV_df2 <- set2$W %>% as.data.frame()
RUV_df2$Names <- rownames(RUV_df2)
#Check that the names match
#k=2
RUV_df_rm2 <- RUV_df2[RUV_df2$Names %in% annot$Final_sample_name, ]
RUV_2 <- RUV_df_rm2$W_2
#PCA checks
#k=2
prcomp_res_2 <- prcomp(t(set2$normalizedCounts), scale. = FALSE, center = TRUE)
annot_prcomp_res_2 <- prcomp_res_2$x %>% cbind(., annot)
##PC1/2
ggplot2::autoplot(prcomp_res_2,
data = annot,
colour = "Condition",
shape = "Time",
size =4,
x=1,
y=2)+
theme_bw()+
scale_color_manual(values=cond_col)+
ggrepel::geom_text_repel(label= ind_num,
vjust = -0.5,
max.overlaps = 30)+
ggtitle("RUVs Correction k=2 NormCounts")Warning: ggrepel: 1 unlabeled data points (too many overlaps). Consider
increasing max.overlaps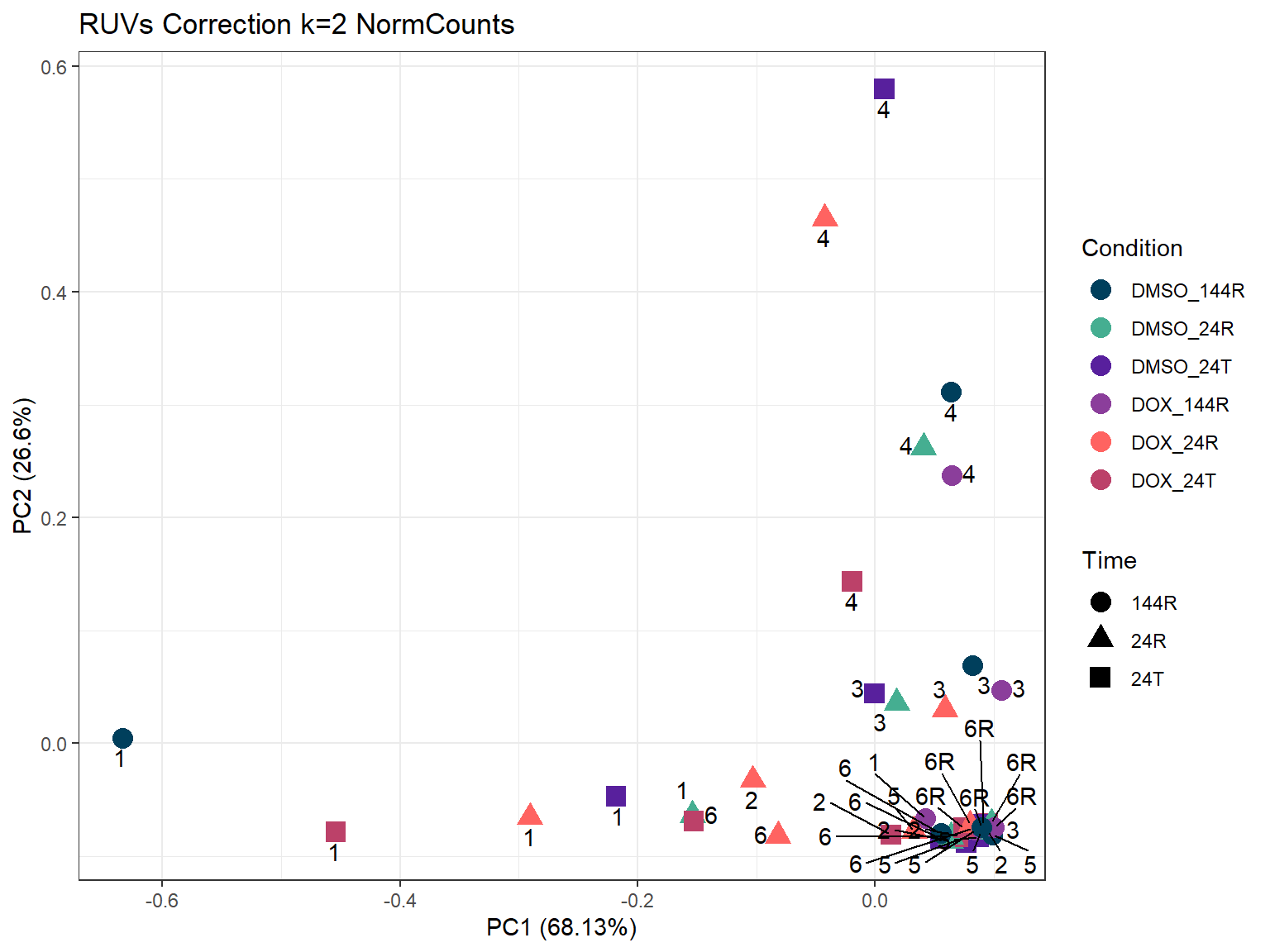
###PC2/3
ggplot2::autoplot(prcomp_res_2,
data = annot,
colour = "Condition",
shape = "Time",
size =4,
x=2,
y=3)+
theme_bw()+
scale_color_manual(values=cond_col)+
ggrepel::geom_text_repel(label= ind_num,
vjust = -0.5,
max.overlaps = 30)+
ggtitle("RUVs Correction k=2 NormCounts")Warning: ggrepel: 13 unlabeled data points (too many overlaps). Consider
increasing max.overlaps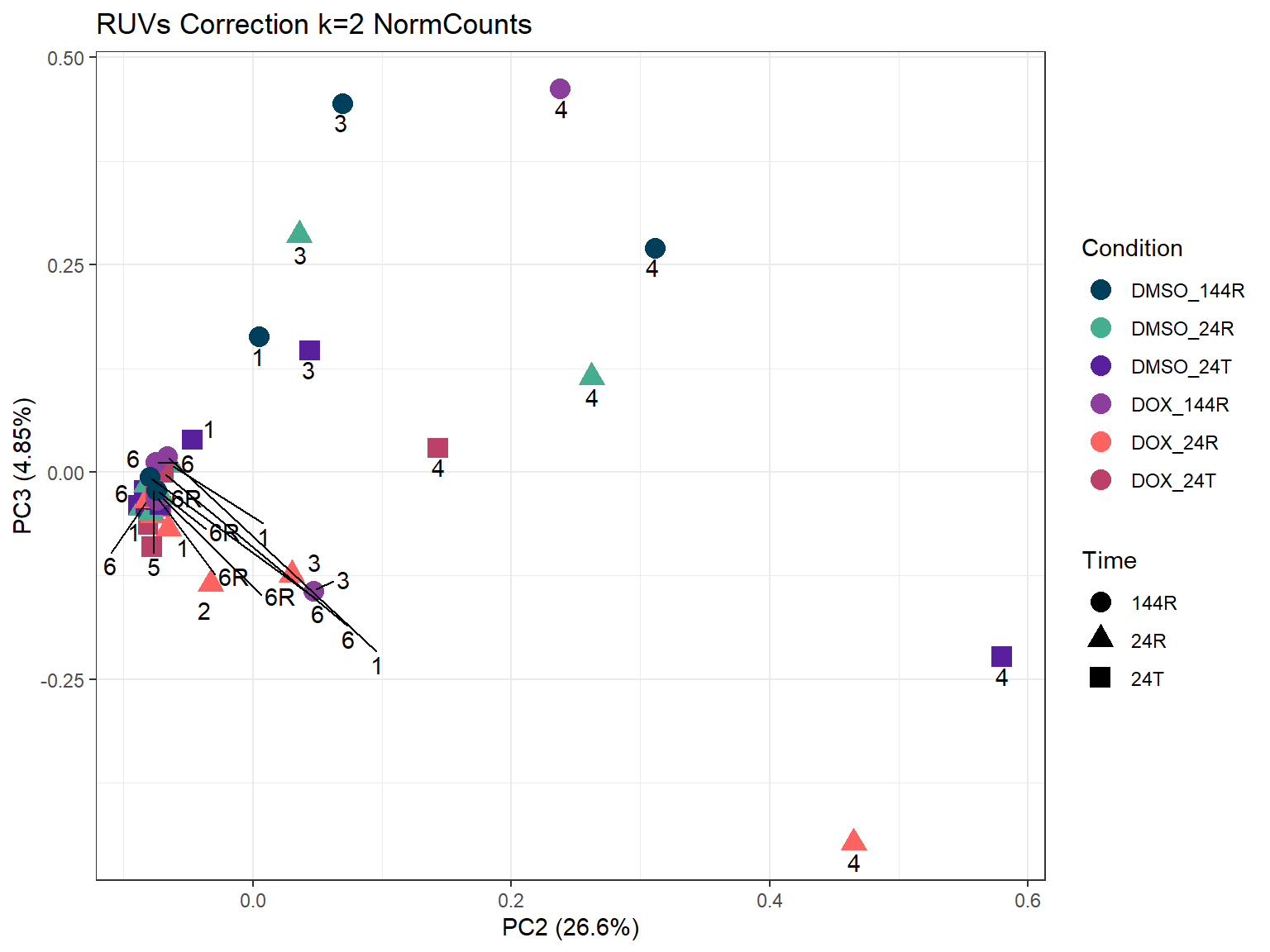
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#now convert this to log2cpm using the normalized counts from set2
RUV_df_rm2_cpm <- cpm(set2$normalizedCounts, log = TRUE)
prcomp_res_2_cpm <- prcomp(t(RUV_df_rm2_cpm), scale. = FALSE, center = TRUE)
annot_prcomp_res_2_cpm <- prcomp_res_2_cpm$x %>% cbind(., annot)
ggplot2::autoplot(prcomp_res_2_cpm,
data = annot,
colour = "Condition",
shape = "Time",
size =4,
x=1,
y=2)+
theme_bw()+
scale_color_manual(values=cond_col)+
ggrepel::geom_text_repel(label= ind_num,
vjust = -0.5,
max.overlaps = 30)+
ggtitle("RUVs Correction k=2 log2cpm")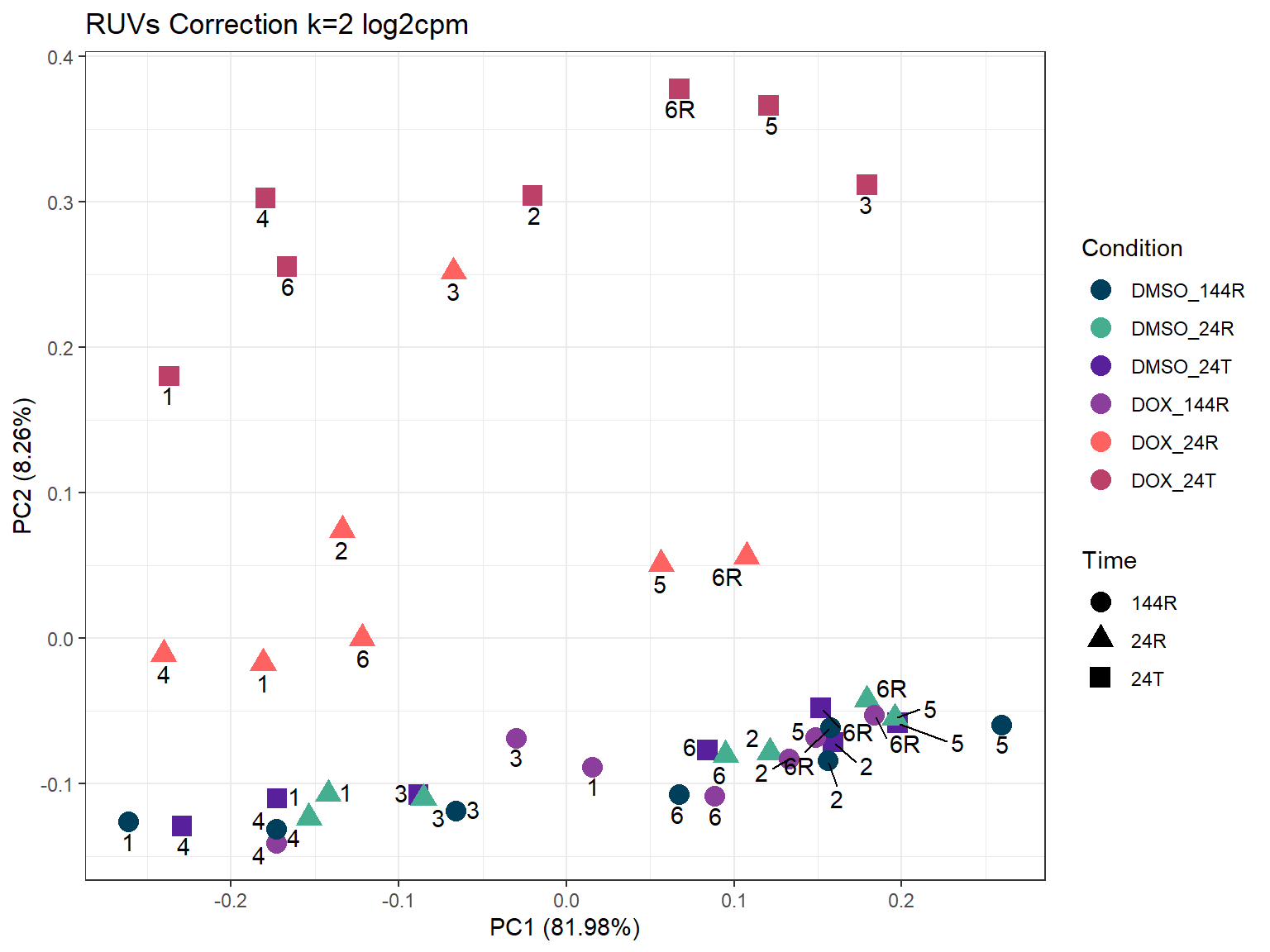
###PC2/3
ggplot2::autoplot(prcomp_res_2_cpm,
data = annot,
colour = "Condition",
shape = "Time",
size =4,
x=2,
y=3)+
theme_bw()+
scale_color_manual(values=cond_col)+
ggrepel::geom_text_repel(label= ind_num,
vjust = -0.5,
max.overlaps = 30)+
ggtitle("RUVs Correction k=2 log2cpm")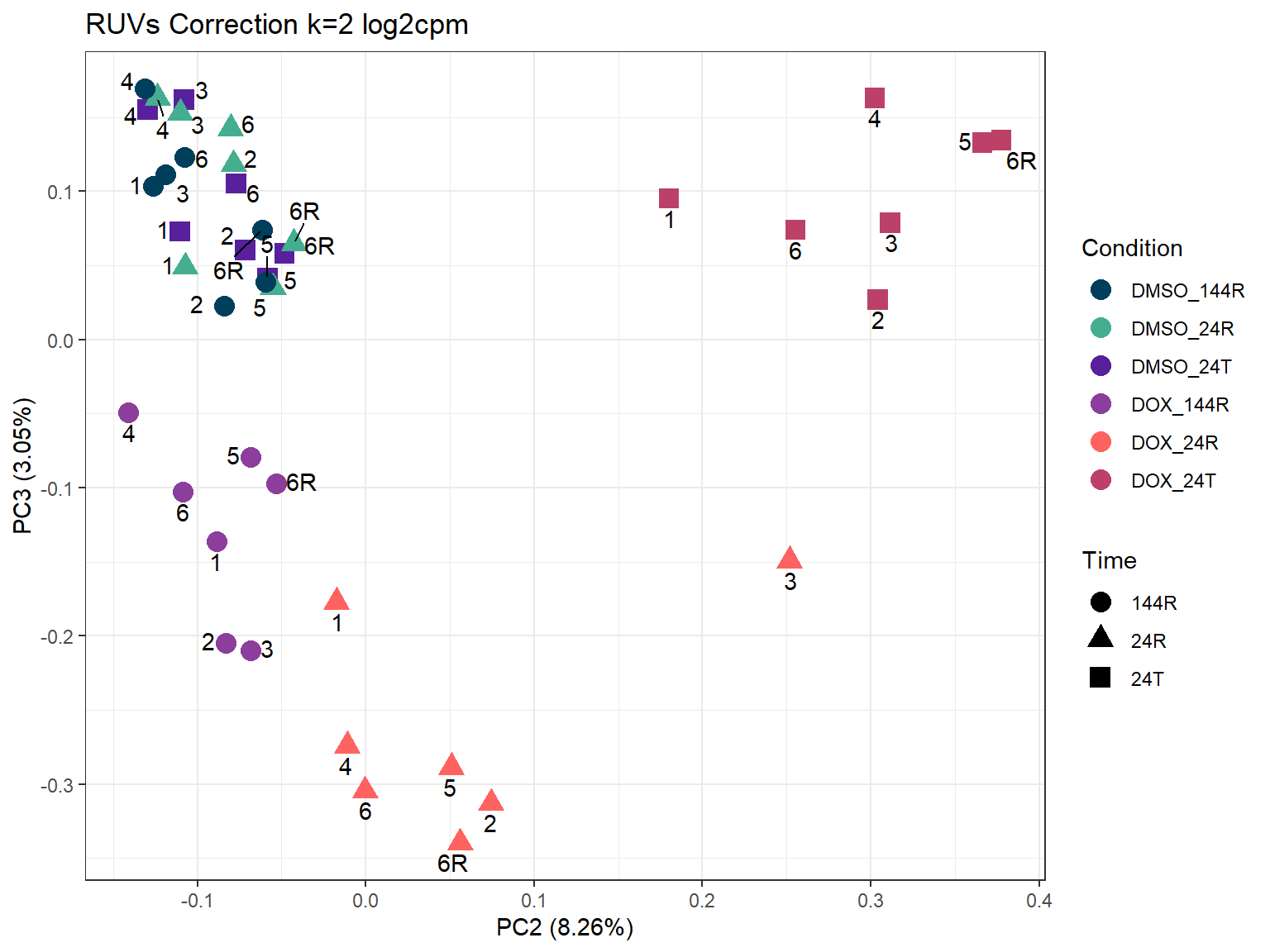
#RUVs with k = 3
#Apply RUVs function from RUVSeq
#"k" will be iteratively adjusted over time depending on your PCA.
set3 <- RUVSeq::RUVs(x = counts, k =3, scIdx = scIdx, isLog = FALSE)
#get the ruv weights to put into the linear model. n weights = k.
#k=3
RUV_df3 <- set3$W %>% as.data.frame()
RUV_df3$Names <- rownames(RUV_df3)
#Check that the names match
#k=3
RUV_df_rm3 <- RUV_df3[RUV_df3$Names %in% annot$Final_sample_name, ]
RUV_3 <- RUV_df_rm3$W_3
#PCA checks
#k=3
prcomp_res_3 <- prcomp(t(set3$normalizedCounts), scale. = FALSE, center = TRUE)
annot_prcomp_res_3 <- prcomp_res_3$x %>% cbind(., annot)
###PC1/2
ggplot2::autoplot(prcomp_res_3,
data = annot,
colour = "Condition",
shape = "Time",
size =4,
x=1,
y=2)+
theme_bw()+
scale_color_manual(values=cond_col)+
ggrepel::geom_text_repel(label= ind_num,
vjust = -0.5,
max.overlaps = 30)+
ggtitle("RUVs Correction k=3 NormCounts")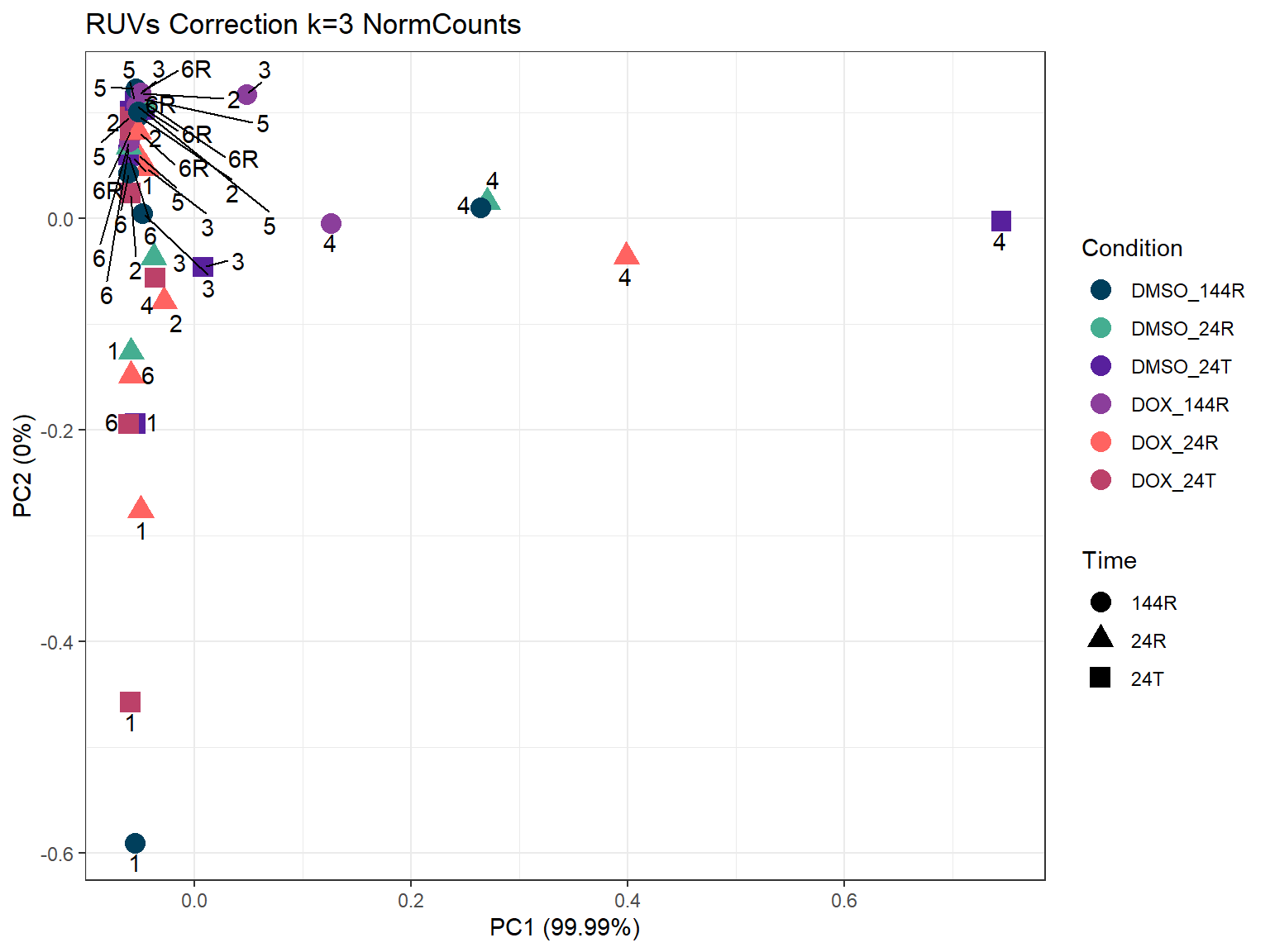
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
###PC2/3
ggplot2::autoplot(prcomp_res_3,
data = annot,
colour = "Condition",
shape = "Time",
size =4,
x=2,
y=3)+
theme_bw()+
scale_color_manual(values=cond_col)+
ggrepel::geom_text_repel(label= ind_num,
vjust = -0.5,
max.overlaps = 30)+
ggtitle("RUVs Correction k=3 NormCounts")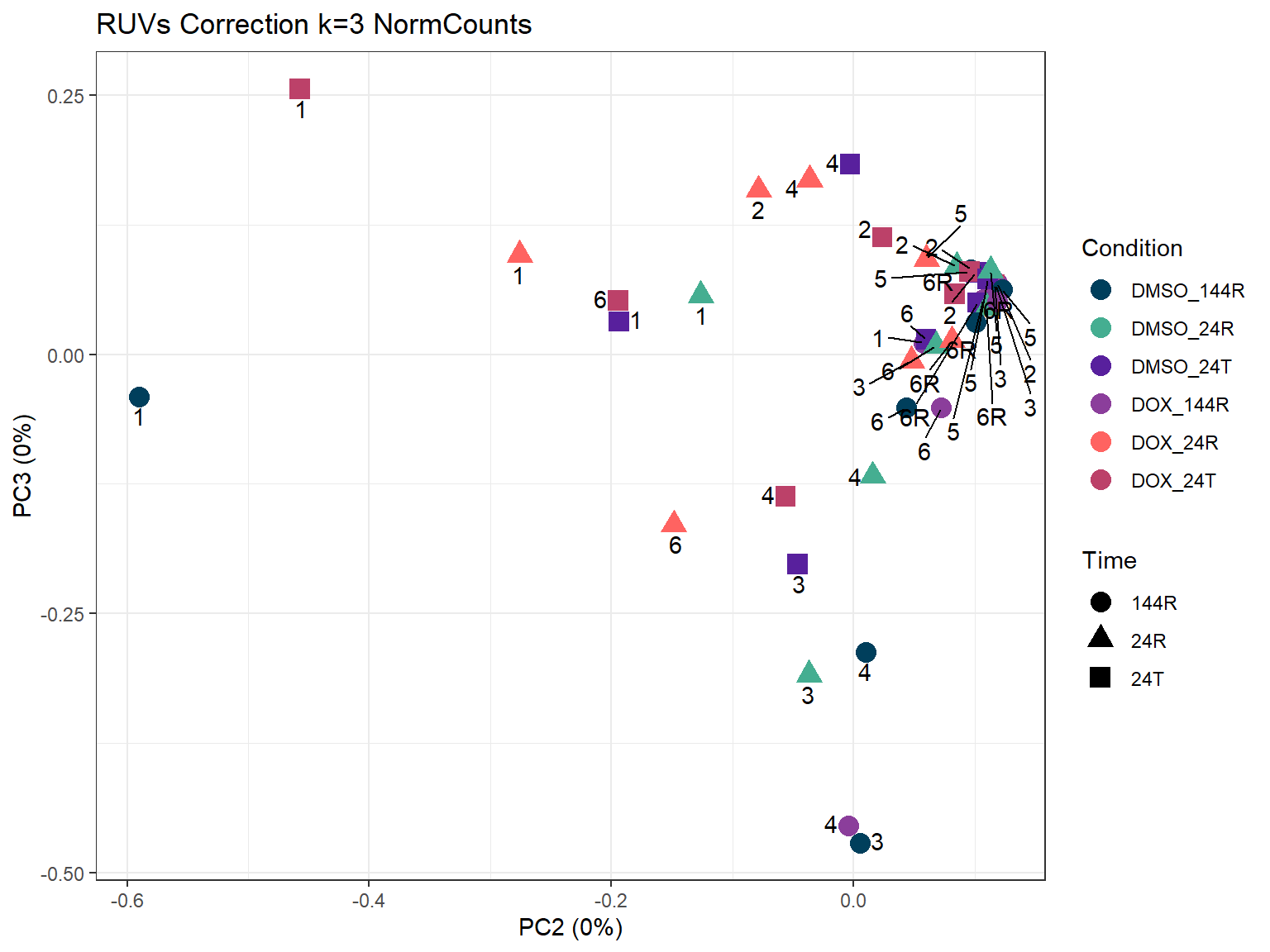
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#now convert this to log2cpm using the normalized counts from set3
RUV_df_rm3_cpm <- cpm(set3$normalizedCounts, log = TRUE)
prcomp_res_3_cpm <- prcomp(t(RUV_df_rm3_cpm), scale. = FALSE, center = TRUE)
annot_prcomp_res_3_cpm <- prcomp_res_3_cpm$x %>% cbind(., annot)
##PC1/2
ggplot2::autoplot(prcomp_res_3_cpm,
data = annot,
colour = "Condition",
shape = "Time",
size =4,
x=1,
y=2)+
theme_bw()+
scale_color_manual(values=cond_col)+
ggrepel::geom_text_repel(label= ind_num,
vjust = -0.5,
max.overlaps = 30)+
ggtitle("RUVs Correction k=3 log2cpm")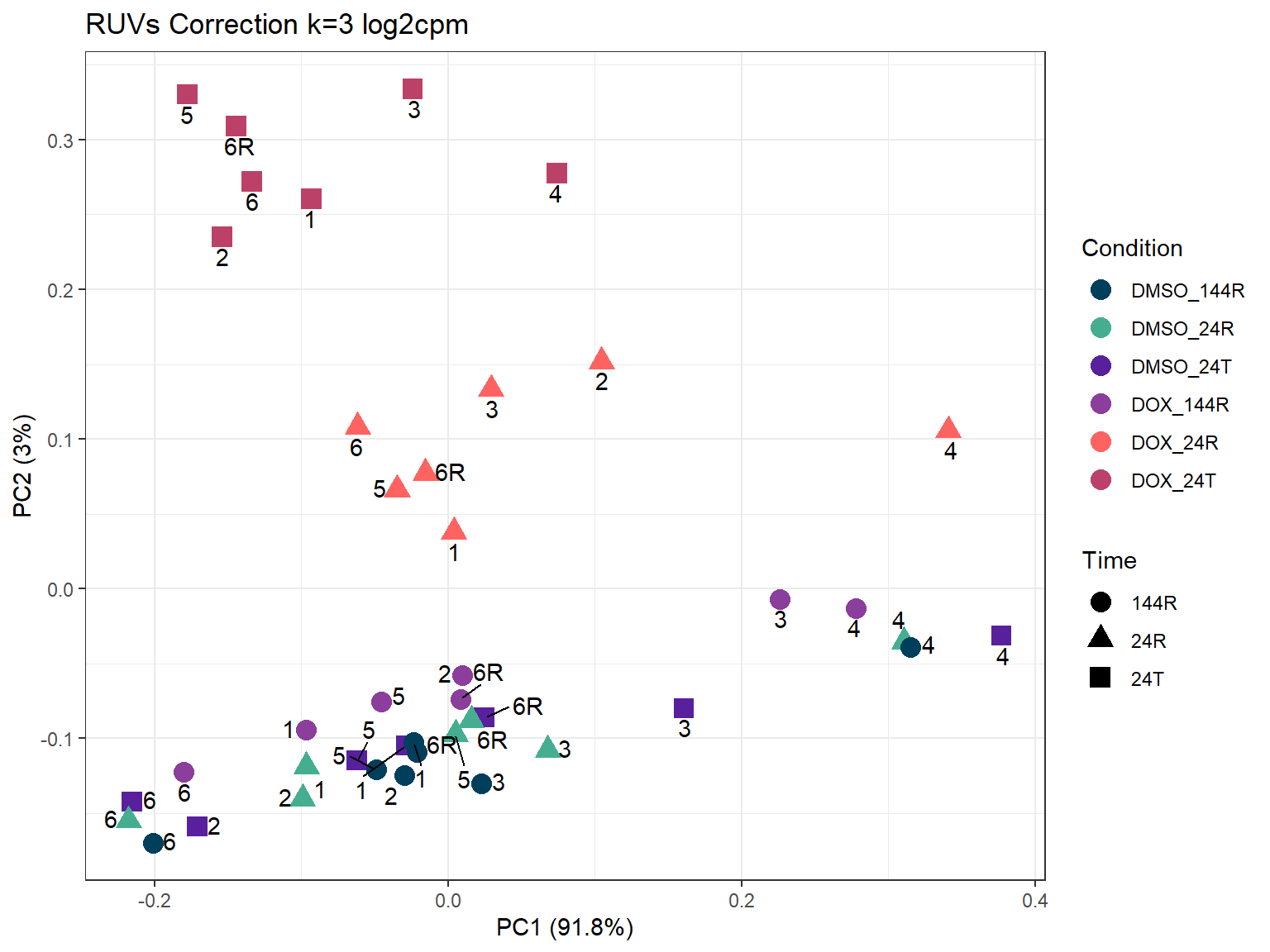
###PC2/3
ggplot2::autoplot(prcomp_res_3_cpm,
data = annot,
colour = "Condition",
shape = "Time",
size =4,
x=2,
y=3)+
theme_bw()+
scale_color_manual(values=cond_col)+
ggrepel::geom_text_repel(label= ind_num,
vjust = -0.5,
max.overlaps = 30)+
ggtitle("RUVs Correction k=3 log2cpm")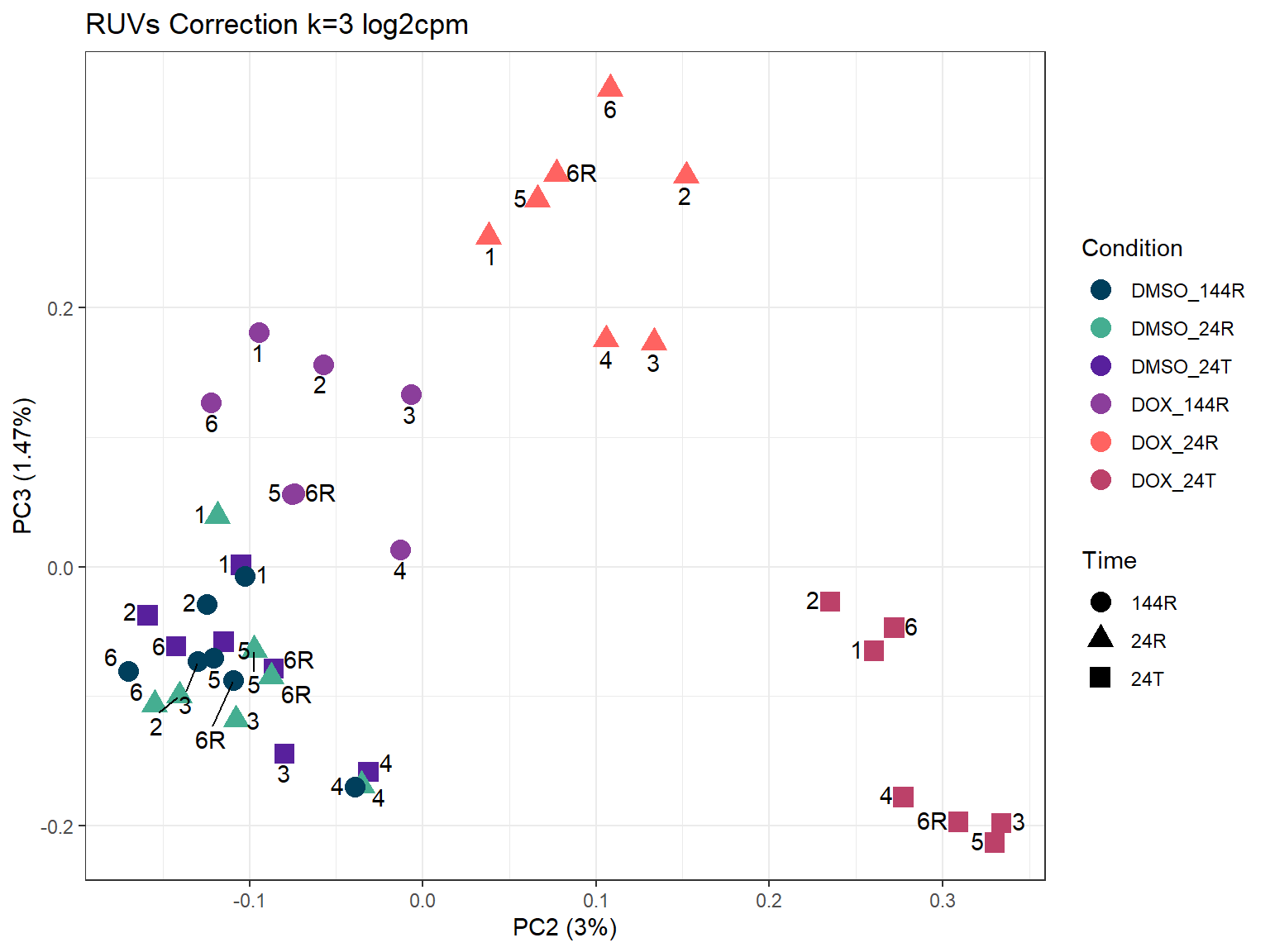
#Plot Spearman Heatmaps for RUVs Normalized Counts Data
#Now that I've put together the PCA plots for both normalized counts and log2cpm
#I want to make these into heatmaps
####RUVs k=1-3
#check to make sure that the column names are correct
dim(RUV_filt_counts)[1] 14319 42dim(set1$normalizedCounts)[1] 14319 42dim(set2$normalizedCounts)[1] 14319 42dim(set3$normalizedCounts)[1] 14319 42#take the normalized counts from k=1 and put together a dataframe with the correct columns
normcounts_k0 <- RUV_filt_counts
normcounts_k1 <- set1$normalizedCounts %>% as.data.frame()
#do the same with k=2 and k=3
normcounts_k2 <- set2$normalizedCounts %>% as.data.frame()
normcounts_k3 <- set3$normalizedCounts %>% as.data.frame()
#do the same with the log2cpm conversion
cpm_k0 <- cpm(normcounts_k0, log = TRUE) %>% as.data.frame()
cpm_k1 <- cpm(set1$normalizedCounts, log = TRUE) %>% as.data.frame()
cpm_k2 <- cpm(set2$normalizedCounts, log = TRUE) %>% as.data.frame()
cpm_k3 <- cpm(set3$normalizedCounts, log = TRUE) %>% as.data.frame()
#compute the correlation matrices for RUVs 1-3 with normalized counts
#k=0
cor_matrix_spmn_k0 <- cor(normcounts_k0,
y = NULL,
use = "everything",
method = "spearman")
#k=1
cor_matrix_spmn_k1 <- cor(normcounts_k1,
y = NULL,
use = "everything",
method = "spearman")
#k=2
cor_matrix_spmn_k2 <- cor(normcounts_k2,
y = NULL,
use = "everything",
method = "spearman")
#k=3
cor_matrix_spmn_k3 <- cor(normcounts_k3,
y = NULL,
use = "everything",
method = "spearman")
#Do the same with the log2cpm converted versions
#k=0
cor_matrix_spmn_k0_cpm <- cor(cpm_k0,
y = NULL,
use = "everything",
method = "spearman")
#k=1
cor_matrix_spmn_k1_cpm <- cor(cpm_k1,
y = NULL,
use = "everything",
method = "spearman")
#k=2
cor_matrix_spmn_k2_cpm <- cor(cpm_k2,
y = NULL,
use = "everything",
method = "spearman")
#k=3
cor_matrix_spmn_k3_cpm <- cor(cpm_k3,
y = NULL,
use = "everything",
method = "spearman")
#extract metadata columns
Individual <- as.character(Metadata$Ind)
Time <- as.character(Metadata$Time)
Treatment <- as.character(Metadata$Drug)
# Define color palettes for annotations
annot_col_cor = list(Treatment = c("DOX" = "#499FBD",
"DMSO" = "#BBBBBC"),
Individuals = c("1" = "#003F5C",
"2" = "#45AE91",
"3" = "#58209D",
"4" = "#8B3E9B",
"5" = "#FF6361",
"6" = "#BC4169",
"6R" = "#FF2362"),
Timepoints = c("24T" = "#238B45",
"24R" = "#74C476",
"144R" = "#C7E9C0"))
tx_colors <- c("DOX" = "#499FBD",
"DMSO" = "#BBBBBC")
ind_colors <- c("1" = "red",
"2" = "orange",
"3" = "yellow",
"4" = "green",
"5" = "blue",
"6" = "violet",
"6R" = "purple")
time_colors <- c("24T" = "#238B45",
"24R" = "#74C476",
"144R" = "#C7E9C0")
# Create annotations
top_annotation <- HeatmapAnnotation(
Individual = Individual,
Time = Time,
Treatment = Treatment,
col = list(
Individual = ind_colors,
Time = time_colors,
Treatment = tx_colors
)
)
####ANNOTATED HEATMAPS####
###Spearman Heatmap k=0 ####
heatmap_spmn_k0 <- Heatmap(cor_matrix_spmn_k0,
name = "Spearman",
top_annotation = top_annotation,
show_row_names = TRUE,
show_column_names = TRUE,
cluster_rows = TRUE,
cluster_columns = TRUE,
border = TRUE,
column_title = "Filtered Counts no RUVs")
# Draw the heatmap k=0
draw(heatmap_spmn_k0)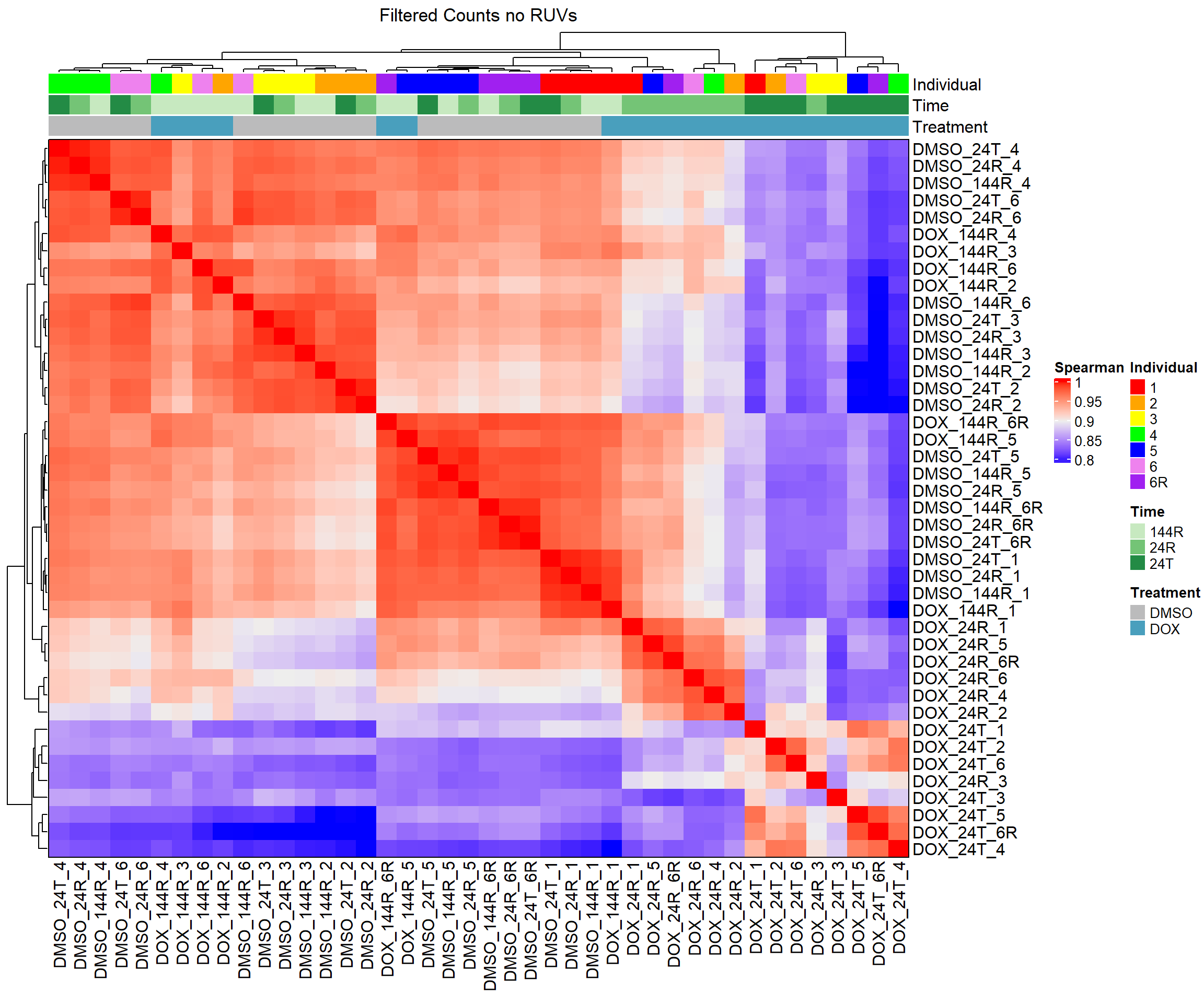
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
####Spearman Heatmap k=1 ####
heatmap_spmn_k1 <- Heatmap(cor_matrix_spmn_k1,
name = "Spearman",
top_annotation = top_annotation,
show_row_names = TRUE,
show_column_names = TRUE,
cluster_rows = TRUE,
cluster_columns = TRUE,
border = TRUE,
column_title = "Normalized Counts k=1")
# Draw the heatmap k=1
draw(heatmap_spmn_k1)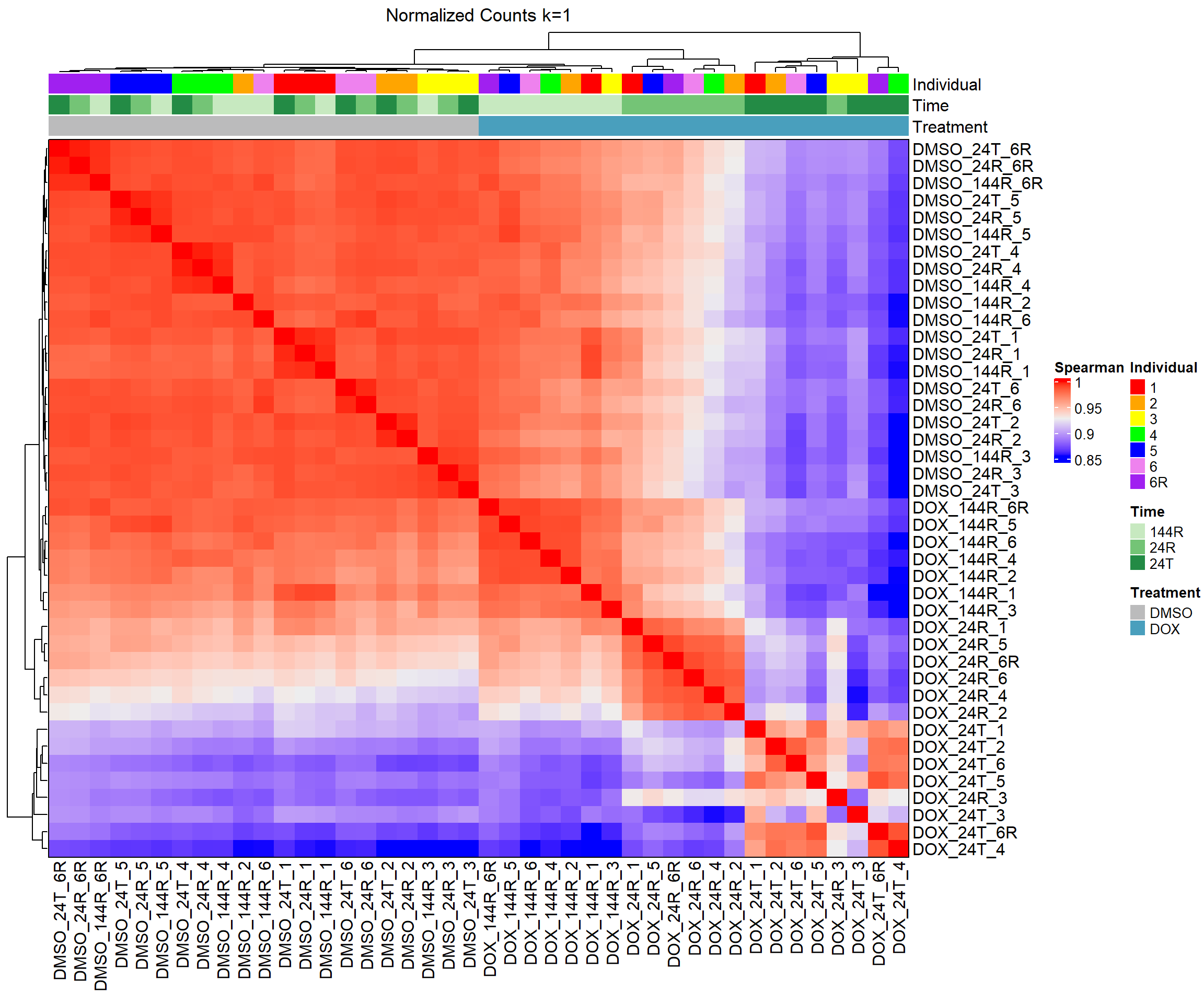
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
####Spearman Heatmap k=2####
heatmap_spmn_k2 <- Heatmap(cor_matrix_spmn_k2,
name = "Spearman",
top_annotation = top_annotation,
show_row_names = TRUE,
show_column_names = TRUE,
cluster_rows = TRUE,
cluster_columns = TRUE,
border = TRUE,
column_title = "Normalized Counts k=2")
# Draw the heatmap k=2
draw(heatmap_spmn_k2)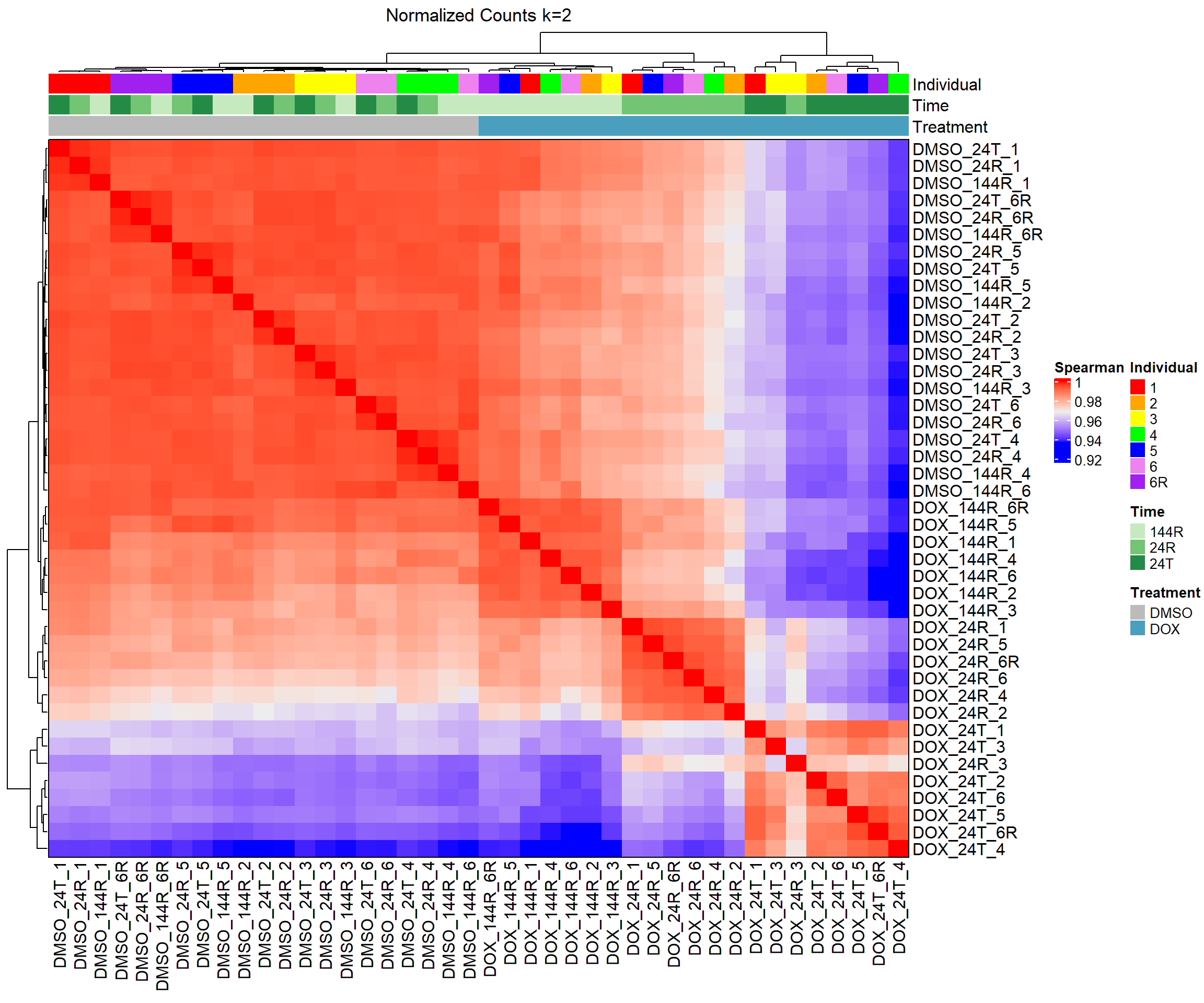
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
####Spearman Heatmap k=3####
heatmap_spmn_k3 <- Heatmap(cor_matrix_spmn_k3,
name = "Spearman",
top_annotation = top_annotation,
show_row_names = TRUE,
show_column_names = TRUE,
cluster_rows = TRUE,
cluster_columns = TRUE,
border = TRUE,
column_title = "Normalized Counts k=3")
# Draw the heatmap k=3
draw(heatmap_spmn_k3)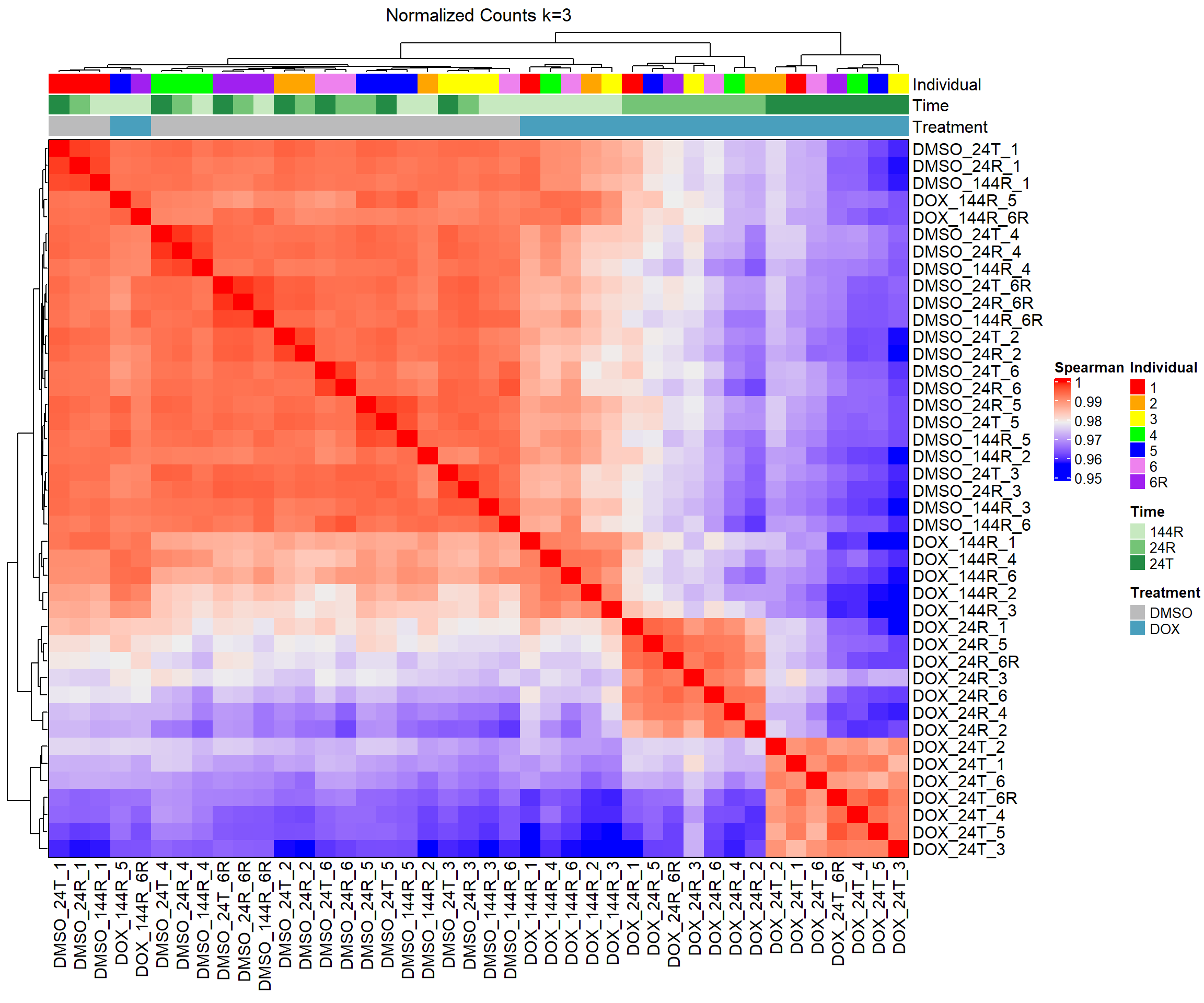
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
####Spearman Heatmap k=0 log2cpm####
heatmap_spmn_k0_cpm <- Heatmap(cor_matrix_spmn_k0_cpm,
name = "Spearman",
top_annotation = top_annotation,
show_row_names = TRUE,
show_column_names = TRUE,
cluster_rows = TRUE,
cluster_columns = TRUE,
border = TRUE,
column_title = "log2cpm of Filtered Counts no RUVs")
# Draw the heatmap k=0 log2cpm
draw(heatmap_spmn_k0_cpm)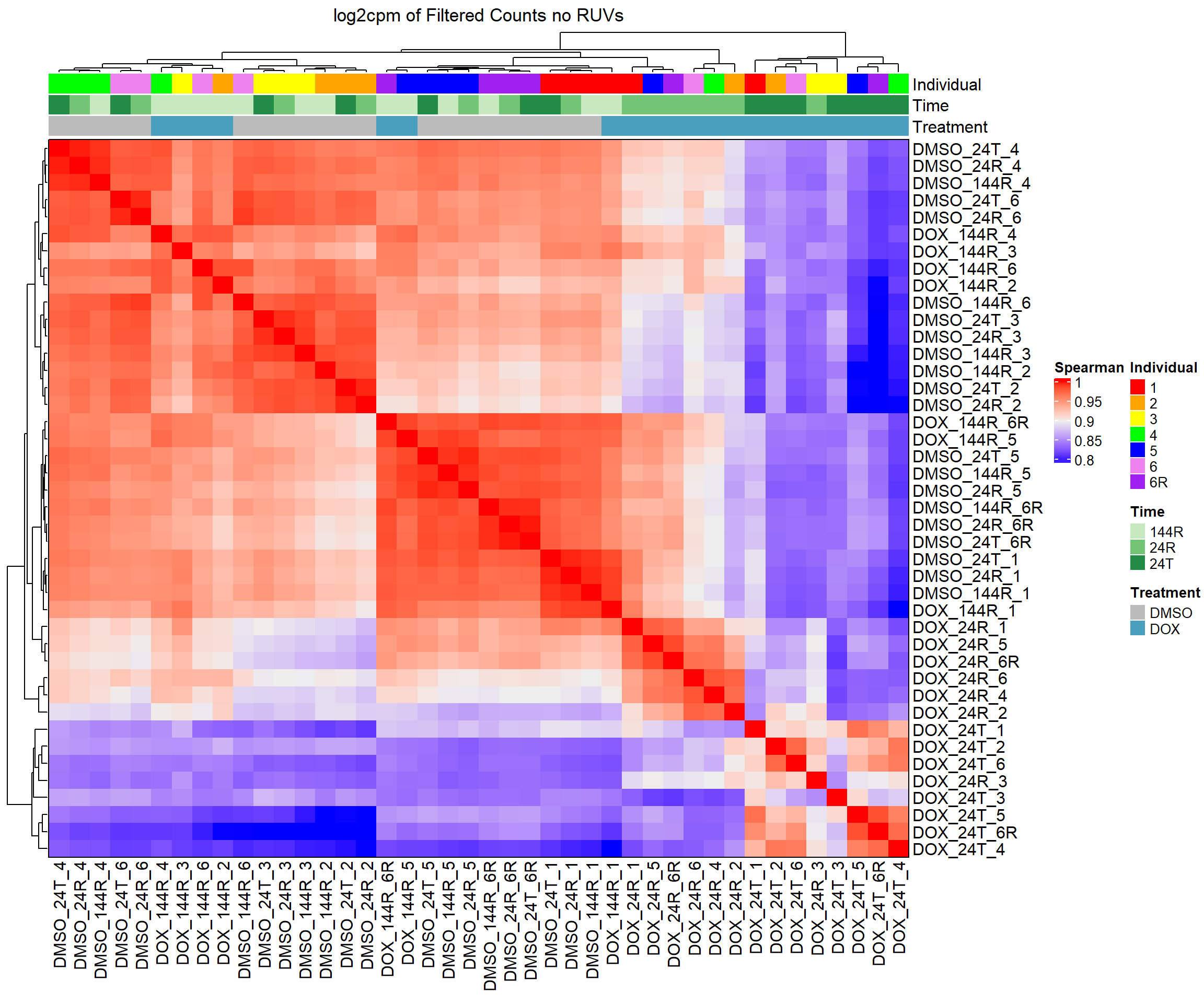
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
####Spearman Heatmap k=1 log2cpm####
heatmap_spmn_k1_cpm <- Heatmap(cor_matrix_spmn_k1_cpm,
name = "Spearman",
top_annotation = top_annotation,
show_row_names = TRUE,
show_column_names = TRUE,
cluster_rows = TRUE,
cluster_columns = TRUE,
border = TRUE,
column_title = "log2cpm of Normalized Counts k=1")
# Draw the heatmap k=1 log2cpm
draw(heatmap_spmn_k1_cpm)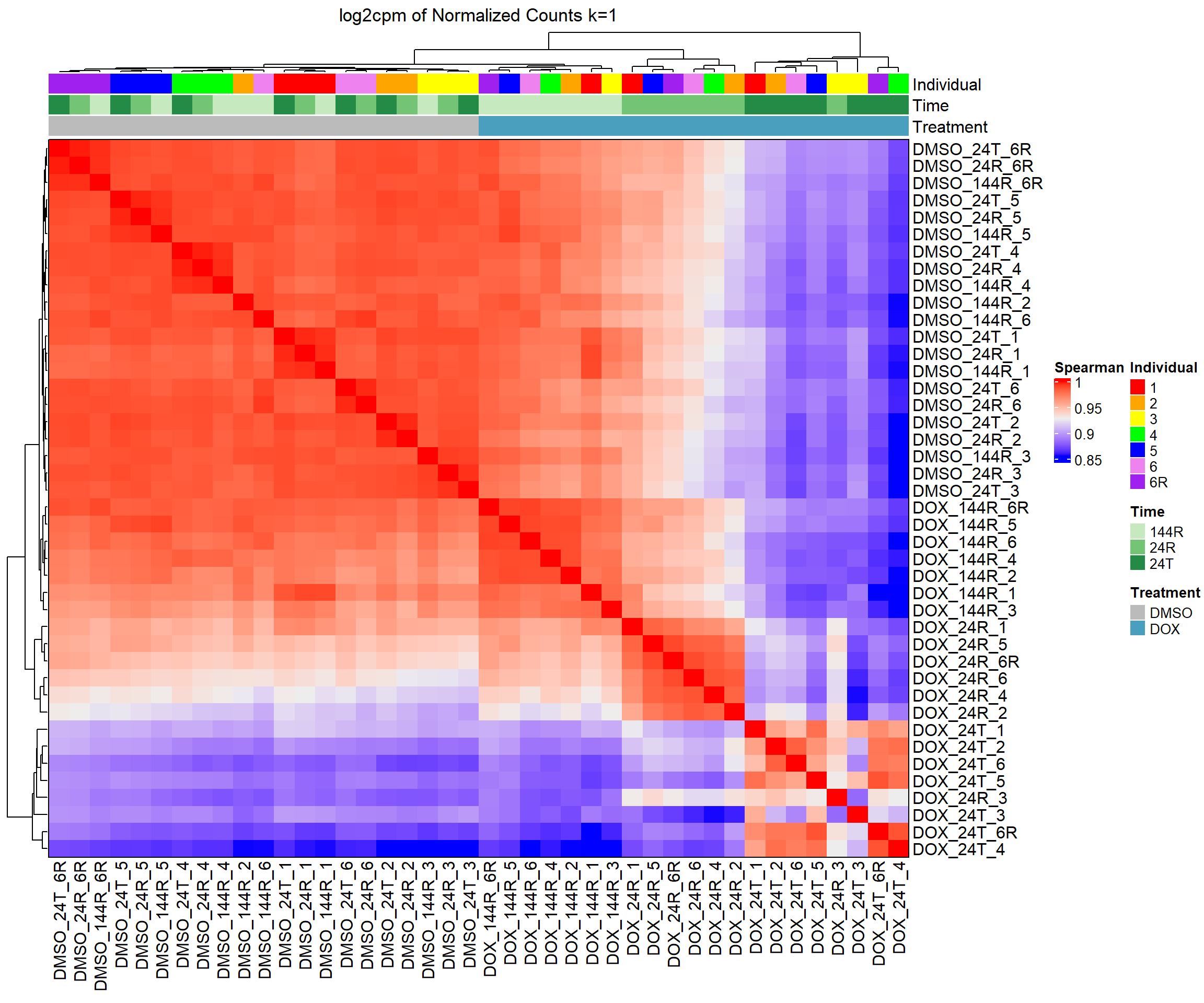
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
####Spearman Heatmap k=2 log2cpm####
heatmap_spmn_k2_cpm <- Heatmap(cor_matrix_spmn_k2_cpm,
name = "Spearman",
top_annotation = top_annotation,
show_row_names = TRUE,
show_column_names = TRUE,
cluster_rows = TRUE,
cluster_columns = TRUE,
border = TRUE,
column_title = "log2cpm of Normalized Counts k=2")
# Draw the heatmap k=2 log2cpm
draw(heatmap_spmn_k2_cpm)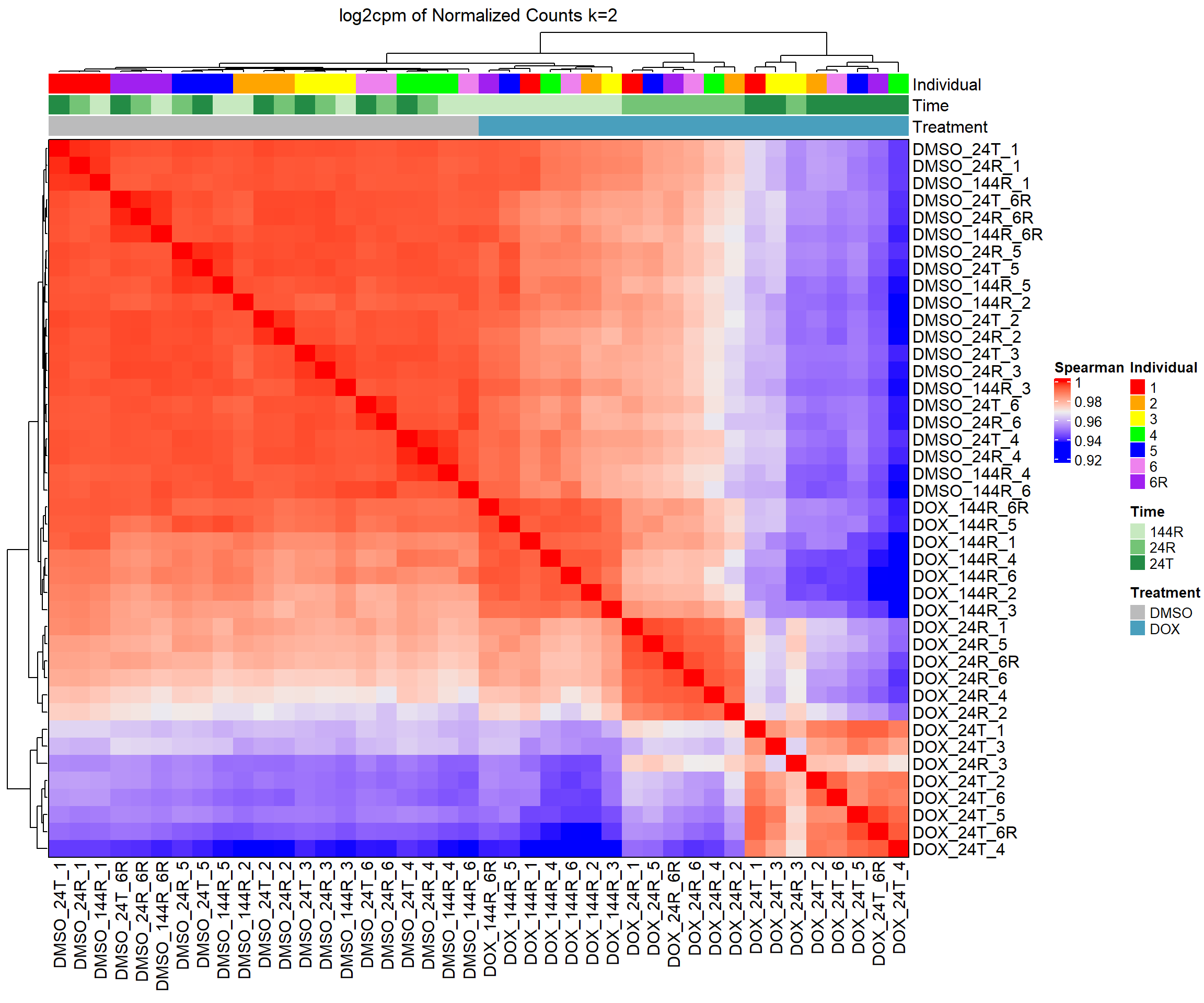
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
####Spearman Heatmap k=3 log2cpm####
heatmap_spmn_k3_cpm <- Heatmap(cor_matrix_spmn_k3_cpm,
name = "Spearman",
top_annotation = top_annotation,
show_row_names = TRUE,
show_column_names = TRUE,
cluster_rows = TRUE,
cluster_columns = TRUE,
border = TRUE,
column_title = "log2cpm of Normalized Counts k=3")
# Draw the heatmap k=3 log2cpm
draw(heatmap_spmn_k3_cpm)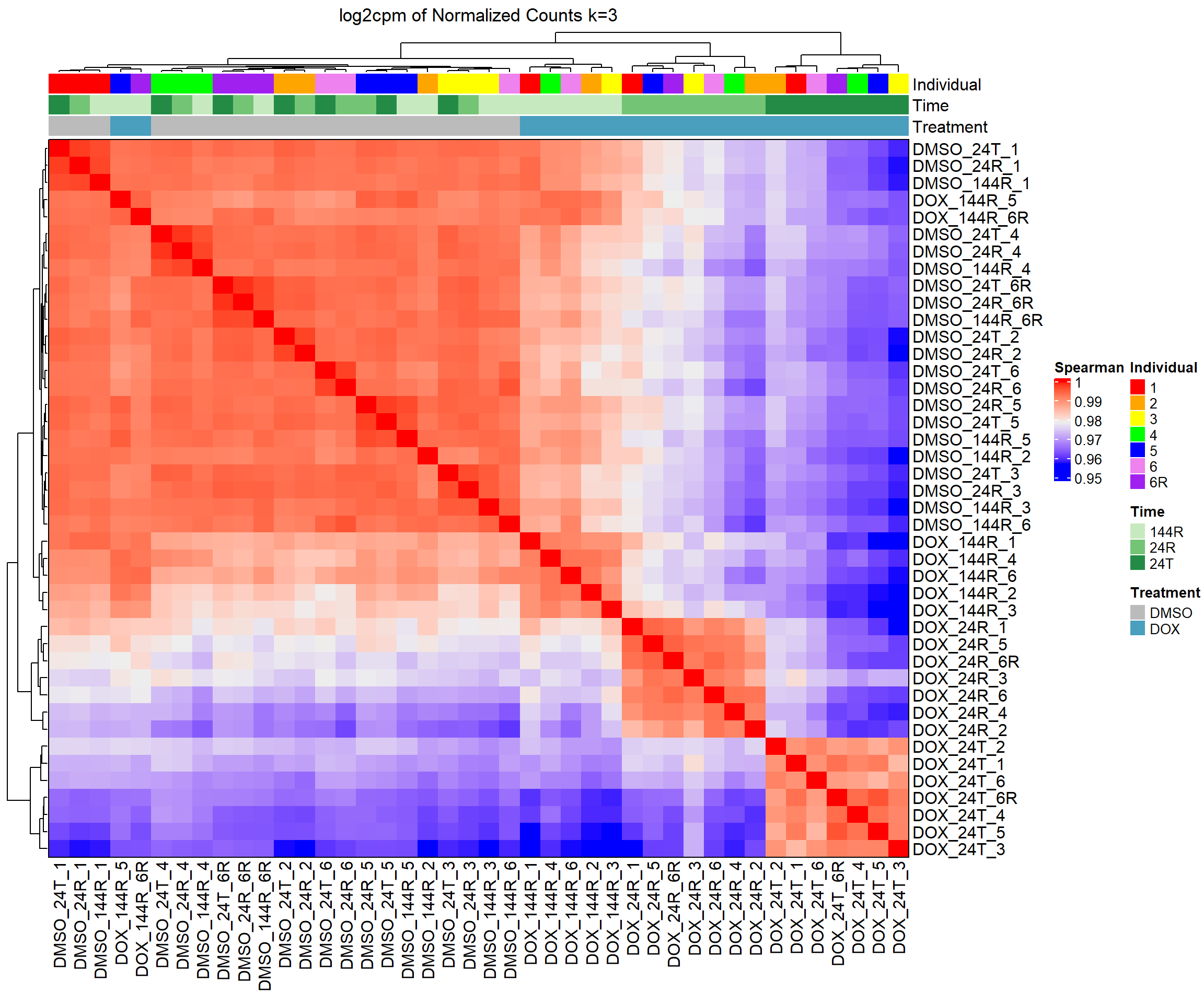
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#Perform Differential Expression Analysis with k = 1 RUVs Correction
#same DGEList object as before
dge <- readRDS("data/new/dge_matrix.RDS")
#check normalization factors from TMM normalization of LIBRARIES
dge$samples group lib.size norm.factors
84-1_DOX_24 DOX_24T 23393931 0.9745263
84-1_DMSO_24 DMSO_24T 22853195 0.9565797
84-1_DOX_24+24 DOX_24R 23846995 1.1659432
84-1_DMSO_24+24 DMSO_24R 21299355 0.9649641
84-1_DOX_24+144 DOX_144R 18222568 0.9913625
84-1_DMSO_24+144 DMSO_144R 28115884 0.9653464
87-1_DOX_24 DOX_24T 19935097 1.0526605
87-1_DMSO_24 DMSO_24T 21302879 0.9773889
87-1_DOX_24+24 DOX_24R 25636959 1.0751043
87-1_DMSO_24+24 DMSO_24R 26319662 0.9940323
87-1_DOX_24+144 DOX_144R 23463426 0.9003102
87-1_DMSO_24+144 DMSO_144R 25840938 0.9888449
78-1_DOX_24 DOX_24T 23085807 0.7676077
78-1_DMSO_24 DMSO_24T 25610495 1.0077383
78-1_DOX_24+24 DOX_24R 18083930 1.1682704
78-1_DMSO_24+24 DMSO_24R 24331177 0.9906872
78-1_DOX_24+144 DOX_144R 19754391 0.9941834
78-1_DMSO_24+144 DMSO_144R 22641509 1.0010734
75-1_DOX_24 DOX_24T 20583626 1.0676786
75-1_DMSO_24 DMSO_24T 28166198 1.0031906
75-1_DOX_24+24 DOX_24R 25831427 1.1530208
75-1_DMSO_24+24 DMSO_24R 26081158 1.0058953
75-1_DOX_24+144 DOX_144R 24659898 0.9261599
75-1_DMSO_24+144 DMSO_144R 25412931 0.9703454
17-3_DOX_24 DOX_24T 22518848 0.9766893
17-3_DMSO_24 DMSO_24T 24589534 0.9612345
17-3_DOX_24+24 DOX_24R 24797547 1.1703079
17-3_DMSO_24+24 DMSO_24R 25977536 0.9509690
17-3_DOX_24+144 DOX_144R 27447106 0.9422729
17-3_DMSO_24+144 DMSO_144R 24893583 0.9356377
90-1_DOX_24 DOX_24T 25187428 1.0311957
90-1_DMSO_24 DMSO_24T 25630519 1.0283437
90-1_DOX_24+24 DOX_24R 26138399 1.1183471
90-1_DMSO_24+24 DMSO_24R 24430396 0.9988688
90-1_DOX_24+144 DOX_144R 23323463 0.9496884
90-1_DMSO_24+144 DMSO_144R 25424152 0.9872926#read in my covariate information RUV_1 dataframe + remove 6R
RUV_df_rm1 <- readRDS("data/new/RUV_df_rm1.RDS")
#filter out 6R for DE analysis
RUV_1_df_filt <- RUV_df_rm1[!grepl("6R$", rownames(RUV_df_rm1)), , drop = FALSE]
Metadata_2$W_1 <- RUV_1_df_filt$W_1
#now ensure that RUV_1 has the right number of cols after removing rep
length(RUV_1_df_filt$W_1)[1] 36#36
#now make this into a list
RUV_1_DE <- RUV_1_df_filt$W_1
#saveRDS(RUV_1_DE, "data/new/RUV_1_DE.RDS")
#create my design matrix for DE + RUVs covariate k=1
design1 <- model.matrix(~0 + RUV_1_DE + Metadata_2$Condition)
colnames(design1) <- gsub("Metadata_2\\$Condition", "", colnames(design1))
#take care that the matrix automatically sorts cols alphabetically
##currently DMSO144R, DMSO24R, DMSO24T, DOX144R, DOX24R, DOX24T
#run duplicate correlation for individual effect
corfit1 <- duplicateCorrelation(object = dge$counts, design = design1, block = Metadata_2$Ind)
#voom transformation and plot
v1 <- voom(dge, design1, block = Metadata_2$Ind, correlation = corfit1$consensus.correlation, plot = TRUE)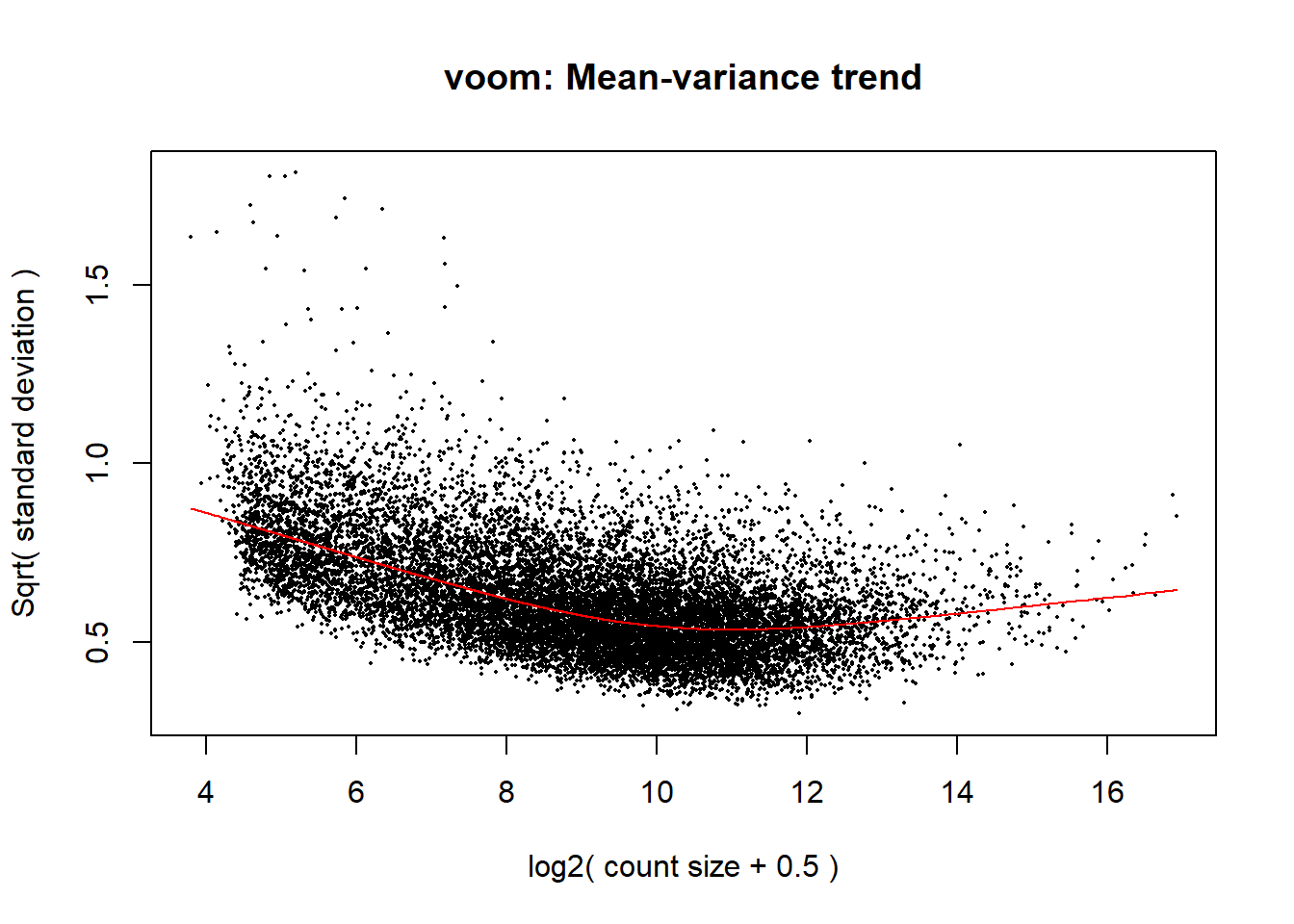
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#fit my linear model
fit1 <- lmFit(v1, design1, block = Metadata_2$Ind, correlation = corfit1$consensus.correlation)
#make my contrast matrix to compare across tx and veh
contrast_matrix_RUV <- makeContrasts(
V.D24T = DOX_24T - DMSO_24T,
V.D24R = DOX_24R - DMSO_24R,
V.D144R = DOX_144R - DMSO_144R,
RUV_1_24T = RUV_1_DE - DMSO_24T,
RUV_1_24R = RUV_1_DE - DMSO_24R,
RUV_1_144R = RUV_1_DE - DMSO_144R,
levels = design1
)
#apply these contrasts to compare DOX to DMSO VEH
fit_RUV <- contrasts.fit(fit1, contrast_matrix_RUV)
fit_RUV <- eBayes(fit_RUV)
#plot the mean-variance trend
plotSA(fit_RUV, main = "Mean-Variance Trend, RUVs k=1")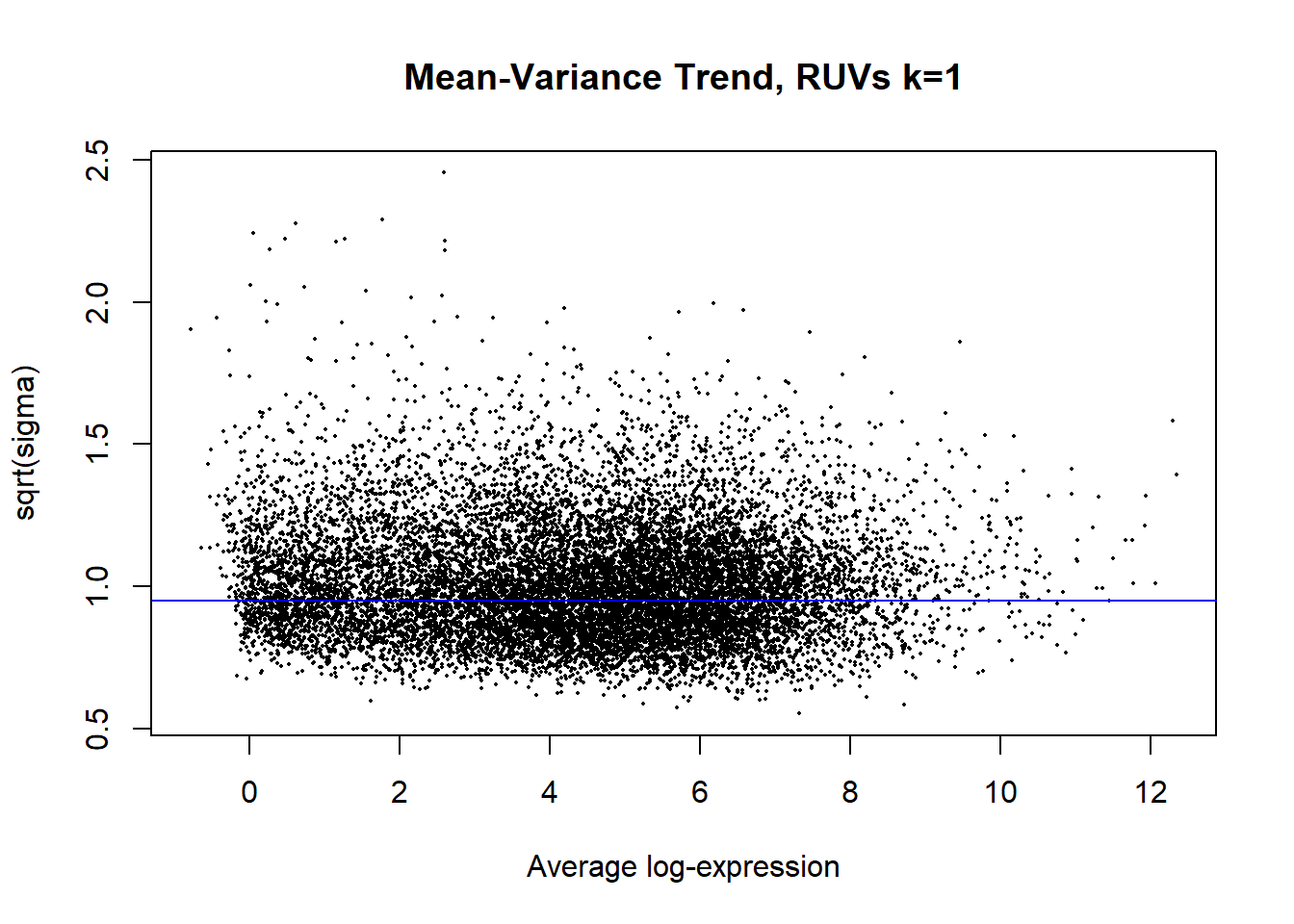
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#look at the summary of your results
##this tells you the number of DEGs in each condition
results_summary1 <- decideTests(fit_RUV, adjust.method = "BH", p.value = 0.05)
summary(results_summary1) V.D24T V.D24R V.D144R RUV_1_24T RUV_1_24R RUV_1_144R
Down 4866 3727 466 13212 13224 13168
NotSig 4899 7107 13659 1014 1002 1058
Up 4554 3485 194 93 93 93# V.D24T V.D24R V.D144R RUV_1_24T RUV_1_24R RUV_1_144R
# Down 4866 3727 466 13212 13224 13168
# NotSig 4899 7107 13659 1014 1002 1058
# Up 4554 3485 194 93 93 93
#compare this to my previous DEGs I found
summary(results_summary) V.D24T V.D24R V.D144R
Down 4723 3593 359
NotSig 5076 7151 13810
Up 4520 3575 150# V.D24T V.D24R V.D144R
# Down 4723 3593 359
# NotSig 5076 7151 13810
# Up 4520 3575 150
#overall, there are more DEGs found after RUVs k=1 correction
#this was an expected result as it increases tx effect w/ correction##Generate Toptables RUVs
# Generate Top Table for Specific Comparisons
Toptable_V.D24T_k1 <- topTable(fit = fit_RUV, coef = "V.D24T", number = nrow(x), adjust.method = "BH", p.value = 1, sort.by = "none")
#write.csv(Toptable_V.D24T_k1, "data/new/DEGs/Toptable_V.D24T_k1.csv")
Toptable_V.D24R_k1 <- topTable(fit = fit_RUV, coef = "V.D24R", number = nrow(x), adjust.method = "BH", p.value = 1, sort.by = "none")
#write.csv(Toptable_V.D24R_k1, "data/new/DEGs/Toptable_V.D24R_k1.csv")
Toptable_V.D144R_k1 <- topTable(fit = fit_RUV, coef = "V.D144R", number = nrow(x), adjust.method = "BH", p.value = 1, sort.by = "none")
#write.csv(Toptable_V.D144R_k1, "data/new/DEGs/Toptable_V.D144R_k1.csv")
# save all of these toptables as R objects
# saveRDS(list(
# V.D24T_k1 = Toptable_V.D24T_k1,
# V.D24R_k1 = Toptable_V.D24R_k1,
# V.D144R_k1 = Toptable_V.D144R_k1
# ), file = "data/new/Toptable_list_RUVk1.RDS")
Toptable_list_RUVk1 <- readRDS("data/new/Toptable_list_RUVk1.RDS")
#################################################################
#this section is commented out since it only needs to run once
#kept the code for posterity
# #I want to add the hgnc symbols to my toptables as well
# ####D24T####
# #load in data
# sample_toptab_24T <- read_csv("data/new/DEGs/Toptable_V.D24T_k1.csv", show_col_types = FALSE)
# # ----------------- Ensure Entrez_ID is Present and in Character Format -----------------
# # Check column names
# print(colnames(sample_toptab_24T))
# # Rename if needed (adjust if the column name is not exactly 'Entrez_ID')
# colnames(sample_toptab_24T)[1] <- "Entrez_ID"
# # Convert Entrez_ID to character
# sample_toptab_24T <- sample_toptab_24T %>%
# mutate(Entrez_ID = as.character(Entrez_ID))
# # ----------------- Map Entrez_ID to Gene Symbol -----------------
# gene_symbols1 <- AnnotationDbi::select(
# org.Hs.eg.db,
# keys = sample_toptab_24T$Entrez_ID,
# columns = c("SYMBOL"),
# keytype = "ENTREZID"
# )
# # ----------------- Join Back to Main Data -----------------
# Toptable_RUV_24T <- left_join(sample_toptab_24T, gene_symbols1, by = c("Entrez_ID" = "ENTREZID"))
# Toptable_RUV_24T %>% column_to_rownames(., var = "Entrez_ID")
# # ----------------- Save Annotated Output -----------------
# #write_csv(Toptable_RUV_24T, "data/new/DEGs/Toptable_RUV_24T.csv")
#
# #now do this again for my other two toptables
#
# ####24R####
# #load in data
# sample_toptab_24R <- read_csv("data/new/DEGs/Toptable_V.D24R_k1.csv", show_col_types = FALSE)
# # ----------------- Ensure Entrez_ID is Present and in Character Format -----------------
# # Check column names
# print(colnames(sample_toptab_24R))
# # Rename if needed (adjust if the column name is not exactly 'Entrez_ID')
# colnames(sample_toptab_24R)[1] <- "Entrez_ID"
# # Convert Entrez_ID to character
# sample_toptab_24R <- sample_toptab_24R %>%
# mutate(Entrez_ID = as.character(Entrez_ID))
# # ----------------- Map Entrez_ID to Gene Symbol -----------------
# gene_symbols2 <- AnnotationDbi::select(
# org.Hs.eg.db,
# keys = sample_toptab_24R$Entrez_ID,
# columns = c("SYMBOL"),
# keytype = "ENTREZID"
# )
# # ----------------- Join Back to Main Data -----------------
# Toptable_RUV_24R <- left_join(sample_toptab_24R, gene_symbols2, by = c("Entrez_ID" = "ENTREZID"))
# Toptable_RUV_24R %>% column_to_rownames(., var = "Entrez_ID")
# # ----------------- Save Annotated Output -----------------
# #I'll make these symbols into rownames later for my volcano plots
# #write_csv(Toptable_RUV_24R, "data/new/DEGs/Toptable_RUV_24R.csv")
#
#
# ####D144R####
# #load in data
# sample_toptab_144R <- read_csv("data/new/DEGs/Toptable_V.D144R_k1.csv", show_col_types = FALSE)
# # ----------------- Ensure Entrez_ID is Present and in Character Format -----------------
# # Check column names
# print(colnames(sample_toptab_144R))
# # Rename if needed (adjust if the column name is not exactly 'Entrez_ID')
# colnames(sample_toptab_144R)[1] <- "Entrez_ID"
# # Convert Entrez_ID to character
# sample_toptab_144R <- sample_toptab_144R %>%
# mutate(Entrez_ID = as.character(Entrez_ID))
# # ----------------- Map Entrez_ID to Gene Symbol -----------------
# gene_symbols3 <- AnnotationDbi::select(
# org.Hs.eg.db,
# keys = sample_toptab_144R$Entrez_ID,
# columns = c("SYMBOL"),
# keytype = "ENTREZID"
# )
# # ----------------- Join Back to Main Data -----------------
# Toptable_RUV_144R <- left_join(sample_toptab_144R, gene_symbols3, by = c("Entrez_ID" = "ENTREZID"))
# Toptable_RUV_144R %>% column_to_rownames(., var = "Entrez_ID")
# # ----------------- Save Annotated Output -----------------
# #I'll make these symbols into rownames later for my volcano plots
# #write_csv(Toptable_RUV_144R, "data/new/DEGs/Toptable_RUV_144R.csv")
# write.csv(Toptable_RUV_24T, "data/new/DEGs/Toptable_RUV_24T_final.csv")
# write.csv(Toptable_RUV_24R, "data/new/DEGs/Toptable_RUV_24R_final.csv")
# write.csv(Toptable_RUV_144R, "data/new/DEGs/Toptable_RUV_144R_final.csv")
Toptable_RUV_24T <- read.csv("data/new/DEGs/Toptable_RUV_24T_final.csv")
Toptable_RUV_24R <- read.csv("data/new/DEGs/Toptable_RUV_24R_final.csv")
Toptable_RUV_144R <- read.csv("data/new/DEGs/Toptable_RUV_144R_final.csv")
# save all of these toptables as R objects
# saveRDS(list(
# V.D24T_RUV = Toptable_RUV_24T,
# V.D24R_RUV = Toptable_RUV_24R,
# V.D144R_RUV = Toptable_RUV_144R
# ), file = "data/new/Toptable_list_RUVk1_Symbols.RDS")
Toptable_list_RUVk1_symbols <- readRDS("data/new/Toptable_list_RUVk1_Symbols.RDS")##Volcano Plots of Original DEGs
#make a function to generate volcano plots + add gene numbers
generate_volcano_plot <- function(toptable, title) {
#make significance labels
toptable$Significance <- "Not Significant"
toptable$Significance[toptable$logFC > 0 & toptable$adj.P.Val < 0.05] <- "Upregulated"
toptable$Significance[toptable$logFC < 0 & toptable$adj.P.Val < 0.05] <- "Downregulated"
#add number of genes for each significance label
upgenes <- toptable %>% filter(Significance == "Upregulated") %>% nrow()
nsgenes <- toptable %>% filter(Significance == "Not Significant") %>% nrow()
downgenes <- toptable %>% filter(Significance == "Downregulated") %>% nrow()
#make legend labels for no of genes
legend_lab <- c(
str_c("Upregulated: ", upgenes),
str_c("Not Significant: ", nsgenes),
str_c("Downregulated: ", downgenes)
)
#specify the colors for the legend
legend_col <- c(
str_c("Upregulated: " = "blue"),
str_c("Not Significant: " = "gray"),
str_c("Downregulated: " = "red")
)
#generate volcano plot w/ legend
ggplot(toptable, aes(x = logFC,
y = -log10(P.Value),
color = Significance)) +
geom_point(alpha = 0.4, size = 2) +
scale_color_manual(values = c("Upregulated" = "blue",
"Not Significant" = "gray",
"Downregulated" = "red"),
labels = legend_lab) +
xlim(-10, 10) +
labs(title = title,
x = expression(x = "log"[2]*"FC"),
y = expression(y = "-log"[10]*"P-value")) +
theme_bw()+
guides(color = guide_legend(override.aes = list(color = legend_col)))+
theme(legend.position = "right",
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = rel(1.25)))
}
#generate volcano plots across each comparison
volcano_plots <- list(
"V.D24T" = generate_volcano_plot(Toptable_V.D24T, "Volcano Plot DOX 24hr (adj P-val<0.05)"),
"V.D24R" = generate_volcano_plot(Toptable_V.D24R, "Volcano Plot DOX 24hr Recovery (adj P-val<0.05)"),
"V.D144R" = generate_volcano_plot(Toptable_V.D144R, "Volcano Plot DOX 144hr Recovery (adj P-val<0.05)")
)
# Display each volcano plot
for (plot_name in names(volcano_plots)) {
print(volcano_plots[[plot_name]])
}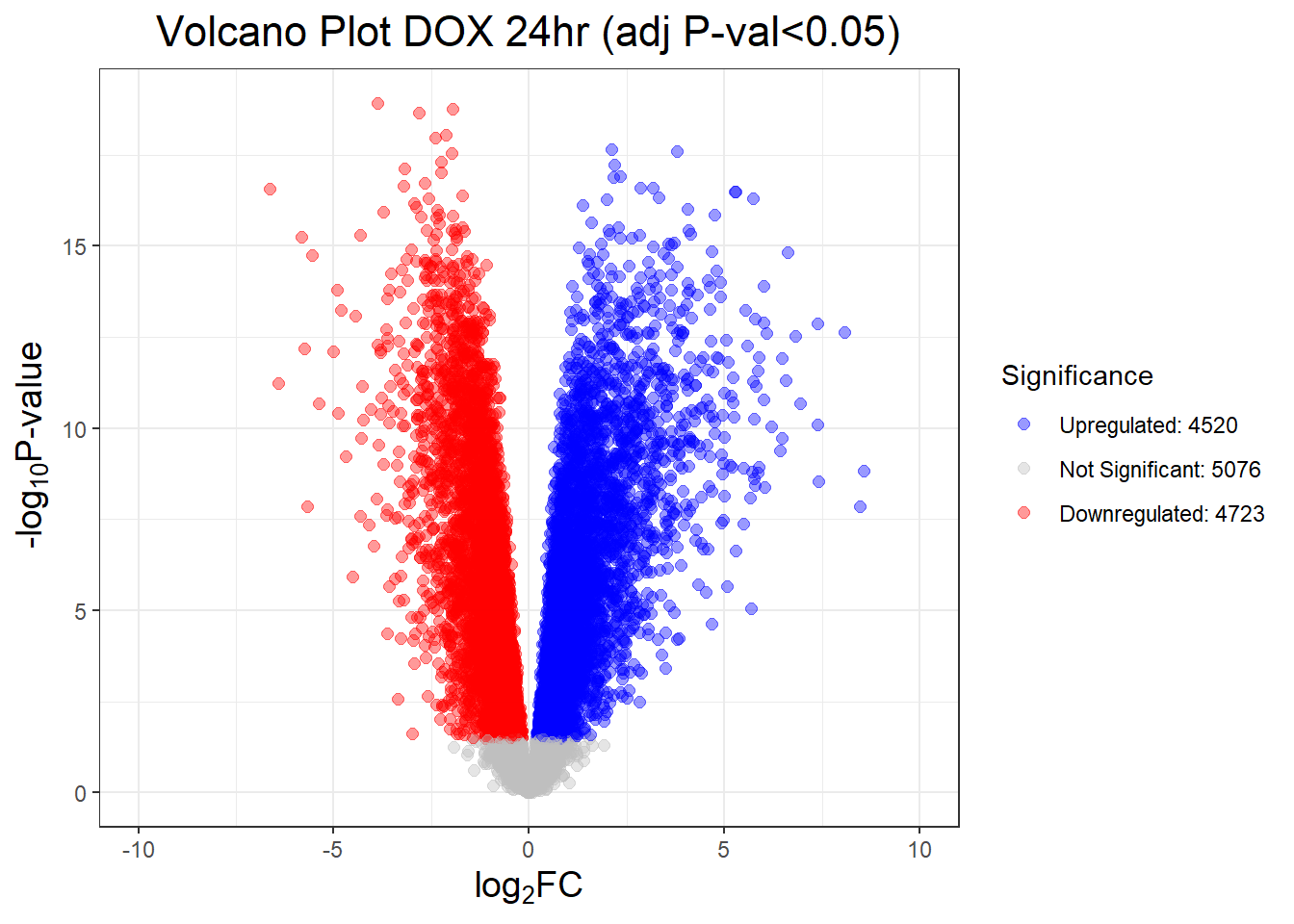
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
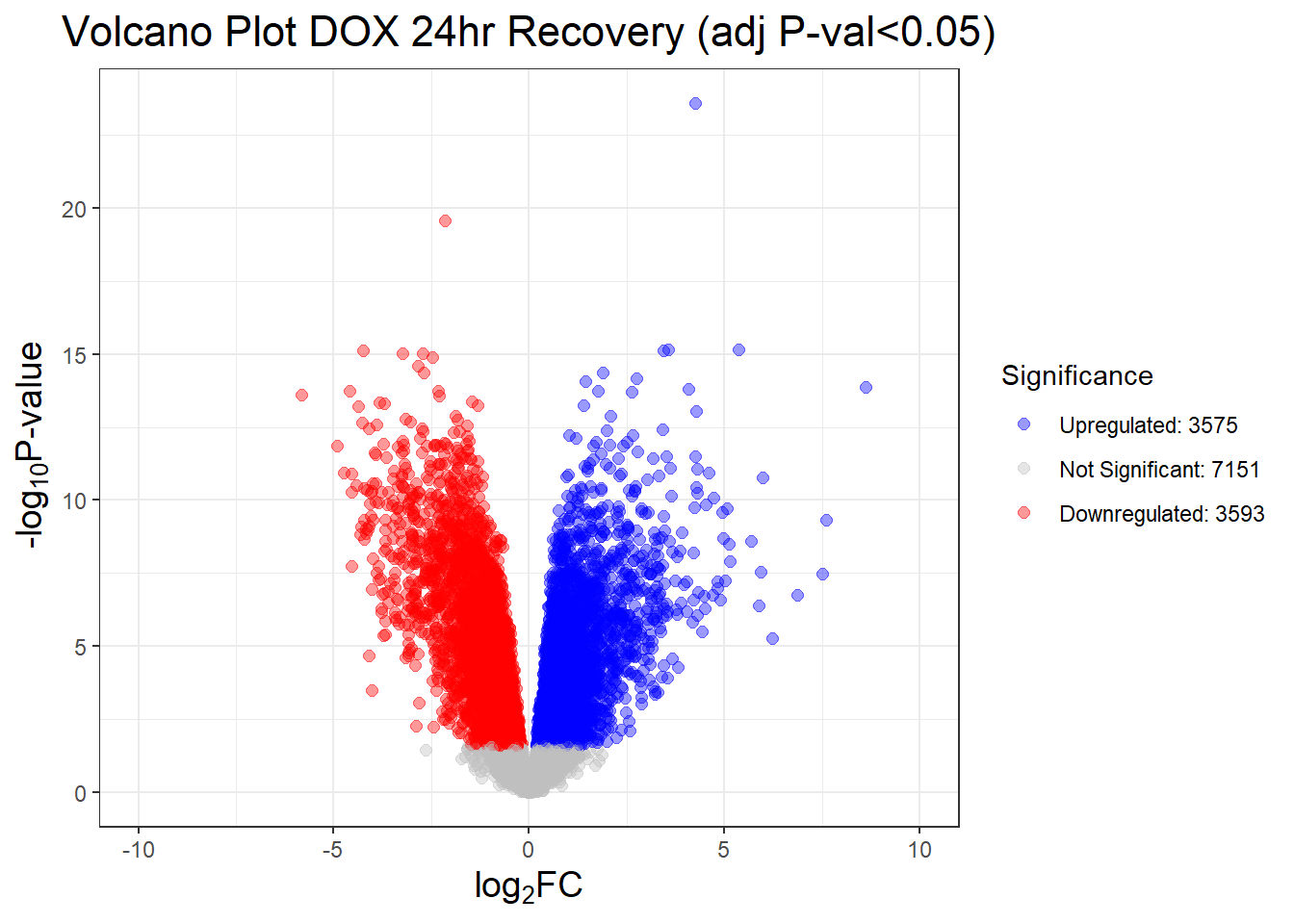
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
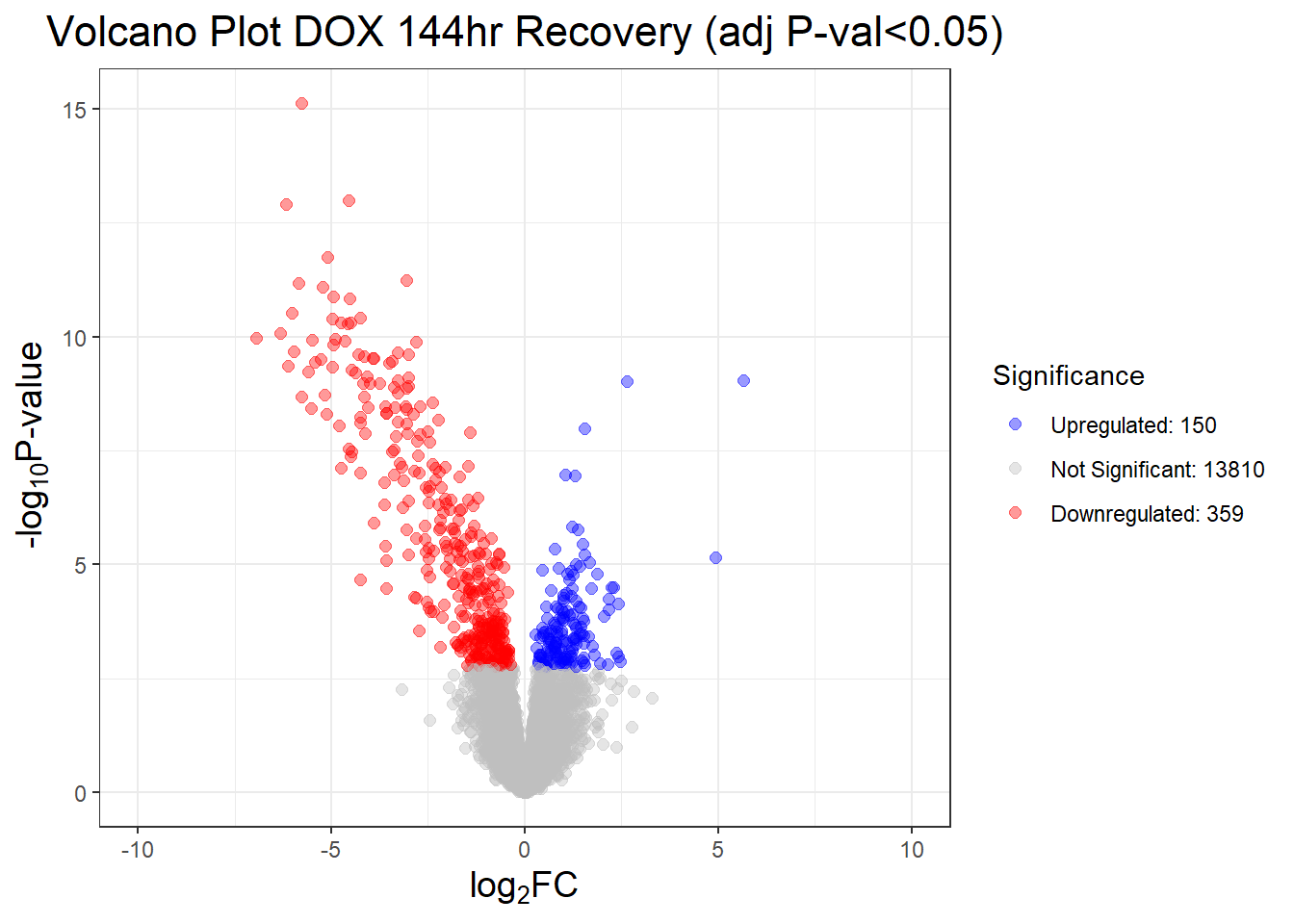
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
##Top 5 DEGs after RUVs k=1 Correction
#use your three toptables so I can pull out top 5 genes from each based on adj. p val
top5_D24T_1 <- Toptable_RUV_24T[order(Toptable_RUV_24T$adj.P.Val), ][1:5,]
top5_D24R_1 <- Toptable_RUV_24R[order(Toptable_RUV_24R$adj.P.Val), ][1:5,]
top5_D144R_1 <- Toptable_RUV_144R[order(Toptable_RUV_144R$adj.P.Val), ][1:5,]
#now that I've pulled the top 5 DEGs from each, make a list to pull them from my log2cpm data
boxplot1 <- read.csv("data/new/filcpm_final_matrix.csv") %>%
as.data.frame()
#Define gene list
#these are the top 5 genes pulled from my toptables
top5_D24T_geneslist_1 <- c(top5_D24T_1$Entrez_ID)
top5_D24R_geneslist_1 <- c(top5_D24R_1$Entrez_ID)
top5_D144R_geneslist_1 <- c(top5_D144R_1$Entrez_ID)
#Add more gene symbols as needed or add more categories
#now pull these from my log2cpm matrix
top5_D24T_genes_1 <- boxplot1[boxplot1$Entrez_ID %in% top5_D24T_geneslist_1,]
dim(top5_D24T_genes_1)[1] 5 44#5 genes in 44 cols
top5_D24R_genes_1 <- boxplot1[boxplot1$Entrez_ID %in% top5_D24R_geneslist_1,]
dim(top5_D24R_genes_1)[1] 5 44#5 genes in 44 cols
top5_D144R_genes_1 <- boxplot1[boxplot1$Entrez_ID %in% top5_D144R_geneslist_1,]
dim(top5_D144R_genes_1)[1] 5 44#5 genes in 44 cols
#Now put in the function I want to use to generate boxplots of genes
#####D24T#####
process_top5_D24T_1 <- function(gene) {
gene_data <- top5_D24T_genes_1 %>% filter(Entrez_ID == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2cpm") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6R$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#Generate Boxplots from the above function using our gene list above
for (gene in top5_D24T_geneslist_1) {
gene_data <- process_top5_D24T_1(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2cpm, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2cpm", gene, "top5DEGs D24T RUVs")) +
labs(x = "Treatment", y = "log2cpm") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}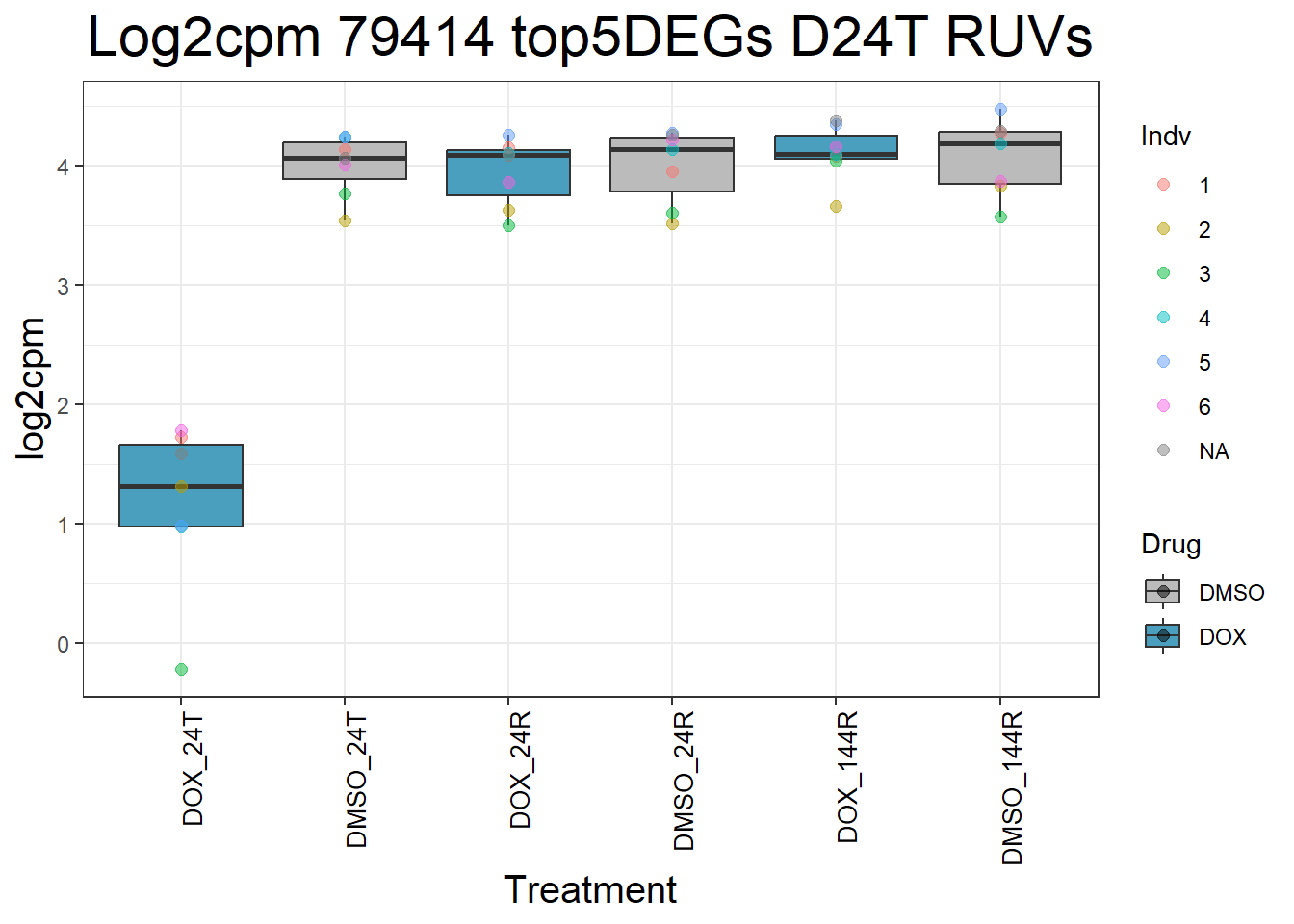
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
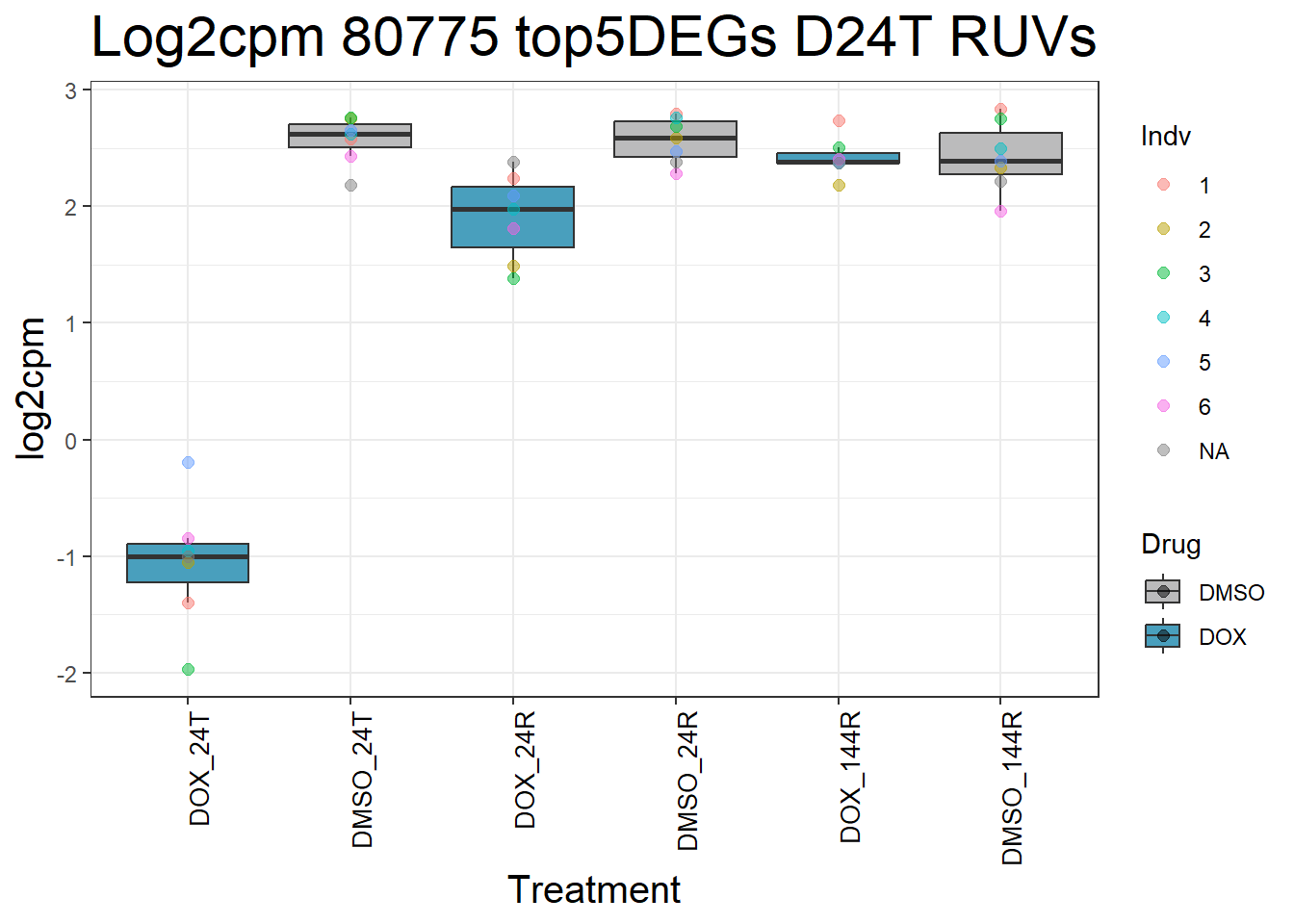
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
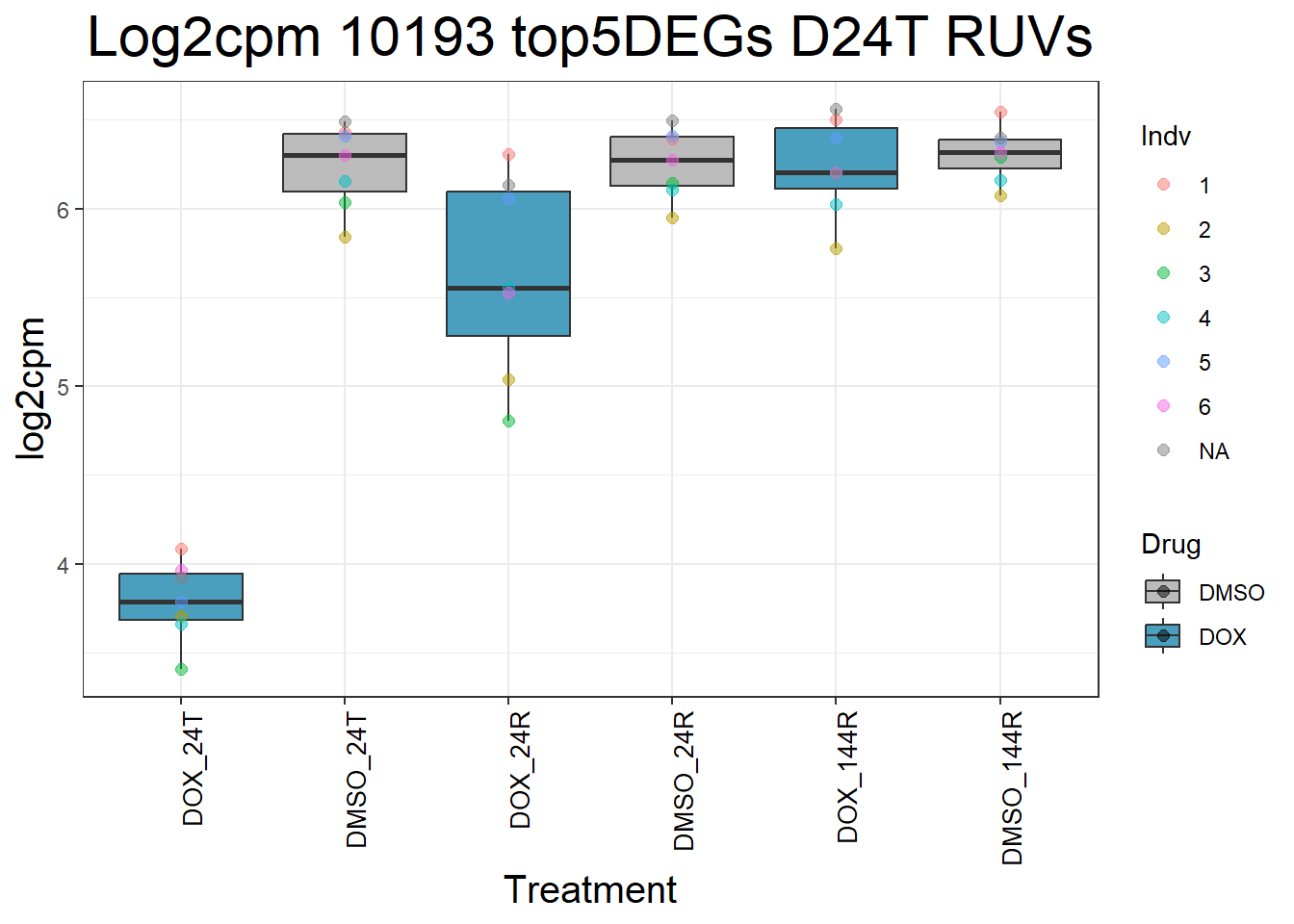
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
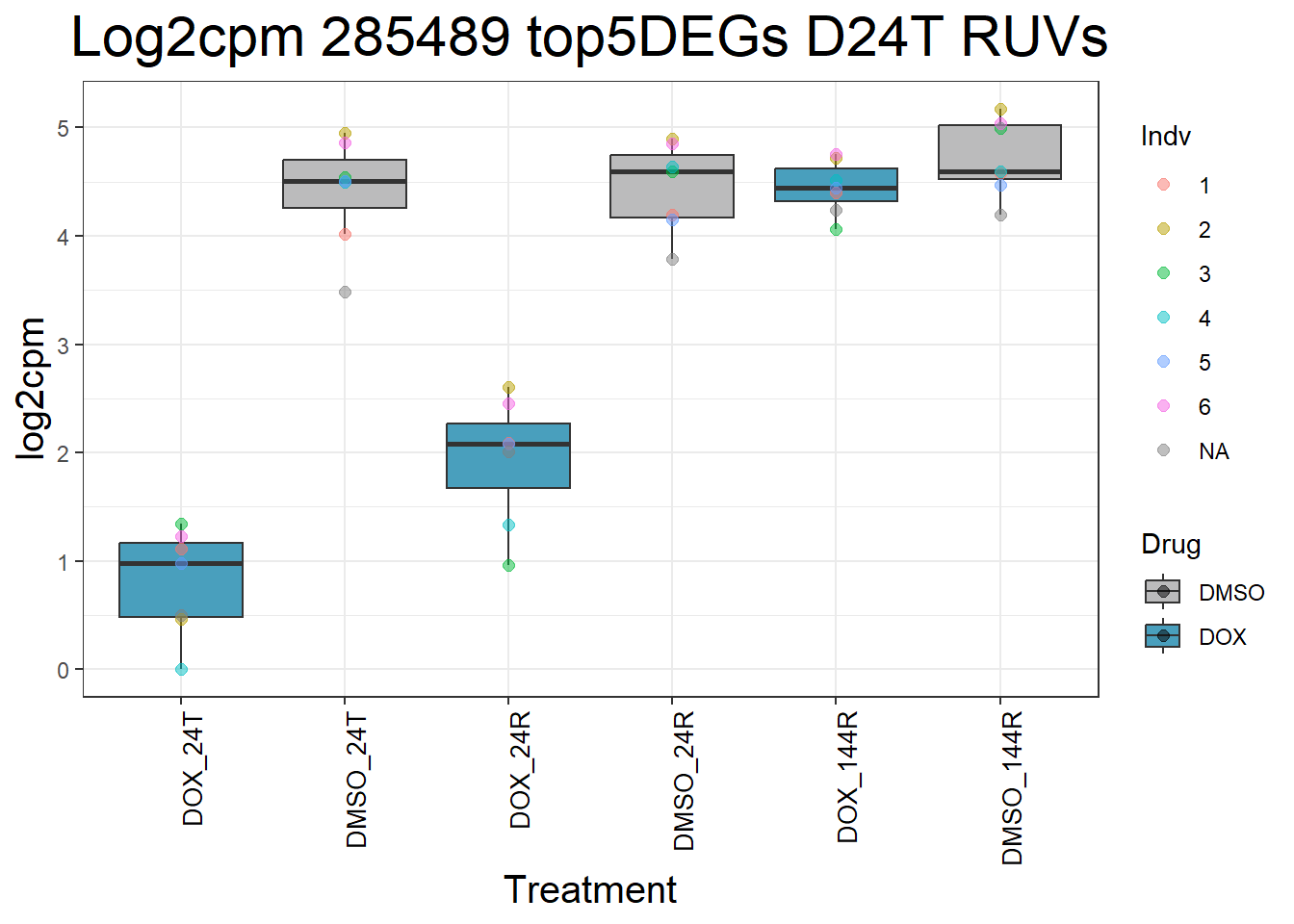
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
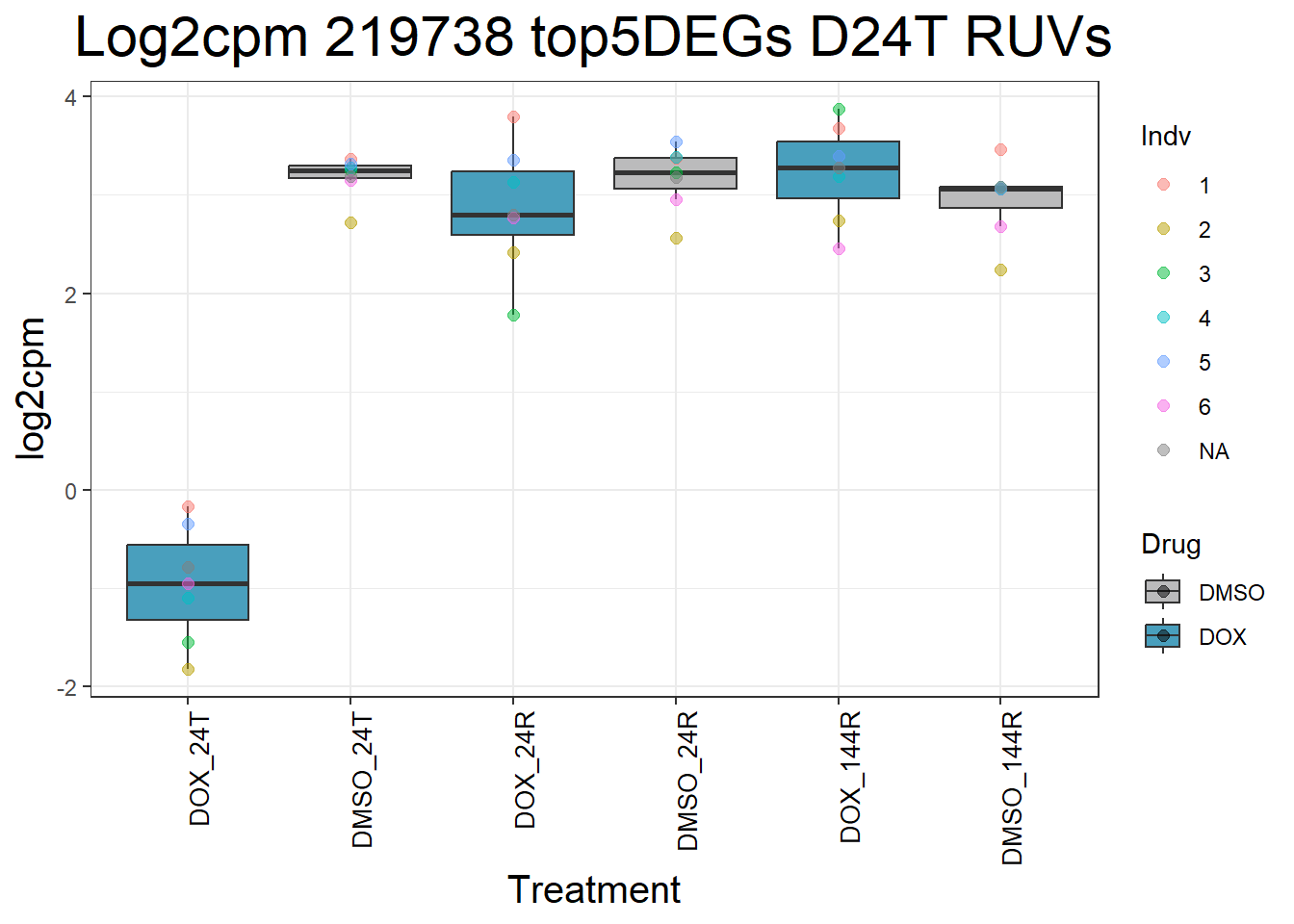
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####D24R#####
process_top5_D24R_1 <- function(gene) {
gene_data <- top5_D24R_genes_1 %>% filter(Entrez_ID == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2cpm") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6R$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#Generate Boxplots from the above function using our gene list above
for (gene in top5_D24R_geneslist_1) {
gene_data <- process_top5_D24R_1(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2cpm, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2cpm", gene, "top5DEGs D24R RUVs")) +
labs(x = "Treatment", y = "log2cpm") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}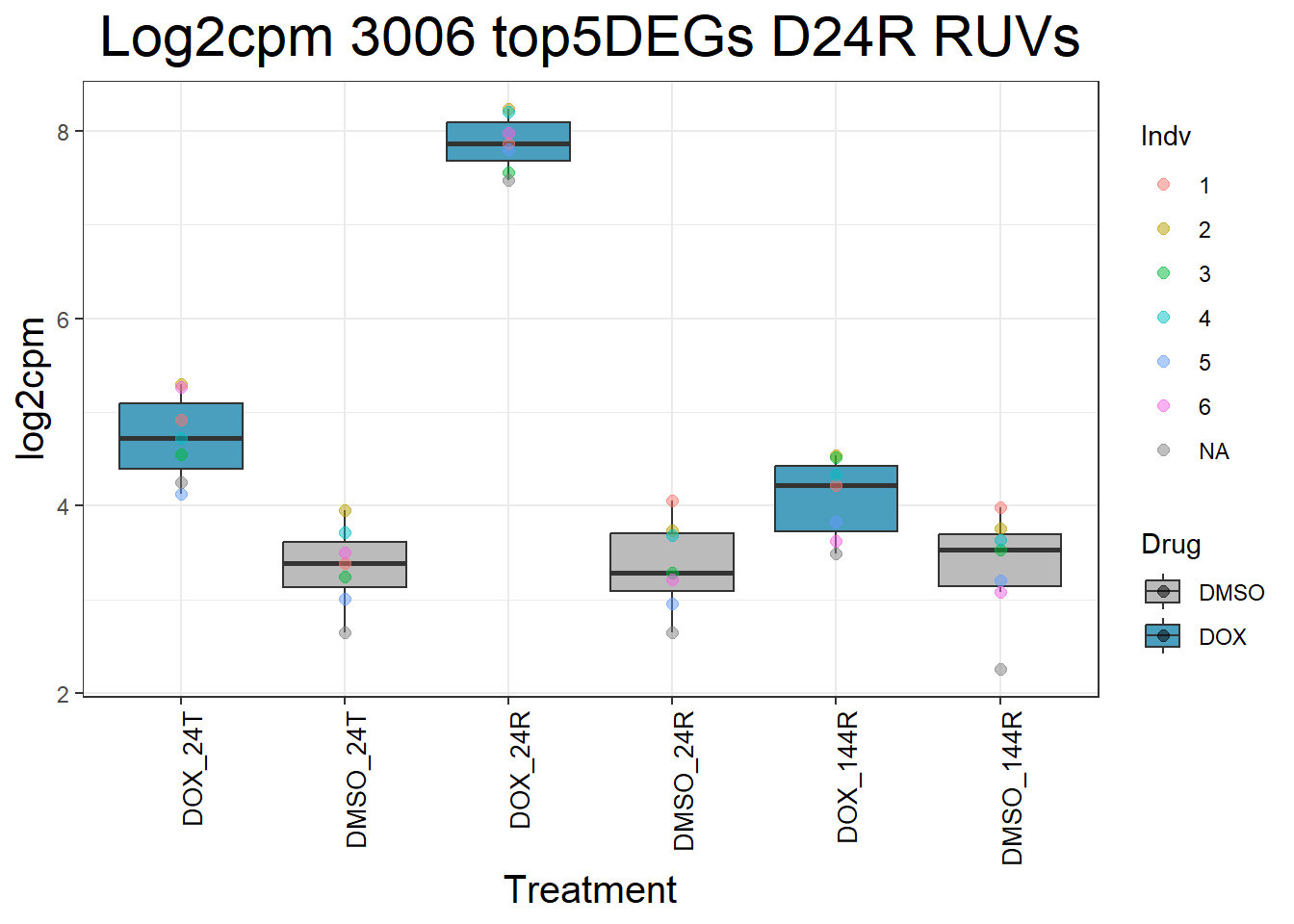
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
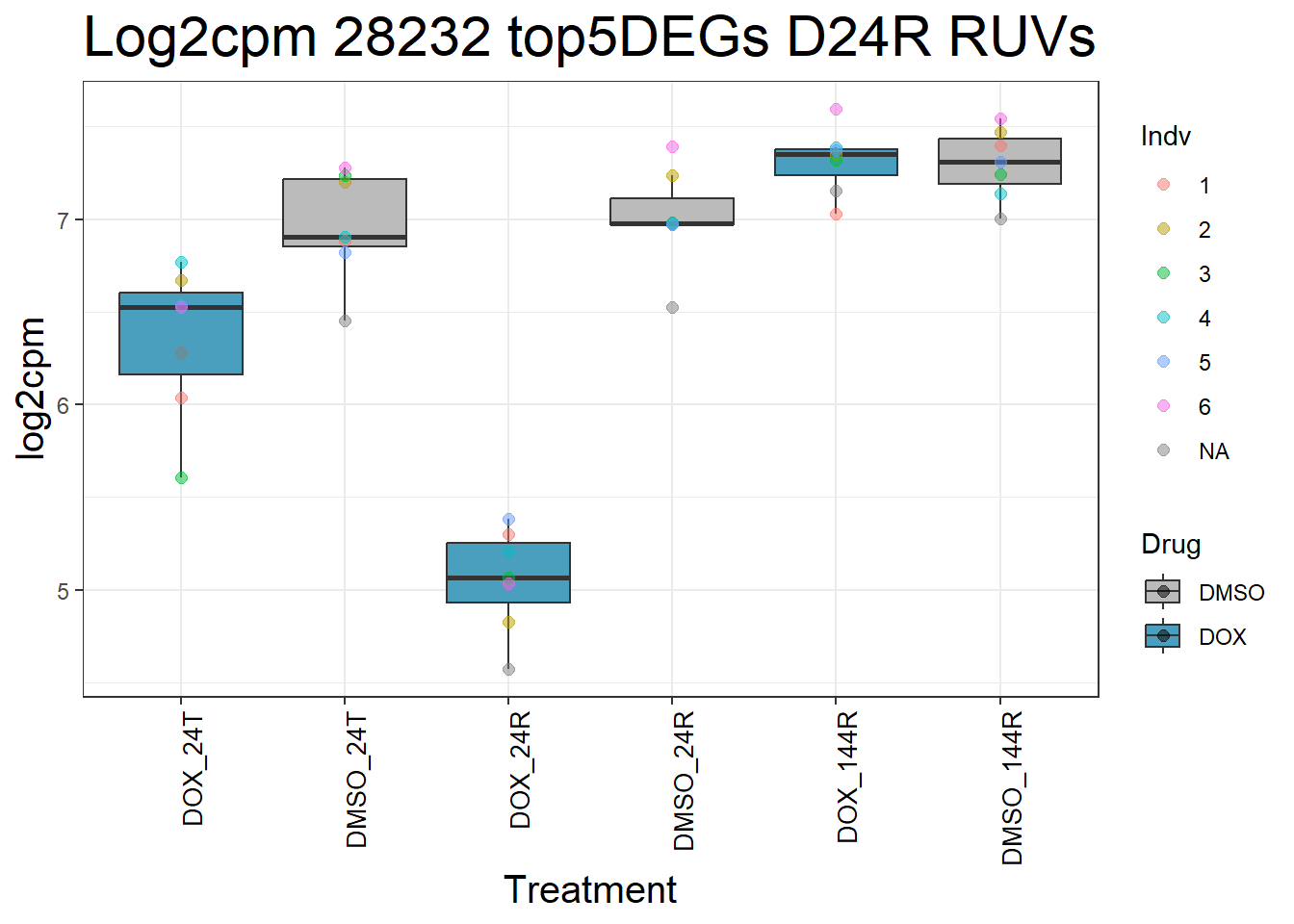
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
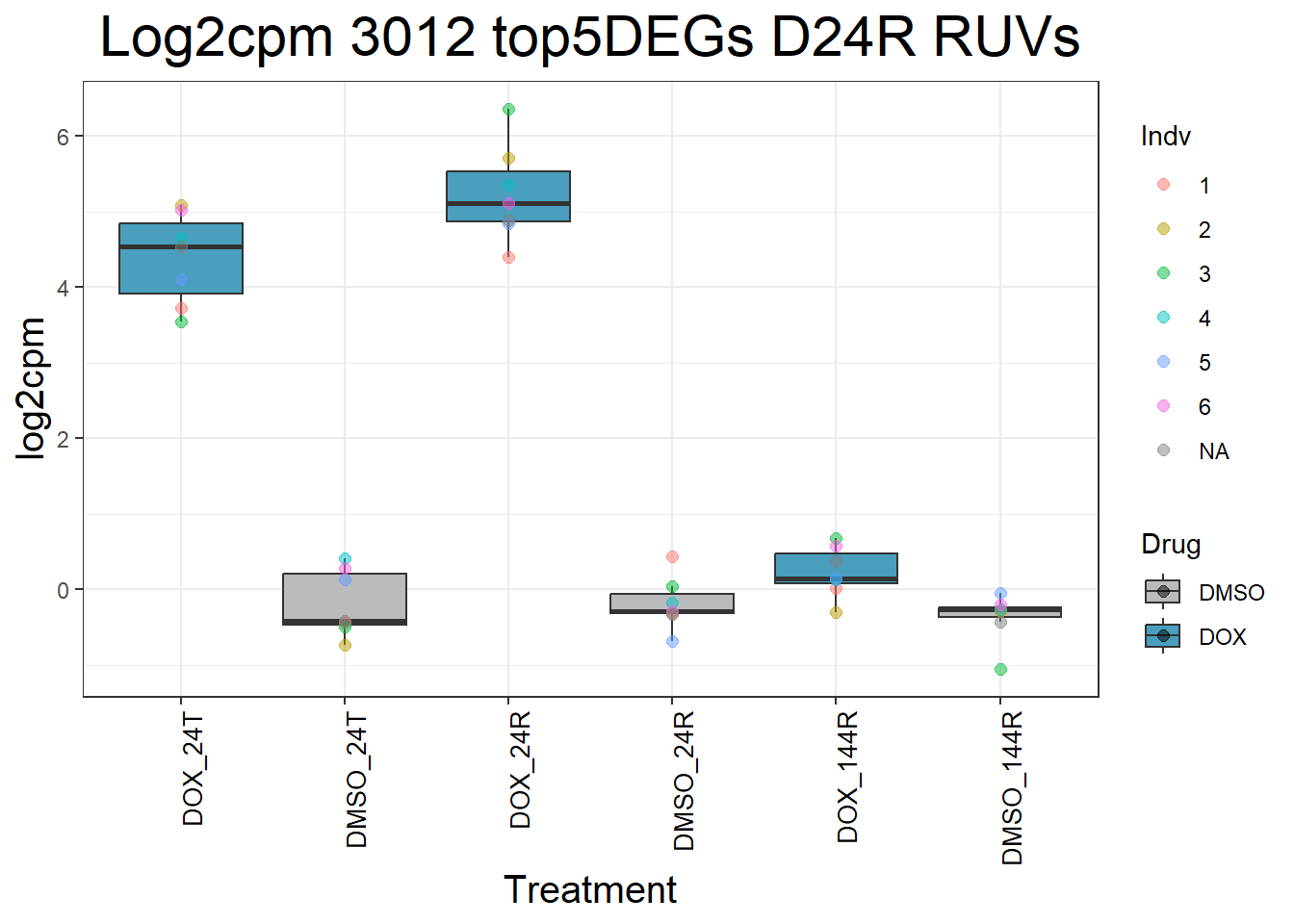
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
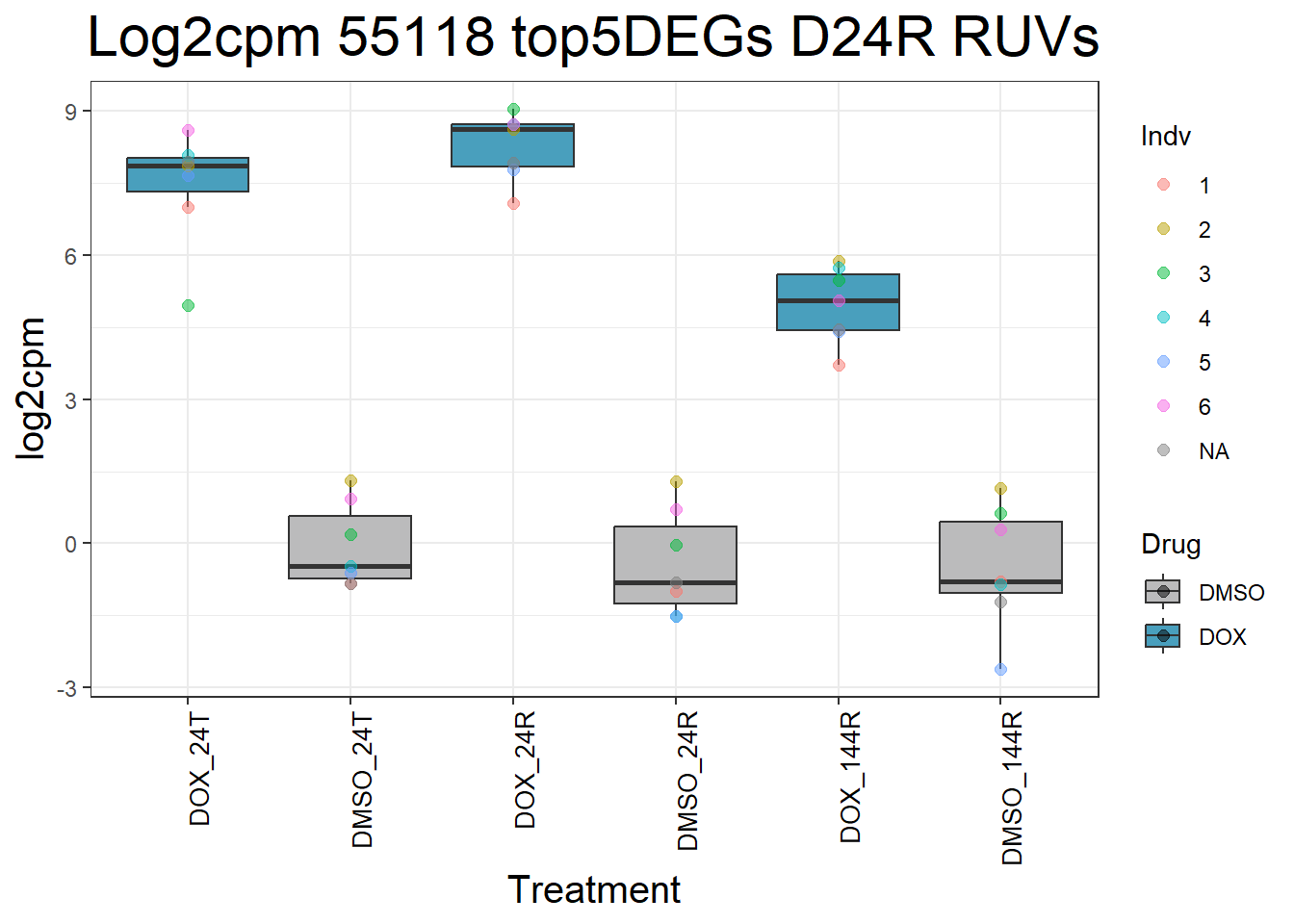
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
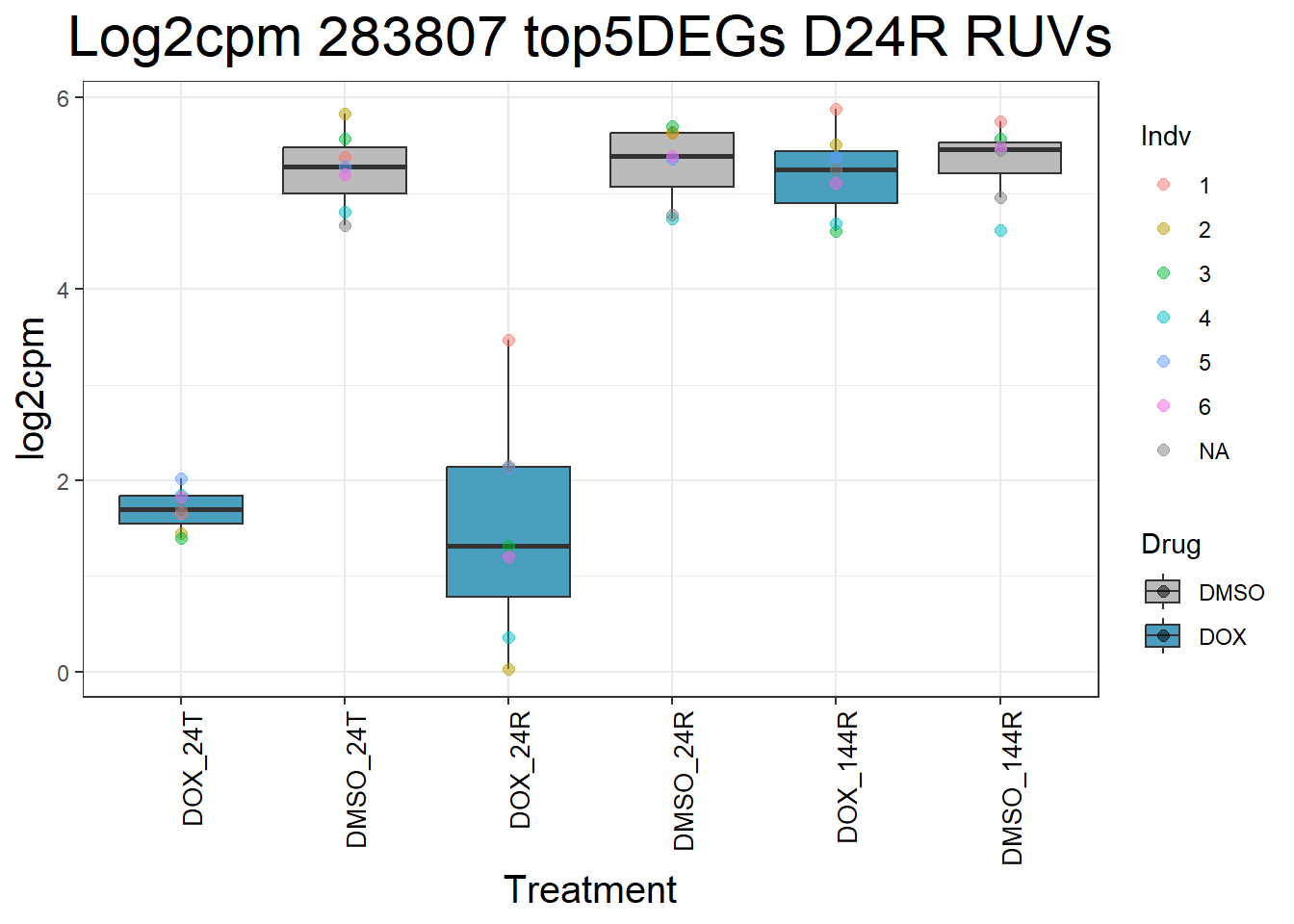
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####D144R#####
process_top5_D144R_1 <- function(gene) {
gene_data <- top5_D144R_genes_1 %>% filter(Entrez_ID == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2cpm") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6R$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#Generate Boxplots from the above function using our gene list above
for (gene in top5_D144R_geneslist_1) {
gene_data <- process_top5_D144R_1(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2cpm, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2cpm", gene, "top5DEGs D144R RUVs")) +
labs(x = "Treatment", y = "log2cpm") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}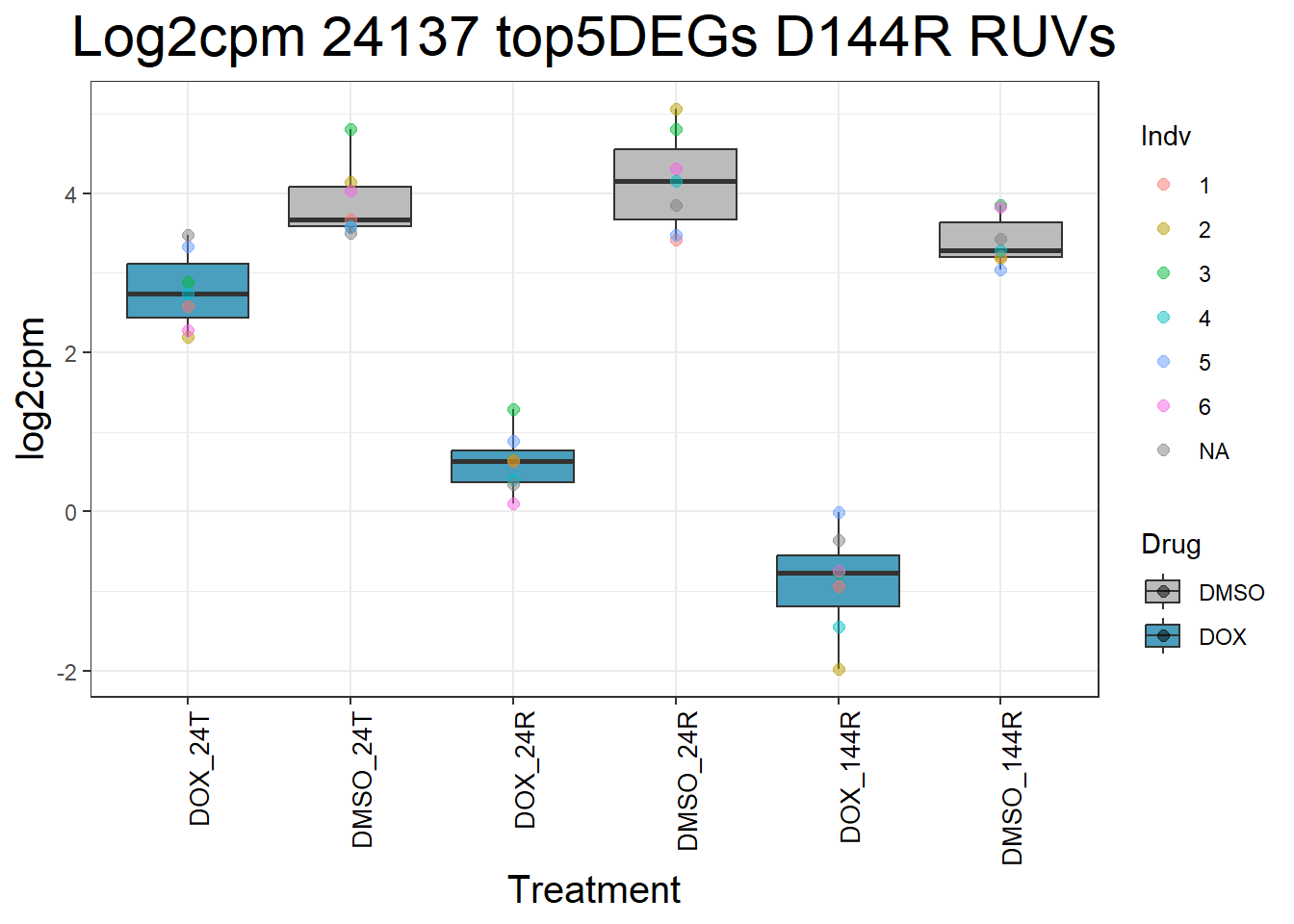
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
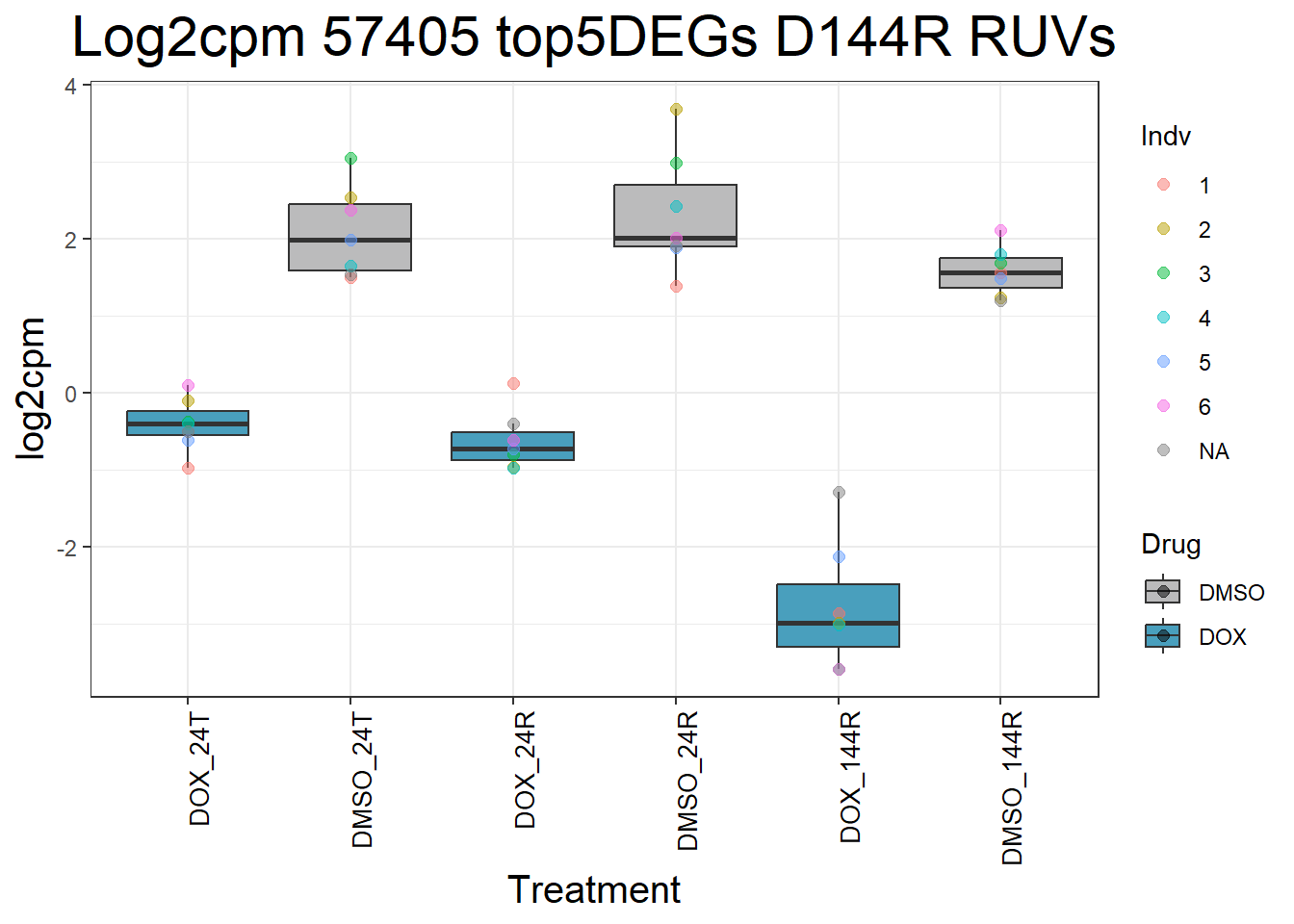
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
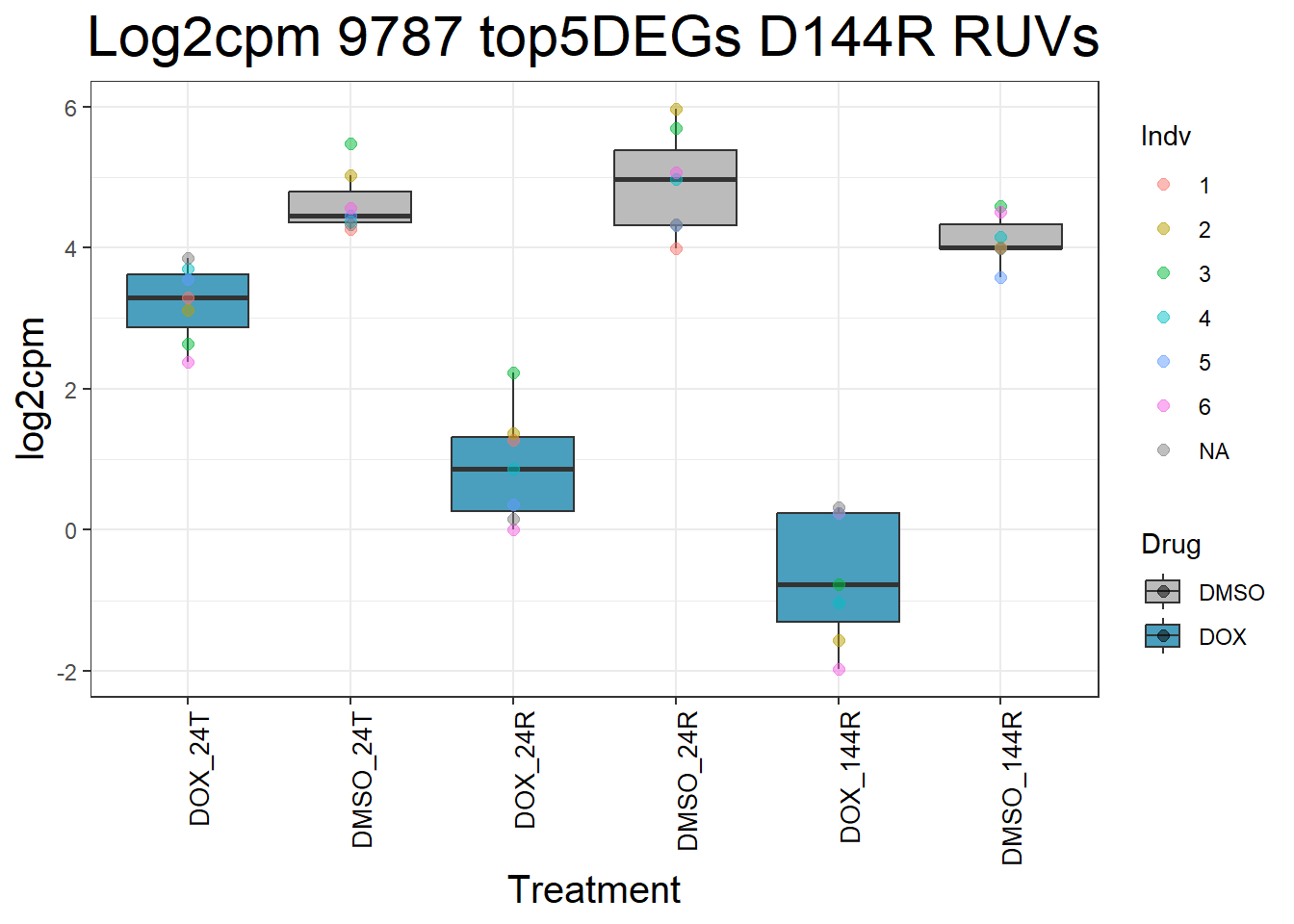
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
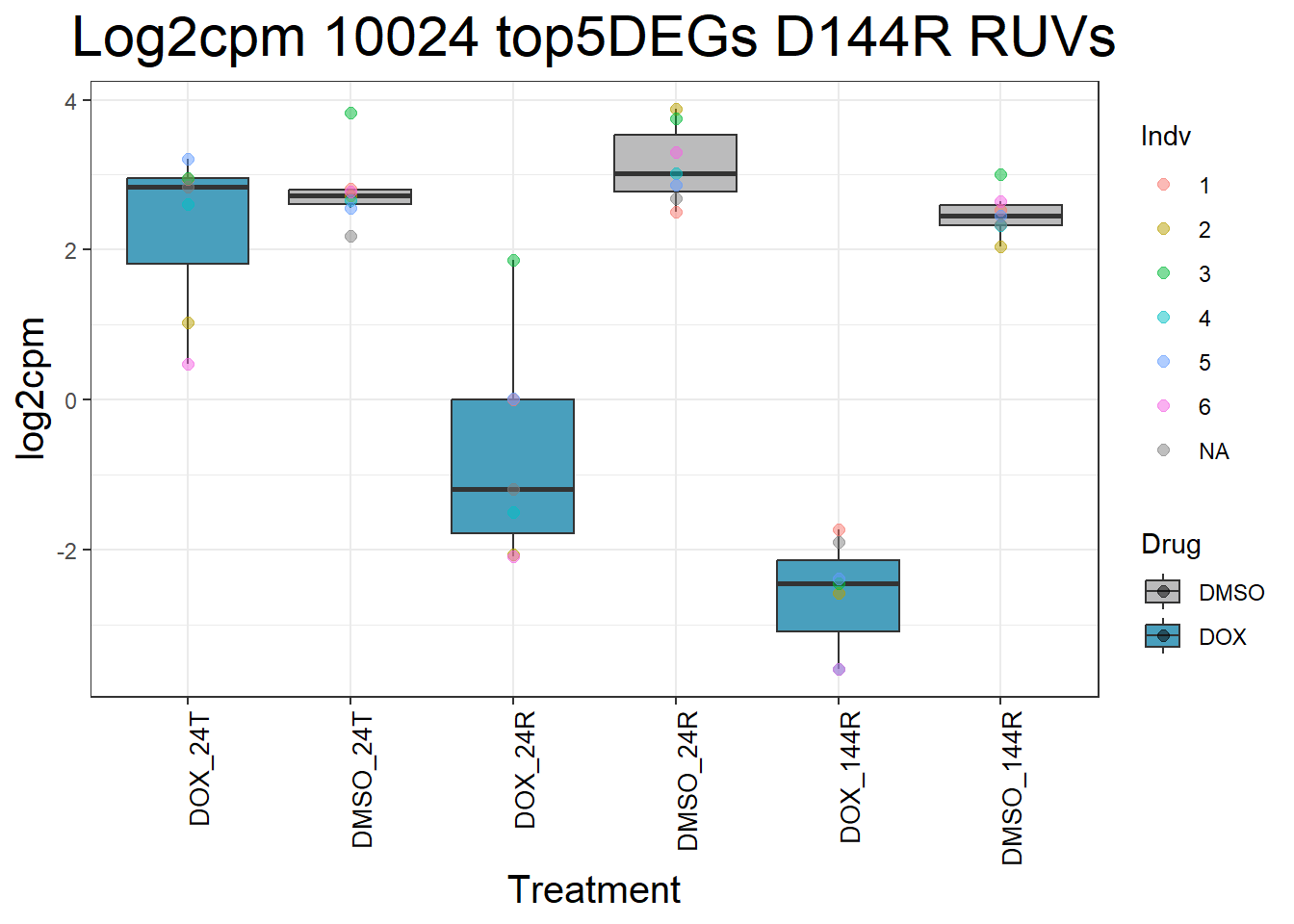
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
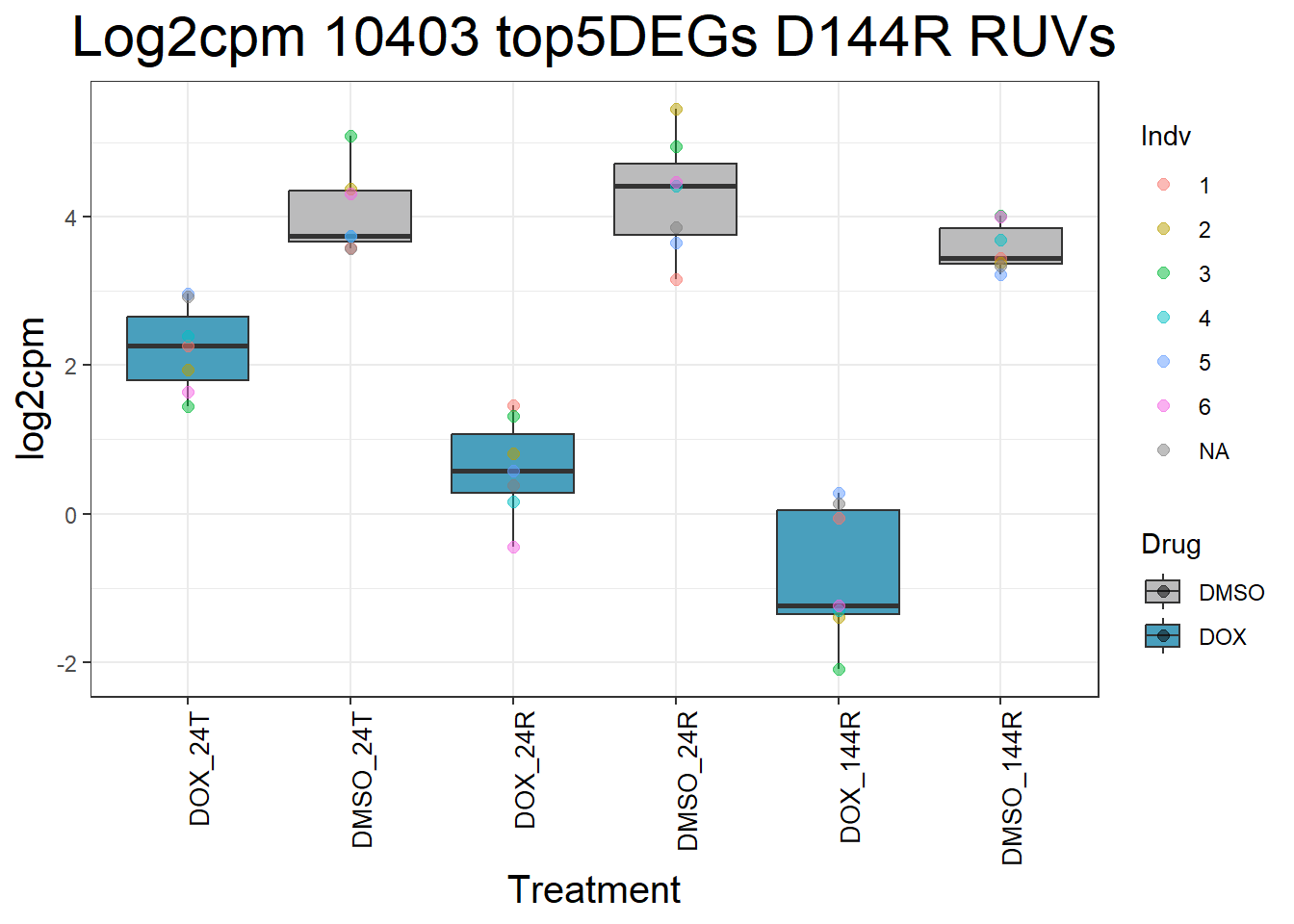
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
##LogFC DEGs RUVs k=1 All Conditions
# Toptable_RUV_24T
# Toptable_RUV_24R
# Toptable_RUV_144R
# Load DEGs Data
DOX_24T_1 <- read.csv("data/new/DEGs/Toptable_RUV_24T_final.csv")
DOX_24R_1 <- read.csv("data/new/DEGs/Toptable_RUV_24R_final.csv")
DOX_144R_1 <- read.csv("data/new/DEGs/Toptable_RUV_144R_final.csv")
#make a list of all of the genes in this set so I can plot the logFC in other sets
D24T_DEGs_1 <- DOX_24T_1$Entrez_ID[DOX_24T_1$adj.P.Val < 0.05]
length(D24T_DEGs_1)[1] 9420#9243 genes
D24R_DEGs_1 <- DOX_24R_1$Entrez_ID[DOX_24R_1$adj.P.Val < 0.05]
length(D24R_DEGs_1)[1] 7212#7168 genes
D144R_DEGs_1 <- DOX_144R_1$Entrez_ID[DOX_144R_1$adj.P.Val < 0.05]
length(D144R_DEGs_1)[1] 660#509 genes
#plot those full gene sets in logFC
D24T_DEGs_1 <- DOX_24T$Entrez_ID[DOX_24T$adj.P.Val < 0.05]
length(D24T_DEGs)[1] 9243D24R_DEGs_1 <- DOX_24R$Entrez_ID[DOX_24R$adj.P.Val < 0.05]
length(D24R_DEGs)[1] 7168D144R_DEGs_1 <- DOX_144R$Entrez_ID[DOX_144R$adj.P.Val < 0.05]
length(D144R_DEGs)[1] 509#now that I have the full list of genes, I want to plot the logFC across conditions
#Combine the toptables I have from pairwise analysis into a single dataframe
d24_toptable_dxr_1 <- Toptable_RUV_24T %>%
mutate(Time = "24")
d24r_toptable_dxr_1 <- Toptable_RUV_24R %>%
mutate(Time = "24R")
d144r_toptable_dxr_1 <- Toptable_RUV_144R %>%
mutate(Time = "144R")
combined_toptables_dxr_RUV <- bind_rows(
d24_toptable_dxr_1,
d24r_toptable_dxr_1,
d144r_toptable_dxr_1)
#Filter the data based on each motif
filt_toptable_dxr_RUV <- combined_toptables_dxr_RUV %>%
dplyr::filter(Entrez_ID %in% D24T_DEGs_1) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC D24T DEGs Across Conditions RUVs")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#now do the same with the other conditions
filt_toptable_dxr_24r_RUV <- combined_toptables_dxr_RUV %>%
dplyr::filter(Entrez_ID %in% D24R_DEGs_1) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC D24R DEGs Across Conditions RUVs")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#D144R
filt_toptable_dxr_144r_RUV <- combined_toptables_dxr_RUV %>%
dplyr::filter(Entrez_ID %in% D144R_DEGs_1) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC D144R DEGs Across Conditions RUVs")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#now put the names of these graphs to print them
filt_toptable_dxr_RUV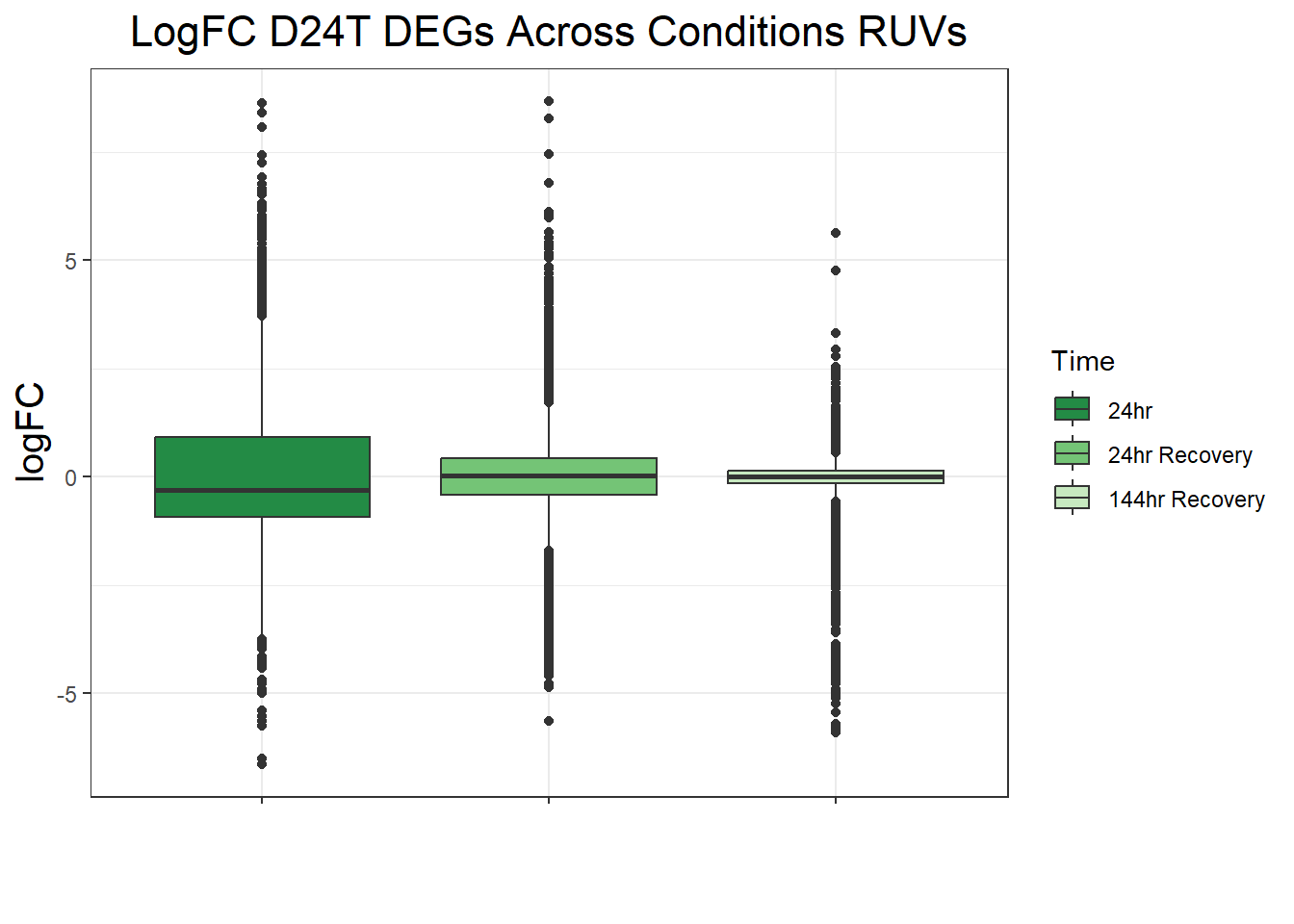
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
filt_toptable_dxr_24r_RUV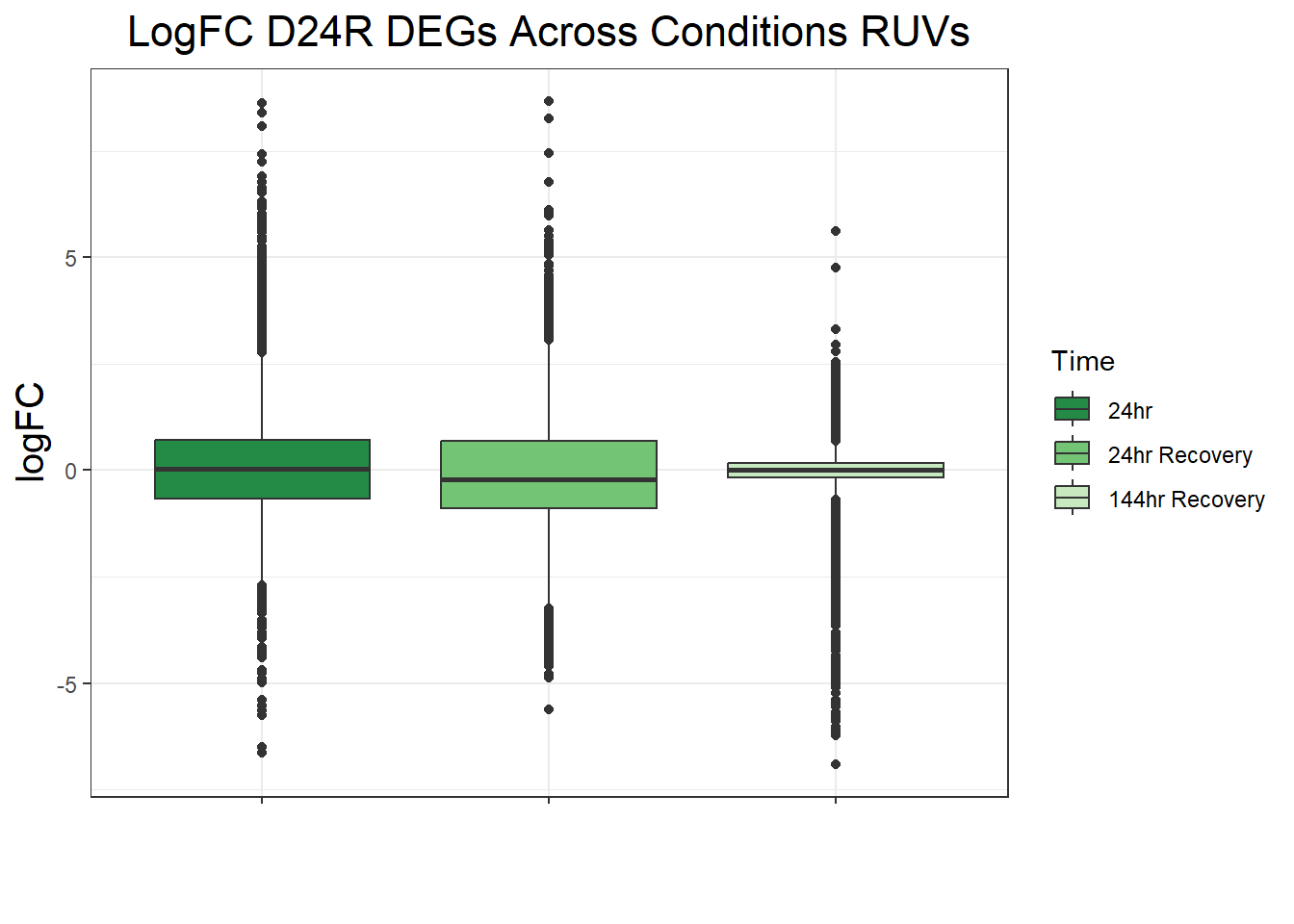
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
filt_toptable_dxr_144r_RUV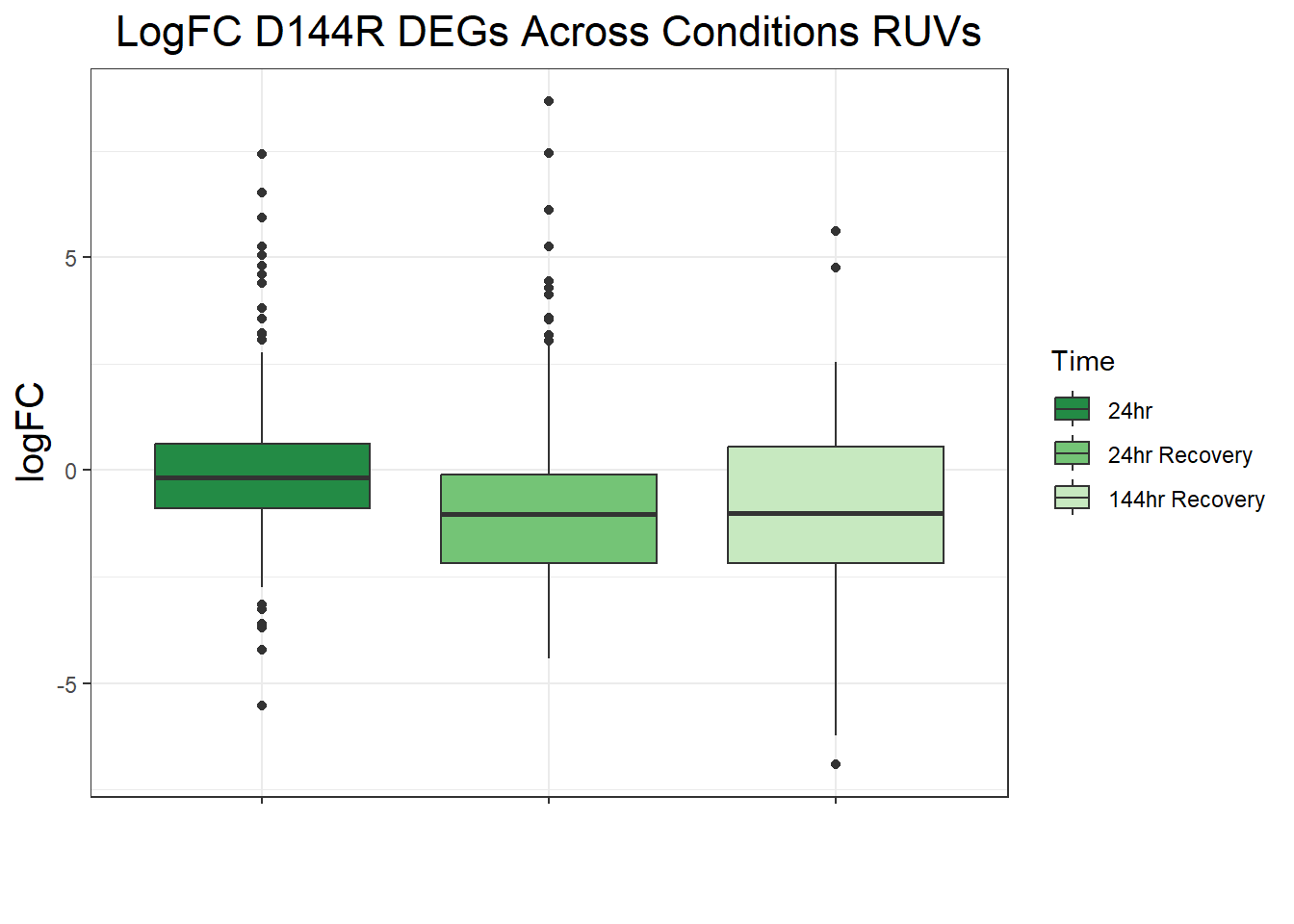
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
##Volcano Plots of RUVs Corrected Data k=1
#make a function to generate volcano plots + add gene numbers
#ensure the gene symbols are in a col so I can plot names of top 15
generate_volcano_plot_RUV <- function(toptable, title) {
#make significance labels
toptable$Significance <- "Not Significant"
toptable$Significance[toptable$logFC > 0 & toptable$adj.P.Val < 0.05] <- "Upregulated"
toptable$Significance[toptable$logFC < 0 & toptable$adj.P.Val < 0.05] <- "Downregulated"
#add number of genes for each significance label
upgenes <- toptable %>% filter(Significance == "Upregulated") %>% nrow()
nsgenes <- toptable %>% filter(Significance == "Not Significant") %>% nrow()
downgenes <- toptable %>% filter(Significance == "Downregulated") %>% nrow()
#make legend labels for no of genes
legend_lab <- c(
str_c("Upregulated: ", upgenes),
str_c("Not Significant: ", nsgenes),
str_c("Downregulated: ", downgenes)
)
#specify the colors for the legend
legend_col <- c(
str_c("Upregulated: " = "blue"),
str_c("Not Significant: " = "gray"),
str_c("Downregulated: " = "red")
)
#add the top 15 genes by adj. p value
top_genes <- toptable %>%
filter(!is.na(SYMBOL)) %>%
arrange(adj.P.Val) %>%
slice_head(n = 15)
#generate volcano plot w/ legend
ggplot(toptable, aes(x = logFC,
y = -log10(P.Value),
color = Significance)) +
geom_point(alpha = 0.4, size = 2) +
scale_color_manual(values = c("Upregulated" = "blue",
"Not Significant" = "gray",
"Downregulated" = "red"),
labels = legend_lab) +
xlim(-10, 10) +
labs(title = title,
x = expression(x = "log"[2]*"FC"),
y = expression(y = "-log"[10]*"P-value")) +
theme_bw()+
guides(color = guide_legend(override.aes = list(color = legend_col)))+
theme(legend.position = "right",
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = rel(1.25)))
}
#generate volcano plots across each comparison
volcano_plots_RUV <- list(
"V.D24T_RUV" = generate_volcano_plot(Toptable_RUV_24T, "DOX 24T RUVs k=1 (adj P-val<0.05)"),
"V.D24R_RUV" = generate_volcano_plot(Toptable_RUV_24R, "DOX 24R RUVs k=1 (adj P-val<0.05)"),
"V.D144R_RUV" = generate_volcano_plot(Toptable_RUV_144R, "DOX 144R RUVs k=1 (adj P-val<0.05)")
)
# Display each volcano plot
for (plot_name in names(volcano_plots_RUV)) {
print(volcano_plots_RUV[[plot_name]])
}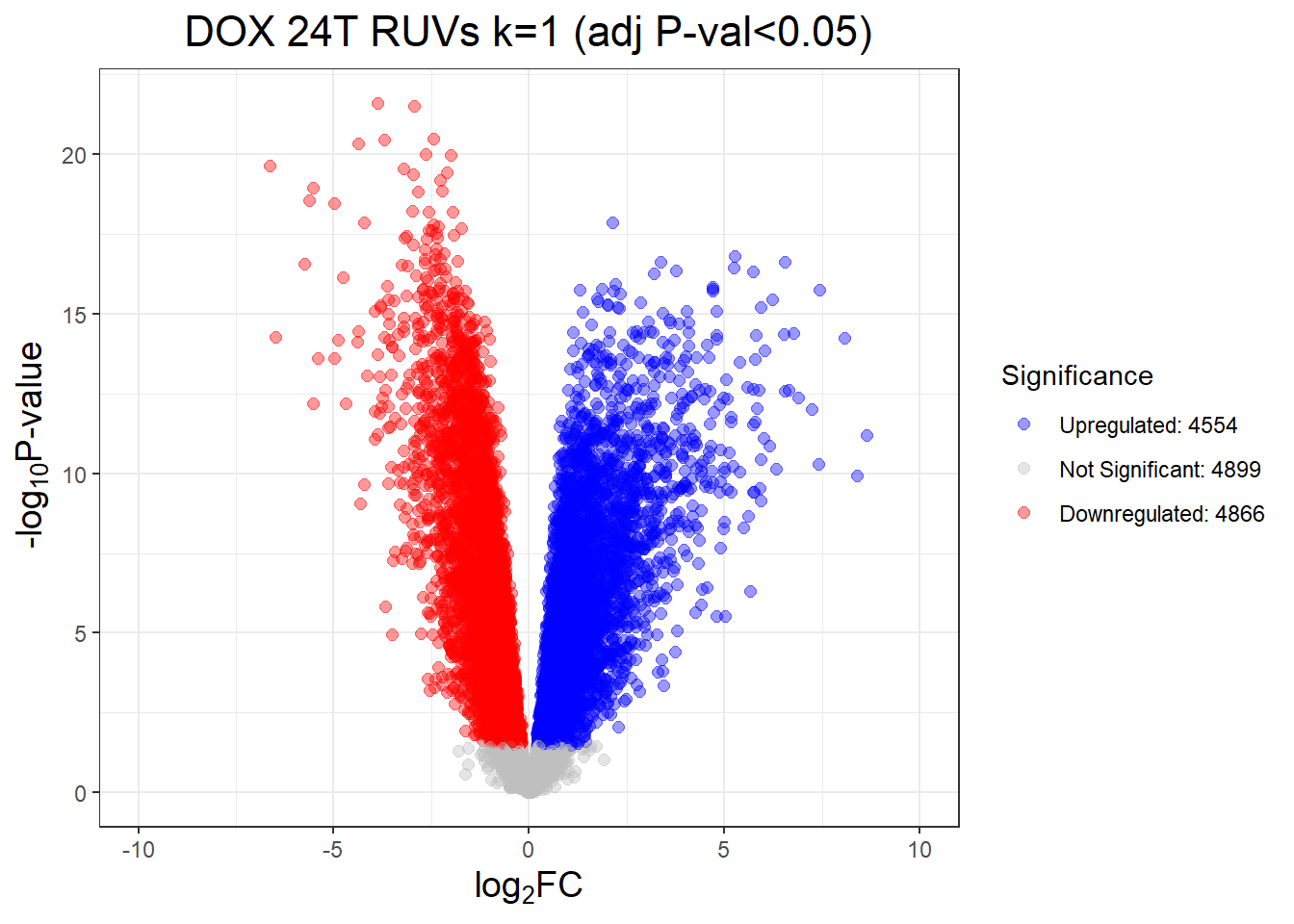
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
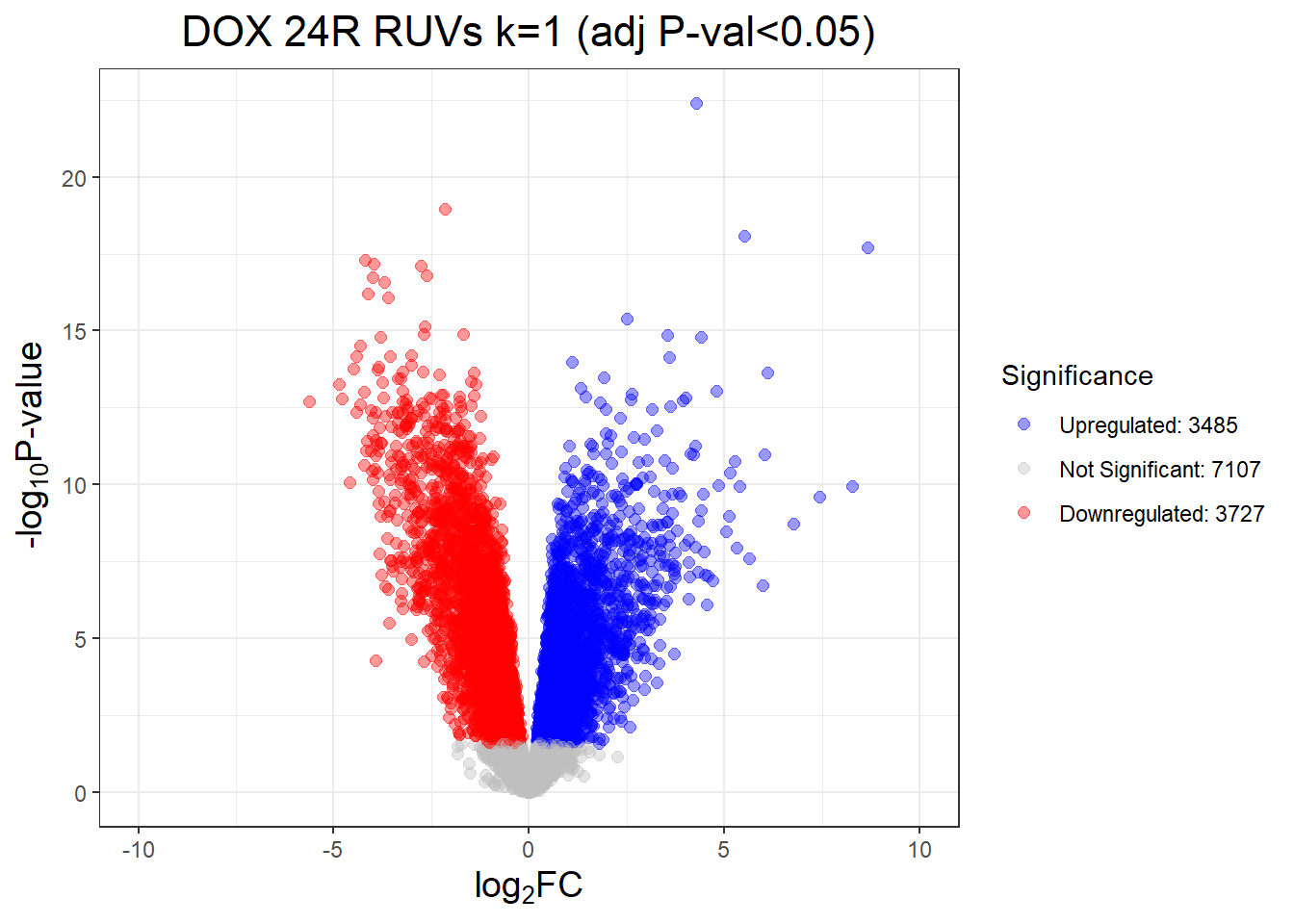
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
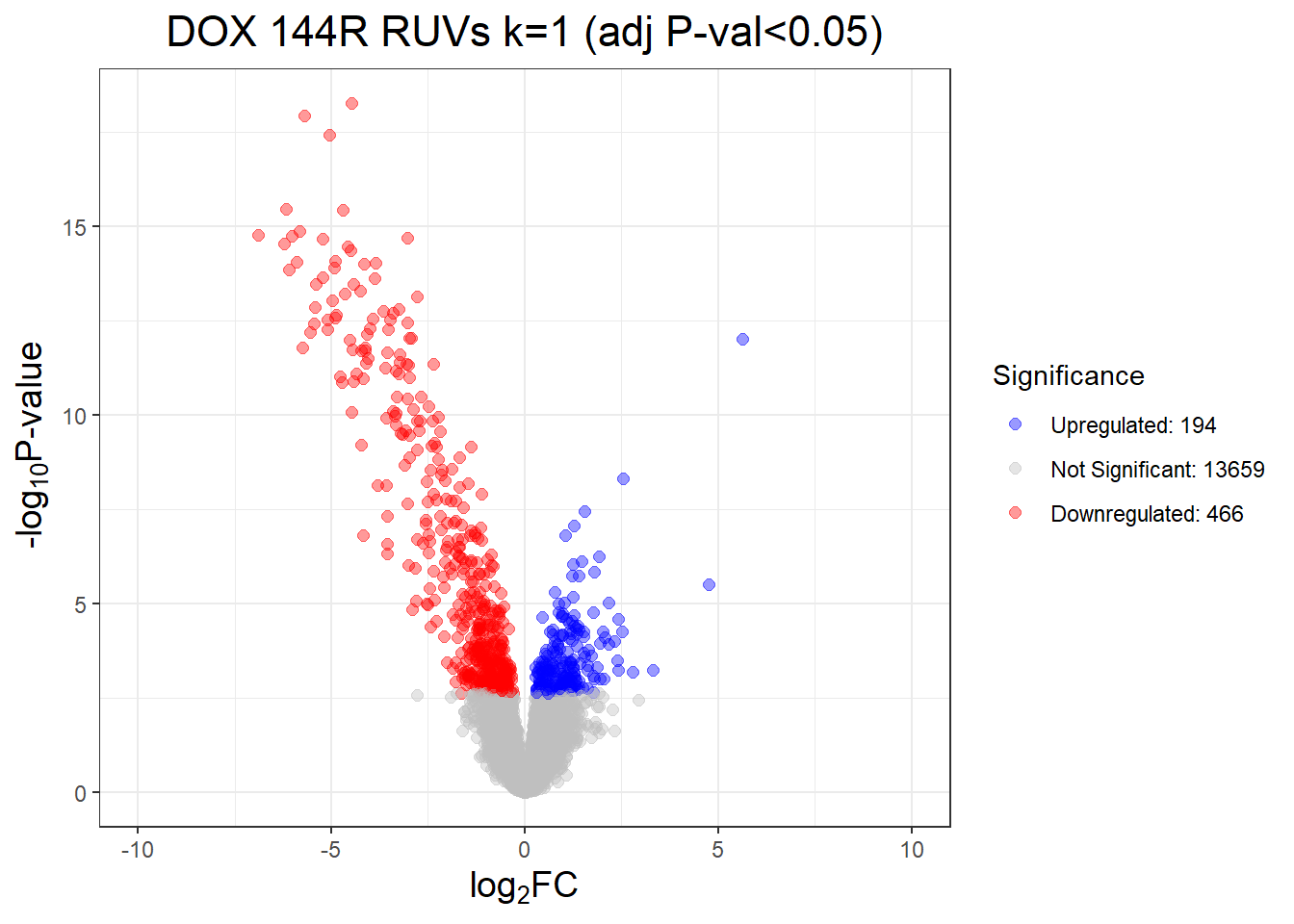
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
##Plot DNA Damage Response Genes (DDR) with Original Data
#DDR Gene Expression Heatmap — DOX Over Recovery Time (68 genes, with categories)
# Load libraries
# library(circlize)
# library(grid)
# library(reshape2)
# Load DEG files
load_deg <- function(path) read.csv(path)
DOX_24T <- load_deg("data/new/DEGs/Toptable_V.D24T.csv")
DOX_24R <- load_deg("data/new/DEGs/Toptable_V.D24R.csv")
DOX_144R <- load_deg("data/new/DEGs/Toptable_V.D144R.csv")
# Final Entrez IDs and categories (68 genes)
entrez_category_DDR <- tribble(
~ENTREZID, ~Category,
317, "Apoptosis", 355, "Apoptosis", 581, "Apoptosis", 637, "Apoptosis",
836, "Apoptosis", 841, "Apoptosis", 842, "Apoptosis", 27113, "Apoptosis",
5366, "Apoptosis", 54205, "Apoptosis", 55367, "Apoptosis", 8795, "Apoptosis",
1026, "Cell Cycle / Checkpoint", 1027, "Cell Cycle / Checkpoint", 595, "Cell Cycle / Checkpoint",
894, "Cell Cycle / Checkpoint", 896, "Cell Cycle / Checkpoint", 898, "Cell Cycle / Checkpoint",
9133, "Cell Cycle / Checkpoint", 9134, "Cell Cycle / Checkpoint", 891, "Cell Cycle / Checkpoint",
983, "Cell Cycle / Checkpoint", 1017, "Cell Cycle / Checkpoint", 1019, "Cell Cycle / Checkpoint",
1020, "Cell Cycle / Checkpoint", 1021, "Cell Cycle / Checkpoint", 993, "Cell Cycle / Checkpoint",
995, "Cell Cycle / Checkpoint", 1869, "Cell Cycle / Checkpoint", 4609, "Cell Cycle / Checkpoint",
5925, "Cell Cycle / Checkpoint", 9874, "Cell Cycle / Checkpoint", 11011, "Cell Cycle / Checkpoint",
1385, "Cell Cycle / Checkpoint",
472, "Damage Sensors / Signal Transducers", 545, "Damage Sensors / Signal Transducers",
5591, "Damage Sensors / Signal Transducers", 5810, "Damage Sensors / Signal Transducers",
5883, "Damage Sensors / Signal Transducers", 5884, "Damage Sensors / Signal Transducers",
6118, "Damage Sensors / Signal Transducers", 4361, "Damage Sensors / Signal Transducers",
10111, "Damage Sensors / Signal Transducers", 4683, "Damage Sensors / Signal Transducers",
84126, "Damage Sensors / Signal Transducers", 3014, "Damage Sensors / Signal Transducers",
672, "DNA Repair", 2177, "DNA Repair", 5888, "DNA Repair", 5893, "DNA Repair",
1647, "DNA Repair", 4616, "DNA Repair", 10912, "DNA Repair", 1111, "DNA Repair",
11200, "DNA Repair", 1643, "DNA Repair", 8243, "DNA Repair", 5981, "DNA Repair",
7157, "p53 Regulators / Targets", 4193, "p53 Regulators / Targets", 5371, "p53 Regulators / Targets",
27244, "p53 Regulators / Targets", 50484, "p53 Regulators / Targets",
5916, "DOX Cardiotoxicity", 7799, "DOX Cardiotoxicity", 4292, "DOX Cardiotoxicity",
207, "Miscellaneous / Broad", 25, "Miscellaneous / Broad"
)
entrez_ids_DDR <- entrez_category_DDR$ENTREZID
# Extract relevant DEG values
extract_data_DDR <- function(df, name) {
df %>%
filter(Entrez_ID %in% entrez_ids_DDR) %>%
mutate(
Gene = mapIds(org.Hs.eg.db, as.character(Entrez_ID),
column = "SYMBOL", keytype = "ENTREZID", multiVals = "first"),
Condition = name,
Signif = ifelse(adj.P.Val < 0.05, "*", "")
)
}
# DEG list
deg_list <- list("DOX_24T" = DOX_24T,
"DOX_24R" = DOX_24R,
"DOX_144R" = DOX_144R
)
# Combine all DEGs and annotate
all_data_DDR <- bind_rows(mapply(extract_data_DDR, deg_list, names(deg_list), SIMPLIFY = FALSE)) %>%
left_join(entrez_category_DDR, by = c("Entrez_ID" = "ENTREZID"))'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns# Create matrices
logFC_matddr <- acast(all_data_DDR, Gene ~ Condition, value.var = "logFC")
signif_matddr <- acast(all_data_DDR, Gene ~ Condition, value.var = "Signif")
# Set desired order
desired_order <- c("DOX_24T",
"DOX_24R",
"DOX_144R")
logFC_mat_DDR <- logFC_matddr[, desired_order, drop = FALSE]
signif_mat_DDR <- signif_matddr[, desired_order, drop = FALSE]
# Column annotation
meta_DDR <- str_split_fixed(colnames(logFC_mat_DDR), "_", 2)
col_annot <- HeatmapAnnotation(
Drug = meta_DDR[, 1],
Time = meta_DDR[, 2],
col = list(
Drug = c("DOX" = "#499FBD"),
Time = c("24T" = "#238B45",
"24R" = "#74C476",
"144R" = "#C7E9C0")
),
annotation_height = unit(c(1, 1, 1), "cm")
)
# Row annotation
gene_order_df_DDR <- all_data_DDR %>%
distinct(Gene, Category) %>%
arrange(factor(Category, levels = sort(unique(entrez_category_DDR$Category))), Gene)
ordered_genes_DDR <- gene_order_df_DDR$Gene
logFC_mat_DDR <- logFC_mat_DDR[ordered_genes_DDR, ]
signif_mat_DDR <- signif_mat_DDR[ordered_genes_DDR, ]
category_colors_DDR <- structure(
c("darkorange", "steelblue", "darkgreen", "firebrick", "gold", "mediumpurple", "gray60"),
names = sort(unique(entrez_category_DDR$Category))
)
ha_left_DDR <- rowAnnotation(
Category = gene_order_df_DDR$Category,
col = list(Category = category_colors_DDR),
annotation_name_side = "top"
)
# Final Heatmap
Heatmap(logFC_mat_DDR,
name = "logFC",
top_annotation = col_annot,
left_annotation = ha_left_DDR,
cluster_columns = FALSE,
cluster_rows = FALSE,
show_row_names = TRUE,
show_column_names = FALSE,
row_names_gp = gpar(fontsize = 10),
column_title = "DDR Gene Expression Response (n = 68)\n DOX Recovery",
column_title_gp = gpar(fontsize = 14, fontface = "bold"),
cell_fun = function(j, i, x, y, width, height, fill) {
grid.text(signif_mat_DDR[i, j], x, y, gp = gpar(fontsize = 9))
}
)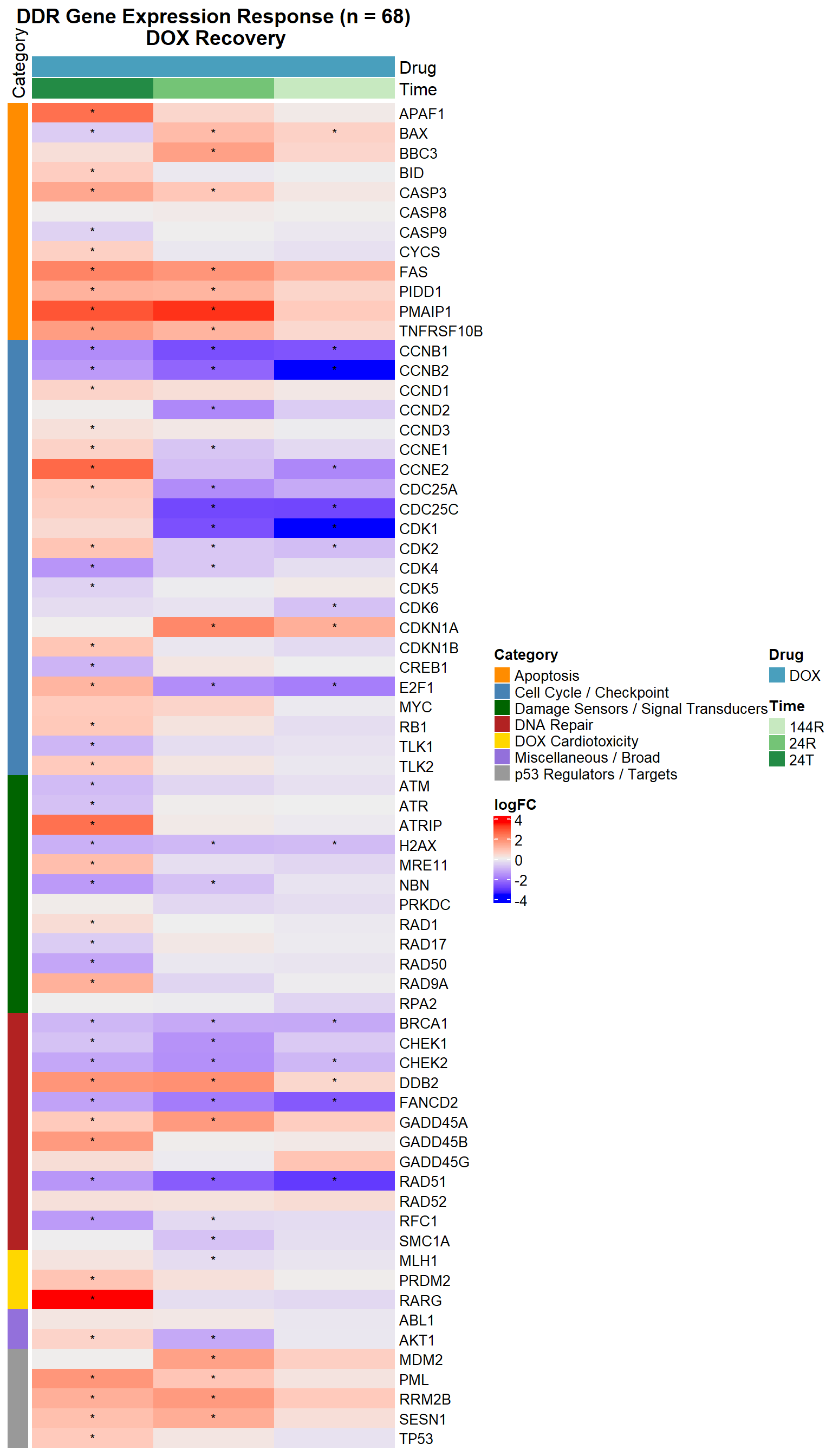
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
##Plot DOX Cardiotox Genes logFC HM Original Data
#plot a list of 29 functionally validated DIC genes
# Load DEG files
load_deg <- function(path) read.csv(path)
DOX_24T <- load_deg("data/new/DEGs/Toptable_V.D24T.csv")
DOX_24R <- load_deg("data/new/DEGs/Toptable_V.D24R.csv")
DOX_144R <- load_deg("data/new/DEGs/Toptable_V.D144R.csv")
#the data I have is in hgnc_symbols, I want to convert this to entrez_id like my df
DIC_genes <- tribble(
~SYMBOL, ~Category,
"CAT", "ROS Generation / Handling",
"CBR1", "ROS Generation / Handling",
"CBR3", "ROS Generation / Handling",
"ERBB2", "ROS Generation / Handling",
"GPX3", "ROS Generation / Handling",
"GSTM1", "ROS Generation / Handling",
"GSTP", "ROS Generation / Handling",
"HAS3", "ROS Generation / Handling",
"NOS3", "ROS Generation / Handling",
"PLCE1", "ROS Generation / Handling",
"RAC2", "ROS Generation / Handling",
"SPG7", "ROS Generation / Handling",
"PRDM2", "DNA Damage",
"MLH1", "DNA Damage",
"RARG", "DNA Damage",
"HFE", "Iron Uptake & Homeostasis",
"SLC22A17", "DOX Uptake",
"SLC28A1", "DOX Uptake",
"SLC28A3", "DOX Uptake",
"ABCB4", "DOX Efflux",
"ABCC2", "DOX Efflux",
"ABCC5", "DOX Efflux",
"ABCC9", "DOX Efflux",
"ABCC10", "DOX Efflux",
"CELF4", "Calcium Handling",
"MYH7", "Calcium Handling",
"CYP2J2", "Cardiac Electrical Activity",
"RIN3", "Cardiac Electrical Activity",
"ZFN521", "Cardiac Electrical Activity")
gene_df_DIC <- tibble(HGNC = DIC_genes)
gene_df_DIC <- gene_df_DIC %>%
mutate(Entrez_ID = mapIds(org.Hs.eg.db,
keys = DIC_genes$SYMBOL,
column = "ENTREZID",
keytype = "SYMBOL",
multiVals = "first")) %>%
unnest_wider(HGNC) %>%
mutate(Entrez_ID = as.character(Entrez_ID))'select()' returned 1:1 mapping between keys and columns#now I've put together a dataframe with the HGNC, Category, and Entrez_ID
#plus I've ensured that Entrez_ID is a character for later joining
entrez_ids_DIC <- gene_df_DIC$Entrez_ID
# saveRDS(entrez_ids_DIC, "data/new/RUV/DIC_genes_entrezid.RDS")
# Extract relevant DEG values
extract_data_DIC <- function(df, name) {
df %>%
filter(Entrez_ID %in% entrez_ids_DIC) %>%
mutate(
Gene = mapIds(org.Hs.eg.db, as.character(Entrez_ID),
column = "SYMBOL", keytype = "ENTREZID", multiVals = "first"),
Condition = name,
Signif = ifelse(adj.P.Val < 0.05, "*", "")
)
}
# DEG list
deg_list <- list("DOX_24T" = DOX_24T,
"DOX_24R" = DOX_24R,
"DOX_144R" = DOX_144R
)
# Combine all DEGs and annotate
all_data_DIC <- bind_rows(mapply(extract_data_DIC,
deg_list,
names(deg_list),
SIMPLIFY = FALSE)) %>%
mutate(Entrez_ID = as.character(Entrez_ID)) %>%
left_join(gene_df_DIC,
by = "Entrez_ID")'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns#I've also made sure here that Entrez_ID is a character and not an integer
# Create matrices
logFC_matdic <- acast(all_data_DIC, Gene ~ Condition, value.var = "logFC")
signif_matdic <- acast(all_data_DIC, Gene ~ Condition, value.var = "Signif")
# Set desired order
desired_order <- c("DOX_24T",
"DOX_24R",
"DOX_144R")
logFC_mat_DIC <- logFC_matdic[, desired_order, drop = FALSE]
signif_mat_DIC <- signif_matdic[, desired_order, drop = FALSE]
# Column annotation
meta_DIC <- str_split_fixed(colnames(logFC_mat_DIC), "_", 3)
meta_DIC <- str_split_fixed(colnames(logFC_mat_DIC), "_", 2)
col_annot <- HeatmapAnnotation(
Drug = meta_DIC[, 1],
Time = meta_DIC[, 2],
col = list(
Drug = c("DOX" = "#499FBD"),
Time = c("24T" = "#238B45",
"24R" = "#74C476",
"144R" = "#C7E9C0")
),
annotation_height = unit(c(2, 2, 2), "cm")
)
#for this I can leave off the DMSO as it's already from a pairwise comparison
#make an extra object with my category order as well
category_order_DIC <- c(
"ROS Generation / Handling",
"DNA Damage",
"Calcium Handling",
"DOX Uptake",
"Iron Uptake & Homeostasis",
"DOX Efflux",
"Cardiac Electrical Activity"
)
# Row annotation
gene_order_df_DIC <- all_data_DIC %>%
distinct(Gene, Category) %>%
mutate(Category = factor(Category, levels = category_order_DIC)) %>%
arrange(Category, Gene)
ordered_genes_DIC <- gene_order_df_DIC$Gene
logFC_mat_DIC <- logFC_mat_DIC[ordered_genes_DIC, ]
signif_mat_DIC <- signif_mat_DIC[ordered_genes_DIC, ]
#add in your colors for each category
category_colors_DIC <- structure(
c("darkorange", "steelblue", "darkgreen", "firebrick", "gold", "mediumpurple", "gray60"),
names = category_order_DIC
)
ha_left_DIC <- rowAnnotation(
Category = gene_order_df_DIC$Category,
col = list(Category = category_colors_DIC),
annotation_name_side = "top"
)
# Draw heatmap
Heatmap(logFC_mat_DIC,
name = "logFC",
top_annotation = col_annot,
left_annotation = ha_left_DIC,
cluster_columns = FALSE,
cluster_rows = FALSE,
show_row_names = TRUE,
show_column_names = FALSE,
cell_fun = function(j, i, x, y, width, height, fill) {
grid.text(signif_mat_DIC[i, j], x, y, gp = gpar(fontsize = 9))
},
column_title = "DIC Genes Expression (n=29)\nDOX Recovery",
column_title_gp = gpar(fontsize = 14, fontface = "bold")
)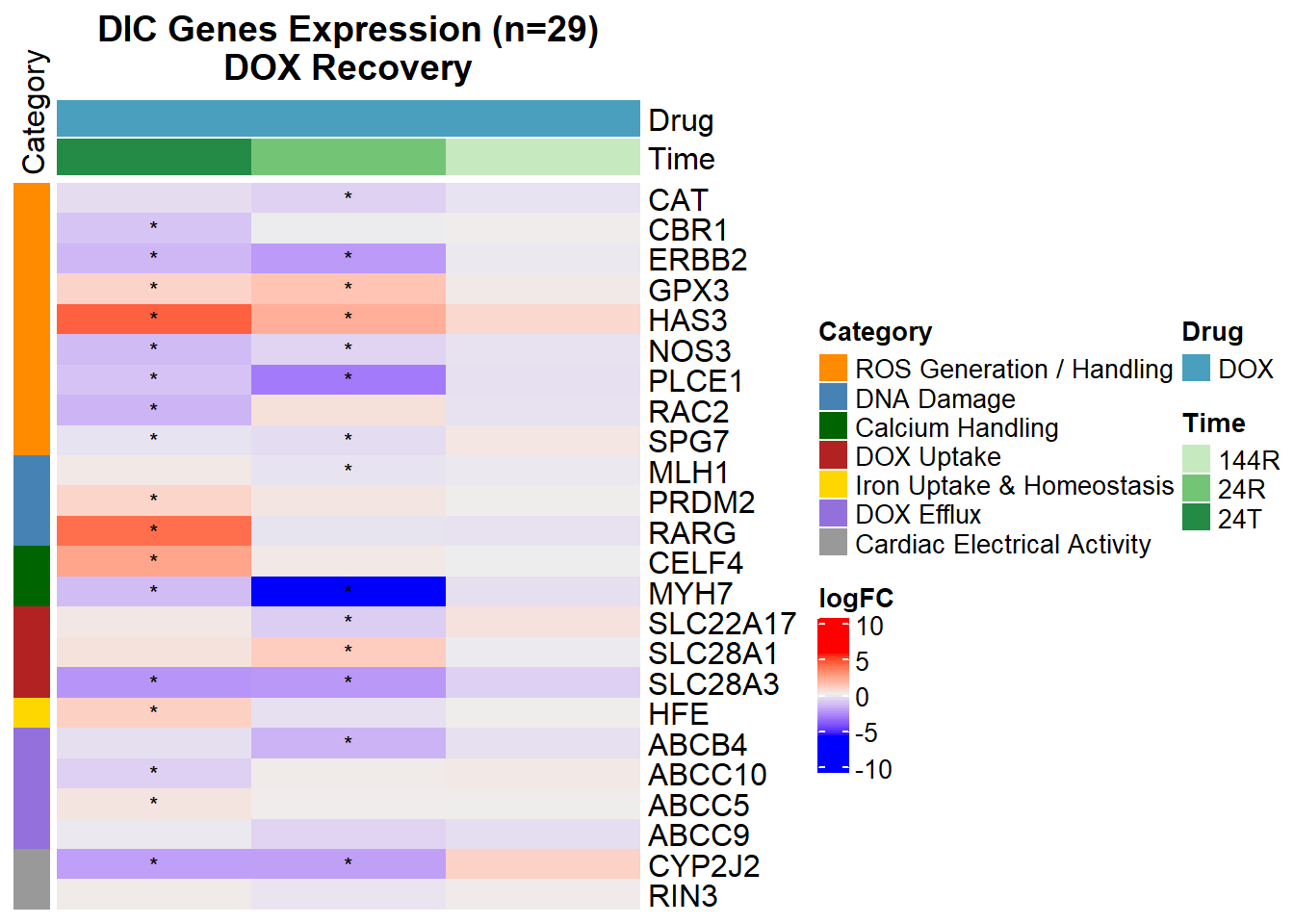
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
##Plot p53 Target Genes logFC HM Original Data
# Load libraries
library(tidyverse)
library(ComplexHeatmap)
library(circlize)
library(grid)
library(org.Hs.eg.db)
library(reshape2)
# Load DEG files
load_deg <- function(path) read.csv(path)
DOX_24T <- load_deg("data/new/DEGs/Toptable_V.D24T.csv")
DOX_24R <- load_deg("data/new/DEGs/Toptable_V.D24R.csv")
DOX_144R <- load_deg("data/new/DEGs/Toptable_V.D144R.csv")
#P53 target Entrez_ID
entrez_ids_p53 <- c(1026,50484,4193,9766,9518,7832,1643,1647,1263,57103,51065,8795,51499,64393,581,
5228,5429,8493,55959,7508,64782,282991,355,53836,4814,10769,9050,27244,9540,94241,
26154,57763,900,26999,55332,26263,23479,23612,29950,9618,10346,8824,134147,55294,
22824,4254,6560,467,27113,60492,8444,60401,1969,220965,2232,3976,55191,84284,93129,
5564,7803,83667,7779,132671,7039,51768,137695,93134,7633,10973,340485,307,27350,
23245,3732,29965,1363,1435,196513,8507,8061,2517,51278,53354,54858,23228,5366,5912,
6236,51222,26152,59,1907,50650,91012,780,9249,11072,144455,64787,116151,27165,2876,
57822,55733,57722,121457,375449,85377,4851,5875,127544,29901,84958,8797,8793,441631,
220001,54541,5889,5054,25816,25987,5111,98,317,598,604,10904,1294,80315,53944,
1606,2770,3628,3675,3985,4035,4163,84552,29085,55367,5371,5791,54884,5980,8794,
1462,50808,220,583,694,1056,9076,10978,54677,1612,55040,114907,2274,127707,4000,
8079,4646,4747,27445,5143,80055,79156,5360,5364,23654,5565,5613,5625,10076,56963,
6004,390,255488,6326,6330,23513,7869,283130,204962,83959,6548,6774,9263,10228,
22954,10475,85363,494514,10142,79714,1006,8446,9648,79828,5507,55240,63874,25841,
9289,84883,154810,51321,421,8553,655,119032,84280,10950,824,839,57828,857,8812,
8837,94027,113189,22837,132864,10898,3300,81704,1847,1849,1947,9538,24139,5168,
147965,115548,9873,23768,2632,2817,3280,3265,23308,3490,51477,182,3856,8844,144811,
9404,4043,9848,2872,23041,740,343263,4638,26509,4792,22861,57523,55214,80025,164091,
57060,64065,51090,5453,8496,333926,55671,5900,55544,23179,8601,389,6223,55800,6385,
4088,6643,122809,257397,285343,7011,54790,374618,55362,51754,7157,9537,22906,7205,
80705,219699,55245,83719,7748,25946,118738)
# p53_genes <- saveRDS(entrez_ids_p53, "data/new/RUV/p53_genelist_entrezid.RDS")
# Function to extract relevant data
extract_data_p53 <- function(df, name) {
df %>%
filter(Entrez_ID %in% entrez_ids_p53) %>%
mutate(Gene = mapIds(org.Hs.eg.db, as.character(Entrez_ID),
column = "SYMBOL", keytype = "ENTREZID", multiVals = "first"),
Condition = name,
Signif = ifelse(adj.P.Val < 0.05, "*", ""))
}
# Collect all data
# DEG list
deg_list <- list("DOX_24T" = DOX_24T,
"DOX_24R" = DOX_24R,
"DOX_144R" = DOX_144R
)
# Combine all DEGs and annotate
all_data_p53 <- bind_rows(mapply(extract_data_p53, deg_list, names(deg_list), SIMPLIFY = FALSE)) 'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns#unnecessary to join again as there are not categories on this list of entrezids
# %>%
# left_join(entrez_ids, by = c("Entrez_ID" = "ENTREZID"))
# Create matrices
logFC_mat53 <- acast(all_data_p53, Gene ~ Condition, value.var = "logFC")
signif_mat53 <- acast(all_data_p53, Gene ~ Condition, value.var = "Signif")
# Desired column order
desired_order <- c("DOX_24T",
"DOX_24R",
"DOX_144R")
logFC_mat_p53 <- logFC_mat53[, desired_order]
signif_mat_p53 <- signif_mat53[, desired_order]
# Column annotation
meta_p53 <- str_split_fixed(colnames(logFC_mat_p53), "_", 3)
meta_p53 <- str_split_fixed(colnames(logFC_mat_p53), "_", 2)
col_annot <- HeatmapAnnotation(
Drug = meta_p53[, 1],
Time = meta_p53[, 2],
col = list(
Drug = c("DOX" = "#499FBD"),
Time = c("24T" = "#238B45",
"24R" = "#74C476",
"144R" = "#C7E9C0")
),
annotation_height = unit(c(1, 1, 1), "cm")
)
# Draw heatmap
Heatmap(logFC_mat_p53,
name = "logFC",
top_annotation = col_annot,
cluster_columns = FALSE,
cluster_rows = TRUE,
show_row_names = TRUE,
show_column_names = FALSE,
layer_fun = function(j, i, x, y, width, height, fill) {
grid.text(signif_mat_p53[cbind(i, j)], x, y, gp = gpar(fontsize = 9))
},
column_title = "P53 Target Genes Expression\nDOX Recovery",
column_title_gp = gpar(fontsize = 14, fontface = "bold")
)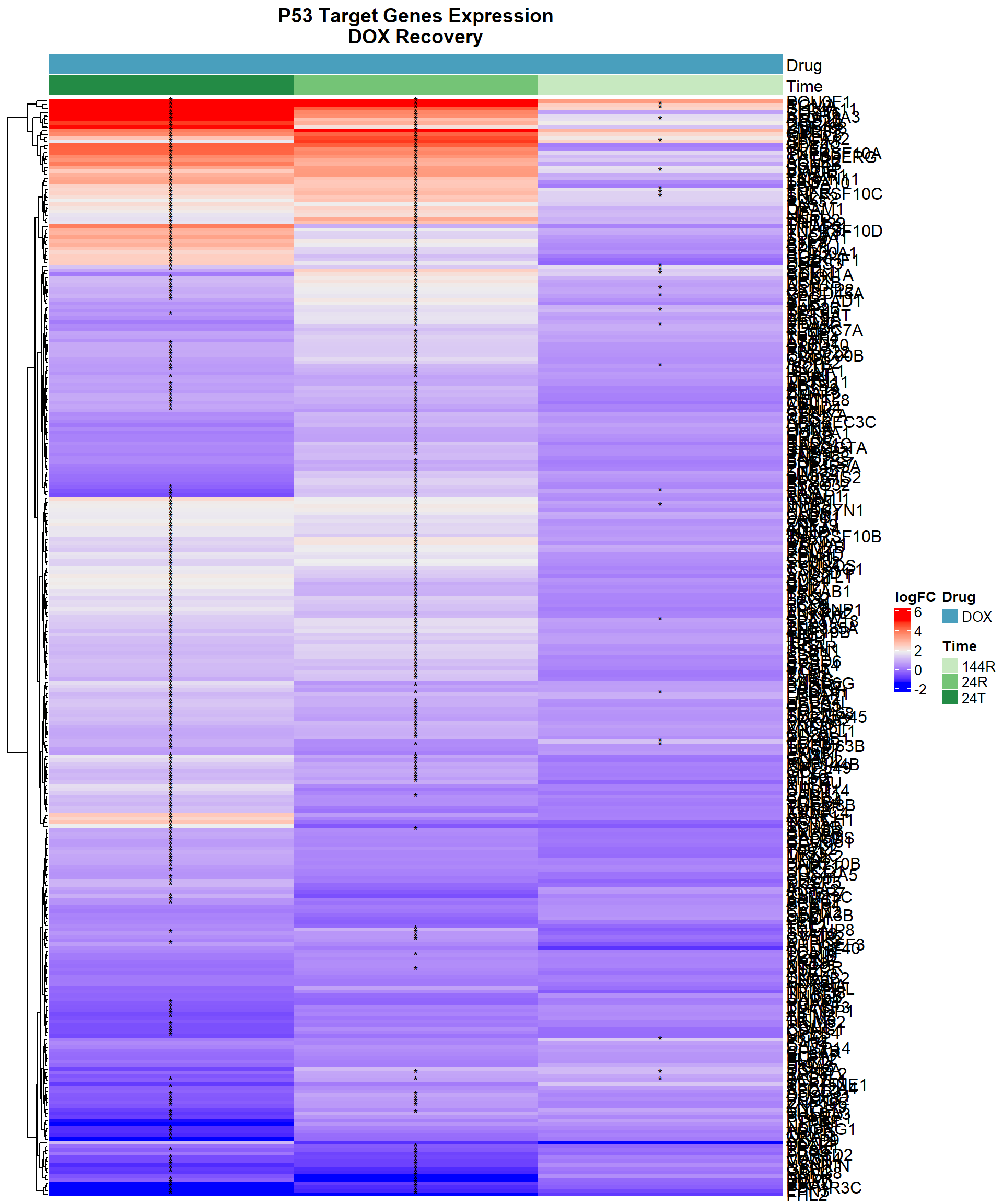
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
##Create a Venn Diagram of DEGs from Original DEA
#plot a venn diagram with all of your conditions from your toptables
# Load DEGs Data
DOX_24T <- read.csv("data/new/DEGs/Toptable_V.D24T.csv")
DOX_24R <- read.csv("data/new/DEGs/Toptable_V.D24R.csv")
DOX_144R <- read.csv("data/new/DEGs/Toptable_V.D144R.csv")
# Extract Significant DEGs
DEG1 <- DOX_24T$Entrez_ID[DOX_24T$adj.P.Val < 0.05]
DEG2 <- DOX_24R$Entrez_ID[DOX_24R$adj.P.Val < 0.05]
DEG3 <- DOX_144R$Entrez_ID[DOX_144R$adj.P.Val < 0.05]
venntest <- list(DEG1, DEG2, DEG3)
ggVennDiagram(
venntest,
category.names = c("DOX_24T", "DOX_24R", "DOX_144R")
) + ggtitle("DXR Specific and Shared DEGs")+
theme(
plot.title = element_text(size = 16, face = "bold"), # Increase title size
text = element_text(size = 16) # Increase text size globally
)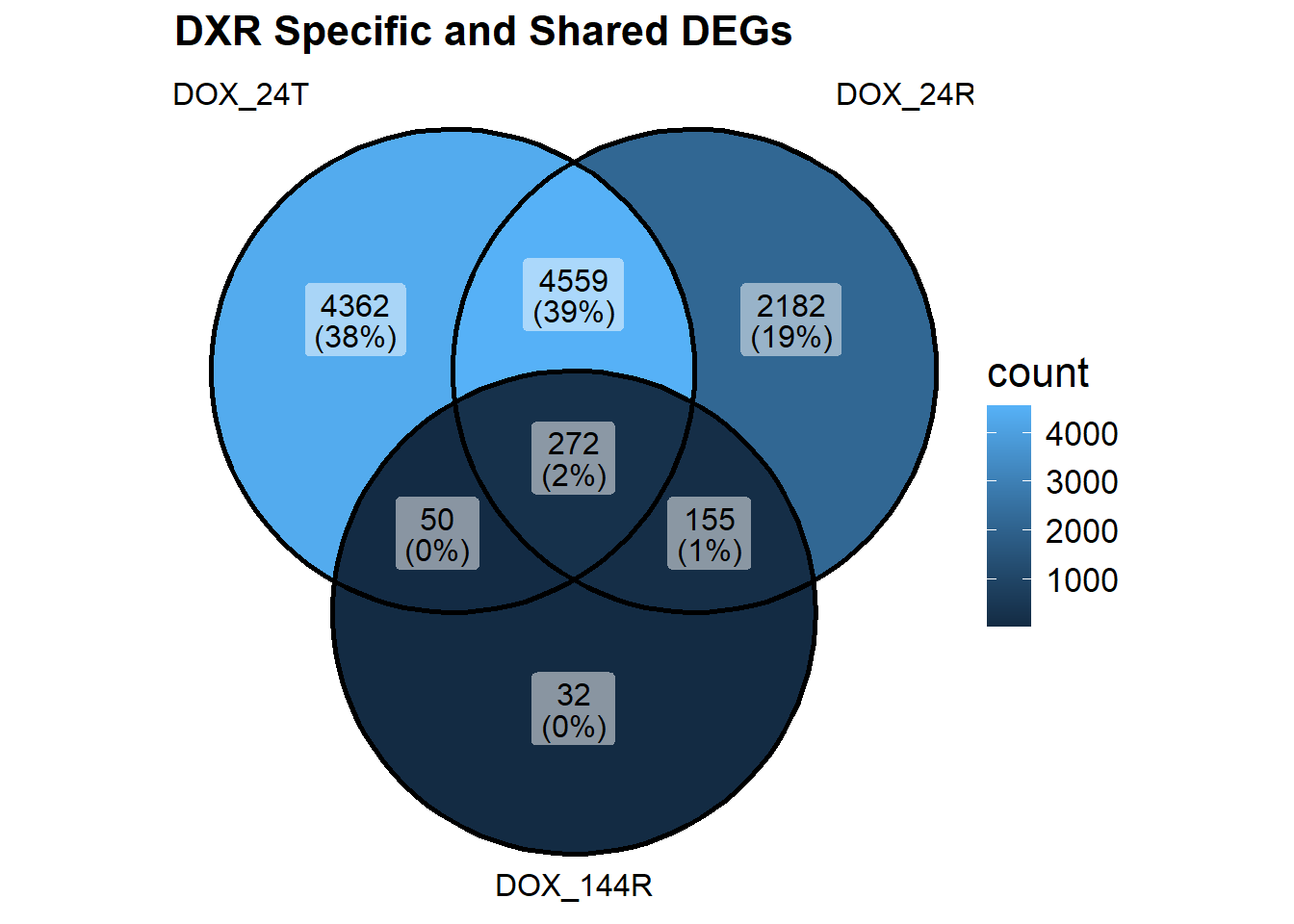
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#Now that I've made my venn diagram, I want to compare these DEGs
#set 1 : 4362 DOX24T specific genes
#set2 : 4362 + 4550 + 50 + 272 genes shared across DOX24T (all genes)
#how many of these are downregulated and how many are upregulated?
# Extract Significant DEGs
# Create a list of DEGs for each sample
# Example gene sets
DEG1 <- DOX_24T$Entrez_ID[DOX_24T$adj.P.Val < 0.05]
DEG2 <- DOX_24R$Entrez_ID[DOX_24R$adj.P.Val < 0.05]
DEG3 <- DOX_144R$Entrez_ID[DOX_144R$adj.P.Val < 0.05]
#try and use the VennDetail package to extract the genes from each condition
#Set 1 - DOX_24T only genes
#Set 2 - DOX_24T shared genes
plot.new()
venn_test <- venndetail(venntest)
plot(venn_test)
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
detail(venn_test)=== Here is the detail of Venndiagram ===
Total results: 11612 x 2
Total sets is: 7
Subset Detail
1 Shared 643837
2 Shared 1870
3 Shared 84958
4 Shared 2537
5 Shared 192670
6 Shared 1298
... with 11606 more rows ...#now that I have the genes subsetted by condition, I can pull out the genes I want to look at
venn_DOX_24T <- getSet(object = venn_test, subset = c("Group 1"))Warning: `filter_()` was deprecated in dplyr 0.7.0.
ℹ Please use `filter()` instead.
ℹ See vignette('programming') for more help
ℹ The deprecated feature was likely used in the VennDetail package.
Please report the issue to the authors.
This warning is displayed once every 8 hours.
Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
generated.dim(venn_DOX_24T)[1] 4362 2#4362 genes in DOX_24T only
venn_DOX_shared <- getSet(object = venn_test, subset = c("Shared", "Group 1", "Group 1_Group 2", "Group 1_Group 3"))
dim(venn_DOX_shared)[1] 9243 2#total of 9243 genes:
#4559 DOX_24T only
#272 shared all
#50 DOX_24T + DOX_144R
#4362 DOX_24T + DOX_24R
venn_DOX144R_shared <- getSet(object = venn_test, subset = c("Shared", "Group 3", "Group 2_Group 3", "Group 1_Group 3"))
dim(venn_DOX144R_shared)[1] 509 2#total of 509 genes:
#272 shared all
#32 DOX144R Specific
#50 DOX24T + DOX144R
#155 DOX24R + DOX144R
#now I can look at these sets to see which ones are up and down regulated in each
#after that, run GO analysis
venn_shared_DEGs <- venn_DOX_shared %>%
as.data.frame() %>%
column_to_rownames(., var = "Detail")
venn_sharedD144R_DEGs <- venn_DOX144R_shared %>%
as.data.frame() %>%
column_to_rownames(., var = "Detail")
venn_DOX24T_DEGs <- venn_DOX_24T %>%
as.data.frame() %>%
column_to_rownames(., var = "Detail")
#pull these genes out of my DEG matrix with logFC
DOX_24T_DEGs <- Toptable_list$V.D24T[row.names(venn_DOX24T_DEGs),]
DOX_24T_shared_DEGs <- Toptable_list$V.D24T[row.names(venn_shared_DEGs),]
DOX_144R_shared_DEGs <- Toptable_list$V.D144R[row.names(venn_sharedD144R_DEGs),]
#I want to go ahead and do this for every condition so I can see the genes in there
#I also want to filter these by their logFC being up or down for GO/KEGG
DOX24T_DEGs_GO <- DOX_24T_DEGs %>%
dplyr::filter(., adj.P.Val < 0.05)
DOX24T_DEGs_GO_up <- DOX24T_DEGs_GO %>%
dplyr::filter(., logFC > 0) %>%
rownames_to_column(., var = "entrezgene_ID") %>%
dplyr::select("entrezgene_ID")
#has 4029 genes
DOX24T_DEGs_GO_down <- DOX24T_DEGs_GO %>%
dplyr::filter(., logFC < 0) %>%
rownames_to_column(., var = "entrezgene_ID") %>%
dplyr::select("entrezgene_ID")
#has 4305 genes
#shared genes venn diagram
DOX24Tshare_DEGs_GO <- DOX_24T_shared_DEGs %>%
dplyr::filter(., adj.P.Val < 0.05)
DOX24T_share_DEGs_GO_plot <- DOX24Tshare_DEGs_GO %>%
rownames_to_column(., var = "Entrez_ID") %>%
dplyr::select("Entrez_ID")
DOX24Tshare_DEGs_GO_up <- DOX24Tshare_DEGs_GO %>%
dplyr::filter(., logFC > 0) %>%
rownames_to_column(., var = "entrezgene_ID") %>%
dplyr::select("entrezgene_ID")
#has 4731 genes
DOX24Tshare_DEGs_GO_down <- DOX24Tshare_DEGs_GO %>%
dplyr::filter(., logFC < 0) %>%
rownames_to_column(., var = "entrezgene_ID") %>%
dplyr::select("entrezgene_ID")
DOX144Rshare_DEGs_GO <- DOX_144R_shared_DEGs %>%
dplyr::filter(., adj.P.Val < 0.05)
DOX144Rshare_DEGs_GO_plot <- DOX144Rshare_DEGs_GO %>%
rownames_to_column(., var = "Entrez_ID") %>%
dplyr::select("Entrez_ID")
#now go ahead and do this for each condition as well
#DOX24T
venn_DOX_24T <- getSet(object = venn_test, subset = c("Group 1"))
dim(venn_DOX_24T)[1] 4362 2#4362 genes in DOX_24T only
venn_DOX24T_DEGs <- venn_DOX_24T %>%
as.data.frame() %>%
column_to_rownames(., var = "Detail")
#pull these genes out of my DEG matrix with logFC
DOX_24T_DEGs <- Toptable_list$V.D24T[row.names(venn_DOX24T_DEGs),]
DOX24T_DEGs_GO <- DOX_24T_DEGs %>%
rownames_to_column(., var = "Entrez_ID") %>%
dplyr::filter(., adj.P.Val < 0.05) %>%
dplyr::select("Entrez_ID")
#DOX24R
venn_DOX_24R <- getSet(object = venn_test, subset = c("Group 2"))
dim(venn_DOX_24R)[1] 2182 2#2182 genes in DOX_24R only
venn_DOX24R_DEGs <- venn_DOX_24R %>%
as.data.frame() %>%
column_to_rownames(., var = "Detail")
#pull these genes out of my DEG matrix with logFC
DOX_24R_DEGs <- Toptable_list$V.D24R[row.names(venn_DOX24R_DEGs),]
DOX24R_DEGs_GO <- DOX_24R_DEGs %>%
rownames_to_column(., var = "Entrez_ID") %>%
dplyr::filter(., adj.P.Val < 0.05) %>%
dplyr::select("Entrez_ID")
#DOX144R
venn_DOX_144R <- getSet(object = venn_test, subset = c("Group 3"))
dim(venn_DOX_144R)[1] 32 2#32 genes in DOX_144R only
venn_DOX144R_DEGs <- venn_DOX_144R %>%
as.data.frame() %>%
column_to_rownames(., var = "Detail")
#pull these genes out of my DEG matrix with logFC
DOX_144R_DEGs <- Toptable_list$V.D144R[row.names(venn_DOX144R_DEGs),]
DOX144R_DEGs_GO <- DOX_144R_DEGs %>%
rownames_to_column(., var = "Entrez_ID") %>%
dplyr::filter(., adj.P.Val < 0.05) %>%
dplyr::select("Entrez_ID")
#this set doesn't yield a GO/KEGG plot as there are too few genes
#instead, I pulled all genes associated with DOX144R##Venn Diagrams of DEG Overlap after RUVs Correction k=1
#plot a venn diagram with all of your conditions from your toptables
# Load DEGs Data
DOX_24T_1 <- read.csv("data/new/DEGs/TTBL_RUV_24T_OLD.csv")
DOX_24R_1 <- read.csv("data/new/DEGs/TTBL_RUV_24R_OLD.csv")
DOX_144R_1 <- read.csv("data/new/DEGs/TTBL_RUV_144R_OLD.csv")
# Extract Significant DEGs
DEG1_RUV <- DOX_24T_1$Entrez_ID[DOX_24T_1$adj.P.Val < 0.05]
DEG2_RUV <- DOX_24R_1$Entrez_ID[DOX_24R_1$adj.P.Val < 0.05]
DEG3_RUV <- DOX_144R_1$Entrez_ID[DOX_144R_1$adj.P.Val < 0.05]
venntest_RUV <- list(DEG1_RUV, DEG2_RUV, DEG3_RUV)
ggVennDiagram(
venntest_RUV,
category.names = c("DOX_24T", "DOX_24R", "DOX_144R")
) + ggtitle("DXR Specific and Shared DEGs RUVs")+
theme(
plot.title = element_text(size = 16, face = "bold"), # Increase title size
text = element_text(size = 16) # Increase text size globally
)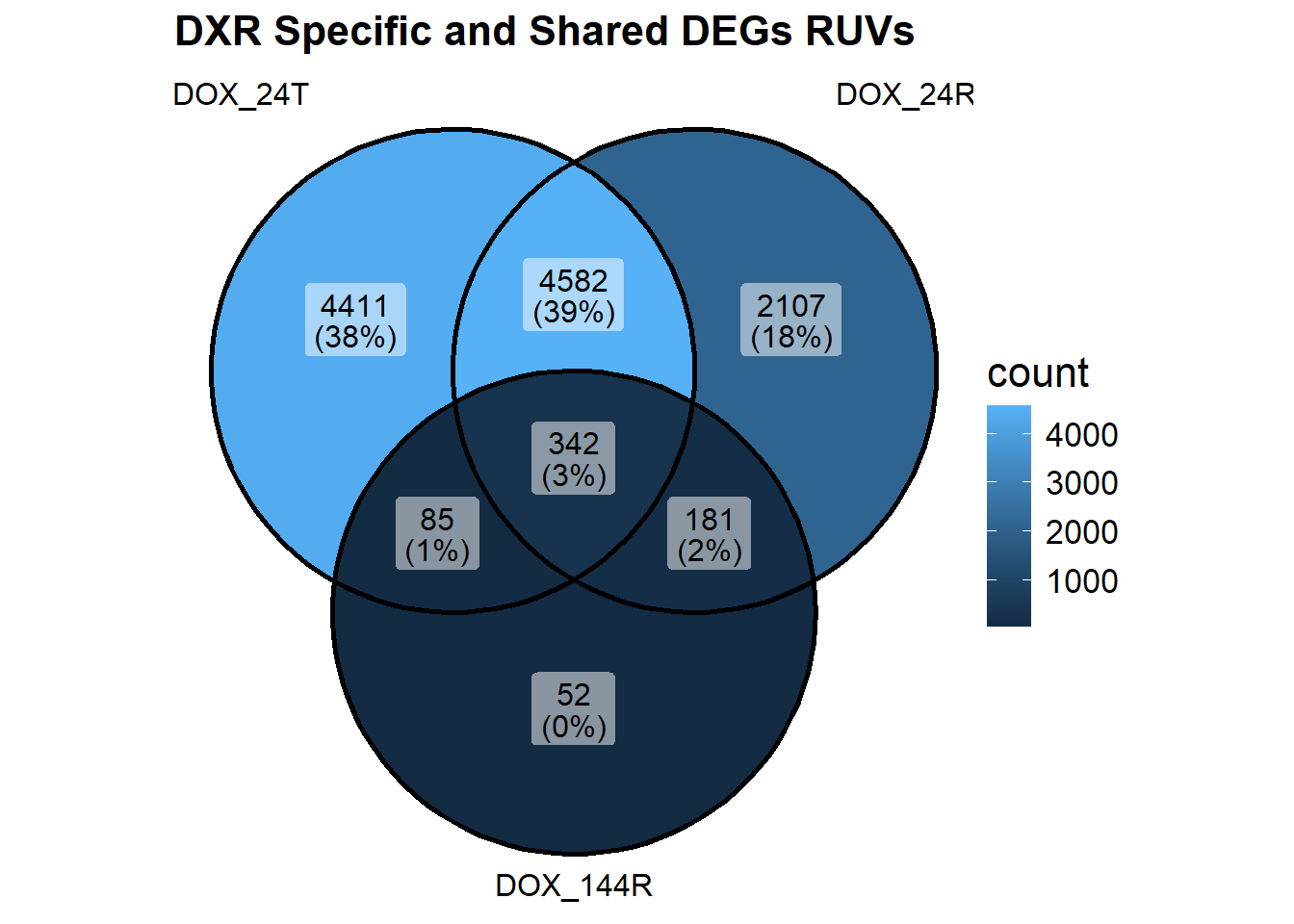
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#try and use the VennDetail package to extract the genes from each condition
#Set 1 - DOX_24T only genes
#Set 2 - DOX_24T shared genes
#Set 3 - DOX_144R shared genes
plot.new()
venn_test_RUV <- venndetail(venntest_RUV)
plot(venn_test_RUV)
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
detail(venn_test_RUV)=== Here is the detail of Venndiagram ===
Total results: 11760 x 2
Total sets is: 7
Subset Detail
1 Shared 643837
2 Shared 60672
3 Shared 55450
4 Shared 1870
5 Shared 57822
6 Shared 84958
... with 11754 more rows ...#now that I have the genes subsetted by condition, I can pull out the genes I want to look at
venn_DOX_24T_RUV <- getSet(object = venn_test_RUV, subset = c("Group 1"))
dim(venn_DOX_24T_RUV)[1] 4411 2#4411 genes in DOX_24T only
venn_DOX_shared_RUV <- getSet(object = venn_test_RUV, subset = c("Shared", "Group 1_Group 2", "Group 1_Group 3"))
dim(venn_DOX_shared_RUV)[1] 5009 2#total of 5009 genes in this set which does not include D24T specific genes
venn_DOX144R_shared_RUV <- getSet(object = venn_test_RUV, subset = c("Shared", "Group 3", "Group 2_Group 3", "Group 1_Group 3"))
dim(venn_DOX144R_shared_RUV)[1] 660 2#total of 660 genes: (original 509)
#342 shared all
#32 DOX144R Specific
#85 DOX24T + DOX144R
#181 DOX24R + DOX144R
#after that, run GO analysis
venn_shared_DEGs_RUV <- venn_DOX_shared_RUV %>%
as.data.frame() %>%
column_to_rownames(., var = "Detail")
venn_DOX24T_DEGs_RUV <- venn_DOX_24T_RUV %>%
as.data.frame() %>%
column_to_rownames(., var = "Detail")
venn_sharedD144R_DEGs_RUV <- venn_DOX144R_shared_RUV %>%
as.data.frame() %>%
column_to_rownames(., var = "Detail")
#pull these genes out of my DEG matrix with logFC
DOX_24T_DEGs_RUV <-
Toptable_list_RUVk1_symbols$V.D24T_RUV[row.names(venn_DOX24T_DEGs_RUV),]
DOX_24T_shared_DEGs_RUV <-
Toptable_list_RUVk1_symbols$V.D24T_RUV[row.names(venn_shared_DEGs_RUV),]
DOX_144R_shared_DEGs_RUV <-
Toptable_list_RUVk1_symbols$V.D144R_RUV[row.names(venn_sharedD144R_DEGs_RUV),]
#next, make the vectors that I need for plotting after cutting off by adj. p value < 0.05
DOX24T_DEGs_GO_RUV <- DOX_24T_DEGs_RUV %>%
dplyr::filter(., adj.P.Val < 0.05)
#shared D24T genes venn diagram
DOX24Tshare_DEGs_GO_RUV <- DOX_24T_shared_DEGs_RUV %>%
dplyr::filter(., adj.P.Val < 0.05)
DOX24T_share_DEGs_GO_plot_RUV <- DOX24Tshare_DEGs_GO_RUV %>%
dplyr::select("Entrez_ID")
#shared D144R genes venn diagram
DOX144Rshare_DEGs_GO_RUV <- DOX_144R_shared_DEGs_RUV %>%
dplyr::filter(., adj.P.Val < 0.05)
DOX144Rshare_DEGs_GO_plot_RUV <- DOX144Rshare_DEGs_GO_RUV %>%
dplyr::select("Entrez_ID")
#now go ahead and do this for each condition as well
#DOX24T
venn_DOX_24T_RUV <- getSet(object = venn_test_RUV, subset = c("Group 1"))
dim(venn_DOX_24T_RUV)[1] 4411 2#4411 genes in DOX_24T only
venn_DOX24T_DEGs_RUV <- venn_DOX_24T_RUV %>%
as.data.frame() %>%
column_to_rownames(., var = "Detail")
#pull these genes out of my DEG matrix with logFC
DOX_24T_DEGs_RUV <- Toptable_list_RUVk1_symbols$V.D24T_RUV[row.names(venn_DOX24T_DEGs_RUV),]
DOX24T_DEGs_GO_RUV <- DOX_24T_DEGs_RUV %>%
dplyr::filter(., adj.P.Val < 0.05) %>%
dplyr::select("Entrez_ID")
#DOX24R
venn_DOX_24R_RUV <- getSet(object = venn_test_RUV, subset = c("Group 2"))
dim(venn_DOX_24R_RUV)[1] 2107 2#2107 genes in DOX_24R only
venn_DOX24R_DEGs_RUV <- venn_DOX_24R_RUV %>%
as.data.frame() %>%
column_to_rownames(., var = "Detail")
#pull these genes out of my DEG matrix with logFC
DOX_24R_DEGs_RUV <- Toptable_list_RUVk1_symbols$V.D24R_RUV[row.names(venn_DOX24R_DEGs_RUV),]
DOX24R_DEGs_GO_RUV <- DOX_24R_DEGs_RUV %>%
dplyr::filter(., adj.P.Val < 0.05) %>%
dplyr::select("Entrez_ID")
#DOX144R
venn_DOX_144R_RUV <- getSet(object = venn_test_RUV, subset = c("Group 3"))
dim(venn_DOX_144R_RUV)[1] 52 2#52 genes in DOX_144R only
venn_DOX144R_DEGs_RUV <- venn_DOX_144R_RUV %>%
as.data.frame() %>%
column_to_rownames(., var = "Detail")
#pull these genes out of my DEG matrix with logFC
DOX_144R_DEGs_RUV <-
Toptable_list_RUVk1_symbols$V.D144R_RUV[row.names(venn_DOX144R_DEGs_RUV),]
DOX144R_DEGs_GO_RUV <- DOX_144R_DEGs_RUV %>%
dplyr::filter(., adj.P.Val < 0.05) %>%
dplyr::select("Entrez_ID")###GO KEGG Overlap of DEGs From Original Data
#####DOX24 Genes#####
library(gprofiler2)
D24_DEGs_mat <- as.matrix(DOX24T_DEGs_GO)
DOX_24_dxr_gene <- gost(query = D24_DEGs_mat,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
DOX_24_gost_genes <- gostplot(DOX_24_dxr_gene, capped = FALSE, interactive = TRUE)
DOX_24_gost_genestable_DOX24_genes <- DOX_24_dxr_gene$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX24_genes %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0080090 | regulation of primary metabolic process | 1355 | 5390 | 2.955e-59 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 1046 | 3990 | 3.142e-51 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 965 | 3687 | 5.969e-46 |
| GO:BP | GO:0006351 | DNA-templated transcription | 930 | 3549 | 3.382e-44 |
| GO:BP | GO:0019538 | protein metabolic process | 1153 | 4721 | 5.625e-41 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 890 | 3428 | 3.256e-40 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 886 | 3409 | 3.256e-40 |
| GO:BP | GO:0043412 | macromolecule modification | 773 | 3030 | 5.277e-31 |
| GO:BP | GO:0006996 | organelle organization | 885 | 3594 | 1.783e-30 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 704 | 2711 | 2.695e-30 |
| GO:BP | GO:0019222 | regulation of metabolic process | 1539 | 7035 | 9.797e-30 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 667 | 2578 | 5.907e-28 |
| GO:BP | GO:0044238 | primary metabolic process | 2457 | 12342 | 2.653e-27 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 1422 | 6492 | 1.244e-26 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1310 | 5920 | 1.249e-25 |
| GO:BP | GO:0036211 | protein modification process | 708 | 2846 | 1.236e-24 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1368 | 6264 | 1.493e-24 |
| GO:BP | GO:0009056 | catabolic process | 659 | 2639 | 4.202e-23 |
| GO:BP | GO:0046907 | intracellular transport | 392 | 1381 | 6.275e-23 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 531 | 2036 | 2.149e-22 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 846 | 3597 | 4.099e-22 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 782 | 3288 | 1.586e-21 |
| GO:BP | GO:0050789 | regulation of biological process | 2420 | 12336 | 3.330e-21 |
| GO:BP | GO:0050794 | regulation of cellular process | 2352 | 11946 | 6.778e-21 |
| GO:BP | GO:0065007 | biological regulation | 2484 | 12743 | 1.666e-20 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 1255 | 5809 | 8.803e-20 |
| GO:BP | GO:0006412 | translation | 232 | 727 | 1.113e-19 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 477 | 1835 | 1.337e-19 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1219 | 5644 | 5.900e-19 |
| GO:BP | GO:0051649 | establishment of localization in cell | 509 | 2010 | 1.547e-18 |
| GO:BP | GO:0033554 | cellular response to stress | 471 | 1830 | 1.968e-18 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 383 | 1417 | 2.239e-18 |
| GO:BP | GO:0010468 | regulation of gene expression | 1192 | 5536 | 6.806e-18 |
| GO:BP | GO:0033036 | macromolecule localization | 748 | 3228 | 2.540e-17 |
| GO:BP | GO:0051641 | cellular localization | 832 | 3661 | 3.002e-17 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 80 | 168 | 4.505e-17 |
| GO:BP | GO:0006325 | chromatin organization | 260 | 884 | 6.229e-17 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 436 | 1697 | 8.309e-17 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 436 | 1699 | 1.019e-16 |
| GO:BP | GO:0043687 | post-translational protein modification | 290 | 1027 | 2.501e-16 |
| GO:BP | GO:0015031 | protein transport | 377 | 1444 | 1.835e-15 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 248 | 853 | 1.909e-15 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 274 | 975 | 4.505e-15 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 610 | 2593 | 4.718e-15 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 629 | 2696 | 8.660e-15 |
| GO:BP | GO:0008104 | protein localization | 643 | 2770 | 1.110e-14 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 645 | 2782 | 1.270e-14 |
| GO:BP | GO:0008152 | metabolic process | 2676 | 14136 | 1.403e-14 |
| GO:BP | GO:0071705 | nitrogen compound transport | 470 | 1923 | 4.476e-14 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 570 | 2433 | 1.473e-13 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 386 | 1529 | 1.765e-13 |
| GO:BP | GO:0030163 | protein catabolic process | 280 | 1030 | 1.765e-13 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 226 | 786 | 1.827e-13 |
| GO:BP | GO:0006886 | intracellular protein transport | 203 | 686 | 2.250e-13 |
| GO:BP | GO:0009894 | regulation of catabolic process | 281 | 1040 | 3.402e-13 |
| GO:BP | GO:0006259 | DNA metabolic process | 273 | 1005 | 4.258e-13 |
| GO:BP | GO:0042254 | ribosome biogenesis | 115 | 323 | 4.644e-13 |
| GO:BP | GO:0035556 | intracellular signal transduction | 670 | 2965 | 8.507e-13 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 191 | 643 | 9.822e-13 |
| GO:BP | GO:0016567 | protein ubiquitination | 218 | 768 | 2.362e-12 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 188 | 638 | 3.636e-12 |
| GO:BP | GO:0006974 | DNA damage response | 248 | 908 | 4.093e-12 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 184 | 628 | 1.200e-11 |
| GO:BP | GO:0032543 | mitochondrial translation | 59 | 130 | 2.039e-11 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 349 | 1400 | 2.520e-11 |
| GO:BP | GO:0006281 | DNA repair | 181 | 621 | 3.090e-11 |
| GO:BP | GO:0006338 | chromatin remodeling | 204 | 725 | 3.859e-11 |
| GO:BP | GO:0023051 | regulation of signaling | 758 | 3478 | 3.859e-11 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 107 | 312 | 5.829e-11 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 854 | 3993 | 6.640e-11 |
| GO:BP | GO:0051179 | localization | 1144 | 5555 | 8.041e-11 |
| GO:BP | GO:0010646 | regulation of cell communication | 757 | 3486 | 8.628e-11 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1157 | 5629 | 9.293e-11 |
| GO:BP | GO:0009966 | regulation of signal transduction | 670 | 3034 | 9.639e-11 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 161 | 543 | 1.299e-10 |
| GO:BP | GO:0006402 | mRNA catabolic process | 93 | 261 | 1.507e-10 |
| GO:BP | GO:0009987 | cellular process | 3598 | 20247 | 1.644e-10 |
| GO:BP | GO:0033043 | regulation of organelle organization | 292 | 1148 | 2.045e-10 |
| GO:BP | GO:0045184 | establishment of protein localization | 453 | 1937 | 2.074e-10 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 436 | 1855 | 2.638e-10 |
| GO:BP | GO:0006403 | RNA localization | 76 | 200 | 4.297e-10 |
| GO:BP | GO:0051169 | nuclear transport | 109 | 334 | 1.044e-09 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 109 | 334 | 1.044e-09 |
| GO:BP | GO:0007005 | mitochondrion organization | 133 | 435 | 1.075e-09 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 156 | 535 | 1.096e-09 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 540 | 2407 | 1.767e-09 |
| GO:BP | GO:0060341 | regulation of cellular localization | 259 | 1013 | 2.043e-09 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 540 | 2410 | 2.139e-09 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 147 | 500 | 2.152e-09 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 151 | 519 | 2.682e-09 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 308 | 1256 | 4.235e-09 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 70 | 187 | 5.937e-09 |
| GO:BP | GO:0006399 | tRNA metabolic process | 76 | 210 | 5.957e-09 |
| GO:BP | GO:0016072 | rRNA metabolic process | 90 | 266 | 7.047e-09 |
| GO:BP | GO:0006810 | transport | 915 | 4407 | 7.401e-09 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1637 | 8393 | 8.414e-09 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1177 | 5834 | 8.512e-09 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 125 | 414 | 9.665e-09 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 155 | 546 | 1.007e-08 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 314 | 1296 | 1.067e-08 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 310 | 1280 | 1.473e-08 |
| GO:BP | GO:0007049 | cell cycle | 387 | 1663 | 1.672e-08 |
| GO:BP | GO:0051168 | nuclear export | 64 | 169 | 1.969e-08 |
| GO:BP | GO:0032880 | regulation of protein localization | 232 | 907 | 1.969e-08 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 165 | 597 | 2.219e-08 |
| GO:BP | GO:0006914 | autophagy | 165 | 597 | 2.219e-08 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 318 | 1326 | 2.573e-08 |
| GO:BP | GO:0051726 | regulation of cell cycle | 268 | 1087 | 4.096e-08 |
| GO:BP | GO:0038202 | TORC1 signaling | 45 | 103 | 4.353e-08 |
| GO:BP | GO:0000209 | protein polyubiquitination | 91 | 280 | 5.172e-08 |
| GO:BP | GO:0006302 | double-strand break repair | 99 | 319 | 1.523e-07 |
| GO:BP | GO:0006401 | RNA catabolic process | 98 | 315 | 1.576e-07 |
| GO:BP | GO:0006364 | rRNA processing | 76 | 225 | 1.901e-07 |
| GO:BP | GO:0023057 | negative regulation of signaling | 335 | 1435 | 1.999e-07 |
| GO:BP | GO:0051236 | establishment of RNA localization | 60 | 163 | 2.334e-07 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 334 | 1436 | 3.186e-07 |
| GO:BP | GO:0006417 | regulation of translation | 112 | 380 | 3.464e-07 |
| GO:BP | GO:0032259 | methylation | 78 | 236 | 3.464e-07 |
| GO:BP | GO:0006282 | regulation of DNA repair | 75 | 225 | 4.441e-07 |
| GO:BP | GO:0008033 | tRNA processing | 53 | 139 | 4.518e-07 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 68 | 198 | 5.697e-07 |
| GO:BP | GO:0031929 | TOR signaling | 60 | 167 | 6.369e-07 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 26 | 48 | 7.306e-07 |
| GO:BP | GO:0050658 | RNA transport | 58 | 160 | 7.544e-07 |
| GO:BP | GO:0050657 | nucleic acid transport | 58 | 160 | 7.544e-07 |
| GO:BP | GO:0051234 | establishment of localization | 992 | 4928 | 8.478e-07 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 2253 | 12048 | 8.593e-07 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 106 | 361 | 1.031e-06 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 63 | 181 | 1.056e-06 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 40 | 96 | 1.713e-06 |
| GO:BP | GO:0141188 | nucleic acid catabolic process | 101 | 343 | 1.879e-06 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 176 | 686 | 1.982e-06 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 377 | 1680 | 2.059e-06 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 430 | 1953 | 2.226e-06 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 77 | 242 | 2.424e-06 |
| GO:BP | GO:0043487 | regulation of RNA stability | 65 | 193 | 2.549e-06 |
| GO:BP | GO:0043414 | macromolecule methylation | 48 | 127 | 2.832e-06 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 1536 | 7992 | 4.411e-06 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 75 | 237 | 4.544e-06 |
| GO:BP | GO:0016073 | snRNA metabolic process | 28 | 58 | 4.722e-06 |
| GO:BP | GO:0000278 | mitotic cell cycle | 217 | 892 | 4.767e-06 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 31 | 68 | 4.989e-06 |
| GO:BP | GO:0061024 | membrane organization | 202 | 821 | 5.548e-06 |
| GO:BP | GO:0051028 | mRNA transport | 48 | 130 | 6.186e-06 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 254 | 1077 | 6.186e-06 |
| GO:BP | GO:0080134 | regulation of response to stress | 316 | 1387 | 6.186e-06 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 92 | 312 | 6.454e-06 |
| GO:BP | GO:0006839 | mitochondrial transport | 60 | 178 | 7.486e-06 |
| GO:BP | GO:0009451 | RNA modification | 59 | 174 | 7.486e-06 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 47 | 127 | 7.535e-06 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 43 | 112 | 7.667e-06 |
| GO:BP | GO:0032879 | regulation of localization | 440 | 2029 | 8.883e-06 |
| GO:BP | GO:0007275 | multicellular organism development | 944 | 4727 | 9.559e-06 |
| GO:BP | GO:0065009 | regulation of molecular function | 336 | 1496 | 9.987e-06 |
| GO:BP | GO:0032790 | ribosome disassembly | 22 | 41 | 1.013e-05 |
| GO:BP | GO:0016043 | cellular component organization | 1565 | 8184 | 1.067e-05 |
| GO:BP | GO:0098727 | maintenance of cell number | 61 | 184 | 1.087e-05 |
| GO:BP | GO:0022402 | cell cycle process | 293 | 1280 | 1.097e-05 |
| GO:BP | GO:0070925 | organelle assembly | 246 | 1046 | 1.164e-05 |
| GO:BP | GO:0000725 | recombinational repair | 63 | 193 | 1.269e-05 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 184 | 745 | 1.424e-05 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 496 | 2331 | 1.443e-05 |
| GO:BP | GO:0051276 | chromosome organization | 148 | 574 | 1.495e-05 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 257 | 1105 | 1.601e-05 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 130 | 492 | 1.894e-05 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 28 | 62 | 2.318e-05 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 61 | 188 | 2.369e-05 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1417 | 7376 | 2.369e-05 |
| GO:BP | GO:0019827 | stem cell population maintenance | 59 | 180 | 2.468e-05 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 47 | 132 | 2.503e-05 |
| GO:BP | GO:0000723 | telomere maintenance | 55 | 164 | 2.600e-05 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 222 | 938 | 2.638e-05 |
| GO:BP | GO:0034504 | protein localization to nucleus | 91 | 318 | 2.819e-05 |
| GO:BP | GO:0006457 | protein folding | 68 | 219 | 3.219e-05 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 688 | 3375 | 4.056e-05 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 50 | 146 | 4.083e-05 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 241 | 1039 | 4.414e-05 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 44 | 123 | 4.752e-05 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 164 | 662 | 4.752e-05 |
| GO:BP | GO:1903008 | organelle disassembly | 30 | 71 | 4.845e-05 |
| GO:BP | GO:0006950 | response to stress | 793 | 3948 | 4.923e-05 |
| GO:BP | GO:0008219 | cell death | 426 | 1991 | 5.646e-05 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 42 | 116 | 5.648e-05 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 229 | 983 | 5.742e-05 |
| GO:BP | GO:0012501 | programmed cell death | 425 | 1987 | 5.949e-05 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 49 | 144 | 6.152e-05 |
| GO:BP | GO:0017004 | cytochrome complex assembly | 21 | 42 | 7.496e-05 |
| GO:BP | GO:0032502 | developmental process | 1263 | 6553 | 7.983e-05 |
| GO:BP | GO:0051604 | protein maturation | 137 | 539 | 8.325e-05 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 127 | 492 | 8.464e-05 |
| GO:BP | GO:0006405 | RNA export from nucleus | 35 | 91 | 8.464e-05 |
| GO:BP | GO:0006508 | proteolysis | 327 | 1486 | 8.760e-05 |
| GO:BP | GO:0044281 | small molecule metabolic process | 383 | 1776 | 8.962e-05 |
| GO:BP | GO:0006400 | tRNA modification | 36 | 95 | 8.988e-05 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 83 | 291 | 9.029e-05 |
| GO:BP | GO:0048856 | anatomical structure development | 1162 | 5997 | 9.554e-05 |
| GO:BP | GO:0006390 | mitochondrial transcription | 14 | 22 | 9.554e-05 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 347 | 1592 | 1.019e-04 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 39 | 107 | 1.025e-04 |
| GO:BP | GO:0050793 | regulation of developmental process | 512 | 2457 | 1.025e-04 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 174 | 720 | 1.049e-04 |
| GO:BP | GO:0033365 | protein localization to organelle | 272 | 1209 | 1.082e-04 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 165 | 677 | 1.116e-04 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 48 | 143 | 1.116e-04 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 36 | 96 | 1.134e-04 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 30 | 74 | 1.202e-04 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 26 | 60 | 1.262e-04 |
| GO:BP | GO:0072344 | rescue of stalled ribosome | 18 | 34 | 1.327e-04 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 47 | 140 | 1.376e-04 |
| GO:BP | GO:0010506 | regulation of autophagy | 99 | 367 | 1.409e-04 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 158 | 646 | 1.411e-04 |
| GO:BP | GO:0023056 | positive regulation of signaling | 385 | 1796 | 1.411e-04 |
| GO:BP | GO:0022411 | cellular component disassembly | 116 | 446 | 1.429e-04 |
| GO:BP | GO:0043009 | chordate embryonic development | 163 | 671 | 1.496e-04 |
| GO:BP | GO:0006915 | apoptotic process | 409 | 1923 | 1.517e-04 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 101 | 377 | 1.520e-04 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 127 | 499 | 1.607e-04 |
| GO:BP | GO:0009790 | embryo development | 256 | 1135 | 1.648e-04 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 118 | 458 | 1.868e-04 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 342 | 1578 | 1.868e-04 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 328 | 1506 | 1.907e-04 |
| GO:BP | GO:0030030 | cell projection organization | 352 | 1631 | 1.978e-04 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 45 | 134 | 2.050e-04 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 45 | 134 | 2.050e-04 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 22 | 48 | 2.220e-04 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 383 | 1795 | 2.237e-04 |
| GO:BP | GO:0006414 | translational elongation | 31 | 80 | 2.301e-04 |
| GO:BP | GO:0031647 | regulation of protein stability | 91 | 335 | 2.362e-04 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 29 | 73 | 2.560e-04 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 162 | 673 | 2.603e-04 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 23 | 52 | 2.788e-04 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 37 | 104 | 3.066e-04 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 26 | 63 | 3.334e-04 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 204 | 884 | 3.373e-04 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 75 | 266 | 3.632e-04 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 165 | 692 | 3.711e-04 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 21 | 46 | 3.727e-04 |
| GO:BP | GO:0016236 | macroautophagy | 96 | 362 | 3.769e-04 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 317 | 1462 | 3.805e-04 |
| GO:BP | GO:0051170 | import into nucleus | 53 | 171 | 4.185e-04 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 68 | 236 | 4.185e-04 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 20 | 43 | 4.210e-04 |
| GO:BP | GO:0016310 | phosphorylation | 289 | 1320 | 4.358e-04 |
| GO:BP | GO:0170036 | import into the mitochondrion | 22 | 50 | 4.628e-04 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 88 | 327 | 4.628e-04 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 36 | 102 | 4.677e-04 |
| GO:BP | GO:0061157 | mRNA destabilization | 36 | 102 | 4.677e-04 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 41 | 122 | 4.708e-04 |
| GO:BP | GO:0050779 | RNA destabilization | 37 | 106 | 4.708e-04 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 97 | 369 | 4.777e-04 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 166 | 701 | 4.950e-04 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 31 | 83 | 5.001e-04 |
| GO:BP | GO:0009267 | cellular response to starvation | 54 | 177 | 5.466e-04 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 401 | 1912 | 6.502e-04 |
| GO:BP | GO:0032200 | telomere organization | 57 | 191 | 6.559e-04 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 34 | 96 | 7.188e-04 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 36 | 104 | 7.345e-04 |
| GO:BP | GO:0006606 | protein import into nucleus | 51 | 166 | 7.425e-04 |
| GO:BP | GO:0010256 | endomembrane system organization | 147 | 613 | 7.575e-04 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 23 | 55 | 7.649e-04 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 9 | 12 | 7.656e-04 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 44 | 137 | 7.973e-04 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 1405 | 7434 | 8.003e-04 |
| GO:BP | GO:0007399 | nervous system development | 530 | 2604 | 8.126e-04 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 47 | 150 | 8.620e-04 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 69 | 246 | 8.692e-04 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 33 | 93 | 8.699e-04 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 131 | 537 | 8.708e-04 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 20 | 45 | 8.749e-04 |
| GO:BP | GO:0050821 | protein stabilization | 64 | 224 | 8.960e-04 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 98 | 380 | 9.194e-04 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 226 | 1011 | 9.206e-04 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 136 | 563 | 1.001e-03 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 14 | 26 | 1.026e-03 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 96 | 372 | 1.061e-03 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 69 | 248 | 1.114e-03 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 18 | 39 | 1.160e-03 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 28 | 75 | 1.186e-03 |
| GO:BP | GO:0007178 | cell surface receptor protein serine/threonine kinase signaling pathway | 94 | 364 | 1.224e-03 |
| GO:BP | GO:0002181 | cytoplasmic translation | 50 | 165 | 1.232e-03 |
| GO:BP | GO:0016197 | endosomal transport | 78 | 290 | 1.263e-03 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 30 | 83 | 1.276e-03 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 91 | 351 | 1.364e-03 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 51 | 170 | 1.371e-03 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 43 | 136 | 1.379e-03 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 107 | 427 | 1.395e-03 |
| GO:BP | GO:0007030 | Golgi organization | 47 | 153 | 1.395e-03 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 336 | 1588 | 1.420e-03 |
| GO:BP | GO:0006310 | DNA recombination | 90 | 347 | 1.445e-03 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 91 | 352 | 1.492e-03 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 32 | 92 | 1.659e-03 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 13 | 24 | 1.725e-03 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 265 | 1223 | 1.812e-03 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 54 | 185 | 1.830e-03 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 546 | 2713 | 1.882e-03 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 120 | 493 | 1.884e-03 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 628 | 3157 | 1.884e-03 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 86 | 331 | 1.919e-03 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 47 | 155 | 1.919e-03 |
| GO:BP | GO:0045727 | positive regulation of translation | 43 | 138 | 1.926e-03 |
| GO:BP | GO:0001510 | RNA methylation | 29 | 81 | 1.961e-03 |
| GO:BP | GO:0007017 | microtubule-based process | 221 | 999 | 2.012e-03 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 69 | 253 | 2.012e-03 |
| GO:BP | GO:0043543 | protein acylation | 31 | 89 | 2.012e-03 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 59 | 208 | 2.026e-03 |
| GO:BP | GO:0006997 | nucleus organization | 45 | 147 | 2.074e-03 |
| GO:BP | GO:0007059 | chromosome segregation | 106 | 427 | 2.115e-03 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 16 | 34 | 2.136e-03 |
| GO:BP | GO:0001701 | in utero embryonic development | 103 | 413 | 2.182e-03 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 38 | 118 | 2.243e-03 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 18 | 41 | 2.350e-03 |
| GO:BP | GO:0031399 | regulation of protein modification process | 230 | 1049 | 2.496e-03 |
| GO:BP | GO:0006413 | translational initiation | 40 | 127 | 2.530e-03 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 44 | 144 | 2.544e-03 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 24 | 63 | 2.583e-03 |
| GO:BP | GO:0042594 | response to starvation | 61 | 219 | 2.632e-03 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 70 | 260 | 2.646e-03 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 70 | 260 | 2.646e-03 |
| GO:BP | GO:0090685 | RNA localization to nucleus | 11 | 19 | 2.681e-03 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 88 | 344 | 2.681e-03 |
| GO:BP | GO:0090670 | RNA localization to Cajal body | 11 | 19 | 2.681e-03 |
| GO:BP | GO:0090671 | telomerase RNA localization to Cajal body | 11 | 19 | 2.681e-03 |
| GO:BP | GO:0090672 | telomerase RNA localization | 11 | 19 | 2.681e-03 |
| GO:BP | GO:0045945 | positive regulation of transcription by RNA polymerase III | 11 | 19 | 2.681e-03 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 42 | 136 | 2.699e-03 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 56 | 197 | 2.699e-03 |
| GO:BP | GO:0018279 | protein N-linked glycosylation via asparagine | 19 | 45 | 2.709e-03 |
| GO:BP | GO:0017148 | negative regulation of translation | 41 | 132 | 2.804e-03 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 96 | 383 | 2.919e-03 |
| GO:BP | GO:0048731 | system development | 789 | 4053 | 2.940e-03 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 76 | 289 | 2.986e-03 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 78 | 299 | 3.225e-03 |
| GO:BP | GO:0007033 | vacuole organization | 65 | 239 | 3.225e-03 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 37 | 116 | 3.225e-03 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 70 | 262 | 3.242e-03 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 73 | 276 | 3.282e-03 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 139 | 594 | 3.282e-03 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 36 | 112 | 3.294e-03 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 105 | 428 | 3.389e-03 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 21 | 53 | 3.411e-03 |
| GO:BP | GO:0051640 | organelle localization | 144 | 620 | 3.565e-03 |
| GO:BP | GO:0051865 | protein autoubiquitination | 27 | 76 | 3.565e-03 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 19 | 46 | 3.664e-03 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 19 | 46 | 3.664e-03 |
| GO:BP | GO:0030518 | nuclear receptor-mediated steroid hormone signaling pathway | 38 | 121 | 3.664e-03 |
| GO:BP | GO:1904874 | positive regulation of telomerase RNA localization to Cajal body | 7 | 9 | 3.723e-03 |
| GO:BP | GO:0042796 | snRNA transcription by RNA polymerase III | 7 | 9 | 3.723e-03 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 105 | 430 | 3.997e-03 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 35 | 109 | 4.037e-03 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 55 | 196 | 4.195e-03 |
| GO:BP | GO:0048255 | mRNA stabilization | 25 | 69 | 4.276e-03 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 42 | 139 | 4.296e-03 |
| GO:BP | GO:0006468 | protein phosphorylation | 262 | 1227 | 4.554e-03 |
| GO:BP | GO:0009301 | snRNA transcription | 11 | 20 | 4.580e-03 |
| GO:BP | GO:0030150 | protein import into mitochondrial matrix | 11 | 20 | 4.580e-03 |
| GO:BP | GO:0031503 | protein-containing complex localization | 59 | 215 | 4.716e-03 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 82 | 322 | 4.834e-03 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 153 | 670 | 4.970e-03 |
| GO:BP | GO:0051049 | regulation of transport | 334 | 1607 | 4.970e-03 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 47 | 162 | 5.163e-03 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 32 | 98 | 5.212e-03 |
| GO:BP | GO:0007010 | cytoskeleton organization | 319 | 1529 | 5.212e-03 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 61 | 225 | 5.212e-03 |
| GO:BP | GO:0072175 | epithelial tube formation | 41 | 136 | 5.226e-03 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 25 | 70 | 5.333e-03 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 25 | 70 | 5.333e-03 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 26 | 74 | 5.416e-03 |
| GO:BP | GO:0044770 | cell cycle phase transition | 125 | 532 | 5.449e-03 |
| GO:BP | GO:0008535 | respiratory chain complex IV assembly | 14 | 30 | 5.706e-03 |
| GO:BP | GO:1904872 | regulation of telomerase RNA localization to Cajal body | 8 | 12 | 5.887e-03 |
| GO:BP | GO:1904867 | protein localization to Cajal body | 8 | 12 | 5.887e-03 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 8 | 12 | 5.887e-03 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 83 | 329 | 5.887e-03 |
| GO:BP | GO:1903405 | protein localization to nuclear body | 8 | 12 | 5.887e-03 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 49 | 172 | 5.911e-03 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 49 | 172 | 5.911e-03 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 70 | 268 | 5.999e-03 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 32 | 99 | 6.167e-03 |
| GO:BP | GO:0048468 | cell development | 569 | 2876 | 6.246e-03 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 13 | 27 | 6.295e-03 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 13 | 27 | 6.295e-03 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 40 | 133 | 6.295e-03 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 79 | 311 | 6.382e-03 |
| GO:BP | GO:0008213 | protein alkylation | 19 | 48 | 6.386e-03 |
| GO:BP | GO:0006479 | protein methylation | 19 | 48 | 6.386e-03 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 30 | 91 | 6.386e-03 |
| GO:BP | GO:0048285 | organelle fission | 118 | 500 | 6.407e-03 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 42 | 142 | 6.507e-03 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 39 | 129 | 6.507e-03 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 27 | 79 | 6.511e-03 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 70 | 269 | 6.511e-03 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 41 | 138 | 6.814e-03 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 41 | 138 | 6.814e-03 |
| GO:BP | GO:0051223 | regulation of protein transport | 106 | 442 | 6.814e-03 |
| GO:BP | GO:0062125 | regulation of mitochondrial gene expression | 15 | 34 | 6.869e-03 |
| GO:BP | GO:0021915 | neural tube development | 46 | 160 | 6.869e-03 |
| GO:BP | GO:0030258 | lipid modification | 54 | 196 | 7.010e-03 |
| GO:BP | GO:0010970 | transport along microtubule | 50 | 178 | 7.010e-03 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 80 | 317 | 7.010e-03 |
| GO:BP | GO:1990928 | response to amino acid starvation | 21 | 56 | 7.204e-03 |
| GO:BP | GO:0090110 | COPII-coated vesicle cargo loading | 10 | 18 | 7.306e-03 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 38 | 126 | 7.909e-03 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 25 | 72 | 7.948e-03 |
| GO:BP | GO:0016226 | iron-sulfur cluster assembly | 14 | 31 | 7.948e-03 |
| GO:BP | GO:0031163 | metallo-sulfur cluster assembly | 14 | 31 | 7.948e-03 |
| GO:BP | GO:0019080 | viral gene expression | 34 | 109 | 7.948e-03 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 16 | 38 | 8.082e-03 |
| GO:BP | GO:0035148 | tube formation | 43 | 148 | 8.091e-03 |
| GO:BP | GO:0000819 | sister chromatid segregation | 62 | 234 | 8.121e-03 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 33 | 105 | 8.203e-03 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 103 | 430 | 8.270e-03 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 53 | 193 | 8.347e-03 |
| GO:BP | GO:0000045 | autophagosome assembly | 36 | 118 | 8.553e-03 |
| GO:BP | GO:0018216 | peptidyl-arginine methylation | 7 | 10 | 8.784e-03 |
| GO:BP | GO:1904871 | positive regulation of protein localization to Cajal body | 7 | 10 | 8.784e-03 |
| GO:BP | GO:1904869 | regulation of protein localization to Cajal body | 7 | 10 | 8.784e-03 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 48 | 171 | 8.905e-03 |
| GO:BP | GO:0044782 | cilium organization | 103 | 431 | 8.905e-03 |
| GO:BP | GO:0140014 | mitotic nuclear division | 72 | 282 | 9.147e-03 |
| GO:BP | GO:0061053 | somite development | 28 | 85 | 9.407e-03 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 15 | 35 | 9.450e-03 |
| GO:BP | GO:0016180 | snRNA processing | 15 | 35 | 9.450e-03 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 15 | 35 | 9.450e-03 |
| GO:BP | GO:0009058 | biosynthetic process | 1952 | 10664 | 9.681e-03 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 42 | 145 | 9.715e-03 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 65 | 250 | 9.968e-03 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 201 | 928 | 1.004e-02 |
| GO:BP | GO:0051098 | regulation of binding | 57 | 213 | 1.008e-02 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 66 | 255 | 1.027e-02 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 31 | 98 | 1.030e-02 |
| GO:BP | GO:0000902 | cell morphogenesis | 214 | 996 | 1.037e-02 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 554 | 2813 | 1.049e-02 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 49 | 177 | 1.062e-02 |
| GO:BP | GO:0141193 | nuclear receptor-mediated signaling pathway | 49 | 177 | 1.062e-02 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 555 | 2819 | 1.062e-02 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 16 | 39 | 1.071e-02 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 16 | 39 | 1.071e-02 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 14 | 32 | 1.101e-02 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 8 | 13 | 1.104e-02 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 33 | 107 | 1.115e-02 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 44 | 155 | 1.115e-02 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 50 | 182 | 1.115e-02 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 96 | 400 | 1.117e-02 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 48 | 173 | 1.118e-02 |
| GO:BP | GO:0016050 | vesicle organization | 91 | 376 | 1.147e-02 |
| GO:BP | GO:0043489 | RNA stabilization | 26 | 78 | 1.147e-02 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 64 | 247 | 1.153e-02 |
| GO:BP | GO:0009191 | ribonucleoside diphosphate catabolic process | 32 | 103 | 1.153e-02 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 9 | 16 | 1.178e-02 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 77 | 309 | 1.200e-02 |
| GO:BP | GO:0043401 | steroid hormone receptor signaling pathway | 40 | 138 | 1.222e-02 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 66 | 257 | 1.225e-02 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 117 | 505 | 1.229e-02 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 100 | 421 | 1.232e-02 |
| GO:BP | GO:0022008 | neurogenesis | 360 | 1771 | 1.232e-02 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 91 | 377 | 1.232e-02 |
| GO:BP | GO:1905037 | autophagosome organization | 37 | 125 | 1.235e-02 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 18 | 47 | 1.251e-02 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 15 | 36 | 1.254e-02 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 64 | 248 | 1.258e-02 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 6 | 8 | 1.258e-02 |
| GO:BP | GO:0051086 | chaperone mediated protein folding independent of cofactor | 6 | 8 | 1.258e-02 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 61 | 234 | 1.259e-02 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 79 | 320 | 1.340e-02 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 85 | 349 | 1.342e-02 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 38 | 130 | 1.344e-02 |
| GO:BP | GO:0140374 | antiviral innate immune response | 20 | 55 | 1.360e-02 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 161 | 729 | 1.361e-02 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 119 | 517 | 1.387e-02 |
| GO:BP | GO:0006998 | nuclear envelope organization | 21 | 59 | 1.387e-02 |
| GO:BP | GO:0009303 | rRNA transcription | 16 | 40 | 1.391e-02 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 44 | 157 | 1.403e-02 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 90 | 374 | 1.407e-02 |
| GO:BP | GO:0033617 | mitochondrial cytochrome c oxidase assembly | 12 | 26 | 1.427e-02 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 12 | 26 | 1.427e-02 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 126 | 553 | 1.468e-02 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 14 | 33 | 1.473e-02 |
| GO:BP | GO:0070085 | glycosylation | 63 | 245 | 1.473e-02 |
| GO:BP | GO:0045995 | regulation of embryonic development | 29 | 92 | 1.486e-02 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 82 | 336 | 1.497e-02 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 306 | 1489 | 1.497e-02 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 139 | 619 | 1.497e-02 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 50 | 185 | 1.529e-02 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 32 | 105 | 1.544e-02 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 18 | 48 | 1.582e-02 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 56 | 213 | 1.582e-02 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 11 | 23 | 1.609e-02 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 39 | 136 | 1.653e-02 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 68 | 270 | 1.667e-02 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 55 | 209 | 1.696e-02 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 36 | 123 | 1.704e-02 |
| GO:BP | GO:0062028 | regulation of stress granule assembly | 7 | 11 | 1.726e-02 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 75 | 304 | 1.726e-02 |
| GO:BP | GO:0018195 | peptidyl-arginine modification | 7 | 11 | 1.726e-02 |
| GO:BP | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening | 13 | 30 | 1.726e-02 |
| GO:BP | GO:0046834 | lipid phosphorylation | 7 | 11 | 1.726e-02 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 75 | 304 | 1.726e-02 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 204 | 956 | 1.732e-02 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 29 | 93 | 1.732e-02 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 538 | 2745 | 1.732e-02 |
| GO:BP | GO:0016482 | cytosolic transport | 40 | 141 | 1.749e-02 |
| GO:BP | GO:0010212 | response to ionizing radiation | 40 | 141 | 1.749e-02 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 64 | 252 | 1.803e-02 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 247 | 1182 | 1.804e-02 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 204 | 957 | 1.811e-02 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 45 | 164 | 1.862e-02 |
| GO:BP | GO:1990173 | protein localization to nucleoplasm | 8 | 14 | 1.882e-02 |
| GO:BP | GO:0098734 | macromolecule depalmitoylation | 8 | 14 | 1.882e-02 |
| GO:BP | GO:0043144 | sno(s)RNA processing | 8 | 14 | 1.882e-02 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 78 | 320 | 1.950e-02 |
| GO:BP | GO:0099111 | microtubule-based transport | 57 | 220 | 1.983e-02 |
| GO:BP | GO:0042073 | intraciliary transport | 18 | 49 | 1.991e-02 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 29 | 94 | 2.036e-02 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 206 | 970 | 2.043e-02 |
| GO:BP | GO:0016925 | protein sumoylation | 23 | 69 | 2.045e-02 |
| GO:BP | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway | 47 | 174 | 2.055e-02 |
| GO:BP | GO:0009134 | nucleoside diphosphate catabolic process | 32 | 107 | 2.057e-02 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 179 | 830 | 2.057e-02 |
| GO:BP | GO:0030097 | hemopoiesis | 207 | 976 | 2.109e-02 |
| GO:BP | GO:0048699 | generation of neurons | 313 | 1536 | 2.113e-02 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 87 | 365 | 2.119e-02 |
| GO:BP | GO:0009179 | purine ribonucleoside diphosphate metabolic process | 34 | 116 | 2.138e-02 |
| GO:BP | GO:0009135 | purine nucleoside diphosphate metabolic process | 34 | 116 | 2.138e-02 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 15 | 38 | 2.151e-02 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 31 | 103 | 2.151e-02 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 54 | 207 | 2.176e-02 |
| GO:BP | GO:0072359 | circulatory system development | 239 | 1145 | 2.212e-02 |
| GO:BP | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 11 | 24 | 2.307e-02 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 11 | 24 | 2.307e-02 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 13 | 31 | 2.338e-02 |
| GO:BP | GO:0030162 | regulation of proteolysis | 95 | 406 | 2.379e-02 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 65 | 260 | 2.409e-02 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 17 | 46 | 2.424e-02 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 23 | 70 | 2.459e-02 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 78 | 323 | 2.462e-02 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 44 | 162 | 2.489e-02 |
| GO:BP | GO:0071396 | cellular response to lipid | 136 | 613 | 2.500e-02 |
| GO:BP | GO:0051099 | positive regulation of binding | 31 | 104 | 2.500e-02 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 92 | 392 | 2.512e-02 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 19 | 54 | 2.515e-02 |
| GO:BP | GO:0060324 | face development | 19 | 54 | 2.515e-02 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 40 | 144 | 2.519e-02 |
| GO:BP | GO:0098732 | macromolecule deacylation | 20 | 58 | 2.519e-02 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 251 | 1212 | 2.554e-02 |
| GO:BP | GO:0009181 | purine ribonucleoside diphosphate catabolic process | 30 | 100 | 2.607e-02 |
| GO:BP | GO:0009137 | purine nucleoside diphosphate catabolic process | 30 | 100 | 2.607e-02 |
| GO:BP | GO:0051607 | defense response to virus | 77 | 319 | 2.609e-02 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 66 | 266 | 2.639e-02 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 54 | 209 | 2.639e-02 |
| GO:BP | GO:0003007 | heart morphogenesis | 66 | 266 | 2.639e-02 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 119 | 528 | 2.654e-02 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 1312 | 7076 | 2.666e-02 |
| GO:BP | GO:0097680 | double-strand break repair via classical nonhomologous end joining | 6 | 9 | 2.672e-02 |
| GO:BP | GO:0031946 | regulation of glucocorticoid biosynthetic process | 6 | 9 | 2.672e-02 |
| GO:BP | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process | 6 | 9 | 2.672e-02 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 114 | 503 | 2.672e-02 |
| GO:BP | GO:0007219 | Notch signaling pathway | 49 | 186 | 2.705e-02 |
| GO:BP | GO:0002218 | activation of innate immune response | 74 | 305 | 2.705e-02 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 12 | 28 | 2.712e-02 |
| GO:BP | GO:0060323 | head morphogenesis | 15 | 39 | 2.744e-02 |
| GO:BP | GO:0070979 | protein K11-linked ubiquitination | 15 | 39 | 2.744e-02 |
| GO:BP | GO:0032205 | negative regulation of telomere maintenance | 15 | 39 | 2.744e-02 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 34 | 118 | 2.744e-02 |
| GO:BP | GO:0007507 | heart development | 134 | 605 | 2.753e-02 |
| GO:BP | GO:0060485 | mesenchyme development | 77 | 320 | 2.769e-02 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 71 | 291 | 2.804e-02 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 31 | 105 | 2.808e-02 |
| GO:BP | GO:0015936 | coenzyme A metabolic process | 9 | 18 | 2.832e-02 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 54 | 210 | 2.854e-02 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 23 | 71 | 2.854e-02 |
| GO:BP | GO:0046034 | ATP metabolic process | 58 | 229 | 2.875e-02 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 121 | 540 | 2.915e-02 |
| GO:BP | GO:0001841 | neural tube formation | 30 | 101 | 2.954e-02 |
| GO:BP | GO:1900152 | negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 7 | 12 | 2.985e-02 |
| GO:BP | GO:0150011 | regulation of neuron projection arborization | 8 | 15 | 2.985e-02 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 7 | 12 | 2.985e-02 |
| GO:BP | GO:0031126 | sno(s)RNA 3’-end processing | 7 | 12 | 2.985e-02 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 18 | 51 | 2.985e-02 |
| GO:BP | GO:0070243 | regulation of thymocyte apoptotic process | 8 | 15 | 2.985e-02 |
| GO:BP | GO:0070986 | left/right axis specification | 8 | 15 | 2.985e-02 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 18 | 51 | 2.985e-02 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 8 | 15 | 2.985e-02 |
| GO:BP | GO:0051081 | nuclear membrane disassembly | 7 | 12 | 2.985e-02 |
| GO:BP | GO:0006220 | pyrimidine nucleotide metabolic process | 19 | 55 | 2.986e-02 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 122 | 546 | 3.038e-02 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 29 | 97 | 3.065e-02 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 140 | 638 | 3.072e-02 |
| GO:BP | GO:0046039 | GTP metabolic process | 11 | 25 | 3.127e-02 |
| GO:BP | GO:0008361 | regulation of cell size | 48 | 183 | 3.136e-02 |
| GO:BP | GO:0035459 | vesicle cargo loading | 14 | 36 | 3.275e-02 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 14 | 36 | 3.275e-02 |
| GO:BP | GO:0006779 | porphyrin-containing compound biosynthetic process | 14 | 36 | 3.275e-02 |
| GO:BP | GO:0033014 | tetrapyrrole biosynthetic process | 14 | 36 | 3.275e-02 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 35 | 124 | 3.302e-02 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 61 | 245 | 3.303e-02 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 30 | 102 | 3.363e-02 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 148 | 681 | 3.397e-02 |
| GO:BP | GO:0023052 | signaling | 1210 | 6515 | 3.400e-02 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 147 | 676 | 3.415e-02 |
| GO:BP | GO:0030488 | tRNA methylation | 15 | 40 | 3.458e-02 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 144 | 661 | 3.494e-02 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 136 | 620 | 3.502e-02 |
| GO:BP | GO:0035239 | tube morphogenesis | 186 | 879 | 3.529e-02 |
| GO:BP | GO:0046785 | microtubule polymerization | 29 | 98 | 3.531e-02 |
| GO:BP | GO:0090114 | COPII-coated vesicle budding | 16 | 44 | 3.574e-02 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 17 | 48 | 3.582e-02 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 92 | 398 | 3.582e-02 |
| GO:BP | GO:0030182 | neuron differentiation | 294 | 1451 | 3.582e-02 |
| GO:BP | GO:0032211 | negative regulation of telomere maintenance via telomerase | 10 | 22 | 3.582e-02 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 10 | 22 | 3.582e-02 |
| GO:BP | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 12 | 29 | 3.582e-02 |
| GO:BP | GO:0140861 | DNA repair-dependent chromatin remodeling | 10 | 22 | 3.582e-02 |
| GO:BP | GO:0071763 | nuclear membrane organization | 17 | 48 | 3.582e-02 |
| GO:BP | GO:0098534 | centriole assembly | 17 | 48 | 3.582e-02 |
| GO:BP | GO:0060271 | cilium assembly | 93 | 403 | 3.582e-02 |
| GO:BP | GO:0006513 | protein monoubiquitination | 17 | 48 | 3.582e-02 |
| GO:BP | GO:0016032 | viral process | 97 | 423 | 3.582e-02 |
| GO:BP | GO:0141137 | positive regulation of gene expression, epigenetic | 19 | 56 | 3.583e-02 |
| GO:BP | GO:0030520 | estrogen receptor signaling pathway | 18 | 52 | 3.595e-02 |
| GO:BP | GO:0014020 | primary neural tube formation | 28 | 94 | 3.654e-02 |
| GO:BP | GO:0035295 | tube development | 228 | 1101 | 3.654e-02 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 55 | 218 | 3.753e-02 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 24 | 77 | 3.759e-02 |
| GO:BP | GO:0045088 | regulation of innate immune response | 102 | 449 | 3.785e-02 |
| GO:BP | GO:0097581 | lamellipodium organization | 27 | 90 | 3.846e-02 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 43 | 162 | 3.880e-02 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 124 | 561 | 3.894e-02 |
| GO:BP | GO:0009218 | pyrimidine ribonucleotide metabolic process | 13 | 33 | 3.894e-02 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 23 | 73 | 3.894e-02 |
| GO:BP | GO:0048869 | cellular developmental process | 838 | 4438 | 3.915e-02 |
| GO:BP | GO:0040031 | snRNA modification | 5 | 7 | 3.944e-02 |
| GO:BP | GO:0007221 | positive regulation of transcription of Notch receptor target | 5 | 7 | 3.944e-02 |
| GO:BP | GO:0046048 | UDP metabolic process | 5 | 7 | 3.944e-02 |
| GO:BP | GO:2000064 | regulation of cortisol biosynthetic process | 5 | 7 | 3.944e-02 |
| GO:BP | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process | 5 | 7 | 3.944e-02 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 29 | 99 | 3.944e-02 |
| GO:BP | GO:0051204 | protein insertion into mitochondrial membrane | 5 | 7 | 3.944e-02 |
| GO:BP | GO:0032459 | regulation of protein oligomerization | 5 | 7 | 3.944e-02 |
| GO:BP | GO:0019364 | pyridine nucleotide catabolic process | 29 | 99 | 3.944e-02 |
| GO:BP | GO:0002084 | protein depalmitoylation | 5 | 7 | 3.944e-02 |
| GO:BP | GO:0006370 | 7-methylguanosine mRNA capping | 5 | 7 | 3.944e-02 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 118 | 531 | 3.955e-02 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 44 | 167 | 3.955e-02 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 88 | 380 | 3.973e-02 |
| GO:BP | GO:0035282 | segmentation | 31 | 108 | 4.019e-02 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 51 | 200 | 4.019e-02 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 14 | 37 | 4.064e-02 |
| GO:BP | GO:0051656 | establishment of organelle localization | 110 | 491 | 4.090e-02 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 166 | 779 | 4.116e-02 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 11 | 26 | 4.137e-02 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 53 | 210 | 4.214e-02 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 15 | 41 | 4.214e-02 |
| GO:BP | GO:0141085 | regulation of inflammasome-mediated signaling pathway | 15 | 41 | 4.214e-02 |
| GO:BP | GO:0030154 | cell differentiation | 837 | 4437 | 4.217e-02 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 140 | 645 | 4.225e-02 |
| GO:BP | GO:0046031 | ADP metabolic process | 30 | 104 | 4.236e-02 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 19 | 57 | 4.241e-02 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 18 | 53 | 4.292e-02 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 32 | 113 | 4.292e-02 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 24 | 78 | 4.292e-02 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 17 | 49 | 4.300e-02 |
| GO:BP | GO:0016074 | sno(s)RNA metabolic process | 8 | 16 | 4.451e-02 |
| GO:BP | GO:0042795 | snRNA transcription by RNA polymerase II | 8 | 16 | 4.451e-02 |
| GO:BP | GO:0032940 | secretion by cell | 180 | 854 | 4.460e-02 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 37 | 136 | 4.499e-02 |
| GO:BP | GO:0035050 | embryonic heart tube development | 26 | 87 | 4.562e-02 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 12 | 30 | 4.593e-02 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 107 | 478 | 4.618e-02 |
| GO:BP | GO:1990144 | intrinsic apoptotic signaling pathway in response to hypoxia | 6 | 10 | 4.697e-02 |
| GO:BP | GO:0070244 | negative regulation of thymocyte apoptotic process | 6 | 10 | 4.697e-02 |
| GO:BP | GO:0071600 | otic vesicle morphogenesis | 6 | 10 | 4.697e-02 |
| GO:BP | GO:0071166 | ribonucleoprotein complex localization | 6 | 10 | 4.697e-02 |
| GO:BP | GO:0015937 | coenzyme A biosynthetic process | 6 | 10 | 4.697e-02 |
| GO:BP | GO:0016559 | peroxisome fission | 6 | 10 | 4.697e-02 |
| GO:BP | GO:0034551 | mitochondrial respiratory chain complex III assembly | 7 | 13 | 4.724e-02 |
| GO:BP | GO:0030397 | membrane disassembly | 7 | 13 | 4.724e-02 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 7 | 13 | 4.724e-02 |
| GO:BP | GO:0017062 | respiratory chain complex III assembly | 7 | 13 | 4.724e-02 |
| GO:BP | GO:0006446 | regulation of translational initiation | 25 | 83 | 4.745e-02 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 56 | 226 | 4.799e-02 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 34 | 123 | 4.799e-02 |
| GO:BP | GO:0006486 | protein glycosylation | 56 | 226 | 4.799e-02 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 10 | 23 | 4.819e-02 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 429 | 2192 | 4.843e-02 |
| GO:BP | GO:0045010 | actin nucleation | 20 | 62 | 4.914e-02 |
| GO:BP | GO:0006611 | protein export from nucleus | 20 | 62 | 4.914e-02 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 27 | 92 | 4.932e-02 |
| GO:BP | GO:0031667 | response to nutrient levels | 113 | 510 | 4.965e-02 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 161 | 506 | 1.414e-08 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 45 | 140 | 2.768e-02 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 52 | 168 | 2.768e-02 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 31 | 84 | 2.768e-02 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 37 | 108 | 2.768e-02 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 21 | 53 | 4.247e-02 |
| KEGG | KEGG:03018 | RNA degradation | 28 | 79 | 4.383e-02 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 96 | 363 | 4.737e-02 |
| KEGG | KEGG:04137 | Mitophagy - animal | 34 | 103 | 4.737e-02 |
| KEGG | KEGG:03082 | ATP-dependent chromatin remodeling | 37 | 116 | 4.974e-02 |
write.csv(table_DOX24_genes, "output/table_DOX24_genes.csv")
#GO:BP
table_DOX24_genes_GOBP <- table_DOX24_genes %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Specific DEGs Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))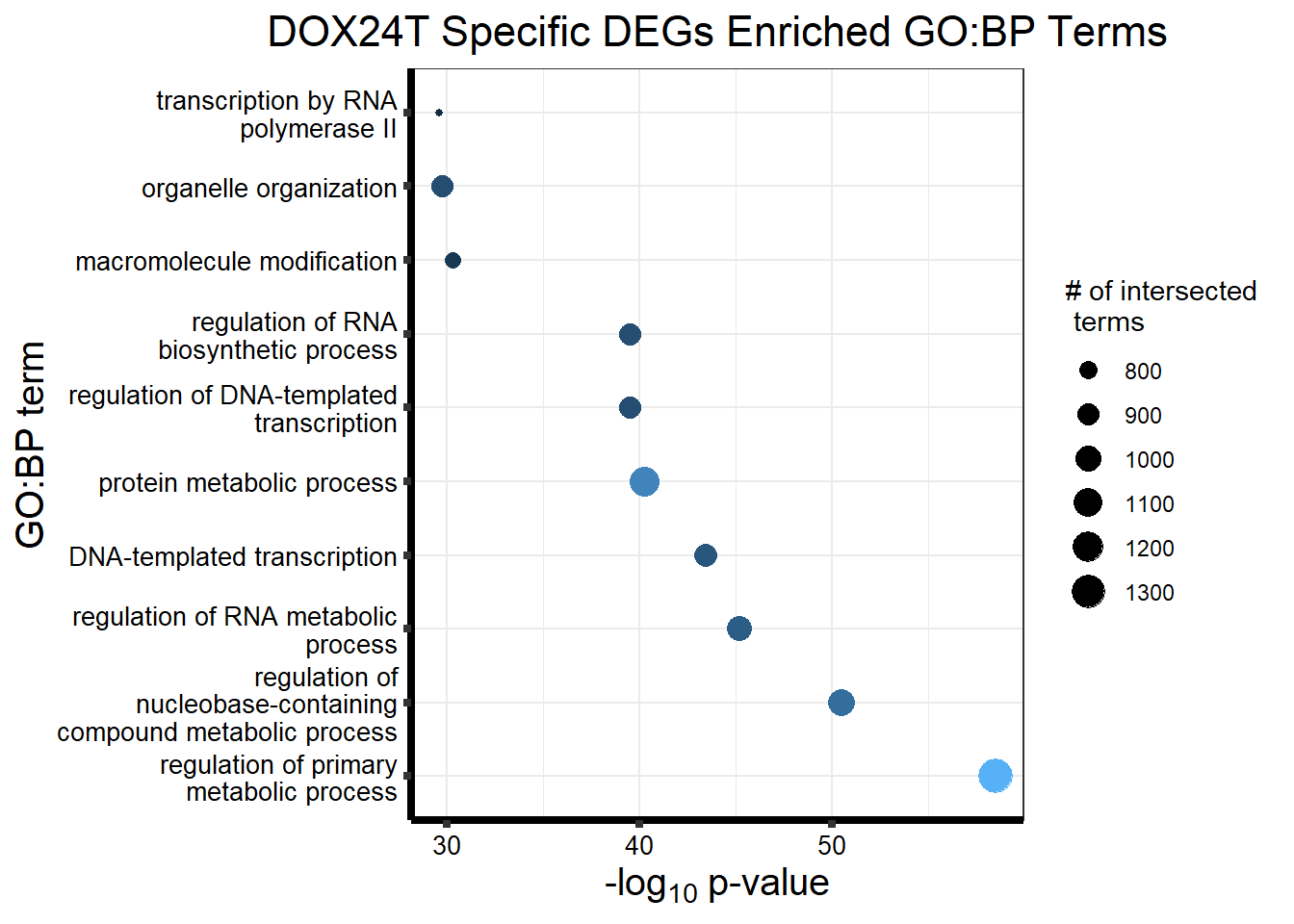
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_DOX24_genes_KEGG <- table_DOX24_genes %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Specific DEGs Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))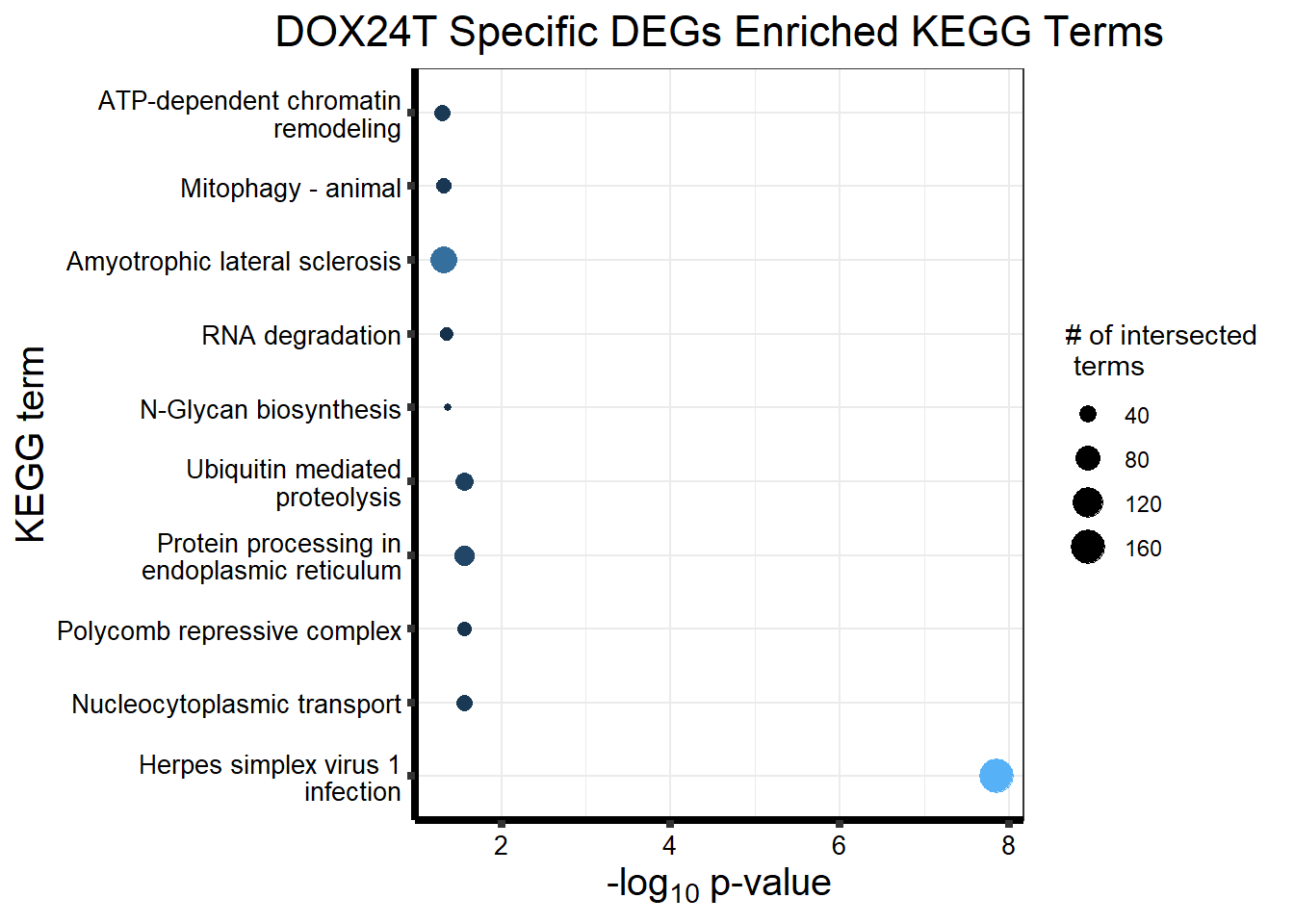
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####DOX24T shared DEGs GO KEGG#####
D24Tshare_DEGs_mat <- as.matrix(DOX24T_share_DEGs_GO_plot)
DOX_24Tshare_dxr_gene <- gost(query = D24Tshare_DEGs_mat,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
DOX_24Tshare_gost_genes <- gostplot(DOX_24Tshare_dxr_gene, capped = FALSE, interactive = TRUE)
DOX_24Tshare_gost_genestable_DOX24Tshare_genes <- DOX_24Tshare_dxr_gene$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX24Tshare_genes %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0080090 | regulation of primary metabolic process | 2739 | 5390 | 1.755e-121 |
| GO:BP | GO:0006996 | organelle organization | 1921 | 3594 | 2.051e-102 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 2049 | 3990 | 1.355e-88 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 2841 | 5920 | 2.002e-87 |
| GO:BP | GO:0019538 | protein metabolic process | 2351 | 4721 | 2.002e-87 |
| GO:BP | GO:0048518 | positive regulation of biological process | 2973 | 6264 | 7.043e-86 |
| GO:BP | GO:0065007 | biological regulation | 5395 | 12743 | 3.303e-81 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 1888 | 3687 | 5.607e-79 |
| GO:BP | GO:0050789 | regulation of biological process | 5240 | 12336 | 6.328e-79 |
| GO:BP | GO:0050794 | regulation of cellular process | 5082 | 11946 | 9.851e-75 |
| GO:BP | GO:0006351 | DNA-templated transcription | 1811 | 3549 | 1.456e-73 |
| GO:BP | GO:0043412 | macromolecule modification | 1586 | 3030 | 6.777e-73 |
| GO:BP | GO:0009056 | catabolic process | 1403 | 2639 | 7.203e-69 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 1729 | 3409 | 5.092e-67 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 1735 | 3428 | 1.890e-66 |
| GO:BP | GO:0051641 | cellular localization | 1829 | 3661 | 3.225e-65 |
| GO:BP | GO:0033554 | cellular response to stress | 1029 | 1830 | 1.246e-64 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 1294 | 2433 | 5.635e-63 |
| GO:BP | GO:0019222 | regulation of metabolic process | 3177 | 7035 | 7.511e-62 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1519 | 2965 | 3.462e-61 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1784 | 3597 | 2.344e-60 |
| GO:BP | GO:0036211 | protein modification process | 1462 | 2846 | 1.701e-59 |
| GO:BP | GO:0051179 | localization | 2578 | 5555 | 1.049e-58 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1091 | 2010 | 5.894e-58 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1636 | 3288 | 1.444e-55 |
| GO:BP | GO:0033036 | macromolecule localization | 1603 | 3228 | 1.599e-53 |
| GO:BP | GO:0046907 | intracellular transport | 790 | 1381 | 1.415e-52 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 2912 | 6492 | 8.338e-51 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 1369 | 2711 | 1.330e-49 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1397 | 2782 | 5.358e-49 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 1074 | 2036 | 7.468e-49 |
| GO:BP | GO:0008104 | protein localization | 1388 | 2770 | 5.618e-48 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 791 | 1417 | 6.371e-47 |
| GO:BP | GO:0033043 | regulation of organelle organization | 666 | 1148 | 1.055e-46 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 1300 | 2578 | 2.341e-46 |
| GO:BP | GO:0007049 | cell cycle | 897 | 1663 | 3.176e-45 |
| GO:BP | GO:0009894 | regulation of catabolic process | 610 | 1040 | 1.127e-44 |
| GO:BP | GO:0023051 | regulation of signaling | 1668 | 3478 | 7.507e-44 |
| GO:BP | GO:0006810 | transport | 2047 | 4407 | 1.218e-43 |
| GO:BP | GO:0010646 | regulation of cell communication | 1669 | 3486 | 2.465e-43 |
| GO:BP | GO:0000278 | mitotic cell cycle | 536 | 892 | 2.989e-43 |
| GO:BP | GO:0007275 | multicellular organism development | 2170 | 4727 | 2.067e-42 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 963 | 1835 | 4.114e-42 |
| GO:BP | GO:0009966 | regulation of signal transduction | 1475 | 3034 | 6.247e-42 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 1210 | 2407 | 6.374e-42 |
| GO:BP | GO:0006974 | DNA damage response | 540 | 908 | 9.179e-42 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 1210 | 2410 | 1.335e-41 |
| GO:BP | GO:0032502 | developmental process | 2879 | 6553 | 4.545e-40 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 455 | 745 | 5.526e-39 |
| GO:BP | GO:0051726 | regulation of cell cycle | 614 | 1087 | 5.990e-38 |
| GO:BP | GO:0048856 | anatomical structure development | 2649 | 5997 | 9.026e-38 |
| GO:BP | GO:0015031 | protein transport | 775 | 1444 | 1.212e-37 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 887 | 1699 | 2.203e-37 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 886 | 1697 | 2.347e-37 |
| GO:BP | GO:0051234 | establishment of localization | 2223 | 4928 | 4.152e-37 |
| GO:BP | GO:0071705 | nitrogen compound transport | 983 | 1923 | 6.126e-37 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1846 | 3993 | 7.522e-37 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 953 | 1855 | 8.137e-37 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 377 | 597 | 1.222e-36 |
| GO:BP | GO:0006914 | autophagy | 377 | 597 | 1.222e-36 |
| GO:BP | GO:0022402 | cell cycle process | 696 | 1280 | 5.677e-36 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 2494 | 5629 | 5.755e-36 |
| GO:BP | GO:0051716 | cellular response to stimulus | 3171 | 7376 | 1.258e-35 |
| GO:BP | GO:0006259 | DNA metabolic process | 569 | 1005 | 1.776e-35 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 1257 | 2593 | 3.510e-34 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 800 | 1529 | 6.884e-34 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 1298 | 2696 | 1.027e-33 |
| GO:BP | GO:0048731 | system development | 1854 | 4053 | 1.078e-33 |
| GO:BP | GO:0048519 | negative regulation of biological process | 2560 | 5834 | 1.769e-33 |
| GO:BP | GO:0070925 | organelle assembly | 582 | 1046 | 1.962e-33 |
| GO:BP | GO:0044238 | primary metabolic process | 5009 | 12342 | 6.407e-33 |
| GO:BP | GO:0030163 | protein catabolic process | 573 | 1030 | 7.242e-33 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 391 | 643 | 8.187e-33 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 979 | 1953 | 1.470e-32 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 2544 | 5809 | 1.853e-32 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 386 | 638 | 1.019e-31 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 455 | 786 | 4.093e-31 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 2471 | 5644 | 4.538e-31 |
| GO:BP | GO:0043687 | post-translational protein modification | 565 | 1027 | 1.187e-30 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 731 | 1400 | 1.994e-30 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 378 | 628 | 2.141e-30 |
| GO:BP | GO:0006281 | DNA repair | 372 | 621 | 2.689e-29 |
| GO:BP | GO:0016236 | macroautophagy | 242 | 362 | 1.151e-28 |
| GO:BP | GO:0006325 | chromatin organization | 493 | 884 | 2.029e-28 |
| GO:BP | GO:0006950 | response to stress | 1783 | 3948 | 2.030e-28 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 533 | 975 | 6.447e-28 |
| GO:BP | GO:0010468 | regulation of gene expression | 2405 | 5536 | 2.817e-27 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 475 | 853 | 3.822e-27 |
| GO:BP | GO:0007399 | nervous system development | 1229 | 2604 | 3.822e-27 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 329 | 546 | 1.868e-26 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 411 | 720 | 2.853e-26 |
| GO:BP | GO:0008219 | cell death | 968 | 1991 | 4.529e-26 |
| GO:BP | GO:0012501 | programmed cell death | 966 | 1987 | 5.279e-26 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 132 | 168 | 5.667e-26 |
| GO:BP | GO:0030030 | cell projection organization | 814 | 1631 | 7.268e-26 |
| GO:BP | GO:0032879 | regulation of localization | 982 | 2029 | 1.237e-25 |
| GO:BP | GO:0006915 | apoptotic process | 937 | 1923 | 1.434e-25 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 660 | 1280 | 2.341e-25 |
| GO:BP | GO:0050793 | regulation of developmental process | 1159 | 2457 | 2.658e-25 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 930 | 1912 | 4.802e-25 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 665 | 1296 | 7.932e-25 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1262 | 2713 | 8.379e-25 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 323 | 543 | 1.273e-24 |
| GO:BP | GO:0045184 | establishment of protein localization | 938 | 1937 | 1.732e-24 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 3483 | 8393 | 1.767e-24 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 791 | 1592 | 2.298e-24 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 789 | 1588 | 2.676e-24 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 297 | 492 | 4.876e-24 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 536 | 1011 | 6.641e-24 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 258 | 414 | 1.178e-23 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 673 | 1326 | 1.580e-23 |
| GO:BP | GO:0051276 | chromosome organization | 332 | 574 | 2.071e-22 |
| GO:BP | GO:0016567 | protein ubiquitination | 421 | 768 | 3.624e-22 |
| GO:BP | GO:0009987 | cellular process | 7650 | 20247 | 3.878e-22 |
| GO:BP | GO:0050896 | response to stimulus | 3692 | 8999 | 5.790e-22 |
| GO:BP | GO:0060341 | regulation of cellular localization | 530 | 1013 | 6.326e-22 |
| GO:BP | GO:0006412 | translation | 401 | 727 | 9.396e-22 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 713 | 1436 | 1.072e-21 |
| GO:BP | GO:0023057 | negative regulation of signaling | 712 | 1435 | 1.459e-21 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 260 | 428 | 1.543e-21 |
| GO:BP | GO:0051640 | organelle localization | 351 | 620 | 1.602e-21 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 202 | 312 | 2.618e-21 |
| GO:BP | GO:0032880 | regulation of protein localization | 480 | 907 | 4.260e-21 |
| GO:BP | GO:0065009 | regulation of molecular function | 736 | 1496 | 4.303e-21 |
| GO:BP | GO:0048468 | cell development | 1308 | 2876 | 4.348e-21 |
| GO:BP | GO:0007010 | cytoskeleton organization | 750 | 1529 | 4.348e-21 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 555 | 1077 | 4.422e-21 |
| GO:BP | GO:0044770 | cell cycle phase transition | 308 | 532 | 7.178e-21 |
| GO:BP | GO:0006338 | chromatin remodeling | 397 | 725 | 8.823e-21 |
| GO:BP | GO:0032543 | mitochondrial translation | 103 | 130 | 9.395e-21 |
| GO:BP | GO:0048869 | cellular developmental process | 1928 | 4438 | 1.751e-20 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 288 | 492 | 1.819e-20 |
| GO:BP | GO:0030154 | cell differentiation | 1927 | 4437 | 2.116e-20 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 630 | 1256 | 2.252e-20 |
| GO:BP | GO:0016310 | phosphorylation | 657 | 1320 | 3.034e-20 |
| GO:BP | GO:0022008 | neurogenesis | 848 | 1771 | 3.387e-20 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 300 | 519 | 3.435e-20 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 384 | 701 | 3.902e-20 |
| GO:BP | GO:0006886 | intracellular protein transport | 377 | 686 | 4.383e-20 |
| GO:BP | GO:0023052 | signaling | 2731 | 6515 | 6.831e-20 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 365 | 662 | 9.512e-20 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 562 | 1105 | 9.656e-20 |
| GO:BP | GO:0043009 | chordate embryonic development | 369 | 671 | 1.020e-19 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 807 | 1680 | 1.258e-19 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 378 | 692 | 1.537e-19 |
| GO:BP | GO:0044281 | small molecule metabolic process | 846 | 1776 | 2.271e-19 |
| GO:BP | GO:0140014 | mitotic nuclear division | 182 | 282 | 5.437e-19 |
| GO:BP | GO:0007005 | mitochondrion organization | 257 | 435 | 5.437e-19 |
| GO:BP | GO:0010506 | regulation of autophagy | 224 | 367 | 6.527e-19 |
| GO:BP | GO:0007154 | cell communication | 2732 | 6540 | 7.798e-19 |
| GO:BP | GO:0042254 | ribosome biogenesis | 202 | 323 | 9.484e-19 |
| GO:BP | GO:0009790 | embryo development | 571 | 1135 | 9.485e-19 |
| GO:BP | GO:0051301 | cell division | 360 | 658 | 9.588e-19 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 287 | 500 | 1.005e-18 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 485 | 938 | 1.005e-18 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 303 | 535 | 1.330e-18 |
| GO:BP | GO:0016072 | rRNA metabolic process | 173 | 266 | 1.422e-18 |
| GO:BP | GO:0032259 | methylation | 157 | 236 | 2.985e-18 |
| GO:BP | GO:0016043 | cellular component organization | 3353 | 8184 | 3.034e-18 |
| GO:BP | GO:0065008 | regulation of biological quality | 1319 | 2947 | 3.315e-18 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 202 | 327 | 6.990e-18 |
| GO:BP | GO:0006302 | double-strand break repair | 198 | 319 | 7.401e-18 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 250 | 427 | 8.915e-18 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 159 | 242 | 1.074e-17 |
| GO:BP | GO:0031399 | regulation of protein modification process | 529 | 1049 | 1.369e-17 |
| GO:BP | GO:0016197 | endosomal transport | 183 | 290 | 1.399e-17 |
| GO:BP | GO:0061024 | membrane organization | 429 | 821 | 1.526e-17 |
| GO:BP | GO:0006399 | tRNA metabolic process | 142 | 210 | 1.828e-17 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 499 | 983 | 2.929e-17 |
| GO:BP | GO:0048699 | generation of neurons | 734 | 1536 | 4.627e-17 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 212 | 352 | 5.807e-17 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 522 | 1039 | 6.083e-17 |
| GO:BP | GO:0006468 | protein phosphorylation | 602 | 1227 | 8.911e-17 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 128 | 187 | 2.163e-16 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 699 | 1462 | 2.945e-16 |
| GO:BP | GO:0000819 | sister chromatid segregation | 152 | 234 | 3.118e-16 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 357 | 670 | 3.416e-16 |
| GO:BP | GO:0001701 | in utero embryonic development | 239 | 413 | 3.859e-16 |
| GO:BP | GO:0043414 | macromolecule methylation | 95 | 127 | 3.859e-16 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 218 | 369 | 4.244e-16 |
| GO:BP | GO:0051656 | establishment of organelle localization | 275 | 491 | 5.285e-16 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 716 | 1506 | 5.609e-16 |
| GO:BP | GO:0006403 | RNA localization | 134 | 200 | 5.740e-16 |
| GO:BP | GO:0006402 | mRNA catabolic process | 165 | 261 | 5.844e-16 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 596 | 1223 | 7.720e-16 |
| GO:BP | GO:0007017 | microtubule-based process | 500 | 999 | 8.098e-16 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 129 | 193 | 3.097e-15 |
| GO:BP | GO:0072359 | circulatory system development | 560 | 1145 | 3.216e-15 |
| GO:BP | GO:0007059 | chromosome segregation | 243 | 427 | 3.216e-15 |
| GO:BP | GO:0030182 | neuron differentiation | 689 | 1451 | 3.417e-15 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 244 | 430 | 4.415e-15 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 145 | 225 | 4.696e-15 |
| GO:BP | GO:0006364 | rRNA processing | 145 | 225 | 4.696e-15 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 219 | 377 | 4.847e-15 |
| GO:BP | GO:0048285 | organelle fission | 276 | 500 | 5.622e-15 |
| GO:BP | GO:0007165 | signal transduction | 2491 | 6002 | 7.400e-15 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 256 | 458 | 9.460e-15 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 739 | 1578 | 1.229e-14 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 701 | 1489 | 1.893e-14 |
| GO:BP | GO:0080134 | regulation of response to stress | 658 | 1387 | 2.272e-14 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 463 | 928 | 2.597e-14 |
| GO:BP | GO:0006508 | proteolysis | 699 | 1486 | 2.605e-14 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 123 | 185 | 2.844e-14 |
| GO:BP | GO:0000902 | cell morphogenesis | 492 | 996 | 2.906e-14 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 276 | 505 | 2.958e-14 |
| GO:BP | GO:0006417 | regulation of translation | 218 | 380 | 3.642e-14 |
| GO:BP | GO:0031175 | neuron projection development | 503 | 1023 | 3.848e-14 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 443 | 884 | 4.385e-14 |
| GO:BP | GO:0051169 | nuclear transport | 196 | 334 | 4.507e-14 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 196 | 334 | 4.507e-14 |
| GO:BP | GO:0007507 | heart development | 320 | 605 | 6.128e-14 |
| GO:BP | GO:0048666 | neuron development | 567 | 1177 | 7.747e-14 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 190 | 323 | 8.822e-14 |
| GO:BP | GO:0051049 | regulation of transport | 746 | 1607 | 9.732e-14 |
| GO:BP | GO:0033365 | protein localization to organelle | 580 | 1209 | 9.732e-14 |
| GO:BP | GO:0006401 | RNA catabolic process | 186 | 315 | 1.027e-13 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 636 | 1343 | 1.084e-13 |
| GO:BP | GO:0007033 | vacuole organization | 149 | 239 | 1.108e-13 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 552 | 1144 | 1.253e-13 |
| GO:BP | GO:0022411 | cellular component disassembly | 247 | 446 | 1.254e-13 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 96 | 136 | 1.254e-13 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 418 | 831 | 1.311e-13 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 173 | 289 | 1.618e-13 |
| GO:BP | GO:0000045 | autophagosome assembly | 86 | 118 | 1.638e-13 |
| GO:BP | GO:0000280 | nuclear division | 249 | 452 | 2.180e-13 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 336 | 646 | 2.422e-13 |
| GO:BP | GO:0008033 | tRNA processing | 97 | 139 | 2.849e-13 |
| GO:BP | GO:0141188 | nucleic acid catabolic process | 198 | 343 | 3.032e-13 |
| GO:BP | GO:0010256 | endomembrane system organization | 321 | 613 | 3.046e-13 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1038 | 2331 | 3.180e-13 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 730 | 1576 | 3.444e-13 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 347 | 673 | 4.159e-13 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 475 | 970 | 5.562e-13 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 277 | 517 | 6.198e-13 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 348 | 677 | 6.533e-13 |
| GO:BP | GO:0023056 | positive regulation of signaling | 818 | 1796 | 8.763e-13 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 339 | 658 | 9.641e-13 |
| GO:BP | GO:0000723 | telomere maintenance | 109 | 164 | 1.165e-12 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 193 | 336 | 1.242e-12 |
| GO:BP | GO:0008152 | metabolic process | 5499 | 14136 | 1.249e-12 |
| GO:BP | GO:0030029 | actin filament-based process | 411 | 825 | 1.409e-12 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 321 | 619 | 1.565e-12 |
| GO:BP | GO:0000209 | protein polyubiquitination | 166 | 280 | 1.809e-12 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 350 | 686 | 2.027e-12 |
| GO:BP | GO:1905037 | autophagosome organization | 88 | 125 | 2.120e-12 |
| GO:BP | GO:0035239 | tube morphogenesis | 433 | 879 | 2.636e-12 |
| GO:BP | GO:0006260 | DNA replication | 167 | 283 | 2.653e-12 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1281 | 2956 | 2.854e-12 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 814 | 1795 | 3.273e-12 |
| GO:BP | GO:0061061 | muscle structure development | 352 | 693 | 3.688e-12 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 178 | 307 | 3.732e-12 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 311 | 600 | 4.121e-12 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 266 | 499 | 4.250e-12 |
| GO:BP | GO:0006839 | mitochondrial transport | 115 | 178 | 4.308e-12 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 118 | 184 | 4.448e-12 |
| GO:BP | GO:0032501 | multicellular organismal process | 2967 | 7322 | 4.478e-12 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 203 | 361 | 4.552e-12 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 329 | 642 | 5.400e-12 |
| GO:BP | GO:0030031 | cell projection assembly | 316 | 613 | 6.303e-12 |
| GO:BP | GO:0051236 | establishment of RNA localization | 107 | 163 | 6.422e-12 |
| GO:BP | GO:0070848 | response to growth factor | 363 | 721 | 6.979e-12 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 112 | 173 | 7.127e-12 |
| GO:BP | GO:0035295 | tube development | 525 | 1101 | 7.169e-12 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 349 | 689 | 7.256e-12 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 149 | 248 | 7.481e-12 |
| GO:BP | GO:0048513 | animal organ development | 1328 | 3085 | 9.127e-12 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 262 | 493 | 9.802e-12 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 169 | 291 | 1.221e-11 |
| GO:BP | GO:0009451 | RNA modification | 112 | 174 | 1.238e-11 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 124 | 198 | 1.254e-11 |
| GO:BP | GO:0060537 | muscle tissue development | 233 | 430 | 1.468e-11 |
| GO:BP | GO:0014706 | striated muscle tissue development | 223 | 408 | 1.468e-11 |
| GO:BP | GO:0006282 | regulation of DNA repair | 137 | 225 | 1.642e-11 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 203 | 365 | 1.827e-11 |
| GO:BP | GO:0044782 | cilium organization | 233 | 431 | 2.008e-11 |
| GO:BP | GO:0006400 | tRNA modification | 70 | 95 | 2.060e-11 |
| GO:BP | GO:0051168 | nuclear export | 109 | 169 | 2.060e-11 |
| GO:BP | GO:0009267 | cellular response to starvation | 113 | 177 | 2.129e-11 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 141 | 234 | 2.308e-11 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 276 | 528 | 2.696e-11 |
| GO:BP | GO:1901698 | response to nitrogen compound | 513 | 1080 | 2.903e-11 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 87 | 127 | 3.057e-11 |
| GO:BP | GO:0050658 | RNA transport | 104 | 160 | 3.309e-11 |
| GO:BP | GO:0050657 | nucleic acid transport | 104 | 160 | 3.309e-11 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 56 | 71 | 3.486e-11 |
| GO:BP | GO:0031647 | regulation of protein stability | 188 | 335 | 4.292e-11 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 148 | 250 | 4.340e-11 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 283 | 546 | 4.839e-11 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 141 | 236 | 5.583e-11 |
| GO:BP | GO:0007178 | cell surface receptor protein serine/threonine kinase signaling pathway | 201 | 364 | 5.619e-11 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 122 | 197 | 5.619e-11 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 90 | 134 | 6.939e-11 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 108 | 170 | 1.044e-10 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 109 | 172 | 1.044e-10 |
| GO:BP | GO:0042592 | homeostatic process | 780 | 1736 | 1.085e-10 |
| GO:BP | GO:0042594 | response to starvation | 132 | 219 | 1.158e-10 |
| GO:BP | GO:0034330 | cell junction organization | 406 | 834 | 1.187e-10 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 176 | 312 | 1.193e-10 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 127 | 209 | 1.316e-10 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 127 | 209 | 1.316e-10 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 150 | 257 | 1.380e-10 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 139 | 234 | 1.410e-10 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 340 | 681 | 1.431e-10 |
| GO:BP | GO:0032200 | telomere organization | 118 | 191 | 1.578e-10 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 175 | 311 | 1.792e-10 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 1431 | 3375 | 1.852e-10 |
| GO:BP | GO:0009888 | tissue development | 898 | 2032 | 1.854e-10 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 457 | 957 | 1.919e-10 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 360 | 729 | 2.018e-10 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 140 | 237 | 2.052e-10 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 337 | 676 | 2.210e-10 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 89 | 134 | 2.245e-10 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 456 | 956 | 2.471e-10 |
| GO:BP | GO:0010970 | transport along microtubule | 111 | 178 | 2.806e-10 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 261 | 503 | 2.887e-10 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 1210 | 2819 | 3.111e-10 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 149 | 257 | 3.149e-10 |
| GO:BP | GO:0016050 | vesicle organization | 204 | 376 | 3.189e-10 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 402 | 830 | 3.455e-10 |
| GO:BP | GO:0060271 | cilium assembly | 216 | 403 | 3.676e-10 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 228 | 430 | 3.996e-10 |
| GO:BP | GO:0043487 | regulation of RNA stability | 118 | 193 | 4.016e-10 |
| GO:BP | GO:0048729 | tissue morphogenesis | 309 | 614 | 4.043e-10 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 91 | 139 | 4.275e-10 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 112 | 181 | 4.468e-10 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 106 | 169 | 4.758e-10 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 300 | 594 | 4.758e-10 |
| GO:BP | GO:0040007 | growth | 443 | 929 | 5.025e-10 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 305 | 606 | 5.342e-10 |
| GO:BP | GO:0006310 | DNA recombination | 190 | 347 | 5.510e-10 |
| GO:BP | GO:0038202 | TORC1 signaling | 72 | 103 | 5.513e-10 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 286 | 563 | 6.207e-10 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 40 | 47 | 6.249e-10 |
| GO:BP | GO:0051028 | mRNA transport | 86 | 130 | 6.582e-10 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 645 | 1418 | 6.703e-10 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 206 | 383 | 6.922e-10 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 49 | 62 | 7.070e-10 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 103 | 164 | 7.976e-10 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 99 | 156 | 8.036e-10 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 260 | 505 | 8.556e-10 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 357 | 729 | 8.693e-10 |
| GO:BP | GO:0000725 | recombinational repair | 117 | 193 | 1.015e-09 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 327 | 660 | 1.070e-09 |
| GO:BP | GO:0042692 | muscle cell differentiation | 215 | 405 | 1.251e-09 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 76 | 112 | 1.456e-09 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 273 | 537 | 1.555e-09 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 187 | 344 | 1.730e-09 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 114 | 188 | 1.742e-09 |
| GO:BP | GO:0031503 | protein-containing complex localization | 127 | 215 | 1.758e-09 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 181 | 331 | 1.820e-09 |
| GO:BP | GO:0006629 | lipid metabolic process | 631 | 1391 | 1.931e-09 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 85 | 130 | 2.063e-09 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 106 | 172 | 2.063e-09 |
| GO:BP | GO:0016073 | snRNA metabolic process | 46 | 58 | 2.252e-09 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 208 | 392 | 2.670e-09 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 953 | 2192 | 2.831e-09 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 124 | 210 | 2.998e-09 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 279 | 554 | 3.372e-09 |
| GO:BP | GO:0031929 | TOR signaling | 103 | 167 | 3.533e-09 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 318 | 645 | 3.930e-09 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 231 | 446 | 5.066e-09 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 77 | 116 | 5.363e-09 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 78 | 118 | 5.545e-09 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 67 | 97 | 5.877e-09 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 101 | 164 | 5.943e-09 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 99 | 160 | 6.135e-09 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 141 | 248 | 6.616e-09 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 169 | 309 | 7.354e-09 |
| GO:BP | GO:0051604 | protein maturation | 271 | 539 | 7.549e-09 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 151 | 270 | 7.844e-09 |
| GO:BP | GO:0040008 | regulation of growth | 299 | 604 | 7.894e-09 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 187 | 349 | 8.006e-09 |
| GO:BP | GO:0003007 | heart morphogenesis | 149 | 266 | 8.754e-09 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 548 | 1199 | 9.123e-09 |
| GO:BP | GO:0001510 | RNA methylation | 58 | 81 | 1.026e-08 |
| GO:BP | GO:0010212 | response to ionizing radiation | 89 | 141 | 1.110e-08 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 31 | 35 | 1.359e-08 |
| GO:BP | GO:0051338 | regulation of transferase activity | 252 | 498 | 1.428e-08 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 200 | 380 | 1.436e-08 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 208 | 398 | 1.442e-08 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 414 | 879 | 1.592e-08 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 372 | 779 | 1.600e-08 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 75 | 114 | 1.642e-08 |
| GO:BP | GO:0016049 | cell growth | 246 | 485 | 1.700e-08 |
| GO:BP | GO:0008283 | cell population proliferation | 876 | 2015 | 1.754e-08 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 148 | 266 | 1.849e-08 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 196 | 372 | 1.861e-08 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 84 | 132 | 1.861e-08 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 295 | 599 | 1.994e-08 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 181 | 339 | 2.136e-08 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 181 | 339 | 2.136e-08 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 70 | 105 | 2.505e-08 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 70 | 105 | 2.505e-08 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 98 | 161 | 2.560e-08 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 96 | 157 | 2.679e-08 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 405 | 861 | 2.910e-08 |
| GO:BP | GO:0099111 | microtubule-based transport | 126 | 220 | 2.957e-08 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 79 | 123 | 3.072e-08 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 79 | 123 | 3.072e-08 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 65 | 96 | 3.698e-08 |
| GO:BP | GO:0007051 | spindle organization | 119 | 206 | 4.168e-08 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 188 | 357 | 4.233e-08 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 58 | 83 | 4.408e-08 |
| GO:BP | GO:0031667 | response to nutrient levels | 255 | 510 | 4.633e-08 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 89 | 144 | 4.932e-08 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 73 | 112 | 4.932e-08 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 158 | 291 | 5.134e-08 |
| GO:BP | GO:0007052 | mitotic spindle organization | 84 | 134 | 5.210e-08 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 61 | 89 | 5.545e-08 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 1322 | 3157 | 5.553e-08 |
| GO:BP | GO:0034504 | protein localization to nucleus | 170 | 318 | 5.887e-08 |
| GO:BP | GO:0007417 | central nervous system development | 486 | 1061 | 6.187e-08 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 137 | 246 | 6.862e-08 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 136 | 244 | 7.269e-08 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 147 | 268 | 7.367e-08 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 141 | 255 | 7.598e-08 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 45 | 60 | 8.069e-08 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 197 | 380 | 8.845e-08 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 196 | 378 | 9.483e-08 |
| GO:BP | GO:0051648 | vesicle localization | 124 | 219 | 9.931e-08 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 60 | 88 | 1.003e-07 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 115 | 200 | 1.081e-07 |
| GO:BP | GO:0030162 | regulation of proteolysis | 208 | 406 | 1.151e-07 |
| GO:BP | GO:0001558 | regulation of cell growth | 208 | 406 | 1.151e-07 |
| GO:BP | GO:0007032 | endosome organization | 65 | 98 | 1.274e-07 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 85 | 138 | 1.375e-07 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 82 | 132 | 1.441e-07 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 141 | 257 | 1.471e-07 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 75 | 118 | 1.498e-07 |
| GO:BP | GO:0006275 | regulation of DNA replication | 76 | 120 | 1.498e-07 |
| GO:BP | GO:0032940 | secretion by cell | 398 | 854 | 1.517e-07 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 730 | 1669 | 1.519e-07 |
| GO:BP | GO:0009303 | rRNA transcription | 33 | 40 | 1.519e-07 |
| GO:BP | GO:0098727 | maintenance of cell number | 107 | 184 | 1.552e-07 |
| GO:BP | GO:0016358 | dendrite development | 134 | 242 | 1.588e-07 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 168 | 317 | 1.588e-07 |
| GO:BP | GO:0019827 | stem cell population maintenance | 105 | 180 | 1.689e-07 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 50 | 70 | 1.692e-07 |
| GO:BP | GO:0007034 | vacuolar transport | 102 | 174 | 1.921e-07 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 62 | 93 | 2.063e-07 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 52 | 74 | 2.096e-07 |
| GO:BP | GO:0016482 | cytosolic transport | 86 | 141 | 2.121e-07 |
| GO:BP | GO:0060322 | head development | 377 | 806 | 2.251e-07 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 115 | 202 | 2.251e-07 |
| GO:BP | GO:0051960 | regulation of nervous system development | 229 | 457 | 2.251e-07 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 67 | 103 | 2.360e-07 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 68 | 105 | 2.397e-07 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 69 | 107 | 2.425e-07 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 70 | 109 | 2.446e-07 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 132 | 239 | 2.467e-07 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 73 | 115 | 2.470e-07 |
| GO:BP | GO:0021915 | neural tube development | 95 | 160 | 2.480e-07 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 165 | 312 | 2.541e-07 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 38 | 49 | 2.615e-07 |
| GO:BP | GO:0050821 | protein stabilization | 125 | 224 | 2.625e-07 |
| GO:BP | GO:0140352 | export from cell | 427 | 928 | 2.815e-07 |
| GO:BP | GO:0099173 | postsynapse organization | 135 | 246 | 2.837e-07 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 144 | 266 | 3.060e-07 |
| GO:BP | GO:0006887 | exocytosis | 186 | 360 | 3.081e-07 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 88 | 146 | 3.102e-07 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 182 | 351 | 3.114e-07 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 273 | 561 | 3.119e-07 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 166 | 315 | 3.119e-07 |
| GO:BP | GO:0006986 | response to unfolded protein | 85 | 140 | 3.354e-07 |
| GO:BP | GO:0072175 | epithelial tube formation | 83 | 136 | 3.522e-07 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 63 | 96 | 3.756e-07 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 66 | 102 | 3.987e-07 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 67 | 104 | 4.031e-07 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 129 | 234 | 4.039e-07 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 36 | 46 | 4.055e-07 |
| GO:BP | GO:0003279 | cardiac septum development | 69 | 108 | 4.072e-07 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 530 | 1182 | 4.084e-07 |
| GO:BP | GO:0048863 | stem cell differentiation | 134 | 245 | 4.099e-07 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 92 | 155 | 4.132e-07 |
| GO:BP | GO:0007030 | Golgi organization | 91 | 153 | 4.282e-07 |
| GO:BP | GO:0009314 | response to radiation | 219 | 437 | 4.394e-07 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 90 | 151 | 4.423e-07 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 212 | 421 | 4.506e-07 |
| GO:BP | GO:0050808 | synapse organization | 273 | 563 | 4.591e-07 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 137 | 252 | 4.634e-07 |
| GO:BP | GO:0006997 | nucleus organization | 88 | 147 | 4.706e-07 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 41 | 55 | 4.841e-07 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 297 | 620 | 4.940e-07 |
| GO:BP | GO:0006479 | protein methylation | 37 | 48 | 5.148e-07 |
| GO:BP | GO:0008213 | protein alkylation | 37 | 48 | 5.148e-07 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 501 | 1113 | 5.402e-07 |
| GO:BP | GO:0060606 | tube closure | 59 | 89 | 5.889e-07 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 79 | 129 | 5.990e-07 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 116 | 207 | 5.990e-07 |
| GO:BP | GO:0006405 | RNA export from nucleus | 60 | 91 | 6.050e-07 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 49 | 70 | 6.237e-07 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 76 | 123 | 6.319e-07 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 63 | 97 | 6.461e-07 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 73 | 117 | 6.569e-07 |
| GO:BP | GO:0060996 | dendritic spine development | 65 | 101 | 6.619e-07 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 68 | 107 | 6.719e-07 |
| GO:BP | GO:0003205 | cardiac chamber development | 98 | 169 | 7.139e-07 |
| GO:BP | GO:0035148 | tube formation | 88 | 148 | 7.139e-07 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 303 | 637 | 8.193e-07 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 82 | 136 | 8.728e-07 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 126 | 230 | 9.065e-07 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 191 | 376 | 9.474e-07 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 381 | 825 | 9.834e-07 |
| GO:BP | GO:0001843 | neural tube closure | 58 | 88 | 1.026e-06 |
| GO:BP | GO:0014020 | primary neural tube formation | 61 | 94 | 1.095e-06 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 61 | 94 | 1.095e-06 |
| GO:BP | GO:1990928 | response to amino acid starvation | 41 | 56 | 1.100e-06 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 63 | 98 | 1.120e-06 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 64 | 100 | 1.127e-06 |
| GO:BP | GO:0001568 | blood vessel development | 342 | 732 | 1.129e-06 |
| GO:BP | GO:0051310 | metaphase chromosome alignment | 66 | 104 | 1.129e-06 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 166 | 320 | 1.169e-06 |
| GO:BP | GO:0048589 | developmental growth | 313 | 663 | 1.203e-06 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 85 | 143 | 1.209e-06 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 49 | 71 | 1.227e-06 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 94 | 162 | 1.257e-06 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 164 | 316 | 1.336e-06 |
| GO:BP | GO:0034329 | cell junction assembly | 245 | 503 | 1.412e-06 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 54 | 81 | 1.621e-06 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 116 | 210 | 1.641e-06 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 75 | 123 | 1.641e-06 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 74 | 121 | 1.675e-06 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 121 | 221 | 1.678e-06 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 115 | 208 | 1.736e-06 |
| GO:BP | GO:0007420 | brain development | 350 | 754 | 1.768e-06 |
| GO:BP | GO:0006457 | protein folding | 120 | 219 | 1.779e-06 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 70 | 113 | 1.796e-06 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 39 | 53 | 1.798e-06 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 85 | 144 | 1.827e-06 |
| GO:BP | GO:0001841 | neural tube formation | 64 | 101 | 1.881e-06 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 63 | 99 | 1.881e-06 |
| GO:BP | GO:0001944 | vasculature development | 353 | 762 | 1.987e-06 |
| GO:BP | GO:0030010 | establishment of cell polarity | 92 | 159 | 2.019e-06 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 47 | 68 | 2.062e-06 |
| GO:BP | GO:0060485 | mesenchyme development | 165 | 320 | 2.075e-06 |
| GO:BP | GO:0060284 | regulation of cell development | 388 | 847 | 2.089e-06 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 132 | 246 | 2.094e-06 |
| GO:BP | GO:0009411 | response to UV | 90 | 155 | 2.202e-06 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 301 | 638 | 2.230e-06 |
| GO:BP | GO:0050807 | regulation of synapse organization | 160 | 309 | 2.248e-06 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 104 | 185 | 2.273e-06 |
| GO:BP | GO:0043434 | response to peptide hormone | 217 | 440 | 2.275e-06 |
| GO:BP | GO:0003015 | heart process | 135 | 253 | 2.301e-06 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 139 | 262 | 2.361e-06 |
| GO:BP | GO:0007169 | cell surface receptor protein tyrosine kinase signaling pathway | 297 | 629 | 2.436e-06 |
| GO:BP | GO:0050000 | chromosome localization | 76 | 126 | 2.447e-06 |
| GO:BP | GO:0044282 | small molecule catabolic process | 187 | 371 | 2.469e-06 |
| GO:BP | GO:0060173 | limb development | 108 | 194 | 2.544e-06 |
| GO:BP | GO:0048736 | appendage development | 108 | 194 | 2.544e-06 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 142 | 269 | 2.552e-06 |
| GO:BP | GO:0006082 | organic acid metabolic process | 415 | 915 | 2.904e-06 |
| GO:BP | GO:0061157 | mRNA destabilization | 64 | 102 | 3.099e-06 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 81 | 137 | 3.181e-06 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 44 | 63 | 3.183e-06 |
| GO:BP | GO:0060429 | epithelium development | 546 | 1238 | 3.200e-06 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 255 | 531 | 3.306e-06 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 189 | 377 | 3.402e-06 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 38 | 52 | 3.402e-06 |
| GO:BP | GO:0046903 | secretion | 446 | 992 | 3.409e-06 |
| GO:BP | GO:0051223 | regulation of protein transport | 217 | 442 | 3.436e-06 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 412 | 909 | 3.474e-06 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 88 | 152 | 3.500e-06 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 168 | 329 | 3.509e-06 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 87 | 150 | 3.653e-06 |
| GO:BP | GO:0099643 | signal release from synapse | 87 | 150 | 3.653e-06 |
| GO:BP | GO:0008016 | regulation of heart contraction | 112 | 204 | 3.873e-06 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 47 | 69 | 3.896e-06 |
| GO:BP | GO:0043549 | regulation of kinase activity | 212 | 431 | 3.918e-06 |
| GO:BP | GO:1903008 | organelle disassembly | 48 | 71 | 4.111e-06 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 49 | 73 | 4.307e-06 |
| GO:BP | GO:0043112 | receptor metabolic process | 49 | 73 | 4.307e-06 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 397 | 874 | 4.319e-06 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 138 | 262 | 4.355e-06 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 50 | 75 | 4.473e-06 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 82 | 140 | 4.518e-06 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 41 | 58 | 4.827e-06 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 402 | 887 | 4.834e-06 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 144 | 276 | 5.014e-06 |
| GO:BP | GO:0030488 | tRNA methylation | 31 | 40 | 5.048e-06 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 56 | 87 | 5.103e-06 |
| GO:BP | GO:0001503 | ossification | 211 | 430 | 5.141e-06 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 155 | 301 | 5.153e-06 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 131 | 247 | 5.195e-06 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 87 | 151 | 5.312e-06 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 86 | 149 | 5.574e-06 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 92 | 162 | 6.026e-06 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 84 | 145 | 6.117e-06 |
| GO:BP | GO:0032868 | response to insulin | 141 | 270 | 6.117e-06 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 71 | 118 | 6.744e-06 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 96 | 171 | 6.950e-06 |
| GO:BP | GO:0098657 | import into cell | 415 | 922 | 7.564e-06 |
| GO:BP | GO:0050779 | RNA destabilization | 65 | 106 | 7.899e-06 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 65 | 106 | 7.899e-06 |
| GO:BP | GO:0060047 | heart contraction | 129 | 244 | 7.908e-06 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 93 | 165 | 8.169e-06 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 86 | 150 | 8.169e-06 |
| GO:BP | GO:0051098 | regulation of binding | 115 | 213 | 8.169e-06 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 23 | 27 | 8.249e-06 |
| GO:BP | GO:0016032 | viral process | 207 | 423 | 8.314e-06 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 52 | 80 | 8.364e-06 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 60 | 96 | 8.478e-06 |
| GO:BP | GO:0051304 | chromosome separation | 54 | 84 | 8.540e-06 |
| GO:BP | GO:0061337 | cardiac conduction | 59 | 94 | 8.540e-06 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 34 | 46 | 9.128e-06 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 117 | 218 | 9.639e-06 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 41 | 59 | 9.639e-06 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 82 | 142 | 9.814e-06 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 82 | 142 | 9.814e-06 |
| GO:BP | GO:0030097 | hemopoiesis | 436 | 976 | 1.036e-05 |
| GO:BP | GO:0051898 | negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 42 | 61 | 1.036e-05 |
| GO:BP | GO:0035107 | appendage morphogenesis | 88 | 155 | 1.054e-05 |
| GO:BP | GO:0035108 | limb morphogenesis | 88 | 155 | 1.054e-05 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 88 | 155 | 1.054e-05 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 407 | 905 | 1.056e-05 |
| GO:BP | GO:0001501 | skeletal system development | 256 | 540 | 1.079e-05 |
| GO:BP | GO:0097352 | autophagosome maturation | 43 | 63 | 1.094e-05 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 43 | 63 | 1.094e-05 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 415 | 925 | 1.108e-05 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 44 | 65 | 1.157e-05 |
| GO:BP | GO:0035265 | organ growth | 92 | 164 | 1.208e-05 |
| GO:BP | GO:0051783 | regulation of nuclear division | 85 | 149 | 1.225e-05 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 64 | 105 | 1.257e-05 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 46 | 69 | 1.261e-05 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 47 | 71 | 1.305e-05 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 462 | 1042 | 1.311e-05 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 107 | 197 | 1.323e-05 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 129 | 246 | 1.344e-05 |
| GO:BP | GO:0016054 | organic acid catabolic process | 129 | 246 | 1.344e-05 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 175 | 351 | 1.360e-05 |
| GO:BP | GO:0009725 | response to hormone | 407 | 907 | 1.367e-05 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 50 | 77 | 1.387e-05 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 51 | 79 | 1.405e-05 |
| GO:BP | GO:0006979 | response to oxidative stress | 196 | 400 | 1.415e-05 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 295 | 635 | 1.415e-05 |
| GO:BP | GO:0035050 | embryonic heart tube development | 55 | 87 | 1.419e-05 |
| GO:BP | GO:0071478 | cellular response to radiation | 100 | 182 | 1.466e-05 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 32 | 43 | 1.469e-05 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 80 | 139 | 1.549e-05 |
| GO:BP | GO:0006301 | postreplication repair | 28 | 36 | 1.572e-05 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 33 | 45 | 1.730e-05 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 97 | 176 | 1.760e-05 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 41 | 60 | 1.839e-05 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 154 | 304 | 1.839e-05 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 154 | 304 | 1.839e-05 |
| GO:BP | GO:0003231 | cardiac ventricle development | 74 | 127 | 2.052e-05 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 196 | 402 | 2.136e-05 |
| GO:BP | GO:0000910 | cytokinesis | 104 | 192 | 2.166e-05 |
| GO:BP | GO:0031032 | actomyosin structure organization | 113 | 212 | 2.208e-05 |
| GO:BP | GO:0051050 | positive regulation of transport | 381 | 847 | 2.251e-05 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 46 | 70 | 2.257e-05 |
| GO:BP | GO:0046777 | protein autophosphorylation | 98 | 179 | 2.289e-05 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 57 | 92 | 2.290e-05 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 717 | 1685 | 2.290e-05 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 47 | 72 | 2.295e-05 |
| GO:BP | GO:0003013 | circulatory system process | 280 | 602 | 2.311e-05 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 86 | 153 | 2.346e-05 |
| GO:BP | GO:0051865 | protein autoubiquitination | 49 | 76 | 2.355e-05 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 54 | 86 | 2.357e-05 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 228 | 478 | 2.367e-05 |
| GO:BP | GO:0007219 | Notch signaling pathway | 101 | 186 | 2.591e-05 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 123 | 235 | 2.608e-05 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 134 | 260 | 2.713e-05 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 134 | 260 | 2.713e-05 |
| GO:BP | GO:0017148 | negative regulation of translation | 76 | 132 | 2.726e-05 |
| GO:BP | GO:0007000 | nucleolus organization | 16 | 17 | 2.794e-05 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 151 | 299 | 2.846e-05 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 38 | 55 | 2.886e-05 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 38 | 55 | 2.886e-05 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 38 | 55 | 2.886e-05 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 64 | 107 | 2.984e-05 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 64 | 107 | 2.984e-05 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 108 | 202 | 3.005e-05 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 160 | 320 | 3.019e-05 |
| GO:BP | GO:0016180 | snRNA processing | 27 | 35 | 3.146e-05 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 40 | 59 | 3.255e-05 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 32 | 44 | 3.258e-05 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 72 | 124 | 3.258e-05 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 527 | 1212 | 3.509e-05 |
| GO:BP | GO:0001649 | osteoblast differentiation | 122 | 234 | 3.625e-05 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 43 | 65 | 3.670e-05 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 84 | 150 | 3.676e-05 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 56 | 91 | 3.695e-05 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 55 | 89 | 3.757e-05 |
| GO:BP | GO:0072665 | protein localization to vacuole | 55 | 89 | 3.757e-05 |
| GO:BP | GO:0016477 | cell migration | 655 | 1534 | 3.809e-05 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 113 | 214 | 3.809e-05 |
| GO:BP | GO:0099536 | synaptic signaling | 363 | 807 | 3.809e-05 |
| GO:BP | GO:0045444 | fat cell differentiation | 125 | 241 | 3.814e-05 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 121 | 232 | 3.837e-05 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 68 | 116 | 3.844e-05 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 47 | 73 | 3.905e-05 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 51 | 81 | 3.910e-05 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 34 | 48 | 4.069e-05 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 352 | 781 | 4.303e-05 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 35 | 50 | 4.467e-05 |
| GO:BP | GO:0071985 | multivesicular body sorting pathway | 35 | 50 | 4.467e-05 |
| GO:BP | GO:0032205 | negative regulation of telomere maintenance | 29 | 39 | 4.624e-05 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 133 | 260 | 4.718e-05 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 96 | 177 | 4.723e-05 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 96 | 177 | 4.723e-05 |
| GO:BP | GO:0007098 | centrosome cycle | 79 | 140 | 4.781e-05 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 62 | 104 | 4.829e-05 |
| GO:BP | GO:0006505 | GPI anchor metabolic process | 25 | 32 | 4.921e-05 |
| GO:BP | GO:0180046 | GPI anchored protein biosynthesis | 25 | 32 | 4.921e-05 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 78 | 138 | 5.032e-05 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 165 | 334 | 5.053e-05 |
| GO:BP | GO:0006390 | mitochondrial transcription | 19 | 22 | 5.085e-05 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 37 | 54 | 5.118e-05 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 37 | 54 | 5.118e-05 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 77 | 136 | 5.288e-05 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 164 | 332 | 5.400e-05 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 69 | 119 | 5.412e-05 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 58 | 96 | 5.449e-05 |
| GO:BP | GO:0106027 | neuron projection organization | 57 | 94 | 5.607e-05 |
| GO:BP | GO:0045995 | regulation of embryonic development | 56 | 92 | 5.758e-05 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 41 | 62 | 6.101e-05 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 41 | 62 | 6.101e-05 |
| GO:BP | GO:0032456 | endocytic recycling | 53 | 86 | 6.157e-05 |
| GO:BP | GO:0062125 | regulation of mitochondrial gene expression | 26 | 34 | 6.169e-05 |
| GO:BP | GO:0140058 | neuron projection arborization | 26 | 34 | 6.169e-05 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 42 | 64 | 6.216e-05 |
| GO:BP | GO:0110154 | RNA decapping | 17 | 19 | 6.216e-05 |
| GO:BP | GO:0045945 | positive regulation of transcription by RNA polymerase III | 17 | 19 | 6.216e-05 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 17 | 19 | 6.216e-05 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 43 | 66 | 6.331e-05 |
| GO:BP | GO:0019080 | viral gene expression | 64 | 109 | 6.689e-05 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 131 | 257 | 6.803e-05 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 705 | 1667 | 6.939e-05 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 84 | 152 | 7.032e-05 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 71 | 124 | 7.142e-05 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 94 | 174 | 7.157e-05 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 62 | 105 | 7.225e-05 |
| GO:BP | GO:0007041 | lysosomal transport | 77 | 137 | 7.461e-05 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 61 | 103 | 7.505e-05 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 133 | 262 | 7.511e-05 |
| GO:BP | GO:0030900 | forebrain development | 202 | 423 | 7.619e-05 |
| GO:BP | GO:0007015 | actin filament organization | 218 | 461 | 7.619e-05 |
| GO:BP | GO:0048732 | gland development | 215 | 454 | 7.811e-05 |
| GO:BP | GO:0000165 | MAPK cascade | 335 | 744 | 7.825e-05 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 34 | 49 | 7.980e-05 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 34 | 49 | 7.980e-05 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 34 | 49 | 7.980e-05 |
| GO:BP | GO:0019725 | cellular homeostasis | 372 | 835 | 8.003e-05 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 59 | 99 | 8.003e-05 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 158 | 320 | 8.007e-05 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 56 | 93 | 8.870e-05 |
| GO:BP | GO:0070482 | response to oxygen levels | 168 | 344 | 9.289e-05 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 79 | 142 | 9.343e-05 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 94 | 175 | 9.617e-05 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 347 | 775 | 9.648e-05 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 347 | 775 | 9.648e-05 |
| GO:BP | GO:0006413 | translational initiation | 72 | 127 | 9.688e-05 |
| GO:BP | GO:0006506 | GPI anchor biosynthetic process | 24 | 31 | 9.759e-05 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 38 | 57 | 9.844e-05 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 29 | 40 | 9.999e-05 |
| GO:BP | GO:0097061 | dendritic spine organization | 51 | 83 | 1.006e-04 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 125 | 245 | 1.013e-04 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 50 | 81 | 1.024e-04 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 41 | 63 | 1.056e-04 |
| GO:BP | GO:0007080 | mitotic metaphase chromosome alignment | 41 | 63 | 1.056e-04 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 48 | 77 | 1.056e-04 |
| GO:BP | GO:0010586 | miRNA metabolic process | 62 | 106 | 1.066e-04 |
| GO:BP | GO:0006664 | glycolipid metabolic process | 62 | 106 | 1.066e-04 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 47 | 75 | 1.066e-04 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 272 | 593 | 1.084e-04 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 18 | 21 | 1.098e-04 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 61 | 104 | 1.110e-04 |
| GO:BP | GO:0048638 | regulation of developmental growth | 153 | 310 | 1.121e-04 |
| GO:BP | GO:0006941 | striated muscle contraction | 100 | 189 | 1.143e-04 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 12 | 12 | 1.150e-04 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 25 | 33 | 1.194e-04 |
| GO:BP | GO:0043247 | telomere maintenance in response to DNA damage | 25 | 33 | 1.194e-04 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 25 | 33 | 1.194e-04 |
| GO:BP | GO:0030258 | lipid modification | 103 | 196 | 1.235e-04 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 318 | 706 | 1.250e-04 |
| GO:BP | GO:0061448 | connective tissue development | 145 | 292 | 1.264e-04 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 255 | 553 | 1.275e-04 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 1149 | 2813 | 1.307e-04 |
| GO:BP | GO:0006836 | neurotransmitter transport | 110 | 212 | 1.312e-04 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 128 | 253 | 1.315e-04 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 32 | 46 | 1.329e-04 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 65 | 113 | 1.360e-04 |
| GO:BP | GO:0090224 | regulation of spindle organization | 33 | 48 | 1.426e-04 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 33 | 48 | 1.426e-04 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 54 | 90 | 1.432e-04 |
| GO:BP | GO:0034502 | protein localization to chromosome | 71 | 126 | 1.432e-04 |
| GO:BP | GO:0034644 | cellular response to UV | 54 | 90 | 1.432e-04 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 53 | 88 | 1.474e-04 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 155 | 316 | 1.474e-04 |
| GO:BP | GO:0010232 | vascular transport | 53 | 88 | 1.474e-04 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 14 | 15 | 1.512e-04 |
| GO:BP | GO:1903509 | liposaccharide metabolic process | 62 | 107 | 1.562e-04 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 154 | 314 | 1.577e-04 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 75 | 135 | 1.610e-04 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 49 | 80 | 1.637e-04 |
| GO:BP | GO:0006414 | translational elongation | 49 | 80 | 1.637e-04 |
| GO:BP | GO:0010761 | fibroblast migration | 36 | 54 | 1.637e-04 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 27 | 37 | 1.637e-04 |
| GO:BP | GO:0051258 | protein polymerization | 141 | 284 | 1.648e-04 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 85 | 157 | 1.655e-04 |
| GO:BP | GO:0043489 | RNA stabilization | 48 | 78 | 1.669e-04 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 19 | 23 | 1.673e-04 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 60 | 103 | 1.689e-04 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 98 | 186 | 1.689e-04 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 98 | 186 | 1.689e-04 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 74 | 133 | 1.689e-04 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 60 | 103 | 1.689e-04 |
| GO:BP | GO:0035329 | hippo signaling | 38 | 58 | 1.709e-04 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 38 | 58 | 1.709e-04 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 39 | 60 | 1.738e-04 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 84 | 155 | 1.746e-04 |
| GO:BP | GO:0002027 | regulation of heart rate | 59 | 101 | 1.758e-04 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 43 | 68 | 1.760e-04 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 178 | 371 | 1.760e-04 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 42 | 66 | 1.763e-04 |
| GO:BP | GO:0007416 | synapse assembly | 130 | 259 | 1.779e-04 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 97 | 184 | 1.791e-04 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 28 | 39 | 1.820e-04 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 78 | 142 | 1.834e-04 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 126 | 250 | 1.860e-04 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 122 | 241 | 1.943e-04 |
| GO:BP | GO:0032509 | endosome transport via multivesicular body sorting pathway | 29 | 41 | 2.020e-04 |
| GO:BP | GO:0032790 | ribosome disassembly | 29 | 41 | 2.020e-04 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 99 | 189 | 2.062e-04 |
| GO:BP | GO:0019395 | fatty acid oxidation | 63 | 110 | 2.134e-04 |
| GO:BP | GO:0072524 | pyridine-containing compound metabolic process | 90 | 169 | 2.143e-04 |
| GO:BP | GO:0007517 | muscle organ development | 175 | 365 | 2.151e-04 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 117 | 230 | 2.157e-04 |
| GO:BP | GO:0051225 | spindle assembly | 75 | 136 | 2.192e-04 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 75 | 136 | 2.192e-04 |
| GO:BP | GO:0035418 | protein localization to synapse | 53 | 89 | 2.215e-04 |
| GO:BP | GO:0043543 | protein acylation | 53 | 89 | 2.215e-04 |
| GO:BP | GO:0043542 | endothelial cell migration | 113 | 221 | 2.217e-04 |
| GO:BP | GO:0060674 | placenta blood vessel development | 24 | 32 | 2.278e-04 |
| GO:BP | GO:0009301 | snRNA transcription | 17 | 20 | 2.339e-04 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 31 | 45 | 2.350e-04 |
| GO:BP | GO:0090148 | membrane fission | 31 | 45 | 2.350e-04 |
| GO:BP | GO:1901875 | positive regulation of post-translational protein modification | 73 | 132 | 2.464e-04 |
| GO:BP | GO:1902414 | protein localization to cell junction | 67 | 119 | 2.466e-04 |
| GO:BP | GO:0048608 | reproductive structure development | 153 | 314 | 2.473e-04 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 92 | 174 | 2.477e-04 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 147 | 300 | 2.481e-04 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 32 | 47 | 2.481e-04 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 32 | 47 | 2.481e-04 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 131 | 263 | 2.576e-04 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 48 | 79 | 2.580e-04 |
| GO:BP | GO:0042073 | intraciliary transport | 33 | 49 | 2.602e-04 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 47 | 77 | 2.646e-04 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 34 | 51 | 2.694e-04 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 34 | 51 | 2.694e-04 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 34 | 51 | 2.694e-04 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 34 | 51 | 2.694e-04 |
| GO:BP | GO:0003281 | ventricular septum development | 45 | 73 | 2.756e-04 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 45 | 73 | 2.756e-04 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 35 | 53 | 2.759e-04 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 35 | 53 | 2.759e-04 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 35 | 53 | 2.759e-04 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 106 | 206 | 2.759e-04 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 35 | 53 | 2.759e-04 |
| GO:BP | GO:0046785 | microtubule polymerization | 57 | 98 | 2.778e-04 |
| GO:BP | GO:0140374 | antiviral innate immune response | 36 | 55 | 2.816e-04 |
| GO:BP | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | 36 | 55 | 2.816e-04 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 43 | 69 | 2.820e-04 |
| GO:BP | GO:0048255 | mRNA stabilization | 43 | 69 | 2.820e-04 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 43 | 69 | 2.820e-04 |
| GO:BP | GO:0001947 | heart looping | 42 | 67 | 2.853e-04 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 70 | 126 | 2.872e-04 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 41 | 65 | 2.873e-04 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 39 | 61 | 2.884e-04 |
| GO:BP | GO:0001822 | kidney development | 156 | 322 | 2.927e-04 |
| GO:BP | GO:0001525 | angiogenesis | 250 | 547 | 3.020e-04 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 69 | 124 | 3.034e-04 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 69 | 124 | 3.034e-04 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 84 | 157 | 3.072e-04 |
| GO:BP | GO:0006606 | protein import into nucleus | 88 | 166 | 3.156e-04 |
| GO:BP | GO:0051259 | protein complex oligomerization | 134 | 271 | 3.169e-04 |
| GO:BP | GO:0072001 | renal system development | 160 | 332 | 3.272e-04 |
| GO:BP | GO:0048568 | embryonic organ development | 215 | 463 | 3.272e-04 |
| GO:BP | GO:0007409 | axonogenesis | 215 | 463 | 3.272e-04 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 27 | 38 | 3.299e-04 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 27 | 38 | 3.299e-04 |
| GO:BP | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 27 | 38 | 3.299e-04 |
| GO:BP | GO:0006897 | endocytosis | 316 | 709 | 3.311e-04 |
| GO:BP | GO:0061458 | reproductive system development | 154 | 318 | 3.332e-04 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 18 | 22 | 3.394e-04 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 18 | 22 | 3.394e-04 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 72 | 131 | 3.514e-04 |
| GO:BP | GO:0019362 | pyridine nucleotide metabolic process | 86 | 162 | 3.576e-04 |
| GO:BP | GO:0046496 | nicotinamide nucleotide metabolic process | 86 | 162 | 3.576e-04 |
| GO:BP | GO:0030334 | regulation of cell migration | 417 | 960 | 3.593e-04 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 28 | 40 | 3.593e-04 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 22 | 29 | 3.603e-04 |
| GO:BP | GO:0001666 | response to hypoxia | 147 | 302 | 3.603e-04 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 22 | 29 | 3.603e-04 |
| GO:BP | GO:0051170 | import into nucleus | 90 | 171 | 3.609e-04 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 71 | 129 | 3.710e-04 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 76 | 140 | 3.743e-04 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 29 | 42 | 3.856e-04 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 1116 | 2745 | 3.875e-04 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 80 | 149 | 3.930e-04 |
| GO:BP | GO:0045727 | positive regulation of translation | 75 | 138 | 3.984e-04 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 84 | 158 | 4.055e-04 |
| GO:BP | GO:1903522 | regulation of blood circulation | 127 | 256 | 4.091e-04 |
| GO:BP | GO:0048511 | rhythmic process | 148 | 305 | 4.093e-04 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 46 | 76 | 4.125e-04 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 46 | 76 | 4.125e-04 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 56 | 97 | 4.182e-04 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 74 | 136 | 4.225e-04 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 31 | 46 | 4.274e-04 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 87 | 165 | 4.389e-04 |
| GO:BP | GO:0071772 | response to BMP | 87 | 165 | 4.389e-04 |
| GO:BP | GO:2000145 | regulation of cell motility | 441 | 1022 | 4.408e-04 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 32 | 48 | 4.426e-04 |
| GO:BP | GO:0071763 | nuclear membrane organization | 32 | 48 | 4.426e-04 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 43 | 70 | 4.430e-04 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 78 | 145 | 4.430e-04 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 155 | 322 | 4.507e-04 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 42 | 68 | 4.519e-04 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 19 | 24 | 4.547e-04 |
| GO:BP | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 19 | 24 | 4.547e-04 |
| GO:BP | GO:0030509 | BMP signaling pathway | 82 | 154 | 4.568e-04 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 41 | 66 | 4.584e-04 |
| GO:BP | GO:0009299 | mRNA transcription | 34 | 52 | 4.623e-04 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 94 | 181 | 4.653e-04 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 105 | 206 | 4.663e-04 |
| GO:BP | GO:0098732 | macromolecule deacylation | 37 | 58 | 4.732e-04 |
| GO:BP | GO:1904505 | regulation of telomere maintenance in response to DNA damage | 16 | 19 | 4.854e-04 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 60 | 106 | 4.880e-04 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 76 | 141 | 5.006e-04 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 80 | 150 | 5.202e-04 |
| GO:BP | GO:0030879 | mammary gland development | 75 | 139 | 5.348e-04 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 25 | 35 | 5.467e-04 |
| GO:BP | GO:0060419 | heart growth | 49 | 83 | 5.649e-04 |
| GO:BP | GO:0006446 | regulation of translational initiation | 49 | 83 | 5.649e-04 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 69 | 126 | 5.694e-04 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 69 | 126 | 5.694e-04 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 69 | 126 | 5.694e-04 |
| GO:BP | GO:0050773 | regulation of dendrite development | 57 | 100 | 5.717e-04 |
| GO:BP | GO:0019985 | translesion synthesis | 20 | 26 | 5.756e-04 |
| GO:BP | GO:0070129 | regulation of mitochondrial translation | 20 | 26 | 5.756e-04 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 20 | 26 | 5.756e-04 |
| GO:BP | GO:0071539 | protein localization to centrosome | 26 | 37 | 5.972e-04 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 26 | 37 | 5.972e-04 |
| GO:BP | GO:0043603 | amide metabolic process | 210 | 455 | 5.990e-04 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 56 | 98 | 5.995e-04 |
| GO:BP | GO:0002200 | somatic diversification of immune receptors | 47 | 79 | 6.056e-04 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 133 | 272 | 6.163e-04 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 89 | 171 | 6.516e-04 |
| GO:BP | GO:0110156 | mRNA methylguanosine-cap decapping | 14 | 16 | 6.599e-04 |
| GO:BP | GO:0045927 | positive regulation of growth | 126 | 256 | 6.607e-04 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 54 | 94 | 6.633e-04 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 10 | 10 | 6.746e-04 |
| GO:BP | GO:1990700 | nucleolar chromatin organization | 10 | 10 | 6.746e-04 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 71 | 131 | 6.842e-04 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 80 | 151 | 6.878e-04 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 88 | 169 | 6.947e-04 |
| GO:BP | GO:0051383 | kinetochore organization | 17 | 21 | 6.978e-04 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 42 | 69 | 7.104e-04 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 298 | 671 | 7.105e-04 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 29 | 43 | 7.147e-04 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 41 | 67 | 7.278e-04 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 59 | 105 | 7.280e-04 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 30 | 45 | 7.407e-04 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 30 | 45 | 7.407e-04 |
| GO:BP | GO:0008406 | gonad development | 118 | 238 | 7.441e-04 |
| GO:BP | GO:0070085 | glycosylation | 121 | 245 | 7.494e-04 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 31 | 47 | 7.599e-04 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 31 | 47 | 7.599e-04 |
| GO:BP | GO:0070914 | UV-damage excision repair | 12 | 13 | 7.787e-04 |
| GO:BP | GO:0006998 | nuclear envelope organization | 37 | 59 | 7.787e-04 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 78 | 147 | 7.787e-04 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 12 | 13 | 7.787e-04 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 36 | 57 | 7.811e-04 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 33 | 51 | 7.811e-04 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 36 | 57 | 7.811e-04 |
| GO:BP | GO:0048878 | chemical homeostasis | 447 | 1043 | 7.811e-04 |
| GO:BP | GO:0031297 | replication fork processing | 33 | 51 | 7.811e-04 |
| GO:BP | GO:0046034 | ATP metabolic process | 114 | 229 | 7.812e-04 |
| GO:BP | GO:0048284 | organelle fusion | 86 | 165 | 7.841e-04 |
| GO:BP | GO:0022406 | membrane docking | 50 | 86 | 7.893e-04 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 120 | 243 | 7.945e-04 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 22 | 30 | 7.972e-04 |
| GO:BP | GO:0006520 | amino acid metabolic process | 143 | 297 | 8.193e-04 |
| GO:BP | GO:0048870 | cell motility | 742 | 1793 | 8.354e-04 |
| GO:BP | GO:0003012 | muscle system process | 202 | 438 | 8.388e-04 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 103 | 204 | 8.444e-04 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 56 | 99 | 8.461e-04 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 72 | 134 | 8.528e-04 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 230 | 506 | 8.557e-04 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 48 | 82 | 8.595e-04 |
| GO:BP | GO:0061564 | axon development | 239 | 528 | 8.647e-04 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 76 | 143 | 8.781e-04 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 23 | 32 | 8.989e-04 |
| GO:BP | GO:0060444 | branching involved in mammary gland duct morphogenesis | 18 | 23 | 9.055e-04 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 321 | 730 | 9.063e-04 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 71 | 132 | 9.063e-04 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 121 | 246 | 9.102e-04 |
| GO:BP | GO:0010720 | positive regulation of cell development | 210 | 458 | 9.218e-04 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 54 | 95 | 9.372e-04 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 45 | 76 | 9.717e-04 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 53 | 93 | 9.858e-04 |
| GO:BP | GO:0072344 | rescue of stalled ribosome | 24 | 34 | 9.858e-04 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 53 | 93 | 9.858e-04 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 53 | 93 | 9.858e-04 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 24 | 34 | 9.858e-04 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 24 | 34 | 9.858e-04 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 44 | 74 | 1.003e-03 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 44 | 74 | 1.003e-03 |
| GO:BP | GO:0071806 | protein transmembrane transport | 44 | 74 | 1.003e-03 |
| GO:BP | GO:0021537 | telencephalon development | 137 | 284 | 1.005e-03 |
| GO:BP | GO:0045926 | negative regulation of growth | 113 | 228 | 1.028e-03 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 64 | 117 | 1.029e-03 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 128 | 263 | 1.032e-03 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 25 | 36 | 1.063e-03 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 25 | 36 | 1.063e-03 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 19 | 25 | 1.116e-03 |
| GO:BP | GO:0007548 | sex differentiation | 141 | 294 | 1.119e-03 |
| GO:BP | GO:0040012 | regulation of locomotion | 455 | 1067 | 1.119e-03 |
| GO:BP | GO:0043010 | camera-type eye development | 170 | 363 | 1.125e-03 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 26 | 38 | 1.130e-03 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 76 | 144 | 1.154e-03 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 140 | 292 | 1.198e-03 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 71 | 133 | 1.209e-03 |
| GO:BP | GO:0033619 | membrane protein proteolysis | 38 | 62 | 1.209e-03 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 66 | 122 | 1.240e-03 |
| GO:BP | GO:0007423 | sensory organ development | 277 | 624 | 1.245e-03 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 48 | 83 | 1.258e-03 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 48 | 83 | 1.258e-03 |
| GO:BP | GO:0008088 | axo-dendritic transport | 48 | 83 | 1.258e-03 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 29 | 44 | 1.261e-03 |
| GO:BP | GO:1901799 | negative regulation of proteasomal protein catabolic process | 35 | 56 | 1.275e-03 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 35 | 56 | 1.275e-03 |
| GO:BP | GO:0006284 | base-excision repair | 30 | 46 | 1.282e-03 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 31 | 48 | 1.291e-03 |
| GO:BP | GO:0001881 | receptor recycling | 31 | 48 | 1.291e-03 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 31 | 48 | 1.291e-03 |
| GO:BP | GO:0006513 | protein monoubiquitination | 31 | 48 | 1.291e-03 |
| GO:BP | GO:0035601 | protein deacylation | 32 | 50 | 1.294e-03 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 32 | 50 | 1.294e-03 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 232 | 514 | 1.297e-03 |
| GO:BP | GO:0036503 | ERAD pathway | 60 | 109 | 1.299e-03 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 54 | 96 | 1.316e-03 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 106 | 213 | 1.325e-03 |
| GO:BP | GO:0060348 | bone development | 109 | 220 | 1.339e-03 |
| GO:BP | GO:2000786 | positive regulation of autophagosome assembly | 16 | 20 | 1.405e-03 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 77 | 147 | 1.407e-03 |
| GO:BP | GO:0001654 | eye development | 191 | 415 | 1.413e-03 |
| GO:BP | GO:0150011 | regulation of neuron projection arborization | 13 | 15 | 1.417e-03 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 45 | 77 | 1.431e-03 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 88 | 172 | 1.438e-03 |
| GO:BP | GO:0060541 | respiratory system development | 111 | 225 | 1.443e-03 |
| GO:BP | GO:0009135 | purine nucleoside diphosphate metabolic process | 63 | 116 | 1.477e-03 |
| GO:BP | GO:0009179 | purine ribonucleoside diphosphate metabolic process | 63 | 116 | 1.477e-03 |
| GO:BP | GO:0034440 | lipid oxidation | 63 | 116 | 1.477e-03 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 21 | 29 | 1.482e-03 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 44 | 75 | 1.488e-03 |
| GO:BP | GO:0061512 | protein localization to cilium | 44 | 75 | 1.488e-03 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 101 | 202 | 1.492e-03 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 51 | 90 | 1.544e-03 |
| GO:BP | GO:0002562 | somatic diversification of immune receptors via germline recombination within a single locus | 43 | 73 | 1.551e-03 |
| GO:BP | GO:0016444 | somatic cell DNA recombination | 43 | 73 | 1.551e-03 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 231 | 513 | 1.567e-03 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 141 | 296 | 1.587e-03 |
| GO:BP | GO:0051402 | neuron apoptotic process | 141 | 296 | 1.587e-03 |
| GO:BP | GO:0003162 | atrioventricular node development | 9 | 9 | 1.621e-03 |
| GO:BP | GO:0000467 | exonucleolytic trimming to generate mature 3’-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 9 | 9 | 1.621e-03 |
| GO:BP | GO:0000183 | rDNA heterochromatin formation | 9 | 9 | 1.621e-03 |
| GO:BP | GO:0042796 | snRNA transcription by RNA polymerase III | 9 | 9 | 1.621e-03 |
| GO:BP | GO:0006006 | glucose metabolic process | 93 | 184 | 1.631e-03 |
| GO:BP | GO:0019082 | viral protein processing | 22 | 31 | 1.631e-03 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 22 | 31 | 1.631e-03 |
| GO:BP | GO:0070536 | protein K63-linked deubiquitination | 22 | 31 | 1.631e-03 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 41 | 69 | 1.669e-03 |
| GO:BP | GO:0098930 | axonal transport | 41 | 69 | 1.669e-03 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 78 | 150 | 1.695e-03 |
| GO:BP | GO:0055006 | cardiac cell development | 49 | 86 | 1.701e-03 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 49 | 86 | 1.701e-03 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 461 | 1087 | 1.713e-03 |
| GO:BP | GO:0044057 | regulation of system process | 247 | 553 | 1.724e-03 |
| GO:BP | GO:0051100 | negative regulation of binding | 55 | 99 | 1.724e-03 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 40 | 67 | 1.725e-03 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 92 | 182 | 1.737e-03 |
| GO:BP | GO:0030518 | nuclear receptor-mediated steroid hormone signaling pathway | 65 | 121 | 1.737e-03 |
| GO:BP | GO:1904872 | regulation of telomerase RNA localization to Cajal body | 11 | 12 | 1.741e-03 |
| GO:BP | GO:0051657 | maintenance of organelle location | 11 | 12 | 1.741e-03 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 117 | 240 | 1.742e-03 |
| GO:BP | GO:0006486 | protein glycosylation | 111 | 226 | 1.744e-03 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 111 | 226 | 1.744e-03 |
| GO:BP | GO:0019318 | hexose metabolic process | 111 | 226 | 1.744e-03 |
| GO:BP | GO:0036257 | multivesicular body organization | 23 | 33 | 1.752e-03 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 17 | 22 | 1.755e-03 |
| GO:BP | GO:0061709 | reticulophagy | 17 | 22 | 1.755e-03 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 39 | 65 | 1.768e-03 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 48 | 84 | 1.768e-03 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 39 | 65 | 1.768e-03 |
| GO:BP | GO:0045185 | maintenance of protein location | 39 | 65 | 1.768e-03 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 38 | 63 | 1.830e-03 |
| GO:BP | GO:0001824 | blastocyst development | 64 | 119 | 1.835e-03 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 84 | 164 | 1.846e-03 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 24 | 35 | 1.856e-03 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 24 | 35 | 1.856e-03 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 76 | 146 | 1.914e-03 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 87 | 171 | 1.915e-03 |
| GO:BP | GO:0060612 | adipose tissue development | 36 | 59 | 1.935e-03 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 36 | 59 | 1.935e-03 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis | 25 | 37 | 1.943e-03 |
| GO:BP | GO:0022616 | DNA strand elongation | 25 | 37 | 1.943e-03 |
| GO:BP | GO:0045324 | late endosome to vacuole transport | 25 | 37 | 1.943e-03 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 35 | 57 | 1.980e-03 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 153 | 326 | 1.989e-03 |
| GO:BP | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 26 | 39 | 2.014e-03 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 75 | 144 | 2.040e-03 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 27 | 41 | 2.065e-03 |
| GO:BP | GO:0051099 | positive regulation of binding | 57 | 104 | 2.079e-03 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 32 | 51 | 2.079e-03 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 32 | 51 | 2.079e-03 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 99 | 199 | 2.079e-03 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 32 | 51 | 2.079e-03 |
| GO:BP | GO:0008089 | anterograde axonal transport | 32 | 51 | 2.079e-03 |
| GO:BP | GO:0031573 | mitotic intra-S DNA damage checkpoint signaling | 14 | 17 | 2.079e-03 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 28 | 43 | 2.084e-03 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 28 | 43 | 2.084e-03 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 30 | 47 | 2.099e-03 |
| GO:BP | GO:0035196 | miRNA processing | 29 | 45 | 2.099e-03 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 30 | 47 | 2.099e-03 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 29 | 45 | 2.099e-03 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 74 | 142 | 2.165e-03 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 43 | 74 | 2.235e-03 |
| GO:BP | GO:1903578 | regulation of ATP metabolic process | 43 | 74 | 2.235e-03 |
| GO:BP | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway | 88 | 174 | 2.241e-03 |
| GO:BP | GO:0046620 | regulation of organ growth | 50 | 89 | 2.241e-03 |
| GO:BP | GO:0045055 | regulated exocytosis | 113 | 232 | 2.244e-03 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 124 | 258 | 2.309e-03 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 55 | 100 | 2.336e-03 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 42 | 72 | 2.336e-03 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 201 | 443 | 2.345e-03 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 145 | 308 | 2.374e-03 |
| GO:BP | GO:0001508 | action potential | 87 | 172 | 2.398e-03 |
| GO:BP | GO:0022038 | corpus callosum development | 19 | 26 | 2.405e-03 |
| GO:BP | GO:0001889 | liver development | 76 | 147 | 2.448e-03 |
| GO:BP | GO:0150063 | visual system development | 191 | 419 | 2.449e-03 |
| GO:BP | GO:0043401 | steroid hormone receptor signaling pathway | 72 | 138 | 2.472e-03 |
| GO:BP | GO:0048041 | focal adhesion assembly | 48 | 85 | 2.499e-03 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 96 | 193 | 2.540e-03 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 99 | 200 | 2.565e-03 |
| GO:BP | GO:0141193 | nuclear receptor-mediated signaling pathway | 89 | 177 | 2.633e-03 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 53 | 96 | 2.633e-03 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 47 | 83 | 2.639e-03 |
| GO:BP | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 20 | 28 | 2.688e-03 |
| GO:BP | GO:0055001 | muscle cell development | 98 | 198 | 2.751e-03 |
| GO:BP | GO:0090672 | telomerase RNA localization | 15 | 19 | 2.759e-03 |
| GO:BP | GO:0090671 | telomerase RNA localization to Cajal body | 15 | 19 | 2.759e-03 |
| GO:BP | GO:0033059 | cellular pigmentation | 38 | 64 | 2.759e-03 |
| GO:BP | GO:0060039 | pericardium development | 15 | 19 | 2.759e-03 |
| GO:BP | GO:0090670 | RNA localization to Cajal body | 15 | 19 | 2.759e-03 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 38 | 64 | 2.759e-03 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 15 | 19 | 2.759e-03 |
| GO:BP | GO:0090685 | RNA localization to nucleus | 15 | 19 | 2.759e-03 |
| GO:BP | GO:0007267 | cell-cell signaling | 559 | 1342 | 2.801e-03 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 37 | 62 | 2.855e-03 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 21 | 30 | 2.912e-03 |
| GO:BP | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening | 21 | 30 | 2.912e-03 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 21 | 30 | 2.912e-03 |
| GO:BP | GO:1901652 | response to peptide | 409 | 962 | 2.925e-03 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 61 | 114 | 2.930e-03 |
| GO:BP | GO:0002520 | immune system development | 100 | 203 | 2.940e-03 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 51 | 92 | 2.942e-03 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 36 | 60 | 2.947e-03 |
| GO:BP | GO:0034097 | response to cytokine | 403 | 947 | 2.949e-03 |
| GO:BP | GO:0007100 | mitotic centrosome separation | 12 | 14 | 2.979e-03 |
| GO:BP | GO:0140718 | facultative heterochromatin formation | 12 | 14 | 2.979e-03 |
| GO:BP | GO:0030278 | regulation of ossification | 65 | 123 | 2.982e-03 |
| GO:BP | GO:0061614 | miRNA transcription | 44 | 77 | 3.066e-03 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 22 | 32 | 3.092e-03 |
| GO:BP | GO:0036258 | multivesicular body assembly | 22 | 32 | 3.092e-03 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 50 | 90 | 3.112e-03 |
| GO:BP | GO:0009408 | response to heat | 55 | 101 | 3.167e-03 |
| GO:BP | GO:0090398 | cellular senescence | 55 | 101 | 3.167e-03 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 72 | 139 | 3.177e-03 |
| GO:BP | GO:0051261 | protein depolymerization | 64 | 121 | 3.182e-03 |
| GO:BP | GO:0055002 | striated muscle cell development | 86 | 171 | 3.198e-03 |
| GO:BP | GO:0043954 | cellular component maintenance | 43 | 75 | 3.216e-03 |
| GO:BP | GO:0006654 | phosphatidic acid biosynthetic process | 23 | 34 | 3.230e-03 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 23 | 34 | 3.230e-03 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 23 | 34 | 3.230e-03 |
| GO:BP | GO:0048880 | sensory system development | 193 | 426 | 3.263e-03 |
| GO:BP | GO:0010332 | response to gamma radiation | 32 | 52 | 3.290e-03 |
| GO:BP | GO:0009615 | response to virus | 188 | 414 | 3.344e-03 |
| GO:BP | GO:0170036 | import into the mitochondrion | 31 | 50 | 3.355e-03 |
| GO:BP | GO:0007019 | microtubule depolymerization | 31 | 50 | 3.355e-03 |
| GO:BP | GO:0032506 | cytokinetic process | 31 | 50 | 3.355e-03 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 42 | 73 | 3.367e-03 |
| GO:BP | GO:0070242 | thymocyte apoptotic process | 16 | 21 | 3.379e-03 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 67 | 128 | 3.404e-03 |
| GO:BP | GO:0098534 | centriole assembly | 30 | 48 | 3.406e-03 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 25 | 38 | 3.408e-03 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 26 | 40 | 3.457e-03 |
| GO:BP | GO:0007249 | canonical NF-kappaB signal transduction | 144 | 308 | 3.466e-03 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 28 | 44 | 3.466e-03 |
| GO:BP | GO:0017004 | cytochrome complex assembly | 27 | 42 | 3.466e-03 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 27 | 42 | 3.466e-03 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 270 | 616 | 3.485e-03 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 58 | 108 | 3.502e-03 |
| GO:BP | GO:0051668 | localization within membrane | 393 | 924 | 3.568e-03 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 216 | 483 | 3.587e-03 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 40 | 69 | 3.698e-03 |
| GO:BP | GO:0045445 | myoblast differentiation | 61 | 115 | 3.858e-03 |
| GO:BP | GO:0034394 | protein localization to cell surface | 39 | 67 | 3.877e-03 |
| GO:BP | GO:0018195 | peptidyl-arginine modification | 10 | 11 | 3.877e-03 |
| GO:BP | GO:0044091 | membrane biogenesis | 39 | 67 | 3.877e-03 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 247 | 560 | 3.892e-03 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 247 | 560 | 3.892e-03 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 83 | 165 | 3.896e-03 |
| GO:BP | GO:0016237 | microautophagy | 17 | 23 | 3.943e-03 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 17 | 23 | 3.943e-03 |
| GO:BP | GO:0044804 | nucleophagy | 17 | 23 | 3.943e-03 |
| GO:BP | GO:0170062 | nutrient storage | 51 | 93 | 4.005e-03 |
| GO:BP | GO:0019915 | lipid storage | 51 | 93 | 4.005e-03 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 38 | 65 | 4.043e-03 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 45 | 80 | 4.092e-03 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 248 | 563 | 4.132e-03 |
| GO:BP | GO:0051293 | establishment of spindle localization | 37 | 63 | 4.230e-03 |
| GO:BP | GO:0072526 | pyridine-containing compound catabolic process | 55 | 102 | 4.245e-03 |
| GO:BP | GO:0042795 | snRNA transcription by RNA polymerase II | 13 | 16 | 4.252e-03 |
| GO:BP | GO:0051607 | defense response to virus | 148 | 319 | 4.330e-03 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 59 | 111 | 4.382e-03 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 232 | 524 | 4.396e-03 |
| GO:BP | GO:0006695 | cholesterol biosynthetic process | 36 | 61 | 4.403e-03 |
| GO:BP | GO:1902653 | secondary alcohol biosynthetic process | 36 | 61 | 4.403e-03 |
| GO:BP | GO:0044090 | positive regulation of vacuole organization | 18 | 25 | 4.433e-03 |
| GO:BP | GO:0070841 | inclusion body assembly | 18 | 25 | 4.433e-03 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 18 | 25 | 4.433e-03 |
| GO:BP | GO:0090257 | regulation of muscle system process | 111 | 231 | 4.471e-03 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 43 | 76 | 4.561e-03 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 35 | 59 | 4.575e-03 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 35 | 59 | 4.575e-03 |
| GO:BP | GO:0007254 | JNK cascade | 87 | 175 | 4.599e-03 |
| GO:BP | GO:0042391 | regulation of membrane potential | 205 | 458 | 4.603e-03 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 96 | 196 | 4.657e-03 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 58 | 109 | 4.657e-03 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 364 | 854 | 4.704e-03 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 34 | 57 | 4.747e-03 |
| GO:BP | GO:0051298 | centrosome duplication | 42 | 74 | 4.795e-03 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 42 | 74 | 4.795e-03 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 42 | 74 | 4.795e-03 |
| GO:BP | GO:0007018 | microtubule-based movement | 213 | 478 | 4.807e-03 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 19 | 27 | 4.822e-03 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 19 | 27 | 4.822e-03 |
| GO:BP | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway | 19 | 27 | 4.822e-03 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 104 | 215 | 4.878e-03 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 33 | 55 | 4.898e-03 |
| GO:BP | GO:0006220 | pyrimidine nucleotide metabolic process | 33 | 55 | 4.898e-03 |
| GO:BP | GO:0009749 | response to glucose | 101 | 208 | 4.915e-03 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 89 | 180 | 4.935e-03 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 98 | 201 | 4.940e-03 |
| GO:BP | GO:0006936 | muscle contraction | 160 | 349 | 5.020e-03 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 41 | 72 | 5.022e-03 |
| GO:BP | GO:0051101 | regulation of DNA binding | 41 | 72 | 5.022e-03 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 138 | 296 | 5.022e-03 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 41 | 72 | 5.022e-03 |
| GO:BP | GO:0001570 | vasculogenesis | 47 | 85 | 5.028e-03 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 47 | 85 | 5.028e-03 |
| GO:BP | GO:0034508 | centromere complex assembly | 20 | 29 | 5.123e-03 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 20 | 29 | 5.123e-03 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 68 | 132 | 5.248e-03 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 40 | 70 | 5.285e-03 |
| GO:BP | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 40 | 70 | 5.285e-03 |
| GO:BP | GO:0090174 | organelle membrane fusion | 64 | 123 | 5.325e-03 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 46 | 83 | 5.331e-03 |
| GO:BP | GO:0097009 | energy homeostasis | 46 | 83 | 5.331e-03 |
| GO:BP | GO:0006497 | protein lipidation | 30 | 49 | 5.336e-03 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 21 | 31 | 5.366e-03 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 51 | 94 | 5.377e-03 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 14 | 18 | 5.382e-03 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 14 | 18 | 5.382e-03 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 14 | 18 | 5.382e-03 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 161 | 352 | 5.415e-03 |
| GO:BP | GO:0006509 | membrane protein ectodomain proteolysis | 29 | 47 | 5.445e-03 |
| GO:BP | GO:0043038 | amino acid activation | 29 | 47 | 5.445e-03 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 81 | 162 | 5.483e-03 |
| GO:BP | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 139 | 299 | 5.484e-03 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 39 | 68 | 5.516e-03 |
| GO:BP | GO:0051653 | spindle localization | 39 | 68 | 5.516e-03 |
| GO:BP | GO:0051149 | positive regulation of muscle cell differentiation | 39 | 68 | 5.516e-03 |
| GO:BP | GO:0051260 | protein homooligomerization | 99 | 204 | 5.556e-03 |
| GO:BP | GO:0022612 | gland morphogenesis | 67 | 130 | 5.561e-03 |
| GO:BP | GO:0046661 | male sex differentiation | 87 | 176 | 5.584e-03 |
| GO:BP | GO:0140029 | exocytic process | 45 | 81 | 5.600e-03 |
| GO:BP | GO:0000154 | rRNA modification | 23 | 35 | 5.600e-03 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 158 | 345 | 5.600e-03 |
| GO:BP | GO:0048489 | synaptic vesicle transport | 27 | 43 | 5.600e-03 |
| GO:BP | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 23 | 35 | 5.600e-03 |
| GO:BP | GO:0043276 | anoikis | 23 | 35 | 5.600e-03 |
| GO:BP | GO:0007099 | centriole replication | 27 | 43 | 5.600e-03 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 63 | 121 | 5.613e-03 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 26 | 41 | 5.643e-03 |
| GO:BP | GO:0003161 | cardiac conduction system development | 24 | 37 | 5.643e-03 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 26 | 41 | 5.643e-03 |
| GO:BP | GO:0035330 | regulation of hippo signaling | 26 | 41 | 5.643e-03 |
| GO:BP | GO:0003230 | cardiac atrium development | 24 | 37 | 5.643e-03 |
| GO:BP | GO:0070979 | protein K11-linked ubiquitination | 25 | 39 | 5.648e-03 |
| GO:BP | GO:0034389 | lipid droplet organization | 25 | 39 | 5.648e-03 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 25 | 39 | 5.648e-03 |
| GO:BP | GO:0097120 | receptor localization to synapse | 44 | 79 | 5.902e-03 |
| GO:BP | GO:0017157 | regulation of exocytosis | 89 | 181 | 5.960e-03 |
| GO:BP | GO:0070555 | response to interleukin-1 | 58 | 110 | 5.992e-03 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 37 | 64 | 6.035e-03 |
| GO:BP | GO:0016447 | somatic recombination of immunoglobulin gene segments | 37 | 64 | 6.035e-03 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 11 | 13 | 6.201e-03 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 43 | 77 | 6.258e-03 |
| GO:BP | GO:0019364 | pyridine nucleotide catabolic process | 53 | 99 | 6.282e-03 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 82 | 165 | 6.306e-03 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 36 | 62 | 6.330e-03 |
| GO:BP | GO:0045010 | actin nucleation | 36 | 62 | 6.330e-03 |
| GO:BP | GO:0006098 | pentose-phosphate shunt | 15 | 20 | 6.363e-03 |
| GO:BP | GO:0060716 | labyrinthine layer blood vessel development | 15 | 20 | 6.363e-03 |
| GO:BP | GO:0048588 | developmental cell growth | 108 | 226 | 6.370e-03 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 75 | 149 | 6.532e-03 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 42 | 75 | 6.618e-03 |
| GO:BP | GO:0060420 | regulation of heart growth | 35 | 60 | 6.630e-03 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 78 | 156 | 6.655e-03 |
| GO:BP | GO:0008015 | blood circulation | 228 | 518 | 6.728e-03 |
| GO:BP | GO:0048864 | stem cell development | 47 | 86 | 6.791e-03 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 47 | 86 | 6.791e-03 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 234 | 533 | 6.836e-03 |
| GO:BP | GO:0030166 | proteoglycan biosynthetic process | 41 | 73 | 7.004e-03 |
| GO:BP | GO:0016042 | lipid catabolic process | 161 | 354 | 7.124e-03 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 51 | 95 | 7.145e-03 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 109 | 229 | 7.146e-03 |
| GO:BP | GO:1900542 | regulation of purine nucleotide metabolic process | 46 | 84 | 7.233e-03 |
| GO:BP | GO:0032211 | negative regulation of telomere maintenance via telomerase | 16 | 22 | 7.256e-03 |
| GO:BP | GO:0035493 | SNARE complex assembly | 16 | 22 | 7.256e-03 |
| GO:BP | GO:0044794 | positive regulation by host of viral process | 16 | 22 | 7.256e-03 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 70 | 138 | 7.292e-03 |
| GO:BP | GO:0007040 | lysosome organization | 59 | 113 | 7.302e-03 |
| GO:BP | GO:0080171 | lytic vacuole organization | 59 | 113 | 7.302e-03 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 59 | 113 | 7.302e-03 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 100 | 208 | 7.520e-03 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 32 | 54 | 7.520e-03 |
| GO:BP | GO:0033121 | regulation of purine nucleotide catabolic process | 32 | 54 | 7.520e-03 |
| GO:BP | GO:0030811 | regulation of nucleotide catabolic process | 32 | 54 | 7.520e-03 |
| GO:BP | GO:0008584 | male gonad development | 76 | 152 | 7.614e-03 |
| GO:BP | GO:0009416 | response to light stimulus | 148 | 323 | 7.665e-03 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 94 | 194 | 7.691e-03 |
| GO:BP | GO:0030048 | actin filament-based movement | 69 | 136 | 7.778e-03 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 69 | 136 | 7.778e-03 |
| GO:BP | GO:0016925 | protein sumoylation | 39 | 69 | 7.778e-03 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 85 | 173 | 7.778e-03 |
| GO:BP | GO:0016126 | sterol biosynthetic process | 39 | 69 | 7.778e-03 |
| GO:BP | GO:0006066 | alcohol metabolic process | 164 | 362 | 7.791e-03 |
| GO:BP | GO:0051216 | cartilage development | 102 | 213 | 7.882e-03 |
| GO:BP | GO:0043122 | regulation of canonical NF-kappaB signal transduction | 133 | 287 | 7.885e-03 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 17 | 24 | 7.928e-03 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 17 | 24 | 7.928e-03 |
| GO:BP | GO:0007042 | lysosomal lumen acidification | 17 | 24 | 7.928e-03 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 30 | 50 | 8.084e-03 |
| GO:BP | GO:0014032 | neural crest cell development | 44 | 80 | 8.100e-03 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 96 | 199 | 8.100e-03 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 53 | 100 | 8.277e-03 |
| GO:BP | GO:0140467 | integrated stress response signaling | 29 | 48 | 8.369e-03 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 275 | 637 | 8.372e-03 |
| GO:BP | GO:1904507 | positive regulation of telomere maintenance in response to DNA damage | 12 | 15 | 8.436e-03 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 12 | 15 | 8.436e-03 |
| GO:BP | GO:0015937 | coenzyme A biosynthetic process | 9 | 10 | 8.436e-03 |
| GO:BP | GO:0031125 | rRNA 3’-end processing | 9 | 10 | 8.436e-03 |
| GO:BP | GO:0110025 | DNA strand resection involved in replication fork processing | 9 | 10 | 8.436e-03 |
| GO:BP | GO:0018216 | peptidyl-arginine methylation | 9 | 10 | 8.436e-03 |
| GO:BP | GO:0031167 | rRNA methylation | 18 | 26 | 8.464e-03 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 18 | 26 | 8.464e-03 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 236 | 540 | 8.494e-03 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 98 | 204 | 8.507e-03 |
| GO:BP | GO:0051057 | positive regulation of small GTPase mediated signal transduction | 43 | 78 | 8.569e-03 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 28 | 46 | 8.578e-03 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 111 | 235 | 8.768e-03 |
| GO:BP | GO:0043583 | ear development | 111 | 235 | 8.768e-03 |
| GO:BP | GO:2000300 | regulation of synaptic vesicle exocytosis | 27 | 44 | 8.811e-03 |
| GO:BP | GO:0060338 | regulation of type I interferon-mediated signaling pathway | 27 | 44 | 8.811e-03 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 27 | 44 | 8.811e-03 |
| GO:BP | GO:0009134 | nucleoside diphosphate catabolic process | 56 | 107 | 8.833e-03 |
| GO:BP | GO:0032801 | receptor catabolic process | 19 | 28 | 8.840e-03 |
| GO:BP | GO:0003272 | endocardial cushion formation | 19 | 28 | 8.840e-03 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 19 | 28 | 8.840e-03 |
| GO:BP | GO:0006476 | protein deacetylation | 26 | 42 | 8.998e-03 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 26 | 42 | 8.998e-03 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 26 | 42 | 8.998e-03 |
| GO:BP | GO:0046048 | UDP metabolic process | 7 | 7 | 9.032e-03 |
| GO:BP | GO:0007221 | positive regulation of transcription of Notch receptor target | 7 | 7 | 9.032e-03 |
| GO:BP | GO:2000638 | regulation of SREBP signaling pathway | 7 | 7 | 9.032e-03 |
| GO:BP | GO:0045054 | constitutive secretory pathway | 7 | 7 | 9.032e-03 |
| GO:BP | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process | 7 | 7 | 9.032e-03 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 7 | 7 | 9.032e-03 |
| GO:BP | GO:0035891 | exit from host cell | 20 | 30 | 9.072e-03 |
| GO:BP | GO:0008535 | respiratory chain complex IV assembly | 20 | 30 | 9.072e-03 |
| GO:BP | GO:0019076 | viral release from host cell | 20 | 30 | 9.072e-03 |
| GO:BP | GO:0046660 | female sex differentiation | 63 | 123 | 9.072e-03 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 25 | 40 | 9.112e-03 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 25 | 40 | 9.112e-03 |
| GO:BP | GO:0032535 | regulation of cellular component size | 161 | 356 | 9.162e-03 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 24 | 38 | 9.206e-03 |
| GO:BP | GO:0071108 | protein K48-linked deubiquitination | 21 | 32 | 9.206e-03 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 24 | 38 | 9.206e-03 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 24 | 38 | 9.206e-03 |
| GO:BP | GO:1903955 | positive regulation of protein targeting to mitochondrion | 21 | 32 | 9.206e-03 |
| GO:BP | GO:2000279 | negative regulation of DNA biosynthetic process | 21 | 32 | 9.206e-03 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 76 | 153 | 9.240e-03 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 76 | 153 | 9.240e-03 |
| GO:BP | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion | 23 | 36 | 9.262e-03 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 22 | 34 | 9.262e-03 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 82 | 167 | 9.321e-03 |
| GO:BP | GO:0071709 | membrane assembly | 35 | 61 | 9.396e-03 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 41 | 74 | 9.498e-03 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 41 | 74 | 9.498e-03 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 266 | 616 | 9.520e-03 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 262 | 606 | 9.520e-03 |
| GO:BP | GO:0006140 | regulation of nucleotide metabolic process | 46 | 85 | 9.528e-03 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 69 | 137 | 9.555e-03 |
| GO:BP | GO:0006906 | vesicle fusion | 62 | 121 | 9.626e-03 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 34 | 59 | 9.865e-03 |
| GO:BP | GO:0032608 | interferon-beta production | 34 | 59 | 9.865e-03 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 75 | 151 | 9.865e-03 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 78 | 158 | 9.922e-03 |
| GO:BP | GO:0007029 | endoplasmic reticulum organization | 54 | 103 | 9.922e-03 |
| GO:BP | GO:0009191 | ribonucleoside diphosphate catabolic process | 54 | 103 | 9.922e-03 |
| GO:BP | GO:0002181 | cytoplasmic translation | 81 | 165 | 9.941e-03 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 40 | 72 | 1.004e-02 |
| GO:BP | GO:0038034 | signal transduction in absence of ligand | 40 | 72 | 1.004e-02 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 45 | 83 | 1.013e-02 |
| GO:BP | GO:0060382 | regulation of DNA strand elongation | 13 | 17 | 1.018e-02 |
| GO:BP | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway | 13 | 17 | 1.018e-02 |
| GO:BP | GO:0051382 | kinetochore assembly | 13 | 17 | 1.018e-02 |
| GO:BP | GO:0070131 | positive regulation of mitochondrial translation | 13 | 17 | 1.018e-02 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 33 | 57 | 1.031e-02 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 33 | 57 | 1.031e-02 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 33 | 57 | 1.031e-02 |
| GO:BP | GO:0007623 | circadian rhythm | 100 | 210 | 1.047e-02 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 219 | 500 | 1.058e-02 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 53 | 101 | 1.058e-02 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 39 | 70 | 1.062e-02 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 105 | 222 | 1.069e-02 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 70 | 140 | 1.116e-02 |
| GO:BP | GO:0007422 | peripheral nervous system development | 48 | 90 | 1.120e-02 |
| GO:BP | GO:0051651 | maintenance of location in cell | 99 | 208 | 1.121e-02 |
| GO:BP | GO:0043149 | stress fiber assembly | 56 | 108 | 1.124e-02 |
| GO:BP | GO:0035282 | segmentation | 56 | 108 | 1.124e-02 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 56 | 108 | 1.124e-02 |
| GO:BP | GO:0051646 | mitochondrion localization | 31 | 53 | 1.134e-02 |
| GO:BP | GO:0030100 | regulation of endocytosis | 136 | 297 | 1.163e-02 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 101 | 213 | 1.171e-02 |
| GO:BP | GO:0009746 | response to hexose | 101 | 213 | 1.171e-02 |
| GO:BP | GO:0035720 | intraciliary anterograde transport | 14 | 19 | 1.179e-02 |
| GO:BP | GO:0021692 | cerebellar Purkinje cell layer morphogenesis | 14 | 19 | 1.179e-02 |
| GO:BP | GO:0031053 | primary miRNA processing | 14 | 19 | 1.179e-02 |
| GO:BP | GO:0097320 | plasma membrane tubulation | 14 | 19 | 1.179e-02 |
| GO:BP | GO:0046579 | positive regulation of Ras protein signal transduction | 14 | 19 | 1.179e-02 |
| GO:BP | GO:0051051 | negative regulation of transport | 183 | 412 | 1.184e-02 |
| GO:BP | GO:0001892 | embryonic placenta development | 47 | 88 | 1.190e-02 |
| GO:BP | GO:0014896 | muscle hypertrophy | 47 | 88 | 1.190e-02 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 69 | 138 | 1.190e-02 |
| GO:BP | GO:0051235 | maintenance of location | 103 | 218 | 1.218e-02 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 29 | 49 | 1.235e-02 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 29 | 49 | 1.235e-02 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 118 | 254 | 1.238e-02 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 92 | 192 | 1.240e-02 |
| GO:BP | GO:0061723 | glycophagy | 10 | 12 | 1.267e-02 |
| GO:BP | GO:0003207 | cardiac chamber formation | 10 | 12 | 1.267e-02 |
| GO:BP | GO:0072710 | response to hydroxyurea | 10 | 12 | 1.267e-02 |
| GO:BP | GO:1903671 | negative regulation of sprouting angiogenesis | 10 | 12 | 1.267e-02 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 10 | 12 | 1.267e-02 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 10 | 12 | 1.267e-02 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 10 | 12 | 1.267e-02 |
| GO:BP | GO:0086067 | AV node cell to bundle of His cell communication | 10 | 12 | 1.267e-02 |
| GO:BP | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase | 10 | 12 | 1.267e-02 |
| GO:BP | GO:1903441 | protein localization to ciliary membrane | 10 | 12 | 1.267e-02 |
| GO:BP | GO:0047496 | vesicle transport along microtubule | 28 | 47 | 1.279e-02 |
| GO:BP | GO:0046031 | ADP metabolic process | 54 | 104 | 1.279e-02 |
| GO:BP | GO:0032365 | intracellular lipid transport | 28 | 47 | 1.279e-02 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 50 | 95 | 1.284e-02 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 41 | 75 | 1.289e-02 |
| GO:BP | GO:0007369 | gastrulation | 94 | 197 | 1.293e-02 |
| GO:BP | GO:0099188 | postsynaptic cytoskeleton organization | 15 | 21 | 1.299e-02 |
| GO:BP | GO:0006611 | protein export from nucleus | 35 | 62 | 1.326e-02 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 27 | 45 | 1.328e-02 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 88 | 183 | 1.340e-02 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 45 | 84 | 1.350e-02 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 85 | 176 | 1.358e-02 |
| GO:BP | GO:0072164 | mesonephric tubule development | 53 | 102 | 1.365e-02 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 53 | 102 | 1.365e-02 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 53 | 102 | 1.365e-02 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 40 | 73 | 1.369e-02 |
| GO:BP | GO:1904888 | cranial skeletal system development | 40 | 73 | 1.369e-02 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 49 | 93 | 1.369e-02 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 26 | 43 | 1.370e-02 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 60 | 118 | 1.386e-02 |
| GO:BP | GO:0006740 | NADPH regeneration | 16 | 23 | 1.388e-02 |
| GO:BP | GO:0032401 | establishment of melanosome localization | 16 | 23 | 1.388e-02 |
| GO:BP | GO:0045540 | regulation of cholesterol biosynthetic process | 16 | 23 | 1.388e-02 |
| GO:BP | GO:0003283 | atrial septum development | 16 | 23 | 1.388e-02 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 16 | 23 | 1.388e-02 |
| GO:BP | GO:0000963 | mitochondrial RNA processing | 16 | 23 | 1.388e-02 |
| GO:BP | GO:0106118 | regulation of sterol biosynthetic process | 16 | 23 | 1.388e-02 |
| GO:BP | GO:0051302 | regulation of cell division | 90 | 188 | 1.400e-02 |
| GO:BP | GO:0006739 | NADP metabolic process | 25 | 41 | 1.410e-02 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 123 | 267 | 1.417e-02 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 44 | 82 | 1.431e-02 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 17 | 25 | 1.443e-02 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 24 | 39 | 1.443e-02 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 39 | 71 | 1.443e-02 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 17 | 25 | 1.443e-02 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 17 | 25 | 1.443e-02 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 17 | 25 | 1.443e-02 |
| GO:BP | GO:0007031 | peroxisome organization | 24 | 39 | 1.443e-02 |
| GO:BP | GO:0009137 | purine nucleoside diphosphate catabolic process | 52 | 100 | 1.443e-02 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 24 | 39 | 1.443e-02 |
| GO:BP | GO:0035994 | response to muscle stretch | 17 | 25 | 1.443e-02 |
| GO:BP | GO:0032400 | melanosome localization | 17 | 25 | 1.443e-02 |
| GO:BP | GO:0009181 | purine ribonucleoside diphosphate catabolic process | 52 | 100 | 1.443e-02 |
| GO:BP | GO:0046677 | response to antibiotic | 24 | 39 | 1.443e-02 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 24 | 39 | 1.443e-02 |
| GO:BP | GO:0055088 | lipid homeostasis | 81 | 167 | 1.443e-02 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 194 | 441 | 1.443e-02 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 24 | 39 | 1.443e-02 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 24 | 39 | 1.443e-02 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 17 | 25 | 1.443e-02 |
| GO:BP | GO:0071396 | cellular response to lipid | 263 | 613 | 1.446e-02 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 78 | 160 | 1.452e-02 |
| GO:BP | GO:0048278 | vesicle docking | 33 | 58 | 1.453e-02 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 72 | 146 | 1.454e-02 |
| GO:BP | GO:0016233 | telomere capping | 18 | 27 | 1.486e-02 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 18 | 27 | 1.486e-02 |
| GO:BP | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 22 | 35 | 1.492e-02 |
| GO:BP | GO:0046320 | regulation of fatty acid oxidation | 22 | 35 | 1.492e-02 |
| GO:BP | GO:0060384 | innervation | 19 | 29 | 1.501e-02 |
| GO:BP | GO:0002063 | chondrocyte development | 21 | 33 | 1.501e-02 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 21 | 33 | 1.501e-02 |
| GO:BP | GO:0021680 | cerebellar Purkinje cell layer development | 19 | 29 | 1.501e-02 |
| GO:BP | GO:1902473 | regulation of protein localization to synapse | 19 | 29 | 1.501e-02 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 19 | 29 | 1.501e-02 |
| GO:BP | GO:0051642 | centrosome localization | 21 | 33 | 1.501e-02 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 19 | 29 | 1.501e-02 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 55 | 107 | 1.501e-02 |
| GO:BP | GO:0009218 | pyrimidine ribonucleotide metabolic process | 21 | 33 | 1.501e-02 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 21 | 33 | 1.501e-02 |
| GO:BP | GO:0009165 | nucleotide biosynthetic process | 107 | 229 | 1.502e-02 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 20 | 31 | 1.503e-02 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 20 | 31 | 1.503e-02 |
| GO:BP | GO:0098868 | bone growth | 20 | 31 | 1.503e-02 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 20 | 31 | 1.503e-02 |
| GO:BP | GO:0002204 | somatic recombination of immunoglobulin genes involved in immune response | 32 | 56 | 1.512e-02 |
| GO:BP | GO:0002208 | somatic diversification of immunoglobulins involved in immune response | 32 | 56 | 1.512e-02 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 32 | 56 | 1.512e-02 |
| GO:BP | GO:0045190 | isotype switching | 32 | 56 | 1.512e-02 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 32 | 56 | 1.512e-02 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 32 | 56 | 1.512e-02 |
| GO:BP | GO:0006473 | protein acetylation | 32 | 56 | 1.512e-02 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 32 | 56 | 1.512e-02 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 80 | 165 | 1.522e-02 |
| GO:BP | GO:0006470 | protein dephosphorylation | 71 | 144 | 1.538e-02 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 74 | 151 | 1.538e-02 |
| GO:BP | GO:0043393 | regulation of protein binding | 58 | 114 | 1.545e-02 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 61 | 121 | 1.591e-02 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 42 | 78 | 1.591e-02 |
| GO:BP | GO:0060324 | face development | 31 | 54 | 1.593e-02 |
| GO:BP | GO:0021695 | cerebellar cortex development | 31 | 54 | 1.593e-02 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 31 | 54 | 1.593e-02 |
| GO:BP | GO:0098734 | macromolecule depalmitoylation | 11 | 14 | 1.600e-02 |
| GO:BP | GO:0051775 | response to redox state | 11 | 14 | 1.600e-02 |
| GO:BP | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation | 11 | 14 | 1.600e-02 |
| GO:BP | GO:1901985 | positive regulation of protein acetylation | 11 | 14 | 1.600e-02 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 82 | 170 | 1.606e-02 |
| GO:BP | GO:0030282 | bone mineralization | 64 | 128 | 1.614e-02 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 46 | 87 | 1.621e-02 |
| GO:BP | GO:0061572 | actin filament bundle organization | 79 | 163 | 1.623e-02 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 57 | 112 | 1.652e-02 |
| GO:BP | GO:0003197 | endocardial cushion development | 30 | 52 | 1.673e-02 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 30 | 52 | 1.673e-02 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 41 | 76 | 1.692e-02 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 36 | 65 | 1.693e-02 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 36 | 65 | 1.693e-02 |
| GO:BP | GO:0061053 | somite development | 45 | 85 | 1.735e-02 |
| GO:BP | GO:0060021 | roof of mouth development | 49 | 94 | 1.735e-02 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 138 | 305 | 1.736e-02 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 112 | 242 | 1.740e-02 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 112 | 242 | 1.740e-02 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 66 | 133 | 1.748e-02 |
| GO:BP | GO:0072073 | kidney epithelium development | 75 | 154 | 1.751e-02 |
| GO:BP | GO:0043622 | cortical microtubule organization | 8 | 9 | 1.763e-02 |
| GO:BP | GO:0031946 | regulation of glucocorticoid biosynthetic process | 8 | 9 | 1.763e-02 |
| GO:BP | GO:0062176 | R-loop processing | 8 | 9 | 1.763e-02 |
| GO:BP | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein | 8 | 9 | 1.763e-02 |
| GO:BP | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process | 8 | 9 | 1.763e-02 |
| GO:BP | GO:0140673 | transcription elongation-coupled chromatin remodeling | 8 | 9 | 1.763e-02 |
| GO:BP | GO:0097680 | double-strand break repair via classical nonhomologous end joining | 8 | 9 | 1.763e-02 |
| GO:BP | GO:1904874 | positive regulation of telomerase RNA localization to Cajal body | 8 | 9 | 1.763e-02 |
| GO:BP | GO:0009200 | deoxyribonucleoside triphosphate metabolic process | 8 | 9 | 1.763e-02 |
| GO:BP | GO:0046602 | regulation of mitotic centrosome separation | 8 | 9 | 1.763e-02 |
| GO:BP | GO:0001657 | ureteric bud development | 52 | 101 | 1.818e-02 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 28 | 48 | 1.835e-02 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 28 | 48 | 1.835e-02 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 28 | 48 | 1.835e-02 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 28 | 48 | 1.835e-02 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 44 | 83 | 1.841e-02 |
| GO:BP | GO:0072028 | nephron morphogenesis | 44 | 83 | 1.841e-02 |
| GO:BP | GO:0051899 | membrane depolarization | 48 | 92 | 1.844e-02 |
| GO:BP | GO:0001659 | temperature homeostasis | 88 | 185 | 1.852e-02 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 118 | 257 | 1.872e-02 |
| GO:BP | GO:0048313 | Golgi inheritance | 12 | 16 | 1.884e-02 |
| GO:BP | GO:0048308 | organelle inheritance | 12 | 16 | 1.884e-02 |
| GO:BP | GO:0043328 | protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 12 | 16 | 1.884e-02 |
| GO:BP | GO:0030242 | autophagy of peroxisome | 12 | 16 | 1.884e-02 |
| GO:BP | GO:0051299 | centrosome separation | 12 | 16 | 1.884e-02 |
| GO:BP | GO:1903358 | regulation of Golgi organization | 12 | 16 | 1.884e-02 |
| GO:BP | GO:0098693 | regulation of synaptic vesicle cycle | 12 | 16 | 1.884e-02 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 12 | 16 | 1.884e-02 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 12 | 16 | 1.884e-02 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 39 | 72 | 1.903e-02 |
| GO:BP | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | 27 | 46 | 1.911e-02 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 27 | 46 | 1.911e-02 |
| GO:BP | GO:0023061 | signal release | 217 | 501 | 1.913e-02 |
| GO:BP | GO:0055085 | transmembrane transport | 639 | 1578 | 1.914e-02 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 79 | 164 | 1.945e-02 |
| GO:BP | GO:0030323 | respiratory tube development | 95 | 202 | 1.952e-02 |
| GO:BP | GO:0097581 | lamellipodium organization | 47 | 90 | 1.964e-02 |
| GO:BP | GO:0008361 | regulation of cell size | 87 | 183 | 1.969e-02 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 64 | 129 | 1.986e-02 |
| GO:BP | GO:0034205 | amyloid-beta formation | 26 | 44 | 1.997e-02 |
| GO:BP | GO:0061462 | protein localization to lysosome | 33 | 59 | 2.000e-02 |
| GO:BP | GO:0061326 | renal tubule development | 54 | 106 | 2.006e-02 |
| GO:BP | GO:0001823 | mesonephros development | 54 | 106 | 2.006e-02 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 84 | 176 | 2.008e-02 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 38 | 70 | 2.021e-02 |
| GO:BP | GO:0048645 | animal organ formation | 38 | 70 | 2.021e-02 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 38 | 70 | 2.021e-02 |
| GO:BP | GO:0002064 | epithelial cell development | 102 | 219 | 2.024e-02 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 50 | 97 | 2.054e-02 |
| GO:BP | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration | 6 | 6 | 2.054e-02 |
| GO:BP | GO:0046032 | ADP catabolic process | 50 | 97 | 2.054e-02 |
| GO:BP | GO:0031999 | negative regulation of fatty acid beta-oxidation | 6 | 6 | 2.054e-02 |
| GO:BP | GO:0014031 | mesenchymal cell development | 6 | 6 | 2.054e-02 |
| GO:BP | GO:1904760 | regulation of myofibroblast differentiation | 6 | 6 | 2.054e-02 |
| GO:BP | GO:1901858 | regulation of mitochondrial DNA metabolic process | 6 | 6 | 2.054e-02 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 50 | 97 | 2.054e-02 |
| GO:BP | GO:0015961 | diadenosine polyphosphate catabolic process | 6 | 6 | 2.054e-02 |
| GO:BP | GO:0046929 | negative regulation of neurotransmitter secretion | 6 | 6 | 2.054e-02 |
| GO:BP | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA | 6 | 6 | 2.054e-02 |
| GO:BP | GO:0003415 | chondrocyte hypertrophy | 6 | 6 | 2.054e-02 |
| GO:BP | GO:0042791 | 5S class rRNA transcription by RNA polymerase III | 6 | 6 | 2.054e-02 |
| GO:BP | GO:0046368 | GDP-L-fucose metabolic process | 6 | 6 | 2.054e-02 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 25 | 42 | 2.062e-02 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 13 | 18 | 2.090e-02 |
| GO:BP | GO:0034727 | piecemeal microautophagy of the nucleus | 13 | 18 | 2.090e-02 |
| GO:BP | GO:0015936 | coenzyme A metabolic process | 13 | 18 | 2.090e-02 |
| GO:BP | GO:0007096 | regulation of exit from mitosis | 13 | 18 | 2.090e-02 |
| GO:BP | GO:0072087 | renal vesicle development | 13 | 18 | 2.090e-02 |
| GO:BP | GO:0032239 | regulation of nucleobase-containing compound transport | 13 | 18 | 2.090e-02 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 66 | 134 | 2.115e-02 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 24 | 40 | 2.140e-02 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 24 | 40 | 2.140e-02 |
| GO:BP | GO:0070266 | necroptotic process | 24 | 40 | 2.140e-02 |
| GO:BP | GO:0006096 | glycolytic process | 49 | 95 | 2.196e-02 |
| GO:BP | GO:0030324 | lung development | 93 | 198 | 2.202e-02 |
| GO:BP | GO:0048286 | lung alveolus development | 31 | 55 | 2.219e-02 |
| GO:BP | GO:0045023 | G0 to G1 transition | 23 | 38 | 2.219e-02 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 31 | 55 | 2.219e-02 |
| GO:BP | GO:0046473 | phosphatidic acid metabolic process | 23 | 38 | 2.219e-02 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 31 | 55 | 2.219e-02 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 110 | 239 | 2.250e-02 |
| GO:BP | GO:0042407 | cristae formation | 14 | 20 | 2.251e-02 |
| GO:BP | GO:0061003 | positive regulation of dendritic spine morphogenesis | 14 | 20 | 2.251e-02 |
| GO:BP | GO:0010866 | regulation of triglyceride biosynthetic process | 14 | 20 | 2.251e-02 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 14 | 20 | 2.251e-02 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 14 | 20 | 2.251e-02 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 68 | 139 | 2.257e-02 |
| GO:BP | GO:0043604 | amide biosynthetic process | 82 | 172 | 2.266e-02 |
| GO:BP | GO:0006904 | vesicle docking involved in exocytosis | 22 | 36 | 2.285e-02 |
| GO:BP | GO:0070316 | regulation of G0 to G1 transition | 22 | 36 | 2.285e-02 |
| GO:BP | GO:0002218 | activation of innate immune response | 137 | 305 | 2.341e-02 |
| GO:BP | GO:0060914 | heart formation | 21 | 34 | 2.341e-02 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 21 | 34 | 2.341e-02 |
| GO:BP | GO:0034405 | response to fluid shear stress | 21 | 34 | 2.341e-02 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 21 | 34 | 2.341e-02 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 21 | 34 | 2.341e-02 |
| GO:BP | GO:0051654 | establishment of mitochondrion localization | 21 | 34 | 2.341e-02 |
| GO:BP | GO:0006734 | NADH metabolic process | 21 | 34 | 2.341e-02 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 21 | 34 | 2.341e-02 |
| GO:BP | GO:0086009 | membrane repolarization | 30 | 53 | 2.341e-02 |
| GO:BP | GO:0019083 | viral transcription | 30 | 53 | 2.341e-02 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 161 | 364 | 2.343e-02 |
| GO:BP | GO:0032402 | melanosome transport | 15 | 22 | 2.343e-02 |
| GO:BP | GO:0009083 | branched-chain amino acid catabolic process | 15 | 22 | 2.343e-02 |
| GO:BP | GO:0016082 | synaptic vesicle priming | 15 | 22 | 2.343e-02 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 15 | 22 | 2.343e-02 |
| GO:BP | GO:0000212 | meiotic spindle organization | 15 | 22 | 2.343e-02 |
| GO:BP | GO:0021756 | striatum development | 15 | 22 | 2.343e-02 |
| GO:BP | GO:0008090 | retrograde axonal transport | 15 | 22 | 2.343e-02 |
| GO:BP | GO:0003228 | atrial cardiac muscle tissue development | 15 | 22 | 2.343e-02 |
| GO:BP | GO:0005977 | glycogen metabolic process | 44 | 84 | 2.352e-02 |
| GO:BP | GO:0006970 | response to osmotic stress | 44 | 84 | 2.352e-02 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 20 | 32 | 2.375e-02 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 20 | 32 | 2.375e-02 |
| GO:BP | GO:0009306 | protein secretion | 167 | 379 | 2.375e-02 |
| GO:BP | GO:0099022 | vesicle tethering | 20 | 32 | 2.375e-02 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 20 | 32 | 2.375e-02 |
| GO:BP | GO:0035751 | regulation of lysosomal lumen pH | 20 | 32 | 2.375e-02 |
| GO:BP | GO:0060966 | regulation of gene silencing by regulatory ncRNA | 20 | 32 | 2.375e-02 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 16 | 24 | 2.392e-02 |
| GO:BP | GO:1901673 | regulation of mitotic spindle assembly | 16 | 24 | 2.392e-02 |
| GO:BP | GO:0051905 | establishment of pigment granule localization | 16 | 24 | 2.392e-02 |
| GO:BP | GO:1902993 | positive regulation of amyloid precursor protein catabolic process | 16 | 24 | 2.392e-02 |
| GO:BP | GO:0009147 | pyrimidine nucleoside triphosphate metabolic process | 16 | 24 | 2.392e-02 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 16 | 24 | 2.392e-02 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 16 | 24 | 2.392e-02 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 16 | 24 | 2.392e-02 |
| GO:BP | GO:0009266 | response to temperature stimulus | 81 | 170 | 2.392e-02 |
| GO:BP | GO:0060993 | kidney morphogenesis | 51 | 100 | 2.396e-02 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by regulatory ncRNA | 19 | 30 | 2.402e-02 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 19 | 30 | 2.402e-02 |
| GO:BP | GO:0042063 | gliogenesis | 156 | 352 | 2.405e-02 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 17 | 26 | 2.408e-02 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 17 | 26 | 2.408e-02 |
| GO:BP | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated | 18 | 28 | 2.408e-02 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 18 | 28 | 2.408e-02 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 18 | 28 | 2.408e-02 |
| GO:BP | GO:0033617 | mitochondrial cytochrome c oxidase assembly | 17 | 26 | 2.408e-02 |
| GO:BP | GO:0009081 | branched-chain amino acid metabolic process | 18 | 28 | 2.408e-02 |
| GO:BP | GO:0003416 | endochondral bone growth | 18 | 28 | 2.408e-02 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 17 | 26 | 2.408e-02 |
| GO:BP | GO:0047497 | mitochondrion transport along microtubule | 18 | 28 | 2.408e-02 |
| GO:BP | GO:0051875 | pigment granule localization | 17 | 26 | 2.408e-02 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 29 | 51 | 2.419e-02 |
| GO:BP | GO:0062028 | regulation of stress granule assembly | 9 | 11 | 2.432e-02 |
| GO:BP | GO:0071028 | nuclear mRNA surveillance | 9 | 11 | 2.432e-02 |
| GO:BP | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA | 9 | 11 | 2.432e-02 |
| GO:BP | GO:0030953 | astral microtubule organization | 9 | 11 | 2.432e-02 |
| GO:BP | GO:0031943 | regulation of glucocorticoid metabolic process | 9 | 11 | 2.432e-02 |
| GO:BP | GO:0010807 | regulation of synaptic vesicle priming | 9 | 11 | 2.432e-02 |
| GO:BP | GO:0016311 | dephosphorylation | 101 | 218 | 2.432e-02 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 9 | 11 | 2.432e-02 |
| GO:BP | GO:0046710 | GDP metabolic process | 9 | 11 | 2.432e-02 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 9 | 11 | 2.432e-02 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 9 | 11 | 2.432e-02 |
| GO:BP | GO:0046834 | lipid phosphorylation | 9 | 11 | 2.432e-02 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 54 | 107 | 2.433e-02 |
| GO:BP | GO:0040011 | locomotion | 512 | 1255 | 2.456e-02 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 43 | 82 | 2.467e-02 |
| GO:BP | GO:0045088 | regulation of innate immune response | 195 | 449 | 2.468e-02 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 57 | 114 | 2.478e-02 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 168 | 382 | 2.479e-02 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 34 | 62 | 2.495e-02 |
| GO:BP | GO:0009161 | ribonucleoside monophosphate metabolic process | 34 | 62 | 2.495e-02 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 28 | 49 | 2.536e-02 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 28 | 49 | 2.536e-02 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 28 | 49 | 2.536e-02 |
| GO:BP | GO:0050890 | cognition | 146 | 328 | 2.562e-02 |
| GO:BP | GO:0009743 | response to carbohydrate | 112 | 245 | 2.584e-02 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 53 | 105 | 2.607e-02 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 46 | 89 | 2.607e-02 |
| GO:BP | GO:0000423 | mitophagy | 38 | 71 | 2.608e-02 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 42 | 80 | 2.637e-02 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 42 | 80 | 2.637e-02 |
| GO:BP | GO:0043124 | negative regulation of canonical NF-kappaB signal transduction | 33 | 60 | 2.657e-02 |
| GO:BP | GO:0008360 | regulation of cell shape | 68 | 140 | 2.672e-02 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 27 | 47 | 2.675e-02 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 49 | 96 | 2.716e-02 |
| GO:BP | GO:0007272 | ensheathment of neurons | 76 | 159 | 2.742e-02 |
| GO:BP | GO:0008366 | axon ensheathment | 76 | 159 | 2.742e-02 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 45 | 87 | 2.790e-02 |
| GO:BP | GO:0006879 | intracellular iron ion homeostasis | 37 | 69 | 2.790e-02 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 52 | 103 | 2.790e-02 |
| GO:BP | GO:2000177 | regulation of neural precursor cell proliferation | 52 | 103 | 2.790e-02 |
| GO:BP | GO:0060349 | bone morphogenesis | 52 | 103 | 2.790e-02 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 73 | 152 | 2.790e-02 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 394 | 954 | 2.790e-02 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 26 | 45 | 2.814e-02 |
| GO:BP | GO:0018279 | protein N-linked glycosylation via asparagine | 26 | 45 | 2.814e-02 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 41 | 78 | 2.817e-02 |
| GO:BP | GO:0001101 | response to acid chemical | 70 | 145 | 2.822e-02 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 70 | 145 | 2.822e-02 |
| GO:BP | GO:0098739 | import across plasma membrane | 91 | 195 | 2.849e-02 |
| GO:BP | GO:0043200 | response to amino acid | 64 | 131 | 2.875e-02 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 192 | 443 | 2.875e-02 |
| GO:BP | GO:0031347 | regulation of defense response | 337 | 809 | 2.892e-02 |
| GO:BP | GO:0042552 | myelination | 75 | 157 | 2.925e-02 |
| GO:BP | GO:0007595 | lactation | 25 | 43 | 2.953e-02 |
| GO:BP | GO:0072528 | pyrimidine-containing compound biosynthetic process | 25 | 43 | 2.953e-02 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 25 | 43 | 2.953e-02 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 25 | 43 | 2.953e-02 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 25 | 43 | 2.953e-02 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 36 | 67 | 2.966e-02 |
| GO:BP | GO:0061668 | mitochondrial ribosome assembly | 10 | 13 | 2.997e-02 |
| GO:BP | GO:0006054 | N-acetylneuraminate metabolic process | 10 | 13 | 2.997e-02 |
| GO:BP | GO:0009304 | tRNA transcription | 10 | 13 | 2.997e-02 |
| GO:BP | GO:0006264 | mitochondrial DNA replication | 10 | 13 | 2.997e-02 |
| GO:BP | GO:0061795 | Golgi lumen acidification | 10 | 13 | 2.997e-02 |
| GO:BP | GO:0010992 | ubiquitin recycling | 10 | 13 | 2.997e-02 |
| GO:BP | GO:0034982 | mitochondrial protein processing | 10 | 13 | 2.997e-02 |
| GO:BP | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation | 10 | 13 | 2.997e-02 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 54 | 108 | 3.032e-02 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 164 | 374 | 3.063e-02 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 57 | 115 | 3.063e-02 |
| GO:BP | GO:0043473 | pigmentation | 57 | 115 | 3.063e-02 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 63 | 129 | 3.064e-02 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 60 | 122 | 3.070e-02 |
| GO:BP | GO:0051046 | regulation of secretion | 261 | 617 | 3.070e-02 |
| GO:BP | GO:0033260 | nuclear DNA replication | 24 | 41 | 3.078e-02 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 24 | 41 | 3.078e-02 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 24 | 41 | 3.078e-02 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 24 | 41 | 3.078e-02 |
| GO:BP | GO:0072210 | metanephric nephron development | 24 | 41 | 3.078e-02 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 24 | 41 | 3.078e-02 |
| GO:BP | GO:0009791 | post-embryonic development | 47 | 92 | 3.086e-02 |
| GO:BP | GO:0034605 | cellular response to heat | 35 | 65 | 3.149e-02 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 30 | 54 | 3.161e-02 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 71 | 148 | 3.161e-02 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 30 | 54 | 3.161e-02 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 30 | 54 | 3.161e-02 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 43 | 83 | 3.170e-02 |
| GO:BP | GO:0043648 | dicarboxylic acid metabolic process | 50 | 99 | 3.174e-02 |
| GO:BP | GO:0007616 | long-term memory | 23 | 39 | 3.225e-02 |
| GO:BP | GO:0035264 | multicellular organism growth | 76 | 160 | 3.247e-02 |
| GO:BP | GO:0034284 | response to monosaccharide | 101 | 220 | 3.255e-02 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 81 | 172 | 3.299e-02 |
| GO:BP | GO:0048592 | eye morphogenesis | 81 | 172 | 3.299e-02 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 137 | 308 | 3.307e-02 |
| GO:BP | GO:0010171 | body morphogenesis | 29 | 52 | 3.356e-02 |
| GO:BP | GO:0031648 | protein destabilization | 29 | 52 | 3.356e-02 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 34 | 63 | 3.356e-02 |
| GO:BP | GO:0042255 | ribosome assembly | 34 | 63 | 3.356e-02 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 34 | 63 | 3.356e-02 |
| GO:BP | GO:0010001 | glial cell differentiation | 119 | 264 | 3.359e-02 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 22 | 37 | 3.363e-02 |
| GO:BP | GO:0060972 | left/right pattern formation | 70 | 146 | 3.374e-02 |
| GO:BP | GO:0090737 | telomere maintenance via telomere trimming | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0090656 | t-circle formation | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0008298 | intracellular mRNA localization | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0046831 | regulation of RNA export from nucleus | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0140042 | lipid droplet formation | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0050708 | regulation of protein secretion | 121 | 269 | 3.376e-02 |
| GO:BP | GO:0033750 | ribosome localization | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0043248 | proteasome assembly | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 42 | 81 | 3.376e-02 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0070986 | left/right axis specification | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0071027 | nuclear RNA surveillance | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0000054 | ribosomal subunit export from nucleus | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0001325 | formation of extrachromosomal circular DNA | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0070243 | regulation of thymocyte apoptotic process | 11 | 15 | 3.376e-02 |
| GO:BP | GO:0007610 | behavior | 283 | 674 | 3.387e-02 |
| GO:BP | GO:0008585 | female gonad development | 52 | 104 | 3.434e-02 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 21 | 35 | 3.474e-02 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 21 | 35 | 3.474e-02 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 33 | 61 | 3.552e-02 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 33 | 61 | 3.552e-02 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 33 | 61 | 3.552e-02 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 169 | 388 | 3.608e-02 |
| GO:BP | GO:0009123 | nucleoside monophosphate metabolic process | 41 | 79 | 3.609e-02 |
| GO:BP | GO:1904354 | negative regulation of telomere capping | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0061511 | centriole elongation | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0007172 | signal complex assembly | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0003149 | membranous septum morphogenesis | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0060509 | type I pneumocyte differentiation | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0006384 | transcription initiation at RNA polymerase III promoter | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0006290 | pyrimidine dimer repair | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 7 | 8 | 3.613e-02 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0003344 | pericardium morphogenesis | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0003211 | cardiac ventricle formation | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0072033 | renal vesicle formation | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0051086 | chaperone mediated protein folding independent of cofactor | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0071051 | poly(A)-dependent snoRNA 3’-end processing | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0034475 | U4 snRNA 3’-end processing | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0015959 | diadenosine polyphosphate metabolic process | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0150012 | positive regulation of neuron projection arborization | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0031077 | post-embryonic camera-type eye development | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0018202 | peptidyl-histidine modification | 7 | 8 | 3.613e-02 |
| GO:BP | GO:0043117 | positive regulation of vascular permeability | 12 | 17 | 3.631e-02 |
| GO:BP | GO:0090205 | positive regulation of cholesterol metabolic process | 12 | 17 | 3.631e-02 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 12 | 17 | 3.631e-02 |
| GO:BP | GO:0006991 | response to sterol depletion | 12 | 17 | 3.631e-02 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 12 | 17 | 3.631e-02 |
| GO:BP | GO:0033993 | response to lipid | 383 | 930 | 3.631e-02 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 12 | 17 | 3.631e-02 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 51 | 102 | 3.641e-02 |
| GO:BP | GO:0072080 | nephron tubule development | 51 | 102 | 3.641e-02 |
| GO:BP | GO:0051085 | chaperone cofactor-dependent protein refolding | 19 | 31 | 3.683e-02 |
| GO:BP | GO:0016226 | iron-sulfur cluster assembly | 19 | 31 | 3.683e-02 |
| GO:BP | GO:0070498 | interleukin-1-mediated signaling pathway | 19 | 31 | 3.683e-02 |
| GO:BP | GO:0031163 | metallo-sulfur cluster assembly | 19 | 31 | 3.683e-02 |
| GO:BP | GO:0045747 | positive regulation of Notch signaling pathway | 27 | 48 | 3.699e-02 |
| GO:BP | GO:0044042 | glucan metabolic process | 44 | 86 | 3.719e-02 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 18 | 29 | 3.786e-02 |
| GO:BP | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 18 | 29 | 3.786e-02 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 18 | 29 | 3.786e-02 |
| GO:BP | GO:0099625 | ventricular cardiac muscle cell membrane repolarization | 18 | 29 | 3.786e-02 |
| GO:BP | GO:0051156 | glucose 6-phosphate metabolic process | 18 | 29 | 3.786e-02 |
| GO:BP | GO:0071025 | RNA surveillance | 13 | 19 | 3.797e-02 |
| GO:BP | GO:0006491 | N-glycan processing | 13 | 19 | 3.797e-02 |
| GO:BP | GO:1902004 | positive regulation of amyloid-beta formation | 13 | 19 | 3.797e-02 |
| GO:BP | GO:0034244 | negative regulation of transcription elongation by RNA polymerase II | 13 | 19 | 3.797e-02 |
| GO:BP | GO:0035372 | protein localization to microtubule | 13 | 19 | 3.797e-02 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 13 | 19 | 3.797e-02 |
| GO:BP | GO:1903533 | regulation of protein targeting | 40 | 77 | 3.803e-02 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 36 | 68 | 3.823e-02 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 98 | 214 | 3.824e-02 |
| GO:BP | GO:0009226 | nucleotide-sugar biosynthetic process | 17 | 27 | 3.851e-02 |
| GO:BP | GO:0051043 | regulation of membrane protein ectodomain proteolysis | 17 | 27 | 3.851e-02 |
| GO:BP | GO:0031468 | nuclear membrane reassembly | 17 | 27 | 3.851e-02 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 17 | 27 | 3.851e-02 |
| GO:BP | GO:0016188 | synaptic vesicle maturation | 17 | 27 | 3.851e-02 |
| GO:BP | GO:0006450 | regulation of translational fidelity | 14 | 21 | 3.873e-02 |
| GO:BP | GO:0016556 | mRNA modification | 14 | 21 | 3.873e-02 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 14 | 21 | 3.873e-02 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 14 | 21 | 3.873e-02 |
| GO:BP | GO:0071474 | cellular hyperosmotic response | 14 | 21 | 3.873e-02 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 50 | 100 | 3.873e-02 |
| GO:BP | GO:0140896 | cGAS/STING signaling pathway | 14 | 21 | 3.873e-02 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 50 | 100 | 3.873e-02 |
| GO:BP | GO:0090043 | regulation of tubulin deacetylation | 14 | 21 | 3.873e-02 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 26 | 46 | 3.882e-02 |
| GO:BP | GO:0007020 | microtubule nucleation | 26 | 46 | 3.882e-02 |
| GO:BP | GO:1903241 | U2-type prespliceosome assembly | 16 | 25 | 3.887e-02 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 15 | 23 | 3.891e-02 |
| GO:BP | GO:1905709 | negative regulation of membrane permeability | 15 | 23 | 3.891e-02 |
| GO:BP | GO:0090042 | tubulin deacetylation | 15 | 23 | 3.891e-02 |
| GO:BP | GO:0140112 | extracellular vesicle biogenesis | 15 | 23 | 3.891e-02 |
| GO:BP | GO:0006672 | ceramide metabolic process | 53 | 107 | 3.891e-02 |
| GO:BP | GO:0051904 | pigment granule transport | 15 | 23 | 3.891e-02 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 15 | 23 | 3.891e-02 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 31 | 57 | 3.942e-02 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 31 | 57 | 3.942e-02 |
| GO:BP | GO:0003170 | heart valve development | 39 | 75 | 4.045e-02 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 46 | 91 | 4.053e-02 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 35 | 66 | 4.053e-02 |
| GO:BP | GO:0006903 | vesicle targeting | 35 | 66 | 4.053e-02 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 46 | 91 | 4.053e-02 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 46 | 91 | 4.053e-02 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 35 | 66 | 4.053e-02 |
| GO:BP | GO:0001890 | placenta development | 72 | 152 | 4.092e-02 |
| GO:BP | GO:0048701 | embryonic cranial skeleton morphogenesis | 25 | 44 | 4.093e-02 |
| GO:BP | GO:0009225 | nucleotide-sugar metabolic process | 25 | 44 | 4.093e-02 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 49 | 98 | 4.128e-02 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 69 | 145 | 4.192e-02 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 30 | 55 | 4.205e-02 |
| GO:BP | GO:0038066 | p38MAPK cascade | 30 | 55 | 4.205e-02 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 42 | 82 | 4.221e-02 |
| GO:BP | GO:0006814 | sodium ion transport | 110 | 244 | 4.243e-02 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 66 | 138 | 4.276e-02 |
| GO:BP | GO:0003158 | endothelium development | 66 | 138 | 4.276e-02 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 24 | 42 | 4.334e-02 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 24 | 42 | 4.334e-02 |
| GO:BP | GO:0099068 | postsynapse assembly | 45 | 89 | 4.350e-02 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 173 | 400 | 4.357e-02 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 183 | 425 | 4.358e-02 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 71 | 150 | 4.371e-02 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 60 | 124 | 4.414e-02 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 51 | 103 | 4.474e-02 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 29 | 53 | 4.480e-02 |
| GO:BP | GO:0170034 | L-amino acid biosynthetic process | 29 | 53 | 4.480e-02 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 29 | 53 | 4.480e-02 |
| GO:BP | GO:2001222 | regulation of neuron migration | 29 | 53 | 4.480e-02 |
| GO:BP | GO:0070987 | error-free translesion synthesis | 5 | 5 | 4.572e-02 |
| GO:BP | GO:1901908 | diadenosine hexaphosphate metabolic process | 5 | 5 | 4.572e-02 |
| GO:BP | GO:1901910 | adenosine 5’-(hexahydrogen pentaphosphate) metabolic process | 5 | 5 | 4.572e-02 |
| GO:BP | GO:1903724 | positive regulation of centriole elongation | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0071543 | diphosphoinositol polyphosphate metabolic process | 5 | 5 | 4.572e-02 |
| GO:BP | GO:1901909 | diadenosine hexaphosphate catabolic process | 5 | 5 | 4.572e-02 |
| GO:BP | GO:1901907 | diadenosine pentaphosphate catabolic process | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0061431 | cellular response to methionine | 5 | 5 | 4.572e-02 |
| GO:BP | GO:1901911 | adenosine 5’-(hexahydrogen pentaphosphate) catabolic process | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0060922 | atrioventricular node cell differentiation | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0019042 | viral latency | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0048790 | maintenance of presynaptic active zone structure | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0090199 | regulation of release of cytochrome c from mitochondria | 23 | 40 | 4.572e-02 |
| GO:BP | GO:0098708 | D-glucose import across plasma membrane | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0042350 | GDP-L-fucose biosynthetic process | 5 | 5 | 4.572e-02 |
| GO:BP | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0086045 | membrane depolarization during AV node cell action potential | 5 | 5 | 4.572e-02 |
| GO:BP | GO:1901906 | diadenosine pentaphosphate metabolic process | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0006113 | fermentation | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0019509 | L-methionine salvage from methylthioadenosine | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0048753 | pigment granule organization | 23 | 40 | 4.572e-02 |
| GO:BP | GO:1990592 | protein K69-linked ufmylation | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0033499 | galactose catabolic process via UDP-galactose, Leloir pathway | 5 | 5 | 4.572e-02 |
| GO:BP | GO:1905908 | positive regulation of amyloid fibril formation | 5 | 5 | 4.572e-02 |
| GO:BP | GO:1990564 | protein polyufmylation | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0003431 | growth plate cartilage chondrocyte development | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0018027 | peptidyl-lysine dimethylation | 5 | 5 | 4.572e-02 |
| GO:BP | GO:1904978 | regulation of endosome organization | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0003292 | cardiac septum cell differentiation | 5 | 5 | 4.572e-02 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 37 | 71 | 4.577e-02 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 33 | 62 | 4.579e-02 |
| GO:BP | GO:0009799 | specification of symmetry | 70 | 148 | 4.614e-02 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 70 | 148 | 4.614e-02 |
| GO:BP | GO:0016125 | sterol metabolic process | 75 | 160 | 4.661e-02 |
| GO:BP | GO:0072006 | nephron development | 75 | 160 | 4.661e-02 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 59 | 122 | 4.664e-02 |
| GO:BP | GO:0000492 | box C/D snoRNP assembly | 8 | 10 | 4.695e-02 |
| GO:BP | GO:0086016 | AV node cell action potential | 8 | 10 | 4.695e-02 |
| GO:BP | GO:1904869 | regulation of protein localization to Cajal body | 8 | 10 | 4.695e-02 |
| GO:BP | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum | 8 | 10 | 4.695e-02 |
| GO:BP | GO:0071166 | ribonucleoprotein complex localization | 8 | 10 | 4.695e-02 |
| GO:BP | GO:0086027 | AV node cell to bundle of His cell signaling | 8 | 10 | 4.695e-02 |
| GO:BP | GO:0060052 | neurofilament cytoskeleton organization | 8 | 10 | 4.695e-02 |
| GO:BP | GO:1904871 | positive regulation of protein localization to Cajal body | 8 | 10 | 4.695e-02 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 8 | 10 | 4.695e-02 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 8 | 10 | 4.695e-02 |
| GO:BP | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport | 8 | 10 | 4.695e-02 |
| GO:BP | GO:0071600 | otic vesicle morphogenesis | 8 | 10 | 4.695e-02 |
| GO:BP | GO:2000739 | regulation of mesenchymal stem cell differentiation | 8 | 10 | 4.695e-02 |
| GO:BP | GO:0046545 | development of primary female sexual characteristics | 53 | 108 | 4.710e-02 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 22 | 38 | 4.734e-02 |
| GO:BP | GO:0099172 | presynapse organization | 32 | 60 | 4.883e-02 |
| GO:BP | GO:0002381 | immunoglobulin production involved in immunoglobulin-mediated immune response | 32 | 60 | 4.883e-02 |
| GO:BP | GO:0033014 | tetrapyrrole biosynthetic process | 21 | 36 | 4.989e-02 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 21 | 36 | 4.989e-02 |
| GO:BP | GO:0006779 | porphyrin-containing compound biosynthetic process | 21 | 36 | 4.989e-02 |
| GO:BP | GO:0007035 | vacuolar acidification | 21 | 36 | 4.989e-02 |
| GO:BP | GO:0090630 | activation of GTPase activity | 21 | 36 | 4.989e-02 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 21 | 36 | 4.989e-02 |
| GO:BP | GO:1902003 | regulation of amyloid-beta formation | 21 | 36 | 4.989e-02 |
| KEGG | KEGG:04140 | Autophagy - animal | 111 | 165 | 1.401e-07 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 283 | 506 | 7.029e-07 |
| KEGG | KEGG:04137 | Mitophagy - animal | 72 | 103 | 5.134e-06 |
| KEGG | KEGG:03082 | ATP-dependent chromatin remodeling | 75 | 116 | 2.104e-04 |
| KEGG | KEGG:05212 | Pancreatic cancer | 53 | 76 | 2.104e-04 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 64 | 97 | 3.365e-04 |
| KEGG | KEGG:05210 | Colorectal cancer | 57 | 86 | 7.571e-04 |
| KEGG | KEGG:03018 | RNA degradation | 53 | 79 | 7.789e-04 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 75 | 121 | 1.079e-03 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 68 | 108 | 1.100e-03 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 55 | 84 | 1.150e-03 |
| KEGG | KEGG:04144 | Endocytosis | 138 | 248 | 1.736e-03 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 21 | 26 | 2.922e-03 |
| KEGG | KEGG:04218 | Cellular senescence | 90 | 155 | 2.922e-03 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 90 | 155 | 2.922e-03 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 47 | 72 | 2.922e-03 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 96 | 166 | 2.922e-03 |
| KEGG | KEGG:04115 | p53 signaling pathway | 48 | 74 | 2.922e-03 |
| KEGG | KEGG:04110 | Cell cycle | 91 | 157 | 2.922e-03 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 111 | 198 | 3.600e-03 |
| KEGG | KEGG:05161 | Hepatitis B | 93 | 162 | 3.600e-03 |
| KEGG | KEGG:05131 | Shigellosis | 134 | 246 | 4.346e-03 |
| KEGG | KEGG:04136 | Autophagy - other | 24 | 32 | 4.425e-03 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 81 | 140 | 5.404e-03 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 95 | 168 | 5.404e-03 |
| KEGG | KEGG:05213 | Endometrial cancer | 38 | 58 | 7.595e-03 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 70 | 120 | 8.842e-03 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 30 | 44 | 9.952e-03 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 187 | 363 | 1.158e-02 |
| KEGG | KEGG:03022 | Basal transcription factors | 29 | 43 | 1.437e-02 |
| KEGG | KEGG:05226 | Gastric cancer | 83 | 148 | 1.437e-02 |
| KEGG | KEGG:04330 | Notch signaling pathway | 38 | 60 | 1.615e-02 |
| KEGG | KEGG:01522 | Endocrine resistance | 56 | 95 | 1.721e-02 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 95 | 174 | 1.891e-02 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 28 | 42 | 2.015e-02 |
| KEGG | KEGG:03410 | Base excision repair | 29 | 44 | 2.133e-02 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 38 | 61 | 2.166e-02 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 155 | 301 | 2.379e-02 |
| KEGG | KEGG:05224 | Breast cancer | 81 | 147 | 2.470e-02 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 78 | 141 | 2.481e-02 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 52 | 89 | 2.545e-02 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 107 | 201 | 2.545e-02 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 61 | 107 | 2.550e-02 |
| KEGG | KEGG:04360 | Axon guidance | 97 | 181 | 2.819e-02 |
| KEGG | KEGG:04710 | Circadian rhythm | 23 | 34 | 2.895e-02 |
| KEGG | KEGG:03020 | RNA polymerase | 23 | 34 | 2.895e-02 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 85 | 157 | 3.092e-02 |
| KEGG | KEGG:04931 | Insulin resistance | 61 | 108 | 3.092e-02 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 33 | 53 | 3.157e-02 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 114 | 218 | 3.385e-02 |
| KEGG | KEGG:00310 | Lysine degradation | 38 | 63 | 3.509e-02 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 46 | 79 | 3.808e-02 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 233 | 474 | 3.822e-02 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 66 | 120 | 4.455e-02 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 48 | 84 | 4.849e-02 |
| KEGG | KEGG:03440 | Homologous recombination | 26 | 41 | 4.849e-02 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 74 | 137 | 4.849e-02 |
#GO:BP
table_DOX24Tshare_genes_GOBP <- table_DOX24Tshare_genes %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24Tshare_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Shared DEGs Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))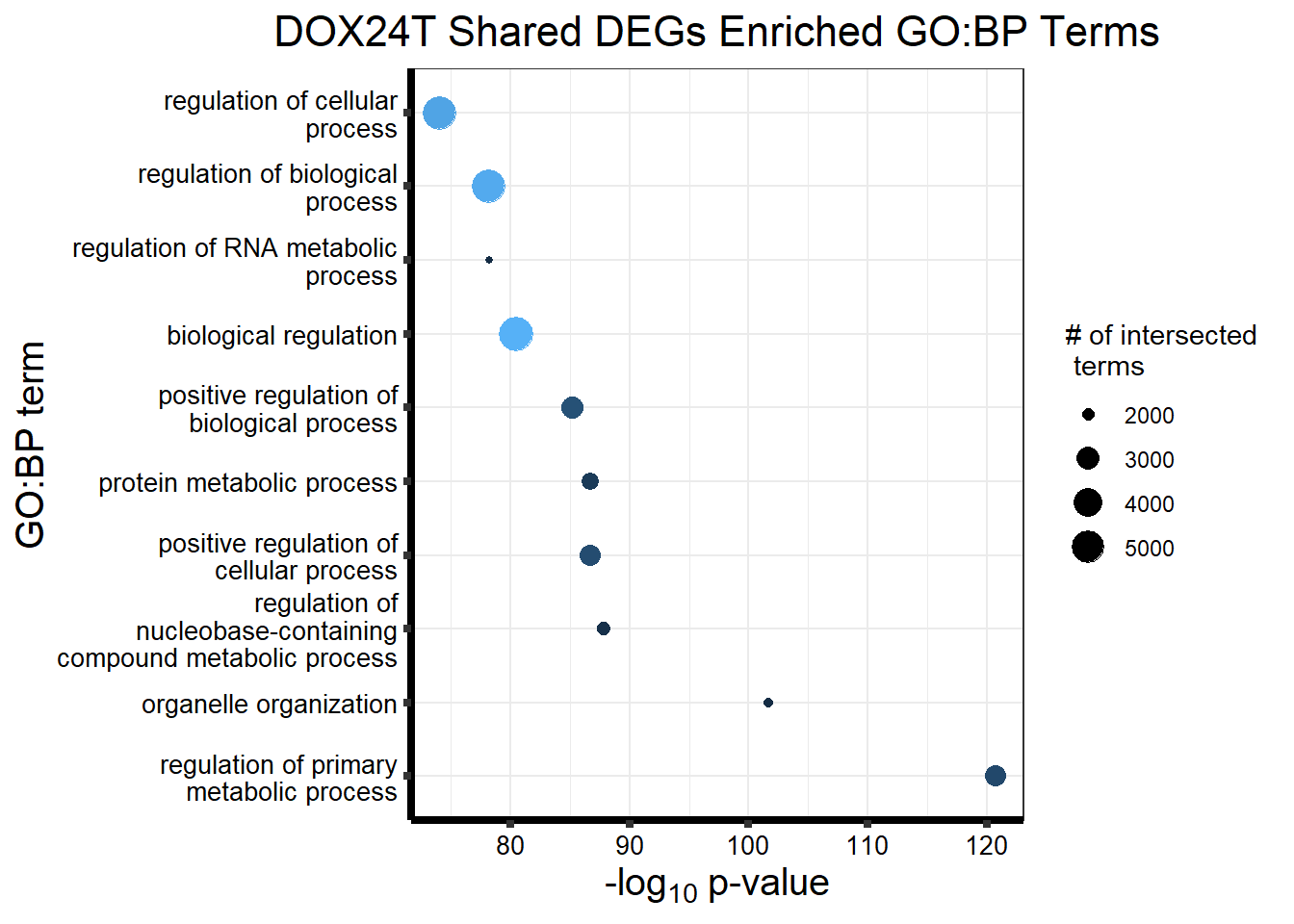
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_DOX24Tshare_genes_KEGG <- table_DOX24Tshare_genes %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24Tshare_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Shared DEGs Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))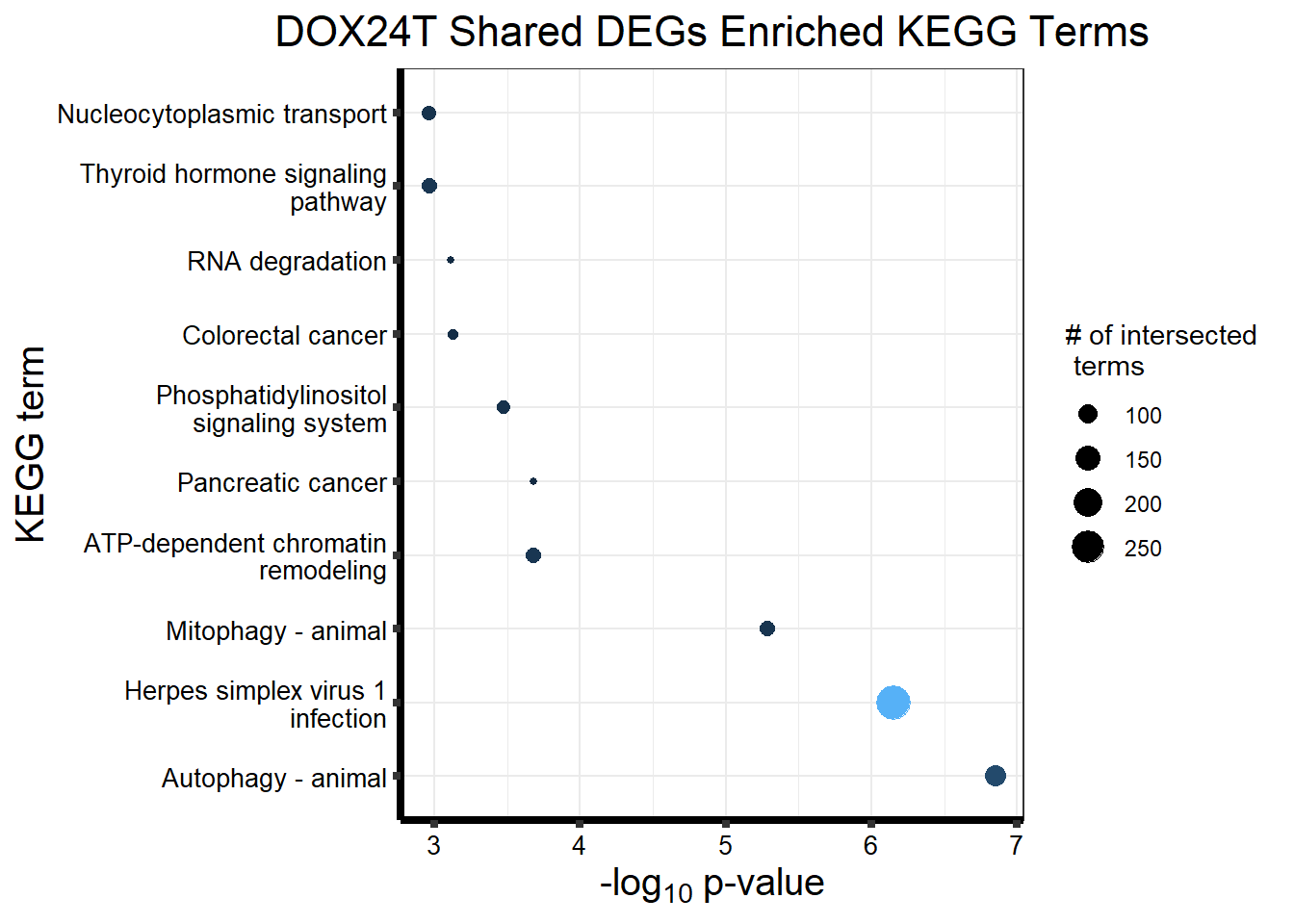
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####DOX24R DEGs GO KEGG#####
D24R_DEGs_mat <- as.matrix(DOX24R_DEGs_GO)
DOX_24R_dxr_gene <- gost(query = D24R_DEGs_mat,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
DOX_24R_gost_genes <- gostplot(DOX_24R_dxr_gene, capped = FALSE, interactive = TRUE)
DOX_24R_gost_genestable_DOX24R_genes <- DOX_24R_dxr_gene$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX24R_genes %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0007275 | multicellular organism development | 578 | 4727 | 1.297e-16 |
| GO:BP | GO:0032502 | developmental process | 754 | 6553 | 1.297e-16 |
| GO:BP | GO:0048856 | anatomical structure development | 699 | 5997 | 2.247e-16 |
| GO:BP | GO:0048731 | system development | 507 | 4053 | 4.867e-16 |
| GO:BP | GO:0006810 | transport | 526 | 4407 | 8.570e-13 |
| GO:BP | GO:0051179 | localization | 637 | 5555 | 8.570e-13 |
| GO:BP | GO:0032501 | multicellular organismal process | 798 | 7322 | 4.356e-12 |
| GO:BP | GO:0006996 | organelle organization | 440 | 3594 | 7.388e-12 |
| GO:BP | GO:0048518 | positive regulation of biological process | 697 | 6264 | 1.037e-11 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 316 | 2407 | 1.931e-11 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 316 | 2410 | 2.074e-11 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 661 | 5920 | 3.346e-11 |
| GO:BP | GO:0010646 | regulation of cell communication | 424 | 3486 | 4.996e-11 |
| GO:BP | GO:0030029 | actin filament-based process | 137 | 825 | 9.458e-11 |
| GO:BP | GO:0007010 | cytoskeleton organization | 218 | 1529 | 9.520e-11 |
| GO:BP | GO:0023051 | regulation of signaling | 421 | 3478 | 1.099e-10 |
| GO:BP | GO:0051234 | establishment of localization | 562 | 4928 | 1.506e-10 |
| GO:BP | GO:0065007 | biological regulation | 1262 | 12743 | 1.745e-10 |
| GO:BP | GO:0007399 | nervous system development | 330 | 2604 | 2.769e-10 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 123 | 729 | 4.020e-10 |
| GO:BP | GO:0048513 | animal organ development | 377 | 3085 | 6.485e-10 |
| GO:BP | GO:0033554 | cellular response to stress | 246 | 1830 | 9.967e-10 |
| GO:BP | GO:0030154 | cell differentiation | 509 | 4437 | 1.062e-09 |
| GO:BP | GO:0048869 | cellular developmental process | 509 | 4438 | 1.062e-09 |
| GO:BP | GO:0019538 | protein metabolic process | 535 | 4721 | 1.682e-09 |
| GO:BP | GO:0003012 | muscle system process | 84 | 438 | 1.846e-09 |
| GO:BP | GO:0051641 | cellular localization | 431 | 3661 | 2.218e-09 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 360 | 2956 | 2.903e-09 |
| GO:BP | GO:0050789 | regulation of biological process | 1218 | 12336 | 3.289e-09 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 335 | 2713 | 3.289e-09 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 462 | 3993 | 3.652e-09 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 217 | 1592 | 4.401e-09 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 134 | 861 | 8.667e-09 |
| GO:BP | GO:0065008 | regulation of biological quality | 356 | 2947 | 9.267e-09 |
| GO:BP | GO:0044281 | small molecule metabolic process | 235 | 1776 | 1.019e-08 |
| GO:BP | GO:0050896 | response to stimulus | 922 | 8999 | 1.620e-08 |
| GO:BP | GO:0009966 | regulation of signal transduction | 363 | 3034 | 1.777e-08 |
| GO:BP | GO:0030334 | regulation of cell migration | 144 | 960 | 2.026e-08 |
| GO:BP | GO:0051716 | cellular response to stimulus | 774 | 7376 | 2.560e-08 |
| GO:BP | GO:0061061 | muscle structure development | 112 | 693 | 3.261e-08 |
| GO:BP | GO:0016477 | cell migration | 206 | 1534 | 4.542e-08 |
| GO:BP | GO:0006936 | muscle contraction | 68 | 349 | 5.944e-08 |
| GO:BP | GO:0072359 | circulatory system development | 162 | 1145 | 1.046e-07 |
| GO:BP | GO:2000145 | regulation of cell motility | 148 | 1022 | 1.201e-07 |
| GO:BP | GO:0032879 | regulation of localization | 256 | 2029 | 1.224e-07 |
| GO:BP | GO:0022008 | neurogenesis | 229 | 1771 | 1.240e-07 |
| GO:BP | GO:0044057 | regulation of system process | 93 | 553 | 1.304e-07 |
| GO:BP | GO:0048519 | negative regulation of biological process | 625 | 5834 | 1.893e-07 |
| GO:BP | GO:0048870 | cell motility | 230 | 1793 | 2.124e-07 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 83 | 478 | 2.124e-07 |
| GO:BP | GO:0030030 | cell projection organization | 213 | 1631 | 2.124e-07 |
| GO:BP | GO:0036211 | protein modification process | 337 | 2846 | 2.478e-07 |
| GO:BP | GO:0040012 | regulation of locomotion | 151 | 1067 | 3.363e-07 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 294 | 2433 | 4.468e-07 |
| GO:BP | GO:0043412 | macromolecule modification | 353 | 3030 | 5.741e-07 |
| GO:BP | GO:0046907 | intracellular transport | 184 | 1381 | 6.118e-07 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 206 | 1588 | 6.118e-07 |
| GO:BP | GO:0051649 | establishment of localization in cell | 250 | 2010 | 6.118e-07 |
| GO:BP | GO:0090257 | regulation of muscle system process | 49 | 231 | 7.325e-07 |
| GO:BP | GO:0040007 | growth | 134 | 929 | 7.664e-07 |
| GO:BP | GO:0007015 | actin filament organization | 79 | 461 | 8.199e-07 |
| GO:BP | GO:0050794 | regulation of cellular process | 1164 | 11946 | 1.124e-06 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 599 | 5629 | 1.269e-06 |
| GO:BP | GO:0009056 | catabolic process | 311 | 2639 | 1.686e-06 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 63 | 345 | 2.366e-06 |
| GO:BP | GO:0009060 | aerobic respiration | 43 | 199 | 3.172e-06 |
| GO:BP | GO:0040011 | locomotion | 167 | 1255 | 3.179e-06 |
| GO:BP | GO:0007155 | cell adhesion | 196 | 1530 | 3.375e-06 |
| GO:BP | GO:0023057 | negative regulation of signaling | 186 | 1435 | 3.375e-06 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 211 | 1680 | 4.469e-06 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 147 | 1077 | 4.469e-06 |
| GO:BP | GO:0009888 | tissue development | 247 | 2032 | 4.696e-06 |
| GO:BP | GO:0034330 | cell junction organization | 120 | 834 | 4.696e-06 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 185 | 1436 | 5.349e-06 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 95 | 619 | 5.970e-06 |
| GO:BP | GO:0050793 | regulation of developmental process | 289 | 2457 | 6.304e-06 |
| GO:BP | GO:0031175 | neuron projection development | 140 | 1023 | 7.533e-06 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 37 | 165 | 9.132e-06 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 227 | 1855 | 9.763e-06 |
| GO:BP | GO:0006950 | response to stress | 433 | 3948 | 9.928e-06 |
| GO:BP | GO:0007507 | heart development | 92 | 605 | 1.384e-05 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 98 | 658 | 1.472e-05 |
| GO:BP | GO:0048699 | generation of neurons | 193 | 1536 | 1.503e-05 |
| GO:BP | GO:0007154 | cell communication | 673 | 6540 | 1.768e-05 |
| GO:BP | GO:0045333 | cellular respiration | 47 | 242 | 1.768e-05 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 86 | 559 | 1.995e-05 |
| GO:BP | GO:0048589 | developmental growth | 98 | 663 | 1.995e-05 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 91 | 603 | 2.113e-05 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 86 | 561 | 2.268e-05 |
| GO:BP | GO:0030182 | neuron differentiation | 183 | 1451 | 2.331e-05 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 317 | 2782 | 2.446e-05 |
| GO:BP | GO:0051049 | regulation of transport | 199 | 1607 | 2.471e-05 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 95 | 642 | 2.727e-05 |
| GO:BP | GO:0033036 | macromolecule localization | 360 | 3228 | 2.802e-05 |
| GO:BP | GO:0043603 | amide metabolic process | 73 | 455 | 2.973e-05 |
| GO:BP | GO:0008104 | protein localization | 315 | 2770 | 3.090e-05 |
| GO:BP | GO:0023052 | signaling | 668 | 6515 | 3.195e-05 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 122 | 887 | 3.254e-05 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 564 | 5390 | 3.369e-05 |
| GO:BP | GO:0009611 | response to wounding | 86 | 568 | 3.469e-05 |
| GO:BP | GO:0048666 | neuron development | 153 | 1177 | 3.612e-05 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 319 | 2819 | 3.824e-05 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 88 | 587 | 3.824e-05 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 135 | 1011 | 3.959e-05 |
| GO:BP | GO:0065009 | regulation of molecular function | 186 | 1496 | 4.215e-05 |
| GO:BP | GO:0060047 | heart contraction | 46 | 244 | 4.649e-05 |
| GO:BP | GO:0006082 | organic acid metabolic process | 124 | 915 | 5.275e-05 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 123 | 909 | 6.284e-05 |
| GO:BP | GO:0006812 | monoatomic cation transport | 140 | 1069 | 6.988e-05 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 184 | 1489 | 6.999e-05 |
| GO:BP | GO:0006979 | response to oxidative stress | 65 | 400 | 7.047e-05 |
| GO:BP | GO:0043500 | muscle adaptation | 26 | 105 | 7.070e-05 |
| GO:BP | GO:0055085 | transmembrane transport | 193 | 1578 | 7.285e-05 |
| GO:BP | GO:0061564 | axon development | 80 | 528 | 7.469e-05 |
| GO:BP | GO:0016310 | phosphorylation | 166 | 1320 | 8.230e-05 |
| GO:BP | GO:0006468 | protein phosphorylation | 156 | 1227 | 9.231e-05 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 391 | 3597 | 9.349e-05 |
| GO:BP | GO:0035556 | intracellular signal transduction | 330 | 2965 | 9.695e-05 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 59 | 356 | 1.081e-04 |
| GO:BP | GO:0003015 | heart process | 46 | 253 | 1.176e-04 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 118 | 877 | 1.251e-04 |
| GO:BP | GO:0050808 | synapse organization | 83 | 563 | 1.350e-04 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 125 | 944 | 1.386e-04 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 87 | 599 | 1.407e-04 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 136 | 1048 | 1.440e-04 |
| GO:BP | GO:0030198 | extracellular matrix organization | 55 | 327 | 1.441e-04 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 165 | 1326 | 1.528e-04 |
| GO:BP | GO:0043062 | extracellular structure organization | 55 | 328 | 1.561e-04 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 55 | 329 | 1.704e-04 |
| GO:BP | GO:0006811 | monoatomic ion transport | 160 | 1282 | 1.803e-04 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 75 | 500 | 2.052e-04 |
| GO:BP | GO:0032535 | regulation of cellular component size | 58 | 356 | 2.072e-04 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 199 | 1667 | 2.147e-04 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 128 | 983 | 2.271e-04 |
| GO:BP | GO:0042060 | wound healing | 67 | 433 | 2.283e-04 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 53 | 317 | 2.434e-04 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 60 | 376 | 2.682e-04 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 94 | 673 | 2.687e-04 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 85 | 593 | 2.781e-04 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 119 | 905 | 3.018e-04 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 68 | 446 | 3.104e-04 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 51 | 304 | 3.168e-04 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 51 | 304 | 3.168e-04 |
| GO:BP | GO:0015031 | protein transport | 175 | 1444 | 3.330e-04 |
| GO:BP | GO:0030239 | myofibril assembly | 20 | 76 | 3.570e-04 |
| GO:BP | GO:0019725 | cellular homeostasis | 111 | 835 | 3.680e-04 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 10 | 22 | 3.763e-04 |
| GO:BP | GO:0000902 | cell morphogenesis | 128 | 996 | 3.972e-04 |
| GO:BP | GO:0098657 | import into cell | 120 | 922 | 4.229e-04 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 90 | 646 | 4.363e-04 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 51 | 308 | 4.379e-04 |
| GO:BP | GO:0055001 | muscle cell development | 37 | 198 | 4.915e-04 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 30 | 146 | 4.973e-04 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 43 | 245 | 4.993e-04 |
| GO:BP | GO:0007165 | signal transduction | 608 | 6002 | 5.230e-04 |
| GO:BP | GO:0007409 | axonogenesis | 69 | 463 | 5.273e-04 |
| GO:BP | GO:0012501 | programmed cell death | 228 | 1987 | 5.510e-04 |
| GO:BP | GO:0042592 | homeostatic process | 203 | 1736 | 5.510e-04 |
| GO:BP | GO:0001706 | endoderm formation | 17 | 60 | 5.510e-04 |
| GO:BP | GO:0051051 | negative regulation of transport | 63 | 412 | 5.510e-04 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 85 | 606 | 5.579e-04 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 112 | 854 | 5.579e-04 |
| GO:BP | GO:0048878 | chemical homeostasis | 132 | 1043 | 5.588e-04 |
| GO:BP | GO:0008015 | blood circulation | 75 | 518 | 5.939e-04 |
| GO:BP | GO:0008219 | cell death | 228 | 1991 | 6.111e-04 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 148 | 1199 | 6.111e-04 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 93 | 681 | 6.261e-04 |
| GO:BP | GO:0055002 | striated muscle cell development | 33 | 171 | 6.677e-04 |
| GO:BP | GO:0048468 | cell development | 314 | 2876 | 6.705e-04 |
| GO:BP | GO:0003013 | circulatory system process | 84 | 602 | 7.249e-04 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 150 | 1223 | 7.255e-04 |
| GO:BP | GO:0016311 | dephosphorylation | 39 | 218 | 7.277e-04 |
| GO:BP | GO:0019318 | hexose metabolic process | 40 | 226 | 7.343e-04 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 43 | 250 | 7.406e-04 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 48 | 292 | 8.406e-04 |
| GO:BP | GO:0001704 | formation of primary germ layer | 27 | 129 | 8.408e-04 |
| GO:BP | GO:0031032 | actomyosin structure organization | 38 | 212 | 8.652e-04 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 44 | 260 | 8.785e-04 |
| GO:BP | GO:0006941 | striated muscle contraction | 35 | 189 | 8.830e-04 |
| GO:BP | GO:0042063 | gliogenesis | 55 | 352 | 9.346e-04 |
| GO:BP | GO:0035987 | endodermal cell differentiation | 15 | 51 | 9.743e-04 |
| GO:BP | GO:0060074 | synapse maturation | 13 | 40 | 1.048e-03 |
| GO:BP | GO:0042692 | muscle cell differentiation | 61 | 405 | 1.048e-03 |
| GO:BP | GO:0060341 | regulation of cellular localization | 127 | 1013 | 1.124e-03 |
| GO:BP | GO:0006974 | DNA damage response | 116 | 908 | 1.124e-03 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 44 | 263 | 1.124e-03 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 121 | 956 | 1.134e-03 |
| GO:BP | GO:0006915 | apoptotic process | 219 | 1923 | 1.166e-03 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 121 | 957 | 1.176e-03 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 91 | 676 | 1.189e-03 |
| GO:BP | GO:0031102 | neuron projection regeneration | 16 | 58 | 1.200e-03 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 91 | 677 | 1.245e-03 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 43 | 257 | 1.316e-03 |
| GO:BP | GO:0048729 | tissue morphogenesis | 84 | 614 | 1.316e-03 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 245 | 2192 | 1.316e-03 |
| GO:BP | GO:0001944 | vasculature development | 100 | 762 | 1.321e-03 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 89 | 660 | 1.321e-03 |
| GO:BP | GO:0051050 | positive regulation of transport | 109 | 847 | 1.351e-03 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 49 | 308 | 1.464e-03 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 52 | 334 | 1.545e-03 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 78 | 563 | 1.572e-03 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 51 | 326 | 1.590e-03 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 19 | 79 | 1.714e-03 |
| GO:BP | GO:0008016 | regulation of heart contraction | 36 | 204 | 1.737e-03 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 14 | 48 | 1.791e-03 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 167 | 1417 | 1.801e-03 |
| GO:BP | GO:0016049 | cell growth | 69 | 485 | 1.834e-03 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 121 | 970 | 1.951e-03 |
| GO:BP | GO:0031399 | regulation of protein modification process | 129 | 1049 | 2.114e-03 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 91 | 689 | 2.205e-03 |
| GO:BP | GO:0007517 | muscle organ development | 55 | 365 | 2.213e-03 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 41 | 247 | 2.213e-03 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 26 | 130 | 2.241e-03 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 106 | 831 | 2.296e-03 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 53 | 349 | 2.399e-03 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 33 | 184 | 2.404e-03 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 182 | 1576 | 2.404e-03 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 175 | 1506 | 2.404e-03 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 95 | 730 | 2.553e-03 |
| GO:BP | GO:0008347 | glial cell migration | 16 | 62 | 2.553e-03 |
| GO:BP | GO:0070848 | response to growth factor | 94 | 721 | 2.590e-03 |
| GO:BP | GO:0030534 | adult behavior | 29 | 154 | 2.621e-03 |
| GO:BP | GO:0071705 | nitrogen compound transport | 216 | 1923 | 2.636e-03 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 72 | 519 | 2.654e-03 |
| GO:BP | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity | 6 | 10 | 2.682e-03 |
| GO:BP | GO:0030199 | collagen fibril organization | 17 | 69 | 2.789e-03 |
| GO:BP | GO:0006886 | intracellular protein transport | 90 | 686 | 2.854e-03 |
| GO:BP | GO:0048588 | developmental cell growth | 38 | 226 | 2.854e-03 |
| GO:BP | GO:0031099 | regeneration | 35 | 202 | 2.889e-03 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 80 | 594 | 2.916e-03 |
| GO:BP | GO:0034329 | cell junction assembly | 70 | 503 | 2.925e-03 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 44 | 276 | 2.935e-03 |
| GO:BP | GO:0040008 | regulation of growth | 81 | 604 | 3.010e-03 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 21 | 97 | 3.153e-03 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 25 | 126 | 3.206e-03 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 48 | 312 | 3.366e-03 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 101 | 794 | 3.366e-03 |
| GO:BP | GO:1903224 | regulation of endodermal cell differentiation | 5 | 7 | 3.366e-03 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 41 | 253 | 3.366e-03 |
| GO:BP | GO:0006897 | endocytosis | 92 | 709 | 3.366e-03 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 26 | 134 | 3.366e-03 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 82 | 616 | 3.366e-03 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 165 | 1418 | 3.366e-03 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 27 | 142 | 3.514e-03 |
| GO:BP | GO:0051209 | release of sequestered calcium ion into cytosol | 25 | 127 | 3.514e-03 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 20 | 91 | 3.514e-03 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 75 | 553 | 3.591e-03 |
| GO:BP | GO:0045184 | establishment of protein localization | 216 | 1937 | 3.659e-03 |
| GO:BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | 23 | 113 | 3.795e-03 |
| GO:BP | GO:0048675 | axon extension | 23 | 113 | 3.795e-03 |
| GO:BP | GO:0006749 | glutathione metabolic process | 14 | 52 | 3.840e-03 |
| GO:BP | GO:0051283 | negative regulation of sequestering of calcium ion | 25 | 128 | 3.885e-03 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 110 | 884 | 3.885e-03 |
| GO:BP | GO:1902600 | proton transmembrane transport | 33 | 190 | 3.924e-03 |
| GO:BP | GO:0030163 | protein catabolic process | 125 | 1030 | 4.013e-03 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 89 | 686 | 4.115e-03 |
| GO:BP | GO:1903522 | regulation of blood circulation | 41 | 256 | 4.116e-03 |
| GO:BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | 23 | 114 | 4.229e-03 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 27 | 144 | 4.247e-03 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 33 | 191 | 4.250e-03 |
| GO:BP | GO:0051651 | maintenance of location in cell | 35 | 208 | 4.623e-03 |
| GO:BP | GO:0006006 | glucose metabolic process | 32 | 184 | 4.670e-03 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 70 | 513 | 4.706e-03 |
| GO:BP | GO:0006629 | lipid metabolic process | 161 | 1391 | 4.788e-03 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 36 | 217 | 4.935e-03 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 70 | 514 | 4.935e-03 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 46 | 301 | 4.951e-03 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 132 | 1105 | 4.974e-03 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 27 | 146 | 5.192e-03 |
| GO:BP | GO:0006816 | calcium ion transport | 61 | 434 | 5.300e-03 |
| GO:BP | GO:1904064 | positive regulation of cation transmembrane transport | 25 | 131 | 5.300e-03 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 23 | 116 | 5.300e-03 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 41 | 260 | 5.424e-03 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 41 | 260 | 5.424e-03 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 34 | 202 | 5.424e-03 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 27 | 147 | 5.673e-03 |
| GO:BP | GO:0019222 | regulation of metabolic process | 688 | 7035 | 5.673e-03 |
| GO:BP | GO:0051648 | vesicle localization | 36 | 219 | 5.675e-03 |
| GO:BP | GO:0090504 | epiboly | 12 | 42 | 5.713e-03 |
| GO:BP | GO:0007169 | cell surface receptor protein tyrosine kinase signaling pathway | 82 | 629 | 5.790e-03 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 18 | 81 | 5.790e-03 |
| GO:BP | GO:0007492 | endoderm development | 19 | 88 | 5.790e-03 |
| GO:BP | GO:0007417 | central nervous system development | 127 | 1061 | 5.790e-03 |
| GO:BP | GO:0051640 | organelle localization | 81 | 620 | 5.876e-03 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 55 | 383 | 5.969e-03 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 343 | 3288 | 6.239e-03 |
| GO:BP | GO:0061024 | membrane organization | 102 | 821 | 6.370e-03 |
| GO:BP | GO:0098869 | cellular oxidant detoxification | 20 | 96 | 6.494e-03 |
| GO:BP | GO:0033275 | actin-myosin filament sliding | 7 | 16 | 6.665e-03 |
| GO:BP | GO:0009790 | embryo development | 134 | 1135 | 6.834e-03 |
| GO:BP | GO:0006551 | L-leucine metabolic process | 5 | 8 | 6.842e-03 |
| GO:BP | GO:0062196 | regulation of lysosome size | 5 | 8 | 6.842e-03 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 30 | 173 | 6.990e-03 |
| GO:BP | GO:0007626 | locomotory behavior | 36 | 222 | 7.048e-03 |
| GO:BP | GO:0010001 | glial cell differentiation | 41 | 264 | 7.048e-03 |
| GO:BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | 23 | 119 | 7.188e-03 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 92 | 729 | 7.274e-03 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 80 | 616 | 7.370e-03 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 37 | 231 | 7.370e-03 |
| GO:BP | GO:0030832 | regulation of actin filament length | 27 | 150 | 7.371e-03 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 107 | 874 | 7.461e-03 |
| GO:BP | GO:0009142 | nucleoside triphosphate biosynthetic process | 24 | 127 | 7.461e-03 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 40 | 257 | 7.725e-03 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 25 | 135 | 7.725e-03 |
| GO:BP | GO:0007610 | behavior | 86 | 674 | 7.778e-03 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 97 | 779 | 7.820e-03 |
| GO:BP | GO:0034767 | positive regulation of monoatomic ion transmembrane transport | 26 | 143 | 7.882e-03 |
| GO:BP | GO:0031103 | axon regeneration | 13 | 50 | 7.968e-03 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 210 | 1912 | 8.633e-03 |
| GO:BP | GO:0006470 | protein dephosphorylation | 26 | 144 | 8.744e-03 |
| GO:BP | GO:0016358 | dendrite development | 38 | 242 | 8.858e-03 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 20 | 99 | 9.131e-03 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 37 | 234 | 9.131e-03 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 47 | 320 | 9.131e-03 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 20 | 99 | 9.131e-03 |
| GO:BP | GO:0006508 | proteolysis | 168 | 1486 | 9.138e-03 |
| GO:BP | GO:0045214 | sarcomere organization | 13 | 51 | 9.504e-03 |
| GO:BP | GO:0071320 | cellular response to cAMP | 13 | 51 | 9.504e-03 |
| GO:BP | GO:0010649 | regulation of cell communication by electrical coupling | 7 | 17 | 9.504e-03 |
| GO:BP | GO:0051235 | maintenance of location | 35 | 218 | 9.521e-03 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 18 | 85 | 9.541e-03 |
| GO:BP | GO:0072524 | pyridine-containing compound metabolic process | 29 | 169 | 9.541e-03 |
| GO:BP | GO:0009152 | purine ribonucleotide biosynthetic process | 28 | 161 | 9.541e-03 |
| GO:BP | GO:1990748 | cellular detoxification | 23 | 122 | 9.541e-03 |
| GO:BP | GO:0030041 | actin filament polymerization | 28 | 161 | 9.541e-03 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 33 | 202 | 9.861e-03 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 51 | 357 | 9.864e-03 |
| GO:BP | GO:0010453 | regulation of cell fate commitment | 12 | 45 | 9.869e-03 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 38 | 244 | 9.888e-03 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 22 | 115 | 1.011e-02 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 70 | 531 | 1.031e-02 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 78 | 606 | 1.036e-02 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 88 | 701 | 1.041e-02 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 46 | 314 | 1.047e-02 |
| GO:BP | GO:0035295 | tube development | 129 | 1101 | 1.052e-02 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 213 | 1953 | 1.053e-02 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 47 | 323 | 1.053e-02 |
| GO:BP | GO:0051928 | positive regulation of calcium ion transport | 22 | 116 | 1.119e-02 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 27 | 155 | 1.122e-02 |
| GO:BP | GO:0046034 | ATP metabolic process | 36 | 229 | 1.122e-02 |
| GO:BP | GO:0099173 | postsynapse organization | 38 | 246 | 1.123e-02 |
| GO:BP | GO:0001568 | blood vessel development | 91 | 732 | 1.126e-02 |
| GO:BP | GO:0009725 | response to hormone | 109 | 907 | 1.136e-02 |
| GO:BP | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol | 6 | 13 | 1.136e-02 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 23 | 124 | 1.136e-02 |
| GO:BP | GO:0032880 | regulation of protein localization | 109 | 907 | 1.136e-02 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 35 | 221 | 1.136e-02 |
| GO:BP | GO:0009078 | pyruvate family amino acid metabolic process | 6 | 13 | 1.136e-02 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 24 | 132 | 1.167e-02 |
| GO:BP | GO:0043604 | amide biosynthetic process | 29 | 172 | 1.186e-02 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 5 | 9 | 1.186e-02 |
| GO:BP | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity | 5 | 9 | 1.186e-02 |
| GO:BP | GO:0001558 | regulation of cell growth | 56 | 406 | 1.187e-02 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 11 | 40 | 1.194e-02 |
| GO:BP | GO:0007611 | learning or memory | 42 | 282 | 1.217e-02 |
| GO:BP | GO:0007369 | gastrulation | 32 | 197 | 1.217e-02 |
| GO:BP | GO:0006281 | DNA repair | 79 | 621 | 1.265e-02 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 30 | 181 | 1.265e-02 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 19 | 95 | 1.273e-02 |
| GO:BP | GO:0042552 | myelination | 27 | 157 | 1.300e-02 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 37 | 240 | 1.301e-02 |
| GO:BP | GO:1901698 | response to nitrogen compound | 126 | 1080 | 1.335e-02 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 14 | 60 | 1.335e-02 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 34 | 215 | 1.335e-02 |
| GO:BP | GO:0048678 | response to axon injury | 17 | 81 | 1.354e-02 |
| GO:BP | GO:0014888 | striated muscle adaptation | 12 | 47 | 1.357e-02 |
| GO:BP | GO:0022900 | electron transport chain | 23 | 126 | 1.357e-02 |
| GO:BP | GO:0032409 | regulation of transporter activity | 30 | 182 | 1.357e-02 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 16 | 74 | 1.358e-02 |
| GO:BP | GO:0051258 | protein polymerization | 42 | 284 | 1.358e-02 |
| GO:BP | GO:1902683 | regulation of receptor localization to synapse | 9 | 29 | 1.384e-02 |
| GO:BP | GO:0035767 | endothelial cell chemotaxis | 9 | 29 | 1.384e-02 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 9 | 29 | 1.384e-02 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 32 | 199 | 1.384e-02 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 27 | 158 | 1.384e-02 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 136 | 1182 | 1.398e-02 |
| GO:BP | GO:0099536 | synaptic signaling | 98 | 807 | 1.404e-02 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 11 | 41 | 1.404e-02 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 11 | 41 | 1.404e-02 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 65 | 493 | 1.415e-02 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 10 | 35 | 1.427e-02 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 37 | 242 | 1.442e-02 |
| GO:BP | GO:0030301 | cholesterol transport | 20 | 104 | 1.465e-02 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 45 | 312 | 1.466e-02 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 32 | 200 | 1.470e-02 |
| GO:BP | GO:0051960 | regulation of nervous system development | 61 | 457 | 1.475e-02 |
| GO:BP | GO:0007272 | ensheathment of neurons | 27 | 159 | 1.476e-02 |
| GO:BP | GO:0008366 | axon ensheathment | 27 | 159 | 1.476e-02 |
| GO:BP | GO:0043462 | regulation of ATP-dependent activity | 14 | 61 | 1.490e-02 |
| GO:BP | GO:0021817 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration | 3 | 3 | 1.491e-02 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 31 | 192 | 1.491e-02 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 58 | 430 | 1.499e-02 |
| GO:BP | GO:0060537 | muscle tissue development | 58 | 430 | 1.499e-02 |
| GO:BP | GO:0071450 | cellular response to oxygen radical | 8 | 24 | 1.540e-02 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 100 | 830 | 1.540e-02 |
| GO:BP | GO:0071451 | cellular response to superoxide | 8 | 24 | 1.540e-02 |
| GO:BP | GO:0008283 | cell population proliferation | 217 | 2015 | 1.540e-02 |
| GO:BP | GO:0009636 | response to toxic substance | 38 | 252 | 1.540e-02 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 105 | 879 | 1.540e-02 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 48 | 340 | 1.540e-02 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 28 | 168 | 1.552e-02 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 126 | 1087 | 1.552e-02 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 23 | 128 | 1.556e-02 |
| GO:BP | GO:0030049 | muscle filament sliding | 6 | 14 | 1.564e-02 |
| GO:BP | GO:0030497 | fatty acid elongation | 6 | 14 | 1.564e-02 |
| GO:BP | GO:0030048 | actin filament-based movement | 24 | 136 | 1.566e-02 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 31 | 193 | 1.566e-02 |
| GO:BP | GO:0046959 | habituation | 4 | 6 | 1.624e-02 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 163 | 1462 | 1.626e-02 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 64 | 488 | 1.651e-02 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 9 | 30 | 1.660e-02 |
| GO:BP | GO:0046621 | negative regulation of organ growth | 9 | 30 | 1.660e-02 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 49 | 351 | 1.693e-02 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 67 | 517 | 1.719e-02 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 30 | 186 | 1.729e-02 |
| GO:BP | GO:0030001 | metal ion transport | 105 | 883 | 1.729e-02 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 36 | 237 | 1.751e-02 |
| GO:BP | GO:1903034 | regulation of response to wounding | 28 | 170 | 1.805e-02 |
| GO:BP | GO:0060322 | head development | 97 | 806 | 1.811e-02 |
| GO:BP | GO:0070303 | negative regulation of stress-activated protein kinase signaling cascade | 5 | 10 | 1.842e-02 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 281 | 2696 | 1.843e-02 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 24 | 138 | 1.843e-02 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 52 | 380 | 1.843e-02 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 24 | 138 | 1.843e-02 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 24 | 138 | 1.843e-02 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 52 | 380 | 1.843e-02 |
| GO:BP | GO:0050921 | positive regulation of chemotaxis | 25 | 146 | 1.843e-02 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 16 | 77 | 1.857e-02 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 14 | 63 | 1.893e-02 |
| GO:BP | GO:0006518 | peptide metabolic process | 14 | 63 | 1.893e-02 |
| GO:BP | GO:0006259 | DNA metabolic process | 117 | 1005 | 1.893e-02 |
| GO:BP | GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | 11 | 43 | 1.898e-02 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 42 | 291 | 1.898e-02 |
| GO:BP | GO:2001259 | positive regulation of cation channel activity | 11 | 43 | 1.898e-02 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 28 | 171 | 1.904e-02 |
| GO:BP | GO:0061572 | actin filament bundle organization | 27 | 163 | 1.931e-02 |
| GO:BP | GO:0034504 | protein localization to nucleus | 45 | 318 | 1.931e-02 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 27 | 163 | 1.931e-02 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 10 | 37 | 1.998e-02 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 38 | 257 | 2.027e-02 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 38 | 257 | 2.027e-02 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 94 | 781 | 2.042e-02 |
| GO:BP | GO:0014037 | Schwann cell differentiation | 12 | 50 | 2.045e-02 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 39 | 266 | 2.050e-02 |
| GO:BP | GO:0001764 | neuron migration | 30 | 189 | 2.100e-02 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 50 | 365 | 2.119e-02 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 182 | 1669 | 2.119e-02 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 13 | 57 | 2.119e-02 |
| GO:BP | GO:0045780 | positive regulation of bone resorption | 7 | 20 | 2.123e-02 |
| GO:BP | GO:0014074 | response to purine-containing compound | 25 | 148 | 2.135e-02 |
| GO:BP | GO:0007267 | cell-cell signaling | 150 | 1342 | 2.141e-02 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 23 | 132 | 2.144e-02 |
| GO:BP | GO:0051956 | negative regulation of amino acid transport | 6 | 15 | 2.144e-02 |
| GO:BP | GO:0051284 | positive regulation of sequestering of calcium ion | 6 | 15 | 2.144e-02 |
| GO:BP | GO:2001028 | positive regulation of endothelial cell chemotaxis | 6 | 15 | 2.144e-02 |
| GO:BP | GO:0015850 | organic hydroxy compound transport | 39 | 267 | 2.144e-02 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 68 | 533 | 2.166e-02 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 86 | 706 | 2.204e-02 |
| GO:BP | GO:0042398 | modified amino acid biosynthetic process | 11 | 44 | 2.204e-02 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 67 | 524 | 2.204e-02 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 799 | 8393 | 2.239e-02 |
| GO:BP | GO:0015918 | sterol transport | 20 | 109 | 2.240e-02 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 68 | 534 | 2.245e-02 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 93 | 775 | 2.259e-02 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 93 | 775 | 2.259e-02 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 31 | 199 | 2.289e-02 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 28 | 174 | 2.330e-02 |
| GO:BP | GO:0032781 | positive regulation of ATP-dependent activity | 10 | 38 | 2.346e-02 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 15 | 72 | 2.352e-02 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 130 | 1144 | 2.357e-02 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 35 | 234 | 2.387e-02 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 19 | 102 | 2.387e-02 |
| GO:BP | GO:0071295 | cellular response to vitamin | 9 | 32 | 2.415e-02 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 29 | 183 | 2.421e-02 |
| GO:BP | GO:0008361 | regulation of cell size | 29 | 183 | 2.421e-02 |
| GO:BP | GO:0021782 | glial cell development | 23 | 134 | 2.508e-02 |
| GO:BP | GO:0032868 | response to insulin | 39 | 270 | 2.530e-02 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 16 | 80 | 2.545e-02 |
| GO:BP | GO:0033273 | response to vitamin | 18 | 95 | 2.545e-02 |
| GO:BP | GO:0010038 | response to metal ion | 48 | 351 | 2.545e-02 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 19 | 103 | 2.641e-02 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 108 | 928 | 2.646e-02 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 28 | 176 | 2.688e-02 |
| GO:BP | GO:0051036 | regulation of endosome size | 5 | 11 | 2.702e-02 |
| GO:BP | GO:0044062 | regulation of excretion | 5 | 11 | 2.702e-02 |
| GO:BP | GO:0072015 | podocyte development | 5 | 11 | 2.702e-02 |
| GO:BP | GO:0014812 | muscle cell migration | 17 | 88 | 2.702e-02 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 7 | 21 | 2.710e-02 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 149 | 1343 | 2.765e-02 |
| GO:BP | GO:0071711 | basement membrane organization | 10 | 39 | 2.779e-02 |
| GO:BP | GO:0045926 | negative regulation of growth | 34 | 228 | 2.797e-02 |
| GO:BP | GO:0007612 | learning | 26 | 160 | 2.797e-02 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 26 | 160 | 2.797e-02 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 50 | 372 | 2.897e-02 |
| GO:BP | GO:0048842 | positive regulation of axon extension involved in axon guidance | 4 | 7 | 2.905e-02 |
| GO:BP | GO:0098877 | neurotransmitter receptor transport to plasma membrane | 6 | 16 | 2.909e-02 |
| GO:BP | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation | 6 | 16 | 2.909e-02 |
| GO:BP | GO:0030167 | proteoglycan catabolic process | 6 | 16 | 2.909e-02 |
| GO:BP | GO:0000303 | response to superoxide | 8 | 27 | 2.913e-02 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 8 | 27 | 2.913e-02 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 27 | 169 | 2.924e-02 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 38 | 264 | 2.927e-02 |
| GO:BP | GO:0032964 | collagen biosynthetic process | 11 | 46 | 2.971e-02 |
| GO:BP | GO:0007599 | hemostasis | 35 | 238 | 3.004e-02 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 14 | 67 | 3.029e-02 |
| GO:BP | GO:0042776 | proton motive force-driven mitochondrial ATP synthesis | 14 | 67 | 3.029e-02 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 12 | 53 | 3.070e-02 |
| GO:BP | GO:0043113 | receptor clustering | 13 | 60 | 3.073e-02 |
| GO:BP | GO:0043542 | endothelial cell migration | 33 | 221 | 3.073e-02 |
| GO:BP | GO:0050890 | cognition | 45 | 328 | 3.104e-02 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 24 | 145 | 3.104e-02 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 68 | 543 | 3.115e-02 |
| GO:BP | GO:0007613 | memory | 22 | 129 | 3.179e-02 |
| GO:BP | GO:0019362 | pyridine nucleotide metabolic process | 26 | 162 | 3.208e-02 |
| GO:BP | GO:0046496 | nicotinamide nucleotide metabolic process | 26 | 162 | 3.208e-02 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 55 | 421 | 3.215e-02 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 268 | 2593 | 3.216e-02 |
| GO:BP | GO:0070925 | organelle assembly | 119 | 1046 | 3.231e-02 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 10 | 40 | 3.231e-02 |
| GO:BP | GO:0046777 | protein autophosphorylation | 28 | 179 | 3.231e-02 |
| GO:BP | GO:0043434 | response to peptide hormone | 57 | 440 | 3.233e-02 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 17 | 90 | 3.238e-02 |
| GO:BP | GO:0098609 | cell-cell adhesion | 112 | 976 | 3.238e-02 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 192 | 1795 | 3.272e-02 |
| GO:BP | GO:0007584 | response to nutrient | 27 | 171 | 3.314e-02 |
| GO:BP | GO:1990138 | neuron projection extension | 27 | 171 | 3.314e-02 |
| GO:BP | GO:0023056 | positive regulation of signaling | 192 | 1796 | 3.337e-02 |
| GO:BP | GO:0098801 | regulation of renal system process | 7 | 22 | 3.375e-02 |
| GO:BP | GO:0021762 | substantia nigra development | 11 | 47 | 3.375e-02 |
| GO:BP | GO:0019430 | removal of superoxide radicals | 7 | 22 | 3.375e-02 |
| GO:BP | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen | 7 | 22 | 3.375e-02 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 50 | 376 | 3.375e-02 |
| GO:BP | GO:0050919 | negative chemotaxis | 11 | 47 | 3.375e-02 |
| GO:BP | GO:0033627 | cell adhesion mediated by integrin | 16 | 83 | 3.412e-02 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 13 | 61 | 3.416e-02 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 13 | 61 | 3.416e-02 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 12 | 54 | 3.428e-02 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 12 | 54 | 3.428e-02 |
| GO:BP | GO:0009081 | branched-chain amino acid metabolic process | 8 | 28 | 3.489e-02 |
| GO:BP | GO:0000305 | response to oxygen radical | 8 | 28 | 3.489e-02 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 24 | 147 | 3.532e-02 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 17 | 91 | 3.532e-02 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 15 | 76 | 3.582e-02 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 57 | 443 | 3.608e-02 |
| GO:BP | GO:0072001 | renal system development | 45 | 332 | 3.676e-02 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 19 | 107 | 3.676e-02 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 50 | 378 | 3.676e-02 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 20 | 115 | 3.687e-02 |
| GO:BP | GO:0042254 | ribosome biogenesis | 44 | 323 | 3.692e-02 |
| GO:BP | GO:0014733 | regulation of skeletal muscle adaptation | 5 | 12 | 3.738e-02 |
| GO:BP | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction | 5 | 12 | 3.738e-02 |
| GO:BP | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway | 5 | 12 | 3.738e-02 |
| GO:BP | GO:1903909 | regulation of receptor clustering | 6 | 17 | 3.738e-02 |
| GO:BP | GO:2000009 | negative regulation of protein localization to cell surface | 5 | 12 | 3.738e-02 |
| GO:BP | GO:0099515 | actin filament-based transport | 5 | 12 | 3.738e-02 |
| GO:BP | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase | 5 | 12 | 3.738e-02 |
| GO:BP | GO:0006094 | gluconeogenesis | 16 | 84 | 3.738e-02 |
| GO:BP | GO:0018126 | protein hydroxylation | 6 | 17 | 3.738e-02 |
| GO:BP | GO:0043589 | skin morphogenesis | 5 | 12 | 3.738e-02 |
| GO:BP | GO:0072310 | glomerular epithelial cell development | 5 | 12 | 3.738e-02 |
| GO:BP | GO:0060414 | aorta smooth muscle tissue morphogenesis | 3 | 4 | 3.817e-02 |
| GO:BP | GO:0042822 | pyridoxal phosphate metabolic process | 3 | 4 | 3.817e-02 |
| GO:BP | GO:0060024 | rhythmic synaptic transmission | 3 | 4 | 3.817e-02 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 17 | 92 | 3.817e-02 |
| GO:BP | GO:0006273 | lagging strand elongation | 3 | 4 | 3.817e-02 |
| GO:BP | GO:1905749 | regulation of endosome to plasma membrane protein transport | 3 | 4 | 3.817e-02 |
| GO:BP | GO:0030207 | chondroitin sulfate proteoglycan catabolic process | 3 | 4 | 3.817e-02 |
| GO:BP | GO:0007420 | brain development | 89 | 754 | 3.817e-02 |
| GO:BP | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process | 3 | 4 | 3.817e-02 |
| GO:BP | GO:0050805 | negative regulation of synaptic transmission | 12 | 55 | 3.817e-02 |
| GO:BP | GO:0061041 | regulation of wound healing | 22 | 132 | 3.834e-02 |
| GO:BP | GO:0032368 | regulation of lipid transport | 22 | 132 | 3.834e-02 |
| GO:BP | GO:0046683 | response to organophosphorus | 22 | 132 | 3.834e-02 |
| GO:BP | GO:0050773 | regulation of dendrite development | 18 | 100 | 3.856e-02 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 242 | 2331 | 3.868e-02 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 9 | 35 | 3.926e-02 |
| GO:BP | GO:0098754 | detoxification | 24 | 149 | 3.983e-02 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 106 | 925 | 3.988e-02 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 14 | 70 | 4.073e-02 |
| GO:BP | GO:0006801 | superoxide metabolic process | 14 | 70 | 4.073e-02 |
| GO:BP | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | 7 | 23 | 4.085e-02 |
| GO:BP | GO:0033043 | regulation of organelle organization | 128 | 1148 | 4.085e-02 |
| GO:BP | GO:0032891 | negative regulation of organic acid transport | 7 | 23 | 4.085e-02 |
| GO:BP | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 7 | 23 | 4.085e-02 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 10 | 42 | 4.227e-02 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 18 | 101 | 4.227e-02 |
| GO:BP | GO:0048144 | fibroblast proliferation | 19 | 109 | 4.251e-02 |
| GO:BP | GO:0007623 | circadian rhythm | 31 | 210 | 4.285e-02 |
| GO:BP | GO:0006935 | chemotaxis | 59 | 467 | 4.285e-02 |
| GO:BP | GO:0015986 | proton motive force-driven ATP synthesis | 15 | 78 | 4.294e-02 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 26 | 167 | 4.299e-02 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 12 | 56 | 4.299e-02 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 12 | 56 | 4.299e-02 |
| GO:BP | GO:0048662 | negative regulation of smooth muscle cell proliferation | 11 | 49 | 4.306e-02 |
| GO:BP | GO:0006164 | purine nucleotide biosynthetic process | 29 | 193 | 4.371e-02 |
| GO:BP | GO:0055006 | cardiac cell development | 16 | 86 | 4.422e-02 |
| GO:BP | GO:0006633 | fatty acid biosynthetic process | 25 | 159 | 4.422e-02 |
| GO:BP | GO:0044042 | glucan metabolic process | 16 | 86 | 4.422e-02 |
| GO:BP | GO:0032456 | endocytic recycling | 16 | 86 | 4.422e-02 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 35 | 246 | 4.422e-02 |
| GO:BP | GO:0035813 | regulation of renal sodium excretion | 4 | 8 | 4.423e-02 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 52 | 402 | 4.423e-02 |
| GO:BP | GO:0017185 | peptidyl-lysine hydroxylation | 4 | 8 | 4.423e-02 |
| GO:BP | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling | 4 | 8 | 4.423e-02 |
| GO:BP | GO:0014052 | regulation of gamma-aminobutyric acid secretion | 4 | 8 | 4.423e-02 |
| GO:BP | GO:0042330 | taxis | 59 | 469 | 4.546e-02 |
| GO:BP | GO:0048638 | regulation of developmental growth | 42 | 310 | 4.546e-02 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 18 | 102 | 4.546e-02 |
| GO:BP | GO:0019058 | viral life cycle | 42 | 310 | 4.546e-02 |
| GO:BP | GO:0072526 | pyridine-containing compound catabolic process | 18 | 102 | 4.546e-02 |
| GO:BP | GO:0006099 | tricarboxylic acid cycle | 9 | 36 | 4.557e-02 |
| GO:BP | GO:0050817 | coagulation | 34 | 238 | 4.600e-02 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 117 | 1042 | 4.607e-02 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 23 | 143 | 4.621e-02 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 152 | 1400 | 4.622e-02 |
| GO:BP | GO:0097120 | receptor localization to synapse | 15 | 79 | 4.640e-02 |
| GO:BP | GO:0099518 | vesicle cytoskeletal trafficking | 13 | 64 | 4.640e-02 |
| GO:BP | GO:0045722 | positive regulation of gluconeogenesis | 6 | 18 | 4.640e-02 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 13 | 64 | 4.640e-02 |
| GO:BP | GO:1902170 | cellular response to reactive nitrogen species | 6 | 18 | 4.640e-02 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 15 | 79 | 4.640e-02 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 12 | 57 | 4.739e-02 |
| GO:BP | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 12 | 57 | 4.739e-02 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 10 | 43 | 4.739e-02 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 12 | 57 | 4.739e-02 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 10 | 43 | 4.739e-02 |
| GO:BP | GO:0061245 | establishment or maintenance of bipolar cell polarity | 12 | 57 | 4.739e-02 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 12 | 57 | 4.739e-02 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 40 | 293 | 4.742e-02 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 16 | 87 | 4.742e-02 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 16 | 87 | 4.742e-02 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 76 | 635 | 4.744e-02 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 39 | 284 | 4.752e-02 |
| GO:BP | GO:0048668 | collateral sprouting | 8 | 30 | 4.779e-02 |
| GO:BP | GO:0006096 | glycolytic process | 17 | 95 | 4.815e-02 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 19 | 111 | 4.838e-02 |
| GO:BP | GO:0009191 | ribonucleoside diphosphate catabolic process | 18 | 103 | 4.841e-02 |
| GO:BP | GO:0042659 | regulation of cell fate specification | 7 | 24 | 4.854e-02 |
| GO:BP | GO:0043501 | skeletal muscle adaptation | 7 | 24 | 4.854e-02 |
| GO:BP | GO:0036119 | response to platelet-derived growth factor | 7 | 24 | 4.854e-02 |
| GO:BP | GO:0044409 | symbiont entry into host | 23 | 144 | 4.854e-02 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 14 | 72 | 4.854e-02 |
| GO:BP | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c | 5 | 13 | 4.902e-02 |
| GO:BP | GO:0010745 | negative regulation of macrophage derived foam cell differentiation | 5 | 13 | 4.902e-02 |
| GO:BP | GO:0007596 | blood coagulation | 33 | 231 | 4.965e-02 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 164 | 1529 | 5.000e-02 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 29 | 87 | 6.023e-06 |
| KEGG | KEGG:01100 | Metabolic pathways | 227 | 1537 | 2.121e-05 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 28 | 89 | 2.121e-05 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 50 | 221 | 2.905e-05 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 46 | 202 | 4.712e-05 |
| KEGG | KEGG:04510 | Focal adhesion | 46 | 202 | 4.712e-05 |
| KEGG | KEGG:01200 | Carbon metabolism | 31 | 115 | 6.136e-05 |
| KEGG | KEGG:05016 | Huntington disease | 61 | 305 | 6.731e-05 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 34 | 134 | 6.731e-05 |
| KEGG | KEGG:05012 | Parkinson disease | 54 | 265 | 1.277e-04 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 27 | 102 | 2.721e-04 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 26 | 97 | 2.854e-04 |
| KEGG | KEGG:04714 | Thermogenesis | 47 | 232 | 4.905e-04 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 80 | 474 | 9.798e-04 |
| KEGG | KEGG:05020 | Prion disease | 51 | 271 | 1.510e-03 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 32 | 153 | 4.572e-03 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 19 | 74 | 5.697e-03 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 61 | 363 | 7.134e-03 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 24 | 107 | 7.951e-03 |
| KEGG | KEGG:01210 | 2-Oxocarboxylic acid metabolism | 11 | 33 | 8.279e-03 |
| KEGG | KEGG:05010 | Alzheimer disease | 63 | 382 | 8.279e-03 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 31 | 154 | 8.362e-03 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 28 | 137 | 1.137e-02 |
| KEGG | KEGG:00630 | Glyoxylate and dicarboxylate metabolism | 10 | 30 | 1.299e-02 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 19 | 84 | 1.921e-02 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 13 | 48 | 1.921e-02 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 54 | 330 | 1.921e-02 |
| KEGG | KEGG:04925 | Aldosterone synthesis and secretion | 21 | 98 | 2.218e-02 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 24 | 118 | 2.234e-02 |
| KEGG | KEGG:00480 | Glutathione metabolism | 14 | 56 | 2.609e-02 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 39 | 227 | 2.934e-02 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 14 | 57 | 2.934e-02 |
| KEGG | KEGG:03050 | Proteasome | 12 | 46 | 3.271e-02 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 9 | 30 | 3.725e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 19 | 92 | 4.297e-02 |
| KEGG | KEGG:05200 | Pathways in cancer | 77 | 527 | 4.297e-02 |
#GO:BP
table_DOX24R_genes_GOBP <- table_DOX24R_genes %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24R_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24R Specific DEGs Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))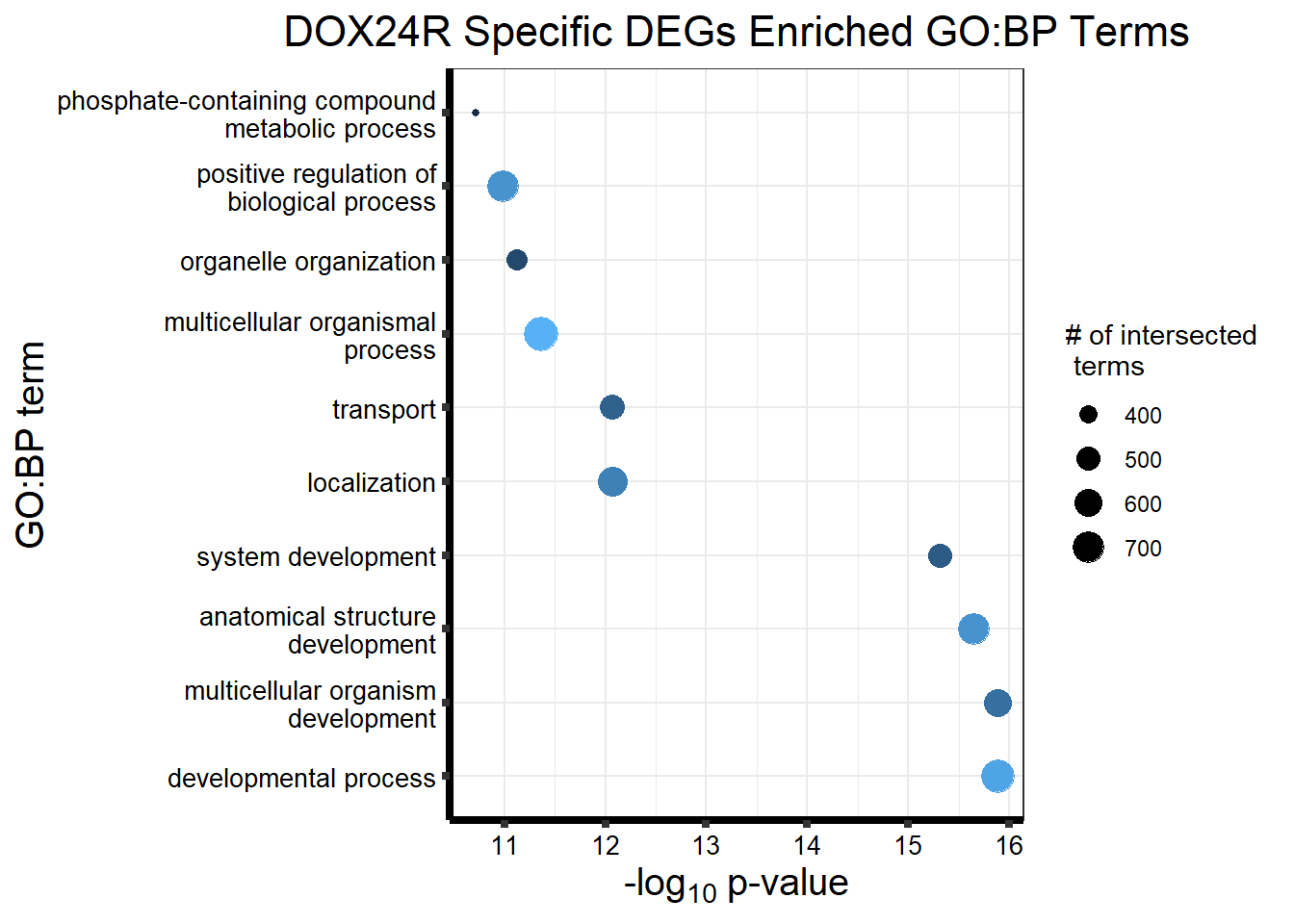
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_DOX24R_genes_KEGG <- table_DOX24R_genes %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24R_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24R DEGs Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))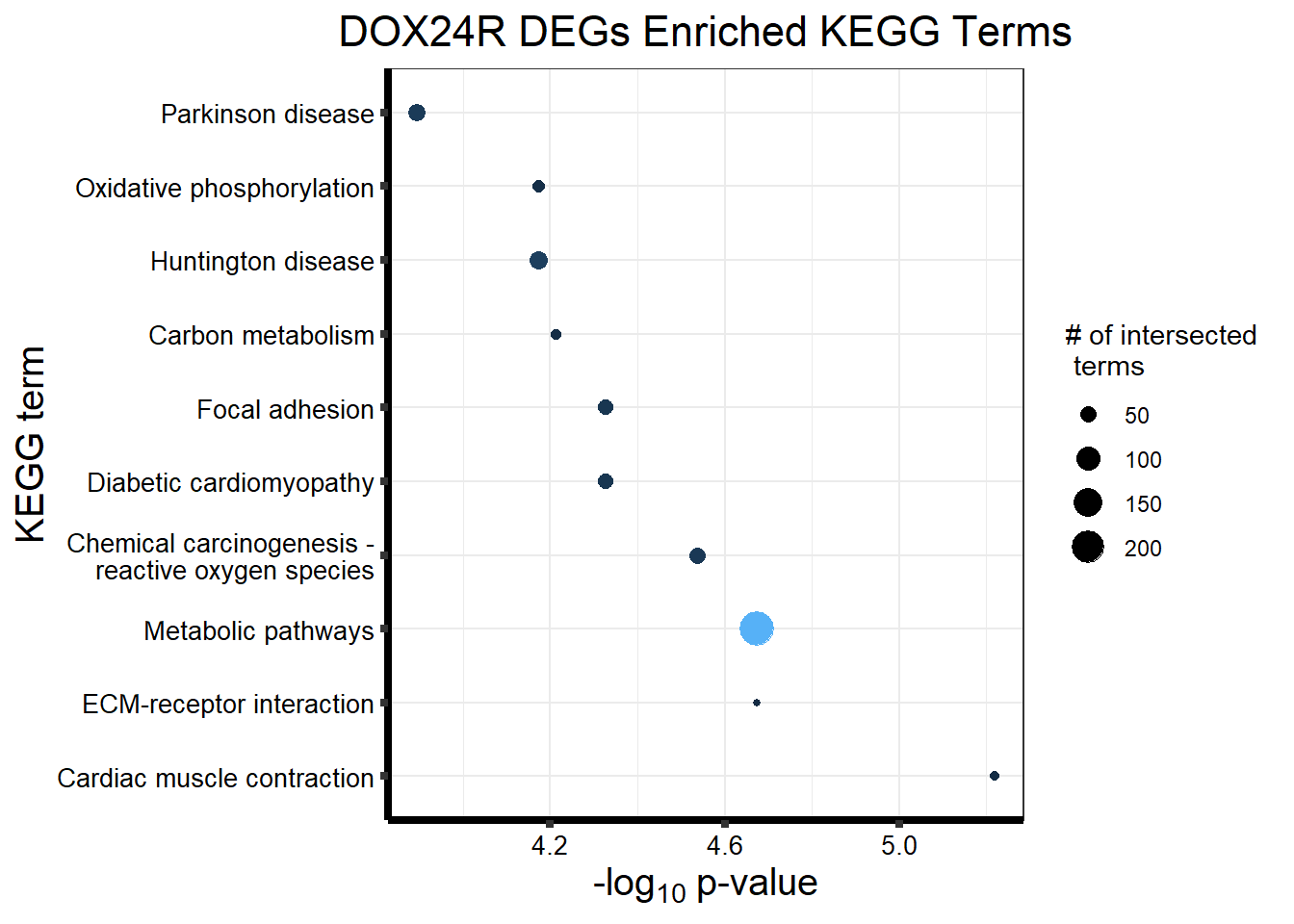
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####DOX144R DEGs GO KEGG#####
# D144R_DEGs_mat <- as.matrix(DOX144R_DEGs_GO)
#
# DOX_144R_dxr_gene <- gost(query = D144R_DEGs_mat,
# organism = "hsapiens",
# ordered_query = FALSE,
# measure_underrepresentation = FALSE,
# evcodes = FALSE,
# user_threshold = 0.05,
# correction_method = c("fdr"),
# sources = c("GO:BP", "KEGG"))
#
# DOX_144R_gost_genes <- gostplot(DOX_144R_dxr_gene, capped = FALSE, interactive = TRUE)
# DOX_144R_gost_genes
#
# table_DOX144R_genes <- DOX_144R_dxr_gene$result %>%
# dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
#
# table_DOX144R_genes %>%
# mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
# kableExtra::kable(.,) %>%
# kableExtra::kable_paper("striped", full_width = FALSE) %>%
# kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
# kableExtra::scroll_box(width = "100%", height = "400px")
#
# #GO:BP
# table_DOX144R_genes_GOBP <- table_DOX144R_genes %>%
# dplyr::filter(source=="GO:BP") %>%
# dplyr::select(p_value, term_name, intersection_size) %>%
# dplyr::slice_min(., n=10, order_by=p_value) %>%
# mutate(log_val = -log10(p_value))
#
# table_DOX144R_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
# geom_point(aes(size = intersection_size)) +
# ggtitle("DOX144R Specific DEGs Enriched GO:BP Terms")+
# xlab(expression("-log"[10]~"p-value"))+
# guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
# ylab("GO:BP term")+
# scale_y_discrete(labels = scales::label_wrap(30))+
# theme_bw()+
# theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
# axis.title = element_text(size = 15, colour = "black"),
# axis.ticks = element_line(linewidth = 1.5),
# axis.line = element_line(linewidth = 1.5),
# axis.text = element_text(size = 10, colour = "black", angle = 0),
# strip.text = element_text(size = 15, colour = "black", face = "bold"))
#
# #KEGG
# table_DOX144R_genes_KEGG <- table_DOX144R_genes %>%
# dplyr::filter(source=="KEGG") %>%
# dplyr::select(p_value, term_name, intersection_size) %>%
# dplyr::slice_min(., n=10, order_by=p_value) %>%
# mutate(log_val = -log10(p_value))
#
# table_DOX144R_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
# geom_point(aes(size = intersection_size)) +
# ggtitle("DOX144R DEGs Enriched KEGG Terms")+
# xlab(expression("-log"[10]~"p-value"))+
# guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
# ylab("KEGG term")+
# scale_y_discrete(labels = scales::label_wrap(30))+
# theme_bw()+
# theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
# axis.title = element_text(size = 15, colour = "black"),
# axis.ticks = element_line(linewidth = 1.5),
# axis.line = element_line(linewidth = 1.5),
# axis.text = element_text(size = 10, colour = "black", angle = 0),
# strip.text = element_text(size = 15, colour = "black", face = "bold"))
#####DOX144R shared DEGs GO KEGG#####
D144Rshare_DEGs_mat <- as.matrix(DOX144Rshare_DEGs_GO_plot)
DOX_144Rshare_dxr_gene <- gost(query = D144Rshare_DEGs_mat,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
DOX_144Rshare_gost_genes <- gostplot(DOX_144Rshare_dxr_gene, capped = FALSE, interactive = TRUE)
DOX_144Rshare_gost_genestable_DOX144Rshare_genes <- DOX_144Rshare_dxr_gene$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX144Rshare_genes %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0022402 | cell cycle process | 169 | 1280 | 8.265e-83 |
| GO:BP | GO:0007049 | cell cycle | 180 | 1663 | 5.991e-75 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 128 | 745 | 1.284e-74 |
| GO:BP | GO:0000278 | mitotic cell cycle | 135 | 892 | 5.412e-72 |
| GO:BP | GO:0007059 | chromosome segregation | 97 | 427 | 4.095e-67 |
| GO:BP | GO:0051301 | cell division | 104 | 658 | 7.689e-56 |
| GO:BP | GO:0000280 | nuclear division | 89 | 452 | 1.399e-55 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 78 | 323 | 1.859e-55 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 107 | 720 | 5.055e-55 |
| GO:BP | GO:0048285 | organelle fission | 90 | 500 | 7.171e-53 |
| GO:BP | GO:0051276 | chromosome organization | 94 | 574 | 1.210e-51 |
| GO:BP | GO:0140014 | mitotic nuclear division | 70 | 282 | 1.752e-50 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 60 | 193 | 2.327e-49 |
| GO:BP | GO:0000819 | sister chromatid segregation | 64 | 234 | 6.805e-49 |
| GO:BP | GO:0051726 | regulation of cell cycle | 120 | 1087 | 3.086e-48 |
| GO:BP | GO:0044770 | cell cycle phase transition | 85 | 532 | 1.429e-45 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 73 | 427 | 8.453e-41 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 72 | 428 | 9.899e-40 |
| GO:BP | GO:0006260 | DNA replication | 60 | 283 | 9.020e-39 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 49 | 185 | 2.536e-36 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 72 | 492 | 1.354e-35 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 53 | 248 | 2.918e-34 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 41 | 132 | 2.543e-33 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 44 | 160 | 2.638e-33 |
| GO:BP | GO:0006259 | DNA metabolic process | 97 | 1005 | 2.876e-33 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 58 | 327 | 5.204e-33 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 55 | 289 | 6.743e-33 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 33 | 77 | 4.239e-32 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 40 | 136 | 1.630e-31 |
| GO:BP | GO:0051310 | metaphase chromosome alignment | 36 | 104 | 3.831e-31 |
| GO:BP | GO:0051304 | chromosome separation | 33 | 84 | 1.299e-30 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 50 | 262 | 6.907e-30 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 37 | 121 | 8.257e-30 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 58 | 377 | 1.216e-29 |
| GO:BP | GO:0050000 | chromosome localization | 37 | 126 | 4.140e-29 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 36 | 118 | 5.703e-29 |
| GO:BP | GO:0006974 | DNA damage response | 86 | 908 | 1.113e-28 |
| GO:BP | GO:0006281 | DNA repair | 71 | 621 | 2.588e-28 |
| GO:BP | GO:0051783 | regulation of nuclear division | 38 | 149 | 1.940e-27 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 44 | 225 | 1.071e-26 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 27 | 62 | 1.854e-26 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 39 | 169 | 1.976e-26 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 27 | 65 | 8.610e-26 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 52 | 351 | 1.042e-25 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 32 | 107 | 2.015e-25 |
| GO:BP | GO:0006996 | organelle organization | 177 | 3594 | 2.448e-25 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 25 | 54 | 2.477e-25 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 24 | 49 | 4.488e-25 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 24 | 49 | 4.488e-25 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 24 | 49 | 4.488e-25 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 38 | 172 | 4.616e-25 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 24 | 50 | 8.130e-25 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 25 | 57 | 1.219e-24 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 24 | 51 | 1.373e-24 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 24 | 51 | 1.373e-24 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 24 | 51 | 1.373e-24 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 24 | 51 | 1.373e-24 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 30 | 97 | 2.236e-24 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 24 | 53 | 4.178e-24 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 24 | 53 | 4.178e-24 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 24 | 53 | 4.178e-24 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 30 | 100 | 5.811e-24 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 29 | 93 | 1.084e-23 |
| GO:BP | GO:0051321 | meiotic cell cycle | 46 | 297 | 1.336e-23 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 29 | 96 | 2.947e-23 |
| GO:BP | GO:0007051 | spindle organization | 39 | 206 | 3.356e-23 |
| GO:BP | GO:0033554 | cellular response to stress | 114 | 1830 | 7.833e-23 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 40 | 224 | 8.005e-23 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 24 | 59 | 9.066e-23 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 41 | 242 | 1.645e-22 |
| GO:BP | GO:0007052 | mitotic spindle organization | 32 | 134 | 3.088e-22 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 24 | 65 | 1.411e-21 |
| GO:BP | GO:0140013 | meiotic nuclear division | 37 | 202 | 1.746e-21 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 33 | 156 | 3.616e-21 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 31 | 139 | 1.389e-20 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 62 | 670 | 7.255e-20 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 25 | 97 | 3.767e-18 |
| GO:BP | GO:0006302 | double-strand break repair | 41 | 319 | 6.401e-18 |
| GO:BP | GO:0006270 | DNA replication initiation | 17 | 37 | 3.948e-17 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 24 | 96 | 4.416e-17 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 46 | 430 | 5.855e-17 |
| GO:BP | GO:0051656 | establishment of organelle localization | 49 | 491 | 7.415e-17 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 26 | 123 | 1.366e-16 |
| GO:BP | GO:0051225 | spindle assembly | 27 | 136 | 1.654e-16 |
| GO:BP | GO:0007080 | mitotic metaphase chromosome alignment | 20 | 63 | 1.906e-16 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 24 | 105 | 3.934e-16 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 27 | 141 | 4.217e-16 |
| GO:BP | GO:0034508 | centromere complex assembly | 15 | 29 | 4.217e-16 |
| GO:BP | GO:0007017 | microtubule-based process | 70 | 999 | 6.111e-16 |
| GO:BP | GO:0006310 | DNA recombination | 40 | 347 | 7.888e-16 |
| GO:BP | GO:0051383 | kinetochore organization | 13 | 21 | 2.587e-15 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 15 | 32 | 2.768e-15 |
| GO:BP | GO:0033043 | regulation of organelle organization | 74 | 1148 | 5.858e-15 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 24 | 118 | 6.366e-15 |
| GO:BP | GO:0051640 | organelle localization | 52 | 620 | 9.313e-15 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 117 | 2433 | 1.266e-14 |
| GO:BP | GO:0000725 | recombinational repair | 29 | 193 | 2.078e-14 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 14 | 30 | 3.157e-14 |
| GO:BP | GO:0051716 | cellular response to stimulus | 248 | 7376 | 4.246e-14 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 28 | 188 | 8.284e-14 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 10 | 12 | 8.310e-14 |
| GO:BP | GO:0007127 | meiosis I | 24 | 133 | 1.031e-13 |
| GO:BP | GO:0034502 | protein localization to chromosome | 23 | 126 | 3.040e-13 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 12 | 23 | 6.528e-13 |
| GO:BP | GO:0035556 | intracellular signal transduction | 128 | 2965 | 1.261e-12 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 12 | 25 | 2.366e-12 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 21 | 114 | 3.693e-12 |
| GO:BP | GO:0033260 | nuclear DNA replication | 14 | 41 | 5.550e-12 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 29 | 241 | 6.872e-12 |
| GO:BP | GO:0000910 | cytokinesis | 26 | 192 | 7.745e-12 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 19 | 93 | 7.745e-12 |
| GO:BP | GO:0007010 | cytoskeleton organization | 81 | 1529 | 8.450e-12 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 19 | 95 | 1.139e-11 |
| GO:BP | GO:0006950 | response to stress | 153 | 3948 | 1.169e-11 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 17 | 74 | 1.835e-11 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 21 | 124 | 1.919e-11 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 14 | 45 | 2.236e-11 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 17 | 76 | 2.860e-11 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 41 | 500 | 2.907e-11 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 20 | 114 | 3.381e-11 |
| GO:BP | GO:0016043 | cellular component organization | 257 | 8184 | 4.377e-11 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 29 | 262 | 5.211e-11 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 14 | 48 | 5.785e-11 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 13 | 40 | 7.637e-11 |
| GO:BP | GO:0006275 | regulation of DNA replication | 20 | 120 | 8.795e-11 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 18 | 94 | 9.552e-11 |
| GO:BP | GO:0050896 | response to stimulus | 274 | 8999 | 1.230e-10 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 33 | 352 | 1.430e-10 |
| GO:BP | GO:0019953 | sexual reproduction | 64 | 1122 | 1.571e-10 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 9 | 15 | 2.056e-10 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 9 | 15 | 2.056e-10 |
| GO:BP | GO:0070925 | organelle assembly | 60 | 1046 | 6.221e-10 |
| GO:BP | GO:0022414 | reproductive process | 79 | 1608 | 6.721e-10 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 257 | 8393 | 9.106e-10 |
| GO:BP | GO:0051382 | kinetochore assembly | 9 | 17 | 9.243e-10 |
| GO:BP | GO:1902969 | mitotic DNA replication | 9 | 17 | 9.243e-10 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 15 | 70 | 1.092e-09 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 19 | 124 | 1.309e-09 |
| GO:BP | GO:0051302 | regulation of cell division | 23 | 188 | 1.309e-09 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 12 | 41 | 1.969e-09 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 10 | 25 | 2.273e-09 |
| GO:BP | GO:0009314 | response to radiation | 35 | 437 | 2.411e-09 |
| GO:BP | GO:0007143 | female meiotic nuclear division | 12 | 42 | 2.645e-09 |
| GO:BP | GO:0051231 | spindle elongation | 8 | 14 | 5.133e-09 |
| GO:BP | GO:0030261 | chromosome condensation | 12 | 45 | 6.387e-09 |
| GO:BP | GO:0051653 | spindle localization | 14 | 68 | 8.293e-09 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 9 | 21 | 9.552e-09 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 7 | 10 | 9.594e-09 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 7 | 10 | 9.594e-09 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 14 | 69 | 9.923e-09 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 21 | 173 | 9.990e-09 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 8 | 15 | 1.009e-08 |
| GO:BP | GO:0034501 | protein localization to kinetochore | 8 | 15 | 1.009e-08 |
| GO:BP | GO:1903083 | protein localization to condensed chromosome | 8 | 15 | 1.009e-08 |
| GO:BP | GO:0010212 | response to ionizing radiation | 19 | 141 | 1.135e-08 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 14 | 71 | 1.426e-08 |
| GO:BP | GO:0050794 | regulation of cellular process | 330 | 11946 | 1.480e-08 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 12 | 50 | 2.237e-08 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 7 | 11 | 2.425e-08 |
| GO:BP | GO:0050789 | regulation of biological process | 337 | 12336 | 2.848e-08 |
| GO:BP | GO:0051293 | establishment of spindle localization | 13 | 63 | 3.211e-08 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 9 | 24 | 3.611e-08 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 9 | 24 | 3.611e-08 |
| GO:BP | GO:0000022 | mitotic spindle elongation | 7 | 12 | 5.535e-08 |
| GO:BP | GO:1905821 | positive regulation of chromosome condensation | 6 | 8 | 9.384e-08 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 8 | 19 | 1.016e-07 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 12 | 57 | 1.071e-07 |
| GO:BP | GO:0065007 | biological regulation | 343 | 12743 | 1.148e-07 |
| GO:BP | GO:0008283 | cell population proliferation | 85 | 2015 | 1.572e-07 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 12 | 59 | 1.599e-07 |
| GO:BP | GO:0000076 | DNA replication checkpoint signaling | 8 | 20 | 1.612e-07 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 34 | 492 | 1.719e-07 |
| GO:BP | GO:0051255 | spindle midzone assembly | 7 | 14 | 2.187e-07 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 56 | 1105 | 2.211e-07 |
| GO:BP | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | 6 | 9 | 2.604e-07 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 22 | 230 | 3.001e-07 |
| GO:BP | GO:0007098 | centrosome cycle | 17 | 140 | 4.203e-07 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 41 | 701 | 5.461e-07 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 12 | 67 | 6.923e-07 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 20 | 202 | 7.680e-07 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 11 | 55 | 7.878e-07 |
| GO:BP | GO:0035825 | homologous recombination | 12 | 68 | 8.104e-07 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 185 | 5920 | 1.444e-06 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 17 | 153 | 1.534e-06 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 18 | 172 | 1.581e-06 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 10 | 47 | 1.760e-06 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 14 | 105 | 2.275e-06 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 9 | 37 | 2.303e-06 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 6 | 12 | 2.493e-06 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 6 | 12 | 2.493e-06 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 5 | 7 | 2.763e-06 |
| GO:BP | GO:0071478 | cellular response to radiation | 18 | 182 | 3.619e-06 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 54 | 1144 | 3.916e-06 |
| GO:BP | GO:0090232 | positive regulation of spindle checkpoint | 6 | 13 | 4.426e-06 |
| GO:BP | GO:1903490 | positive regulation of mitotic cytokinesis | 6 | 13 | 4.426e-06 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 11 | 66 | 5.239e-06 |
| GO:BP | GO:0051781 | positive regulation of cell division | 13 | 98 | 6.639e-06 |
| GO:BP | GO:0048518 | positive regulation of biological process | 190 | 6264 | 7.661e-06 |
| GO:BP | GO:0000212 | meiotic spindle organization | 7 | 22 | 8.096e-06 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 9 | 43 | 8.717e-06 |
| GO:BP | GO:0040007 | growth | 46 | 929 | 8.928e-06 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 6 | 15 | 1.200e-05 |
| GO:BP | GO:0000915 | actomyosin contractile ring assembly | 5 | 9 | 1.496e-05 |
| GO:BP | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis | 5 | 9 | 1.496e-05 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 22 | 291 | 1.631e-05 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 24 | 339 | 1.657e-05 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 24 | 339 | 1.657e-05 |
| GO:BP | GO:1902412 | regulation of mitotic cytokinesis | 6 | 16 | 1.829e-05 |
| GO:BP | GO:0045840 | positive regulation of mitotic nuclear division | 9 | 47 | 1.863e-05 |
| GO:BP | GO:0048589 | developmental growth | 36 | 663 | 2.075e-05 |
| GO:BP | GO:0090224 | regulation of spindle organization | 9 | 48 | 2.227e-05 |
| GO:BP | GO:0048856 | anatomical structure development | 181 | 5997 | 2.508e-05 |
| GO:BP | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition | 6 | 17 | 2.709e-05 |
| GO:BP | GO:0051307 | meiotic chromosome separation | 5 | 10 | 2.787e-05 |
| GO:BP | GO:0044837 | actomyosin contractile ring organization | 5 | 10 | 2.787e-05 |
| GO:BP | GO:0022616 | DNA strand elongation | 8 | 37 | 2.805e-05 |
| GO:BP | GO:0032502 | developmental process | 194 | 6553 | 2.931e-05 |
| GO:BP | GO:0032506 | cytokinetic process | 9 | 50 | 3.095e-05 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 10 | 64 | 3.119e-05 |
| GO:BP | GO:0031297 | replication fork processing | 9 | 51 | 3.651e-05 |
| GO:BP | GO:1902101 | positive regulation of metaphase/anaphase transition of cell cycle | 6 | 18 | 3.842e-05 |
| GO:BP | GO:0007292 | female gamete generation | 16 | 174 | 4.084e-05 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 7 | 28 | 4.468e-05 |
| GO:BP | GO:0009411 | response to UV | 15 | 155 | 4.476e-05 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 44 | 928 | 4.579e-05 |
| GO:BP | GO:0033314 | mitotic DNA replication checkpoint signaling | 5 | 11 | 4.771e-05 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 67 | 1685 | 5.133e-05 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 10 | 68 | 5.312e-05 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 6 | 19 | 5.318e-05 |
| GO:BP | GO:0016310 | phosphorylation | 56 | 1320 | 5.607e-05 |
| GO:BP | GO:0006468 | protein phosphorylation | 53 | 1227 | 6.307e-05 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 8 | 42 | 7.241e-05 |
| GO:BP | GO:0010032 | meiotic chromosome condensation | 4 | 6 | 7.341e-05 |
| GO:BP | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 5 | 12 | 7.722e-05 |
| GO:BP | GO:0007056 | spindle assembly involved in female meiosis | 5 | 12 | 7.722e-05 |
| GO:BP | GO:0048513 | animal organ development | 105 | 3085 | 7.862e-05 |
| GO:BP | GO:0007275 | multicellular organism development | 147 | 4727 | 8.565e-05 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 10 | 72 | 8.669e-05 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 7 | 31 | 8.702e-05 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 7 | 31 | 8.702e-05 |
| GO:BP | GO:0007154 | cell communication | 191 | 6540 | 9.128e-05 |
| GO:BP | GO:0007165 | signal transduction | 178 | 6002 | 9.531e-05 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 8 | 44 | 9.943e-05 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 8 | 44 | 9.943e-05 |
| GO:BP | GO:0023052 | signaling | 190 | 6515 | 1.076e-04 |
| GO:BP | GO:0007144 | female meiosis I | 5 | 13 | 1.177e-04 |
| GO:BP | GO:0140527 | reciprocal homologous recombination | 9 | 60 | 1.318e-04 |
| GO:BP | GO:0007131 | reciprocal meiotic recombination | 9 | 60 | 1.318e-04 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 168 | 5629 | 1.439e-04 |
| GO:BP | GO:0051338 | regulation of transferase activity | 28 | 498 | 1.534e-04 |
| GO:BP | GO:1901994 | negative regulation of meiotic cell cycle phase transition | 4 | 7 | 1.556e-04 |
| GO:BP | GO:0044785 | metaphase/anaphase transition of meiotic cell cycle | 4 | 7 | 1.556e-04 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 5 | 14 | 1.747e-04 |
| GO:BP | GO:0033316 | meiotic spindle assembly checkpoint signaling | 3 | 3 | 2.103e-04 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 10 | 80 | 2.120e-04 |
| GO:BP | GO:0048519 | negative regulation of biological process | 172 | 5834 | 2.158e-04 |
| GO:BP | GO:0006301 | postreplication repair | 7 | 36 | 2.343e-04 |
| GO:BP | GO:0007276 | gamete generation | 39 | 841 | 2.577e-04 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 7 | 37 | 2.813e-04 |
| GO:BP | GO:0009790 | embryo development | 48 | 1135 | 2.944e-04 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 4 | 8 | 2.951e-04 |
| GO:BP | GO:0010332 | response to gamma radiation | 8 | 52 | 3.365e-04 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 14 | 164 | 3.531e-04 |
| GO:BP | GO:0051649 | establishment of localization in cell | 73 | 2010 | 3.594e-04 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 11 | 105 | 4.127e-04 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 71 | 1953 | 4.571e-04 |
| GO:BP | GO:0031573 | mitotic intra-S DNA damage checkpoint signaling | 5 | 17 | 4.843e-04 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 4 | 9 | 5.064e-04 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 4 | 9 | 5.064e-04 |
| GO:BP | GO:0048731 | system development | 126 | 4053 | 5.234e-04 |
| GO:BP | GO:0051447 | negative regulation of meiotic cell cycle | 5 | 18 | 6.486e-04 |
| GO:BP | GO:0010165 | response to X-ray | 6 | 29 | 6.489e-04 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 9 | 74 | 6.801e-04 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 14 | 176 | 7.443e-04 |
| GO:BP | GO:1990086 | lens fiber cell apoptotic process | 3 | 4 | 7.532e-04 |
| GO:BP | GO:0003150 | muscular septum morphogenesis | 3 | 4 | 7.532e-04 |
| GO:BP | GO:0007057 | spindle assembly involved in female meiosis I | 3 | 4 | 7.532e-04 |
| GO:BP | GO:0044779 | meiotic spindle checkpoint signaling | 3 | 4 | 7.532e-04 |
| GO:BP | GO:0043060 | meiotic metaphase I homologous chromosome alignment | 3 | 4 | 7.532e-04 |
| GO:BP | GO:0033313 | meiotic cell cycle checkpoint signaling | 4 | 10 | 7.965e-04 |
| GO:BP | GO:2000241 | regulation of reproductive process | 15 | 203 | 9.123e-04 |
| GO:BP | GO:0071168 | protein localization to chromatin | 8 | 60 | 9.123e-04 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 82 | 2407 | 9.469e-04 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 82 | 2410 | 9.850e-04 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 42 | 1000 | 1.090e-03 |
| GO:BP | GO:0006298 | mismatch repair | 6 | 32 | 1.113e-03 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 12 | 139 | 1.132e-03 |
| GO:BP | GO:0022411 | cellular component disassembly | 24 | 446 | 1.178e-03 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 4 | 11 | 1.191e-03 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 19 | 312 | 1.344e-03 |
| GO:BP | GO:0010224 | response to UV-B | 5 | 21 | 1.357e-03 |
| GO:BP | GO:0065009 | regulation of molecular function | 56 | 1496 | 1.536e-03 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 9 | 83 | 1.580e-03 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 40 | 954 | 1.643e-03 |
| GO:BP | GO:1905133 | negative regulation of meiotic chromosome separation | 3 | 5 | 1.699e-03 |
| GO:BP | GO:1902292 | cell cycle DNA replication initiation | 3 | 5 | 1.699e-03 |
| GO:BP | GO:1905784 | regulation of anaphase-promoting complex-dependent catabolic process | 3 | 5 | 1.699e-03 |
| GO:BP | GO:1902315 | nuclear cell cycle DNA replication initiation | 3 | 5 | 1.699e-03 |
| GO:BP | GO:0051311 | meiotic metaphase chromosome alignment | 3 | 5 | 1.699e-03 |
| GO:BP | GO:1902975 | mitotic DNA replication initiation | 3 | 5 | 1.699e-03 |
| GO:BP | GO:0001832 | blastocyst growth | 5 | 22 | 1.699e-03 |
| GO:BP | GO:1902103 | negative regulation of metaphase/anaphase transition of meiotic cell cycle | 3 | 5 | 1.699e-03 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 17 | 266 | 1.699e-03 |
| GO:BP | GO:0110030 | regulation of G2/MI transition of meiotic cell cycle | 3 | 5 | 1.699e-03 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 6 | 35 | 1.766e-03 |
| GO:BP | GO:0007019 | microtubule depolymerization | 7 | 50 | 1.800e-03 |
| GO:BP | GO:0007548 | sex differentiation | 18 | 294 | 1.818e-03 |
| GO:BP | GO:0009966 | regulation of signal transduction | 97 | 3034 | 1.854e-03 |
| GO:BP | GO:0051641 | cellular localization | 113 | 3661 | 1.874e-03 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 18 | 296 | 1.960e-03 |
| GO:BP | GO:0009416 | response to light stimulus | 19 | 323 | 1.973e-03 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 7 | 51 | 2.001e-03 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 7 | 51 | 2.001e-03 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 27 | 554 | 2.048e-03 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 12 | 150 | 2.143e-03 |
| GO:BP | GO:0023051 | regulation of signaling | 108 | 3478 | 2.153e-03 |
| GO:BP | GO:0008584 | male gonad development | 12 | 152 | 2.413e-03 |
| GO:BP | GO:0048144 | fibroblast proliferation | 10 | 109 | 2.482e-03 |
| GO:BP | GO:0046661 | male sex differentiation | 13 | 176 | 2.521e-03 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 12 | 153 | 2.543e-03 |
| GO:BP | GO:0034644 | cellular response to UV | 9 | 90 | 2.702e-03 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 7 | 54 | 2.810e-03 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 5 | 25 | 2.967e-03 |
| GO:BP | GO:2000105 | positive regulation of DNA-templated DNA replication | 4 | 14 | 3.001e-03 |
| GO:BP | GO:0046385 | deoxyribose phosphate biosynthetic process | 4 | 14 | 3.001e-03 |
| GO:BP | GO:0001833 | inner cell mass cell proliferation | 4 | 14 | 3.001e-03 |
| GO:BP | GO:1900025 | negative regulation of substrate adhesion-dependent cell spreading | 4 | 14 | 3.001e-03 |
| GO:BP | GO:0009263 | deoxyribonucleotide biosynthetic process | 4 | 14 | 3.001e-03 |
| GO:BP | GO:0009265 | 2’-deoxyribonucleotide biosynthetic process | 4 | 14 | 3.001e-03 |
| GO:BP | GO:0036211 | protein modification process | 91 | 2846 | 3.001e-03 |
| GO:BP | GO:0003175 | tricuspid valve development | 3 | 6 | 3.033e-03 |
| GO:BP | GO:1905132 | regulation of meiotic chromosome separation | 3 | 6 | 3.033e-03 |
| GO:BP | GO:0006335 | DNA replication-dependent chromatin assembly | 3 | 6 | 3.033e-03 |
| GO:BP | GO:0016321 | female meiosis chromosome segregation | 3 | 6 | 3.033e-03 |
| GO:BP | GO:1902102 | regulation of metaphase/anaphase transition of meiotic cell cycle | 3 | 6 | 3.033e-03 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 120 | 3993 | 3.058e-03 |
| GO:BP | GO:0030154 | cell differentiation | 131 | 4437 | 3.058e-03 |
| GO:BP | GO:0048869 | cellular developmental process | 131 | 4438 | 3.080e-03 |
| GO:BP | GO:0048638 | regulation of developmental growth | 18 | 310 | 3.176e-03 |
| GO:BP | GO:0040008 | regulation of growth | 28 | 604 | 3.266e-03 |
| GO:BP | GO:0051298 | centrosome duplication | 8 | 74 | 3.379e-03 |
| GO:BP | GO:0010646 | regulation of cell communication | 107 | 3486 | 3.384e-03 |
| GO:BP | GO:0030010 | establishment of cell polarity | 12 | 159 | 3.388e-03 |
| GO:BP | GO:0019985 | translesion synthesis | 5 | 26 | 3.388e-03 |
| GO:BP | GO:0048608 | reproductive structure development | 18 | 314 | 3.648e-03 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 11 | 138 | 3.789e-03 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 7 | 58 | 4.101e-03 |
| GO:BP | GO:0061458 | reproductive system development | 18 | 318 | 4.215e-03 |
| GO:BP | GO:0008406 | gonad development | 15 | 238 | 4.284e-03 |
| GO:BP | GO:0007099 | centriole replication | 6 | 43 | 4.932e-03 |
| GO:BP | GO:0051782 | negative regulation of cell division | 4 | 16 | 4.932e-03 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 6 | 43 | 4.932e-03 |
| GO:BP | GO:0051299 | centrosome separation | 4 | 16 | 4.932e-03 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 15 | 243 | 5.266e-03 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 7 | 61 | 5.478e-03 |
| GO:BP | GO:0007420 | brain development | 32 | 754 | 5.478e-03 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 5 | 29 | 5.576e-03 |
| GO:BP | GO:0006338 | chromatin remodeling | 31 | 725 | 5.899e-03 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 7 | 62 | 6.008e-03 |
| GO:BP | GO:0006915 | apoptotic process | 65 | 1923 | 6.055e-03 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 6 | 45 | 6.189e-03 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 6 | 45 | 6.189e-03 |
| GO:BP | GO:1905325 | regulation of meiosis I spindle assembly checkpoint | 2 | 2 | 6.462e-03 |
| GO:BP | GO:0031619 | homologous chromosome orientation in meiotic metaphase I | 2 | 2 | 6.462e-03 |
| GO:BP | GO:1905318 | meiosis I spindle assembly checkpoint signaling | 2 | 2 | 6.462e-03 |
| GO:BP | GO:0141112 | broken chromosome clustering | 2 | 2 | 6.462e-03 |
| GO:BP | GO:1990426 | mitotic recombination-dependent replication fork processing | 2 | 2 | 6.462e-03 |
| GO:BP | GO:0099607 | lateral attachment of mitotic spindle microtubules to kinetochore | 2 | 2 | 6.462e-03 |
| GO:BP | GO:0110029 | negative regulation of meiosis I | 2 | 2 | 6.462e-03 |
| GO:BP | GO:1990505 | mitotic DNA replication maintenance of fidelity | 2 | 2 | 6.462e-03 |
| GO:BP | GO:1902298 | cell cycle DNA replication maintenance of fidelity | 2 | 2 | 6.462e-03 |
| GO:BP | GO:0140274 | repair of kinetochore microtubule attachment defect | 2 | 2 | 6.462e-03 |
| GO:BP | GO:0140273 | repair of mitotic kinetochore microtubule attachment defect | 2 | 2 | 6.462e-03 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 7 | 64 | 7.012e-03 |
| GO:BP | GO:0007077 | mitotic nuclear membrane disassembly | 3 | 8 | 7.261e-03 |
| GO:BP | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process | 3 | 8 | 7.261e-03 |
| GO:BP | GO:0010458 | exit from mitosis | 5 | 31 | 7.261e-03 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 3 | 8 | 7.261e-03 |
| GO:BP | GO:0097681 | double-strand break repair via alternative nonhomologous end joining | 3 | 8 | 7.261e-03 |
| GO:BP | GO:0007096 | regulation of exit from mitosis | 4 | 18 | 7.423e-03 |
| GO:BP | GO:0060322 | head development | 33 | 806 | 7.781e-03 |
| GO:BP | GO:0098534 | centriole assembly | 6 | 48 | 8.354e-03 |
| GO:BP | GO:0045777 | positive regulation of blood pressure | 5 | 33 | 9.579e-03 |
| GO:BP | GO:0006325 | chromatin organization | 35 | 884 | 9.579e-03 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 5 | 33 | 9.579e-03 |
| GO:BP | GO:0051642 | centrosome localization | 5 | 33 | 9.579e-03 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 5 | 33 | 9.579e-03 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 14 | 234 | 1.011e-02 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 6 | 50 | 1.023e-02 |
| GO:BP | GO:1904776 | regulation of protein localization to cell cortex | 3 | 9 | 1.036e-02 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 3 | 9 | 1.036e-02 |
| GO:BP | GO:0043653 | mitochondrial fragmentation involved in apoptotic process | 3 | 9 | 1.036e-02 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 5 | 34 | 1.083e-02 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 4 | 20 | 1.094e-02 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 8 | 90 | 1.102e-02 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 5 | 35 | 1.232e-02 |
| GO:BP | GO:0043412 | macromolecule modification | 92 | 3030 | 1.261e-02 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 8 | 92 | 1.262e-02 |
| GO:BP | GO:0001701 | in utero embryonic development | 20 | 413 | 1.262e-02 |
| GO:BP | GO:0012501 | programmed cell death | 65 | 1987 | 1.276e-02 |
| GO:BP | GO:0008219 | cell death | 65 | 1991 | 1.339e-02 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 11 | 164 | 1.389e-02 |
| GO:BP | GO:0035265 | organ growth | 11 | 164 | 1.389e-02 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 3 | 10 | 1.408e-02 |
| GO:BP | GO:0045835 | negative regulation of meiotic nuclear division | 3 | 10 | 1.408e-02 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 29 | 706 | 1.469e-02 |
| GO:BP | GO:0007129 | homologous chromosome pairing at meiosis | 6 | 54 | 1.475e-02 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 7 | 74 | 1.537e-02 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 4 | 22 | 1.537e-02 |
| GO:BP | GO:1903475 | mitotic actomyosin contractile ring assembly | 2 | 3 | 1.713e-02 |
| GO:BP | GO:1903479 | mitotic actomyosin contractile ring assembly actin filament organization | 2 | 3 | 1.713e-02 |
| GO:BP | GO:0110032 | positive regulation of G2/MI transition of meiotic cell cycle | 2 | 3 | 1.713e-02 |
| GO:BP | GO:1902407 | assembly of actomyosin apparatus involved in mitotic cytokinesis | 2 | 3 | 1.713e-02 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 10 | 144 | 1.713e-02 |
| GO:BP | GO:0034421 | post-translational protein acetylation | 2 | 3 | 1.713e-02 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 5 | 38 | 1.713e-02 |
| GO:BP | GO:0071163 | DNA replication preinitiation complex assembly | 2 | 3 | 1.713e-02 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 6 | 56 | 1.740e-02 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 4 | 23 | 1.780e-02 |
| GO:BP | GO:0046599 | regulation of centriole replication | 4 | 23 | 1.780e-02 |
| GO:BP | GO:0051261 | protein depolymerization | 9 | 121 | 1.802e-02 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 56 | 1680 | 1.804e-02 |
| GO:BP | GO:1990918 | double-strand break repair involved in meiotic recombination | 3 | 11 | 1.811e-02 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 3 | 11 | 1.811e-02 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 3 | 11 | 1.811e-02 |
| GO:BP | GO:0070601 | centromeric sister chromatid cohesion | 3 | 11 | 1.811e-02 |
| GO:BP | GO:0048468 | cell development | 87 | 2876 | 1.839e-02 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 9 | 122 | 1.876e-02 |
| GO:BP | GO:0033365 | protein localization to organelle | 43 | 1209 | 1.918e-02 |
| GO:BP | GO:0009162 | deoxyribonucleoside monophosphate metabolic process | 4 | 24 | 2.050e-02 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 28 | 692 | 2.078e-02 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 38 | 1038 | 2.139e-02 |
| GO:BP | GO:0031099 | regeneration | 12 | 202 | 2.222e-02 |
| GO:BP | GO:0072710 | response to hydroxyurea | 3 | 12 | 2.305e-02 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 5 | 41 | 2.305e-02 |
| GO:BP | GO:0071493 | cellular response to UV-B | 3 | 12 | 2.305e-02 |
| GO:BP | GO:0051081 | nuclear membrane disassembly | 3 | 12 | 2.305e-02 |
| GO:BP | GO:0009394 | 2’-deoxyribonucleotide metabolic process | 5 | 41 | 2.305e-02 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 3 | 12 | 2.305e-02 |
| GO:BP | GO:0001783 | B cell apoptotic process | 4 | 25 | 2.333e-02 |
| GO:BP | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 4 | 25 | 2.333e-02 |
| GO:BP | GO:0006312 | mitotic recombination | 4 | 25 | 2.333e-02 |
| GO:BP | GO:0060420 | regulation of heart growth | 6 | 60 | 2.349e-02 |
| GO:BP | GO:0048678 | response to axon injury | 7 | 81 | 2.383e-02 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 7 | 81 | 2.383e-02 |
| GO:BP | GO:0003231 | cardiac ventricle development | 9 | 127 | 2.383e-02 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 20 | 441 | 2.444e-02 |
| GO:BP | GO:0009262 | deoxyribonucleotide metabolic process | 5 | 42 | 2.506e-02 |
| GO:BP | GO:0019692 | deoxyribose phosphate metabolic process | 5 | 42 | 2.506e-02 |
| GO:BP | GO:0043009 | chordate embryonic development | 27 | 671 | 2.576e-02 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 4 | 26 | 2.643e-02 |
| GO:BP | GO:0062009 | secondary palate development | 4 | 26 | 2.643e-02 |
| GO:BP | GO:0008156 | negative regulation of DNA replication | 4 | 26 | 2.643e-02 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 7 | 83 | 2.686e-02 |
| GO:BP | GO:0060419 | heart growth | 7 | 83 | 2.686e-02 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 11 | 181 | 2.686e-02 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 6 | 62 | 2.694e-02 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 5 | 43 | 2.730e-02 |
| GO:BP | GO:0051179 | localization | 151 | 5555 | 2.737e-02 |
| GO:BP | GO:0060429 | epithelium development | 43 | 1238 | 2.779e-02 |
| GO:BP | GO:0040015 | negative regulation of multicellular organism growth | 3 | 13 | 2.782e-02 |
| GO:BP | GO:1903251 | multi-ciliated epithelial cell differentiation | 3 | 13 | 2.782e-02 |
| GO:BP | GO:0016446 | somatic hypermutation of immunoglobulin genes | 3 | 13 | 2.782e-02 |
| GO:BP | GO:0030397 | membrane disassembly | 3 | 13 | 2.782e-02 |
| GO:BP | GO:0070914 | UV-damage excision repair | 3 | 13 | 2.782e-02 |
| GO:BP | GO:0007417 | central nervous system development | 38 | 1061 | 2.885e-02 |
| GO:BP | GO:0050793 | regulation of developmental process | 75 | 2457 | 2.912e-02 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 4 | 27 | 2.918e-02 |
| GO:BP | GO:0002339 | B cell selection | 2 | 4 | 2.918e-02 |
| GO:BP | GO:1905007 | positive regulation of epithelial to mesenchymal transition involved in endocardial cushion formation | 2 | 4 | 2.918e-02 |
| GO:BP | GO:2000689 | actomyosin contractile ring assembly actin filament organization | 2 | 4 | 2.918e-02 |
| GO:BP | GO:1905448 | positive regulation of mitochondrial ATP synthesis coupled electron transport | 2 | 4 | 2.918e-02 |
| GO:BP | GO:0072757 | cellular response to camptothecin | 2 | 4 | 2.918e-02 |
| GO:BP | GO:0051296 | establishment of meiotic spindle orientation | 2 | 4 | 2.918e-02 |
| GO:BP | GO:1905005 | regulation of epithelial to mesenchymal transition involved in endocardial cushion formation | 2 | 4 | 2.918e-02 |
| GO:BP | GO:1901995 | positive regulation of meiotic cell cycle phase transition | 2 | 4 | 2.918e-02 |
| GO:BP | GO:1901563 | response to camptothecin | 2 | 4 | 2.918e-02 |
| GO:BP | GO:0048627 | myoblast development | 2 | 4 | 2.918e-02 |
| GO:BP | GO:0007079 | mitotic chromosome movement towards spindle pole | 2 | 4 | 2.918e-02 |
| GO:BP | GO:0051754 | meiotic sister chromatid cohesion, centromeric | 2 | 4 | 2.918e-02 |
| GO:BP | GO:0060400 | negative regulation of growth hormone receptor signaling pathway | 2 | 4 | 2.918e-02 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 22 | 517 | 3.056e-02 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 11 | 186 | 3.128e-02 |
| GO:BP | GO:0023057 | negative regulation of signaling | 48 | 1435 | 3.181e-02 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 22 | 519 | 3.181e-02 |
| GO:BP | GO:0035264 | multicellular organism growth | 10 | 160 | 3.187e-02 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 48 | 1436 | 3.204e-02 |
| GO:BP | GO:0003272 | endocardial cushion formation | 4 | 28 | 3.238e-02 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 45 | 1326 | 3.238e-02 |
| GO:BP | GO:0033262 | regulation of nuclear cell cycle DNA replication | 3 | 14 | 3.285e-02 |
| GO:BP | GO:0007100 | mitotic centrosome separation | 3 | 14 | 3.285e-02 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 3 | 14 | 3.285e-02 |
| GO:BP | GO:0002566 | somatic diversification of immune receptors via somatic mutation | 3 | 14 | 3.285e-02 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 5 | 46 | 3.413e-02 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 17 | 364 | 3.414e-02 |
| GO:BP | GO:0009987 | cellular process | 461 | 20247 | 3.540e-02 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 31 | 830 | 3.552e-02 |
| GO:BP | GO:0046620 | regulation of organ growth | 7 | 89 | 3.636e-02 |
| GO:BP | GO:0001947 | heart looping | 6 | 67 | 3.647e-02 |
| GO:BP | GO:0007399 | nervous system development | 78 | 2604 | 3.647e-02 |
| GO:BP | GO:0000723 | telomere maintenance | 10 | 164 | 3.680e-02 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 10 | 164 | 3.680e-02 |
| GO:BP | GO:0043549 | regulation of kinase activity | 19 | 431 | 3.763e-02 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 7 | 90 | 3.823e-02 |
| GO:BP | GO:0031100 | animal organ regeneration | 6 | 68 | 3.884e-02 |
| GO:BP | GO:0003198 | epithelial to mesenchymal transition involved in endocardial cushion formation | 3 | 15 | 3.919e-02 |
| GO:BP | GO:0044027 | negative regulation of gene expression via chromosomal CpG island methylation | 3 | 15 | 3.919e-02 |
| GO:BP | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process | 3 | 15 | 3.919e-02 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 45 | 1343 | 3.921e-02 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 18 | 402 | 3.972e-02 |
| GO:BP | GO:0071763 | nuclear membrane organization | 5 | 48 | 3.975e-02 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 8 | 115 | 4.015e-02 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 6 | 69 | 4.110e-02 |
| GO:BP | GO:0009888 | tissue development | 63 | 2032 | 4.162e-02 |
| GO:BP | GO:0045666 | positive regulation of neuron differentiation | 7 | 92 | 4.225e-02 |
| GO:BP | GO:0071962 | mitotic sister chromatid cohesion, centromeric | 2 | 5 | 4.361e-02 |
| GO:BP | GO:0098535 | de novo centriole assembly involved in multi-ciliated epithelial cell differentiation | 2 | 5 | 4.361e-02 |
| GO:BP | GO:0098722 | asymmetric stem cell division | 2 | 5 | 4.361e-02 |
| GO:BP | GO:0098728 | germline stem cell asymmetric division | 2 | 5 | 4.361e-02 |
| GO:BP | GO:0090233 | negative regulation of spindle checkpoint | 2 | 5 | 4.361e-02 |
| GO:BP | GO:0003186 | tricuspid valve morphogenesis | 2 | 5 | 4.361e-02 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 6 | 70 | 4.361e-02 |
| GO:BP | GO:0140499 | negative regulation of mitotic spindle assembly checkpoint signaling | 2 | 5 | 4.361e-02 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 2 | 5 | 4.361e-02 |
| GO:BP | GO:0006282 | regulation of DNA repair | 12 | 225 | 4.361e-02 |
| GO:BP | GO:0031536 | positive regulation of exit from mitosis | 2 | 5 | 4.361e-02 |
| GO:BP | GO:0043137 | DNA replication, removal of RNA primer | 2 | 5 | 4.361e-02 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 6 | 70 | 4.361e-02 |
| GO:BP | GO:0003274 | endocardial cushion fusion | 2 | 5 | 4.361e-02 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 9 | 143 | 4.411e-02 |
| GO:BP | GO:0007507 | heart development | 24 | 605 | 4.436e-02 |
| GO:BP | GO:1990166 | protein localization to site of double-strand break | 3 | 16 | 4.497e-02 |
| GO:BP | GO:0048308 | organelle inheritance | 3 | 16 | 4.497e-02 |
| GO:BP | GO:0048313 | Golgi inheritance | 3 | 16 | 4.497e-02 |
| GO:BP | GO:0006334 | nucleosome assembly | 8 | 118 | 4.497e-02 |
| GO:BP | GO:0003214 | cardiac left ventricle morphogenesis | 3 | 16 | 4.497e-02 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 32 | 884 | 4.522e-02 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 10 | 171 | 4.604e-02 |
| GO:BP | GO:0001824 | blastocyst development | 8 | 119 | 4.685e-02 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 13 | 257 | 4.712e-02 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 7 | 95 | 4.785e-02 |
| GO:BP | GO:1904376 | negative regulation of protein localization to cell periphery | 4 | 32 | 4.785e-02 |
| GO:BP | GO:0007018 | microtubule-based movement | 20 | 478 | 4.933e-02 |
| KEGG | KEGG:04110 | Cell cycle | 45 | 157 | 6.713e-33 |
| KEGG | KEGG:03030 | DNA replication | 17 | 36 | 4.517e-16 |
| KEGG | KEGG:04115 | p53 signaling pathway | 14 | 74 | 4.224e-07 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 12 | 54 | 5.928e-07 |
| KEGG | KEGG:04114 | Oocyte meiosis | 17 | 131 | 2.176e-06 |
| KEGG | KEGG:03440 | Homologous recombination | 10 | 41 | 2.687e-06 |
| KEGG | KEGG:03430 | Mismatch repair | 7 | 23 | 4.458e-05 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 12 | 102 | 3.428e-04 |
| KEGG | KEGG:04218 | Cellular senescence | 15 | 155 | 3.428e-04 |
| KEGG | KEGG:03410 | Base excision repair | 8 | 44 | 3.627e-04 |
| KEGG | KEGG:05212 | Pancreatic cancer | 10 | 76 | 5.152e-04 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 9 | 61 | 5.152e-04 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 17 | 218 | 9.973e-04 |
| KEGG | KEGG:04814 | Motor proteins | 15 | 193 | 2.657e-03 |
| KEGG | KEGG:05222 | Small cell lung cancer | 9 | 92 | 1.024e-02 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 8 | 76 | 1.176e-02 |
| KEGG | KEGG:04934 | Cushing syndrome | 11 | 153 | 3.091e-02 |
| KEGG | KEGG:01524 | Platinum drug resistance | 7 | 72 | 3.329e-02 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 7 | 72 | 3.329e-02 |
| KEGG | KEGG:05214 | Glioma | 7 | 75 | 3.990e-02 |
#GO:BP
table_DOX144Rshare_genes_GOBP <- table_DOX144Rshare_genes %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX144Rshare_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX144R Shared DEGs Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))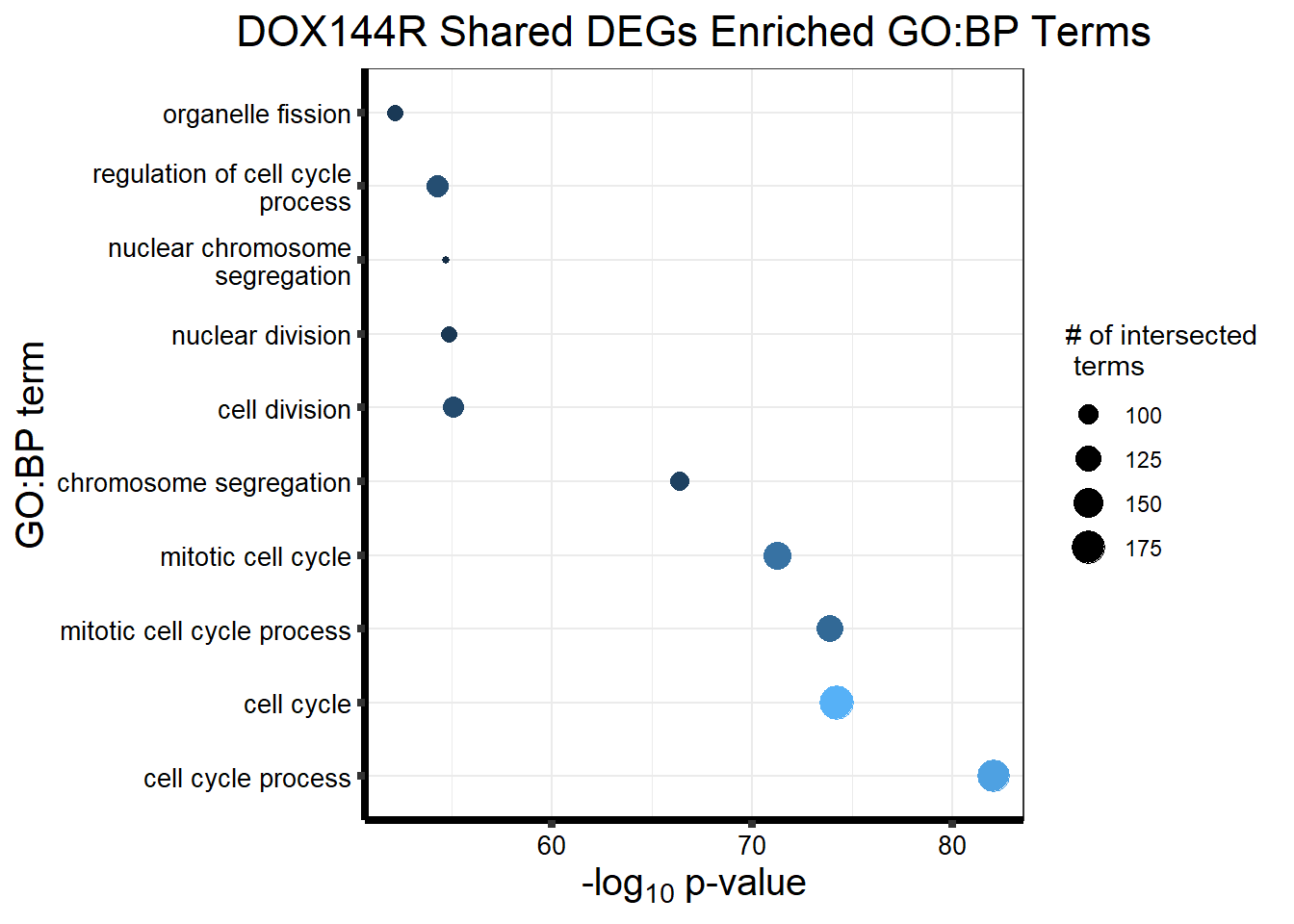
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_DOX144Rshare_genes_KEGG <- table_DOX144Rshare_genes %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX144Rshare_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX144R Shared DEGs Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))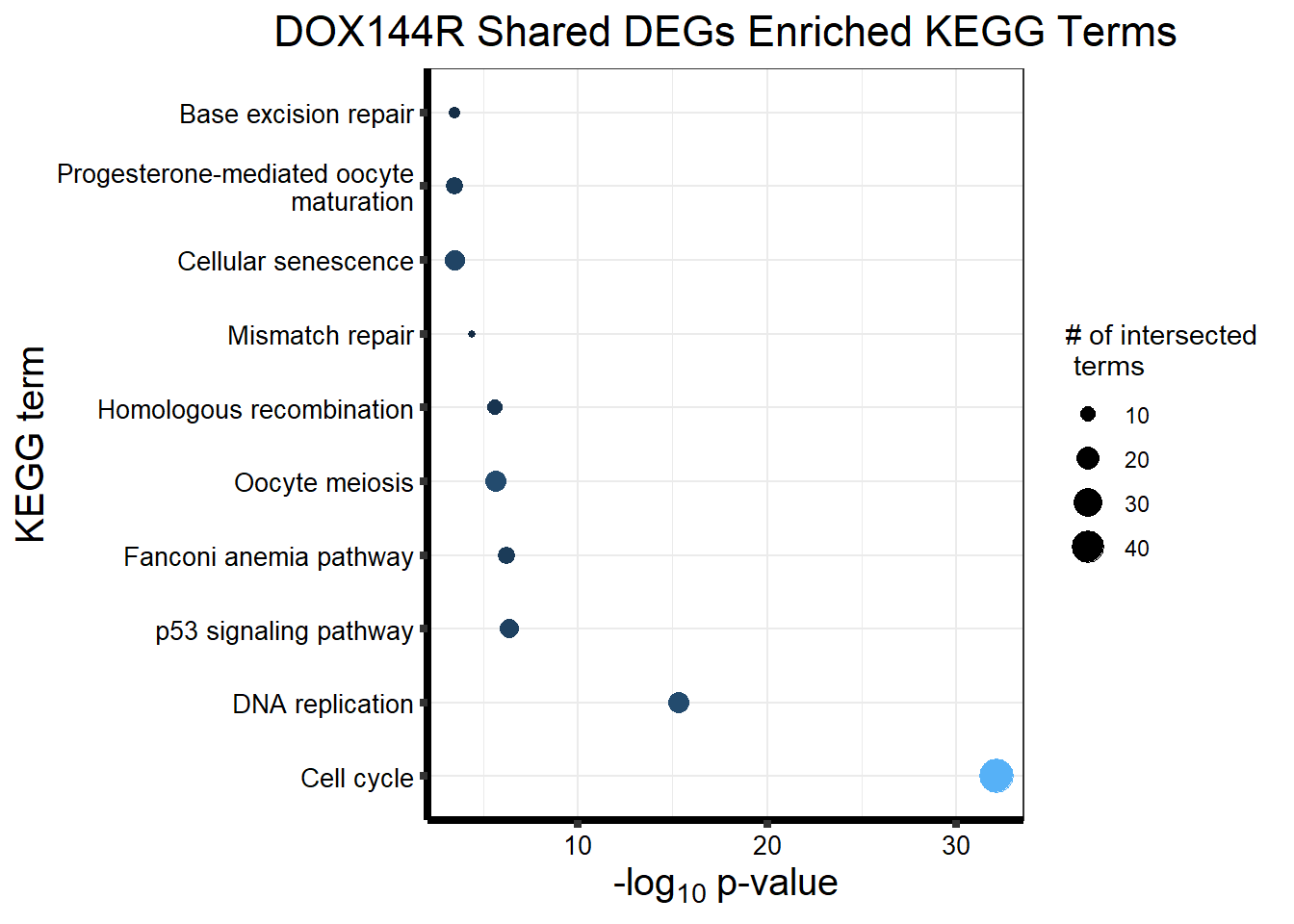
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
###GO/KEGG Analysis of Upreg and Downreg Genes Original Data
# library(gprofiler2)
#####DOX24 Upregulated Genes#####
D24_DEGs_up_mat <- as.matrix(DOX24T_DEGs_GO_up)
DOX_24_up_dxr_gene <- gost(query = D24_DEGs_up_mat,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
DOX_24_up_gost_genes <- gostplot(DOX_24_up_dxr_gene, capped = FALSE, interactive = TRUE)
DOX_24_up_gost_genestable_DOX24_up_genes <- DOX_24_up_dxr_gene$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX24_up_genes %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0080090 | regulation of primary metabolic process | 576 | 5390 | 1.045e-15 |
| GO:BP | GO:0019538 | protein metabolic process | 506 | 4721 | 2.272e-13 |
| GO:BP | GO:0048518 | positive regulation of biological process | 634 | 6264 | 6.684e-13 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 604 | 5920 | 6.884e-13 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 438 | 3990 | 6.884e-13 |
| GO:BP | GO:0065007 | biological regulation | 1136 | 12743 | 3.199e-12 |
| GO:BP | GO:0050789 | regulation of biological process | 1104 | 12336 | 6.167e-12 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 405 | 3687 | 7.025e-12 |
| GO:BP | GO:0050794 | regulation of cellular process | 1074 | 11946 | 7.025e-12 |
| GO:BP | GO:0036211 | protein modification process | 319 | 2846 | 1.022e-09 |
| GO:BP | GO:0023051 | regulation of signaling | 374 | 3478 | 1.999e-09 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 366 | 3409 | 4.296e-09 |
| GO:BP | GO:0007275 | multicellular organism development | 480 | 4727 | 4.661e-09 |
| GO:BP | GO:0010646 | regulation of cell communication | 372 | 3486 | 4.904e-09 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 367 | 3428 | 4.904e-09 |
| GO:BP | GO:0009056 | catabolic process | 296 | 2639 | 4.904e-09 |
| GO:BP | GO:0043412 | macromolecule modification | 331 | 3030 | 5.358e-09 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 415 | 3993 | 7.293e-09 |
| GO:BP | GO:0051716 | cellular response to stimulus | 697 | 7376 | 7.338e-09 |
| GO:BP | GO:0035556 | intracellular signal transduction | 324 | 2965 | 7.338e-09 |
| GO:BP | GO:0006351 | DNA-templated transcription | 376 | 3549 | 7.338e-09 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 178 | 1400 | 7.338e-09 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 190 | 1529 | 9.567e-09 |
| GO:BP | GO:0032502 | developmental process | 629 | 6553 | 9.602e-09 |
| GO:BP | GO:0048856 | anatomical structure development | 583 | 5997 | 9.625e-09 |
| GO:BP | GO:0050793 | regulation of developmental process | 277 | 2457 | 9.625e-09 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 550 | 5629 | 2.235e-08 |
| GO:BP | GO:0009966 | regulation of signal transduction | 327 | 3034 | 2.335e-08 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 168 | 1326 | 2.744e-08 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 201 | 1680 | 5.285e-08 |
| GO:BP | GO:0023057 | negative regulation of signaling | 177 | 1435 | 6.564e-08 |
| GO:BP | GO:0019222 | regulation of metabolic process | 662 | 7035 | 6.564e-08 |
| GO:BP | GO:0051179 | localization | 540 | 5555 | 7.321e-08 |
| GO:BP | GO:0006810 | transport | 443 | 4407 | 7.975e-08 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 176 | 1436 | 1.111e-07 |
| GO:BP | GO:0048519 | negative regulation of biological process | 561 | 5834 | 1.428e-07 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 264 | 2407 | 3.563e-07 |
| GO:BP | GO:0023052 | signaling | 614 | 6515 | 3.674e-07 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 264 | 2410 | 3.819e-07 |
| GO:BP | GO:0048731 | system development | 408 | 4053 | 4.044e-07 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 158 | 1280 | 4.890e-07 |
| GO:BP | GO:0007154 | cell communication | 614 | 6540 | 6.087e-07 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 159 | 1296 | 6.087e-07 |
| GO:BP | GO:0050896 | response to stimulus | 811 | 8999 | 6.087e-07 |
| GO:BP | GO:0035295 | tube development | 140 | 1101 | 6.087e-07 |
| GO:BP | GO:0006325 | chromatin organization | 118 | 884 | 6.705e-07 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 277 | 2578 | 7.967e-07 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 298 | 2819 | 9.410e-07 |
| GO:BP | GO:0046907 | intracellular transport | 166 | 1381 | 1.072e-06 |
| GO:BP | GO:0044281 | small molecule metabolic process | 203 | 1776 | 1.157e-06 |
| GO:BP | GO:0033554 | cellular response to stress | 208 | 1830 | 1.157e-06 |
| GO:BP | GO:0030154 | cell differentiation | 437 | 4437 | 1.180e-06 |
| GO:BP | GO:0048869 | cellular developmental process | 437 | 4438 | 1.193e-06 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 365 | 3597 | 1.233e-06 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 287 | 2711 | 1.478e-06 |
| GO:BP | GO:0007165 | signal transduction | 566 | 6002 | 1.729e-06 |
| GO:BP | GO:0032879 | regulation of localization | 224 | 2029 | 2.987e-06 |
| GO:BP | GO:0043687 | post-translational protein modification | 129 | 1027 | 4.072e-06 |
| GO:BP | GO:0051641 | cellular localization | 367 | 3661 | 4.085e-06 |
| GO:BP | GO:0042592 | homeostatic process | 196 | 1736 | 4.749e-06 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 70 | 458 | 4.826e-06 |
| GO:BP | GO:0033036 | macromolecule localization | 329 | 3228 | 4.873e-06 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 111 | 853 | 5.512e-06 |
| GO:BP | GO:0051234 | establishment of localization | 473 | 4928 | 5.512e-06 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 56 | 336 | 5.512e-06 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 46 | 252 | 5.512e-06 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 53 | 311 | 5.778e-06 |
| GO:BP | GO:0060485 | mesenchyme development | 54 | 320 | 5.986e-06 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 283 | 2713 | 6.310e-06 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 304 | 2956 | 7.053e-06 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 122 | 975 | 9.981e-06 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 599 | 6492 | 1.141e-05 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 35 | 171 | 1.141e-05 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 118 | 938 | 1.181e-05 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 220 | 2036 | 1.694e-05 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 54 | 331 | 1.694e-05 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 45 | 255 | 1.831e-05 |
| GO:BP | GO:0032880 | regulation of protein localization | 114 | 907 | 1.945e-05 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 541 | 5809 | 2.022e-05 |
| GO:BP | GO:0016567 | protein ubiquitination | 100 | 768 | 2.099e-05 |
| GO:BP | GO:0035239 | tube morphogenesis | 111 | 879 | 2.152e-05 |
| GO:BP | GO:0048468 | cell development | 293 | 2876 | 2.683e-05 |
| GO:BP | GO:0007178 | cell surface receptor protein serine/threonine kinase signaling pathway | 57 | 364 | 2.898e-05 |
| GO:BP | GO:0061053 | somite development | 22 | 85 | 3.606e-05 |
| GO:BP | GO:0060341 | regulation of cellular localization | 123 | 1013 | 3.797e-05 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 31 | 150 | 4.104e-05 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 161 | 1418 | 4.417e-05 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 37 | 198 | 4.628e-05 |
| GO:BP | GO:0035282 | segmentation | 25 | 108 | 5.614e-05 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 142 | 1223 | 6.125e-05 |
| GO:BP | GO:0006996 | organelle organization | 352 | 3594 | 6.328e-05 |
| GO:BP | GO:0051649 | establishment of localization in cell | 214 | 2010 | 6.369e-05 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 333 | 3375 | 6.859e-05 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 23 | 96 | 7.805e-05 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 174 | 1576 | 8.306e-05 |
| GO:BP | GO:0032501 | multicellular organismal process | 657 | 7322 | 1.020e-04 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 273 | 2696 | 1.074e-04 |
| GO:BP | GO:0043009 | chordate embryonic development | 87 | 671 | 1.279e-04 |
| GO:BP | GO:0009790 | embryo development | 132 | 1135 | 1.290e-04 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 158 | 1417 | 1.431e-04 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 196 | 1835 | 1.431e-04 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 44 | 268 | 1.431e-04 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 323 | 3288 | 1.431e-04 |
| GO:BP | GO:0006950 | response to stress | 379 | 3948 | 1.431e-04 |
| GO:BP | GO:0065009 | regulation of molecular function | 165 | 1496 | 1.557e-04 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 126 | 1077 | 1.557e-04 |
| GO:BP | GO:0008104 | protein localization | 278 | 2770 | 1.600e-04 |
| GO:BP | GO:0051049 | regulation of transport | 175 | 1607 | 1.612e-04 |
| GO:BP | GO:0071705 | nitrogen compound transport | 203 | 1923 | 1.936e-04 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 81 | 620 | 1.947e-04 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 52 | 344 | 2.085e-04 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 278 | 2782 | 2.177e-04 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 150 | 1343 | 2.202e-04 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 107 | 887 | 2.234e-04 |
| GO:BP | GO:0060429 | epithelium development | 140 | 1238 | 2.433e-04 |
| GO:BP | GO:0044238 | primary metabolic process | 1044 | 12342 | 2.433e-04 |
| GO:BP | GO:0006338 | chromatin remodeling | 91 | 725 | 2.434e-04 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 33 | 181 | 2.468e-04 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 163 | 1489 | 2.494e-04 |
| GO:BP | GO:0008219 | cell death | 208 | 1991 | 2.647e-04 |
| GO:BP | GO:0030097 | hemopoiesis | 115 | 976 | 2.773e-04 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 13 | 39 | 2.963e-04 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 516 | 5644 | 3.176e-04 |
| GO:BP | GO:0012501 | programmed cell death | 207 | 1987 | 3.271e-04 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 87 | 692 | 3.366e-04 |
| GO:BP | GO:0015031 | protein transport | 158 | 1444 | 3.366e-04 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 246 | 2433 | 3.366e-04 |
| GO:BP | GO:0072175 | epithelial tube formation | 27 | 136 | 3.366e-04 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 260 | 2593 | 3.366e-04 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 68 | 503 | 3.366e-04 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 108 | 909 | 3.491e-04 |
| GO:BP | GO:0051169 | nuclear transport | 50 | 334 | 3.524e-04 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 10 | 24 | 3.524e-04 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 50 | 334 | 3.524e-04 |
| GO:BP | GO:0009894 | regulation of catabolic process | 120 | 1040 | 4.203e-04 |
| GO:BP | GO:0034504 | protein localization to nucleus | 48 | 318 | 4.227e-04 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 71 | 537 | 4.242e-04 |
| GO:BP | GO:0009888 | tissue development | 210 | 2032 | 4.242e-04 |
| GO:BP | GO:0006082 | organic acid metabolic process | 108 | 915 | 4.416e-04 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 113 | 970 | 4.875e-04 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 48 | 320 | 4.875e-04 |
| GO:BP | GO:0006629 | lipid metabolic process | 152 | 1391 | 5.029e-04 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 47 | 312 | 5.271e-04 |
| GO:BP | GO:0035148 | tube formation | 28 | 148 | 5.281e-04 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 22 | 102 | 5.487e-04 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 7 | 12 | 5.488e-04 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 223 | 2192 | 5.863e-04 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 306 | 3157 | 6.253e-04 |
| GO:BP | GO:0006606 | protein import into nucleus | 30 | 166 | 6.402e-04 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 108 | 925 | 6.603e-04 |
| GO:BP | GO:0006402 | mRNA catabolic process | 41 | 261 | 6.639e-04 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 276 | 2813 | 7.378e-04 |
| GO:BP | GO:0043487 | regulation of RNA stability | 33 | 193 | 7.803e-04 |
| GO:BP | GO:0046649 | lymphocyte activation | 97 | 814 | 7.938e-04 |
| GO:BP | GO:0016125 | sterol metabolic process | 29 | 160 | 8.084e-04 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 234 | 2331 | 8.154e-04 |
| GO:BP | GO:0010468 | regulation of gene expression | 502 | 5536 | 9.334e-04 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 27 | 145 | 9.334e-04 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 131 | 1182 | 9.938e-04 |
| GO:BP | GO:0048729 | tissue morphogenesis | 77 | 614 | 1.032e-03 |
| GO:BP | GO:0006886 | intracellular protein transport | 84 | 686 | 1.062e-03 |
| GO:BP | GO:0051170 | import into nucleus | 30 | 171 | 1.071e-03 |
| GO:BP | GO:0065008 | regulation of biological quality | 286 | 2947 | 1.080e-03 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 22 | 107 | 1.080e-03 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 83 | 677 | 1.108e-03 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 24 | 123 | 1.151e-03 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 177 | 1697 | 1.191e-03 |
| GO:BP | GO:0072359 | circulatory system development | 127 | 1145 | 1.220e-03 |
| GO:BP | GO:0016310 | phosphorylation | 143 | 1320 | 1.224e-03 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 28 | 156 | 1.237e-03 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 177 | 1699 | 1.244e-03 |
| GO:BP | GO:0007399 | nervous system development | 256 | 2604 | 1.246e-03 |
| GO:BP | GO:0006468 | protein phosphorylation | 134 | 1227 | 1.486e-03 |
| GO:BP | GO:0006915 | apoptotic process | 196 | 1923 | 1.614e-03 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 12 | 40 | 1.614e-03 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 26 | 142 | 1.614e-03 |
| GO:BP | GO:0098727 | maintenance of cell number | 31 | 184 | 1.620e-03 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 195 | 1912 | 1.620e-03 |
| GO:BP | GO:0048732 | gland development | 60 | 454 | 1.658e-03 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 79 | 645 | 1.658e-03 |
| GO:BP | GO:0001756 | somitogenesis | 16 | 67 | 1.915e-03 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 27 | 152 | 1.935e-03 |
| GO:BP | GO:0007219 | Notch signaling pathway | 31 | 186 | 1.952e-03 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 26 | 144 | 1.978e-03 |
| GO:BP | GO:0001568 | blood vessel development | 87 | 732 | 1.996e-03 |
| GO:BP | GO:0030163 | protein catabolic process | 115 | 1030 | 2.027e-03 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 26 | 145 | 2.199e-03 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 66 | 519 | 2.212e-03 |
| GO:BP | GO:0007369 | gastrulation | 32 | 197 | 2.402e-03 |
| GO:BP | GO:0080134 | regulation of response to stress | 147 | 1387 | 2.492e-03 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 21 | 106 | 2.523e-03 |
| GO:BP | GO:0022008 | neurogenesis | 181 | 1771 | 2.604e-03 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 172 | 1669 | 2.604e-03 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 69 | 553 | 2.656e-03 |
| GO:BP | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib | 7 | 15 | 2.696e-03 |
| GO:BP | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway | 7 | 15 | 2.696e-03 |
| GO:BP | GO:0023056 | positive regulation of signaling | 183 | 1796 | 2.736e-03 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 39 | 262 | 2.791e-03 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 33 | 208 | 2.832e-03 |
| GO:BP | GO:0001944 | vasculature development | 89 | 762 | 2.855e-03 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 188 | 1855 | 2.855e-03 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 30 | 182 | 2.880e-03 |
| GO:BP | GO:0009880 | embryonic pattern specification | 17 | 77 | 2.894e-03 |
| GO:BP | GO:1904238 | pericyte cell differentiation | 6 | 11 | 2.984e-03 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 45 | 320 | 3.140e-03 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 171 | 1667 | 3.267e-03 |
| GO:BP | GO:0001775 | cell activation | 123 | 1132 | 3.295e-03 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 196 | 1953 | 3.344e-03 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 24 | 133 | 3.409e-03 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 264 | 2745 | 3.476e-03 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 109 | 983 | 3.718e-03 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 24 | 134 | 3.797e-03 |
| GO:BP | GO:0002520 | immune system development | 32 | 203 | 3.838e-03 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 20 | 102 | 3.870e-03 |
| GO:BP | GO:0048608 | reproductive structure development | 44 | 314 | 3.870e-03 |
| GO:BP | GO:0015914 | phospholipid transport | 20 | 102 | 3.870e-03 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 72 | 593 | 3.943e-03 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 13 | 51 | 3.983e-03 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 76 | 635 | 3.983e-03 |
| GO:BP | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib | 7 | 16 | 3.983e-03 |
| GO:BP | GO:0021915 | neural tube development | 27 | 160 | 3.983e-03 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 13 | 51 | 3.983e-03 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 94 | 825 | 4.043e-03 |
| GO:BP | GO:0051223 | regulation of protein transport | 57 | 442 | 4.062e-03 |
| GO:BP | GO:0060284 | regulation of cell development | 96 | 847 | 4.119e-03 |
| GO:BP | GO:0006839 | mitochondrial transport | 29 | 178 | 4.175e-03 |
| GO:BP | GO:0016032 | viral process | 55 | 423 | 4.232e-03 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 81 | 689 | 4.244e-03 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 39 | 269 | 4.296e-03 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 181 | 1795 | 4.509e-03 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 90 | 786 | 4.569e-03 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 128 | 1199 | 4.569e-03 |
| GO:BP | GO:1904026 | regulation of collagen fibril organization | 5 | 8 | 4.697e-03 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 5 | 8 | 4.697e-03 |
| GO:BP | GO:0061458 | reproductive system development | 44 | 318 | 4.770e-03 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 6 | 12 | 4.819e-03 |
| GO:BP | GO:0019827 | stem cell population maintenance | 29 | 180 | 4.869e-03 |
| GO:BP | GO:0045184 | establishment of protein localization | 193 | 1937 | 4.927e-03 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 34 | 225 | 4.927e-03 |
| GO:BP | GO:0048513 | animal organ development | 291 | 3085 | 4.974e-03 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 45 | 329 | 5.116e-03 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 16 | 74 | 5.116e-03 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 28 | 172 | 5.129e-03 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 52 | 398 | 5.239e-03 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 47 | 349 | 5.239e-03 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 24 | 138 | 5.239e-03 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 119 | 1105 | 5.239e-03 |
| GO:BP | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I | 8 | 22 | 5.314e-03 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 62 | 499 | 5.398e-03 |
| GO:BP | GO:0016054 | organic acid catabolic process | 36 | 246 | 5.816e-03 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 36 | 246 | 5.816e-03 |
| GO:BP | GO:0006979 | response to oxidative stress | 52 | 400 | 5.816e-03 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 45 | 332 | 5.987e-03 |
| GO:BP | GO:0044282 | small molecule catabolic process | 49 | 371 | 5.987e-03 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 23 | 131 | 5.987e-03 |
| GO:BP | GO:0048699 | generation of neurons | 157 | 1536 | 6.194e-03 |
| GO:BP | GO:0070848 | response to growth factor | 83 | 721 | 6.261e-03 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 42 | 304 | 6.261e-03 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 42 | 304 | 6.261e-03 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 73 | 616 | 6.261e-03 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 132 | 1256 | 6.283e-03 |
| GO:BP | GO:0045321 | leukocyte activation | 107 | 980 | 6.283e-03 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 20 | 107 | 6.283e-03 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 56 | 442 | 6.283e-03 |
| GO:BP | GO:0048255 | mRNA stabilization | 15 | 69 | 6.752e-03 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 93 | 831 | 6.752e-03 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 15 | 69 | 6.752e-03 |
| GO:BP | GO:0030182 | neuron differentiation | 149 | 1451 | 6.752e-03 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 78 | 671 | 6.752e-03 |
| GO:BP | GO:0030199 | collagen fibril organization | 15 | 69 | 6.752e-03 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 66 | 546 | 6.752e-03 |
| GO:BP | GO:0002475 | antigen processing and presentation via MHC class Ib | 8 | 23 | 6.752e-03 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 27 | 167 | 6.752e-03 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 65 | 535 | 6.752e-03 |
| GO:BP | GO:0006658 | phosphatidylserine metabolic process | 8 | 23 | 6.752e-03 |
| GO:BP | GO:0048736 | appendage development | 30 | 194 | 6.752e-03 |
| GO:BP | GO:0042632 | cholesterol homeostasis | 18 | 92 | 6.752e-03 |
| GO:BP | GO:0060173 | limb development | 30 | 194 | 6.752e-03 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 112 | 1038 | 6.752e-03 |
| GO:BP | GO:0001525 | angiogenesis | 66 | 547 | 6.994e-03 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 160 | 1578 | 7.173e-03 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 36 | 250 | 7.173e-03 |
| GO:BP | GO:0009987 | cellular process | 1596 | 20247 | 7.498e-03 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 64 | 528 | 7.508e-03 |
| GO:BP | GO:0055092 | sterol homeostasis | 18 | 93 | 7.596e-03 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 75 | 643 | 7.607e-03 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 24 | 143 | 7.864e-03 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 11 | 42 | 7.926e-03 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 83 | 729 | 7.926e-03 |
| GO:BP | GO:0031946 | regulation of glucocorticoid biosynthetic process | 5 | 9 | 7.926e-03 |
| GO:BP | GO:0000209 | protein polyubiquitination | 39 | 280 | 7.926e-03 |
| GO:BP | GO:0048318 | axial mesoderm development | 5 | 9 | 7.926e-03 |
| GO:BP | GO:0001501 | skeletal system development | 65 | 540 | 7.939e-03 |
| GO:BP | GO:0050810 | regulation of steroid biosynthetic process | 16 | 78 | 8.008e-03 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 13 | 56 | 8.225e-03 |
| GO:BP | GO:0060541 | respiratory system development | 33 | 225 | 8.680e-03 |
| GO:BP | GO:0002483 | antigen processing and presentation of endogenous peptide antigen | 8 | 24 | 8.871e-03 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 31 | 207 | 8.944e-03 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 16 | 79 | 9.172e-03 |
| GO:BP | GO:0048863 | stem cell differentiation | 35 | 245 | 9.456e-03 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 47 | 361 | 9.456e-03 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 35 | 245 | 9.456e-03 |
| GO:BP | GO:0030509 | BMP signaling pathway | 25 | 154 | 9.500e-03 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 64 | 534 | 9.560e-03 |
| GO:BP | GO:0015748 | organophosphate ester transport | 27 | 172 | 9.734e-03 |
| GO:BP | GO:0002486 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent | 6 | 14 | 1.054e-02 |
| GO:BP | GO:0001843 | neural tube closure | 17 | 88 | 1.060e-02 |
| GO:BP | GO:0048878 | chemical homeostasis | 111 | 1043 | 1.081e-02 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 152 | 1506 | 1.150e-02 |
| GO:BP | GO:0002181 | cytoplasmic translation | 26 | 165 | 1.150e-02 |
| GO:BP | GO:0019883 | antigen processing and presentation of endogenous antigen | 9 | 31 | 1.154e-02 |
| GO:BP | GO:0033077 | T cell differentiation in thymus | 17 | 89 | 1.194e-02 |
| GO:BP | GO:0060606 | tube closure | 17 | 89 | 1.194e-02 |
| GO:BP | GO:1901698 | response to nitrogen compound | 114 | 1080 | 1.208e-02 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 66 | 561 | 1.253e-02 |
| GO:BP | GO:0035621 | ER to Golgi ceramide transport | 3 | 3 | 1.256e-02 |
| GO:BP | GO:0071403 | cellular response to high density lipoprotein particle stimulus | 3 | 3 | 1.256e-02 |
| GO:BP | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis | 3 | 3 | 1.256e-02 |
| GO:BP | GO:0002085 | inhibition of neuroepithelial cell differentiation | 4 | 6 | 1.280e-02 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 96 | 884 | 1.300e-02 |
| GO:BP | GO:0002521 | leukocyte differentiation | 73 | 636 | 1.300e-02 |
| GO:BP | GO:0030278 | regulation of ossification | 21 | 123 | 1.306e-02 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 36 | 260 | 1.333e-02 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 66 | 563 | 1.333e-02 |
| GO:BP | GO:1990144 | intrinsic apoptotic signaling pathway in response to hypoxia | 5 | 10 | 1.333e-02 |
| GO:BP | GO:0055088 | lipid homeostasis | 26 | 167 | 1.333e-02 |
| GO:BP | GO:0000902 | cell morphogenesis | 106 | 996 | 1.368e-02 |
| GO:BP | GO:0006974 | DNA damage response | 98 | 908 | 1.371e-02 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 73 | 638 | 1.388e-02 |
| GO:BP | GO:0009306 | protein secretion | 48 | 379 | 1.402e-02 |
| GO:BP | GO:0060674 | placenta blood vessel development | 9 | 32 | 1.406e-02 |
| GO:BP | GO:0009798 | axis specification | 18 | 99 | 1.437e-02 |
| GO:BP | GO:0051259 | protein complex oligomerization | 37 | 271 | 1.437e-02 |
| GO:BP | GO:0030030 | cell projection organization | 162 | 1631 | 1.437e-02 |
| GO:BP | GO:0030258 | lipid modification | 29 | 196 | 1.487e-02 |
| GO:BP | GO:0008283 | cell population proliferation | 195 | 2015 | 1.487e-02 |
| GO:BP | GO:0090030 | regulation of steroid hormone biosynthetic process | 6 | 15 | 1.487e-02 |
| GO:BP | GO:0070243 | regulation of thymocyte apoptotic process | 6 | 15 | 1.487e-02 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 75 | 662 | 1.515e-02 |
| GO:BP | GO:0051168 | nuclear export | 26 | 169 | 1.524e-02 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 64 | 546 | 1.542e-02 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 18 | 100 | 1.587e-02 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 49 | 392 | 1.597e-02 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 48 | 382 | 1.603e-02 |
| GO:BP | GO:0006066 | alcohol metabolic process | 46 | 362 | 1.611e-02 |
| GO:BP | GO:0001701 | in utero embryonic development | 51 | 413 | 1.645e-02 |
| GO:BP | GO:1902653 | secondary alcohol biosynthetic process | 13 | 61 | 1.684e-02 |
| GO:BP | GO:0006695 | cholesterol biosynthetic process | 13 | 61 | 1.684e-02 |
| GO:BP | GO:0001841 | neural tube formation | 18 | 101 | 1.758e-02 |
| GO:BP | GO:0051604 | protein maturation | 63 | 539 | 1.775e-02 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 25 | 162 | 1.778e-02 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 10 | 40 | 1.836e-02 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 73 | 646 | 1.837e-02 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 41 | 315 | 1.862e-02 |
| GO:BP | GO:0003007 | heart morphogenesis | 36 | 266 | 1.862e-02 |
| GO:BP | GO:0006401 | RNA catabolic process | 41 | 315 | 1.862e-02 |
| GO:BP | GO:0061157 | mRNA destabilization | 18 | 102 | 1.933e-02 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 18 | 102 | 1.933e-02 |
| GO:BP | GO:0007507 | heart development | 69 | 605 | 1.941e-02 |
| GO:BP | GO:0032940 | secretion by cell | 92 | 854 | 1.941e-02 |
| GO:BP | GO:0014020 | primary neural tube formation | 17 | 94 | 1.989e-02 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 146 | 1462 | 1.990e-02 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 72 | 638 | 2.005e-02 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 69 | 606 | 2.005e-02 |
| GO:BP | GO:0043489 | RNA stabilization | 15 | 78 | 2.023e-02 |
| GO:BP | GO:0032926 | negative regulation of activin receptor signaling pathway | 6 | 16 | 2.025e-02 |
| GO:BP | GO:1900038 | negative regulation of cellular response to hypoxia | 5 | 11 | 2.025e-02 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 9 | 34 | 2.025e-02 |
| GO:BP | GO:0031943 | regulation of glucocorticoid metabolic process | 5 | 11 | 2.025e-02 |
| GO:BP | GO:0038066 | p38MAPK cascade | 12 | 55 | 2.025e-02 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 71 | 628 | 2.025e-02 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 41 | 317 | 2.025e-02 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 9 | 34 | 2.025e-02 |
| GO:BP | GO:0007033 | vacuole organization | 33 | 239 | 2.033e-02 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 48 | 388 | 2.066e-02 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 11 | 48 | 2.094e-02 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 10 | 41 | 2.112e-02 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 25 | 165 | 2.154e-02 |
| GO:BP | GO:0071772 | response to BMP | 25 | 165 | 2.154e-02 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 22 | 138 | 2.154e-02 |
| GO:BP | GO:0035116 | embryonic hindlimb morphogenesis | 8 | 28 | 2.210e-02 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 40 | 309 | 2.218e-02 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 76 | 686 | 2.333e-02 |
| GO:BP | GO:2000064 | regulation of cortisol biosynthetic process | 4 | 7 | 2.333e-02 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 24 | 157 | 2.344e-02 |
| GO:BP | GO:0090183 | regulation of kidney development | 9 | 35 | 2.449e-02 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 9 | 35 | 2.449e-02 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 10 | 42 | 2.508e-02 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 112 | 1087 | 2.579e-02 |
| GO:BP | GO:0050821 | protein stabilization | 31 | 224 | 2.642e-02 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 12 | 57 | 2.668e-02 |
| GO:BP | GO:0032024 | positive regulation of insulin secretion | 16 | 89 | 2.734e-02 |
| GO:BP | GO:0007389 | pattern specification process | 56 | 477 | 2.734e-02 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 38 | 293 | 2.734e-02 |
| GO:BP | GO:0034375 | high-density lipoprotein particle remodeling | 6 | 17 | 2.734e-02 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 8 | 29 | 2.734e-02 |
| GO:BP | GO:0090205 | positive regulation of cholesterol metabolic process | 6 | 17 | 2.734e-02 |
| GO:BP | GO:0050779 | RNA destabilization | 18 | 106 | 2.757e-02 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 15 | 81 | 2.771e-02 |
| GO:BP | GO:0006575 | modified amino acid metabolic process | 26 | 178 | 2.833e-02 |
| GO:BP | GO:0007548 | sex differentiation | 38 | 294 | 2.865e-02 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 7 | 23 | 2.875e-02 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 7 | 23 | 2.875e-02 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 17 | 98 | 2.875e-02 |
| GO:BP | GO:0035137 | hindlimb morphogenesis | 9 | 36 | 2.889e-02 |
| GO:BP | GO:0050657 | nucleic acid transport | 24 | 160 | 2.889e-02 |
| GO:BP | GO:0050658 | RNA transport | 24 | 160 | 2.889e-02 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 20 | 124 | 2.889e-02 |
| GO:BP | GO:0048505 | regulation of timing of cell differentiation | 5 | 12 | 2.911e-02 |
| GO:BP | GO:0019318 | hexose metabolic process | 31 | 226 | 2.911e-02 |
| GO:BP | GO:0022414 | reproductive process | 157 | 1608 | 2.944e-02 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 62 | 543 | 2.951e-02 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 18 | 107 | 2.967e-02 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 33 | 246 | 3.007e-02 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 74 | 673 | 3.057e-02 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 77 | 706 | 3.094e-02 |
| GO:BP | GO:0006259 | DNA metabolic process | 104 | 1005 | 3.094e-02 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 27 | 189 | 3.094e-02 |
| GO:BP | GO:0043648 | dicarboxylic acid metabolic process | 17 | 99 | 3.110e-02 |
| GO:BP | GO:1902930 | regulation of alcohol biosynthetic process | 11 | 51 | 3.188e-02 |
| GO:BP | GO:0003279 | cardiac septum development | 18 | 108 | 3.245e-02 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 16 | 91 | 3.251e-02 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 8 | 30 | 3.260e-02 |
| GO:BP | GO:0006508 | proteolysis | 146 | 1486 | 3.268e-02 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 15 | 83 | 3.359e-02 |
| GO:BP | GO:0042594 | response to starvation | 30 | 219 | 3.366e-02 |
| GO:BP | GO:0006457 | protein folding | 30 | 219 | 3.366e-02 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 26 | 181 | 3.366e-02 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 34 | 258 | 3.366e-02 |
| GO:BP | GO:0003382 | epithelial cell morphogenesis | 9 | 37 | 3.366e-02 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 9 | 37 | 3.366e-02 |
| GO:BP | GO:0006403 | RNA localization | 28 | 200 | 3.369e-02 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 39 | 308 | 3.384e-02 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 48 | 400 | 3.396e-02 |
| GO:BP | GO:1904028 | positive regulation of collagen fibril organization | 3 | 4 | 3.427e-02 |
| GO:BP | GO:0045632 | negative regulation of mechanoreceptor differentiation | 3 | 4 | 3.427e-02 |
| GO:BP | GO:0045608 | negative regulation of inner ear auditory receptor cell differentiation | 3 | 4 | 3.427e-02 |
| GO:BP | GO:2000981 | negative regulation of inner ear receptor cell differentiation | 3 | 4 | 3.427e-02 |
| GO:BP | GO:0003383 | apical constriction | 3 | 4 | 3.427e-02 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 46 | 380 | 3.432e-02 |
| GO:BP | GO:0006417 | regulation of translation | 46 | 380 | 3.432e-02 |
| GO:BP | GO:0019080 | viral gene expression | 18 | 109 | 3.432e-02 |
| GO:BP | GO:0051236 | establishment of RNA localization | 24 | 163 | 3.432e-02 |
| GO:BP | GO:0043691 | reverse cholesterol transport | 6 | 18 | 3.432e-02 |
| GO:BP | GO:0007042 | lysosomal lumen acidification | 7 | 24 | 3.432e-02 |
| GO:BP | GO:0045995 | regulation of embryonic development | 16 | 92 | 3.461e-02 |
| GO:BP | GO:0061024 | membrane organization | 87 | 821 | 3.467e-02 |
| GO:BP | GO:0050708 | regulation of protein secretion | 35 | 269 | 3.467e-02 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 22 | 145 | 3.506e-02 |
| GO:BP | GO:0031175 | neuron projection development | 105 | 1023 | 3.554e-02 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 33 | 250 | 3.610e-02 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 40 | 320 | 3.625e-02 |
| GO:BP | GO:0032868 | response to insulin | 35 | 270 | 3.650e-02 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 14 | 76 | 3.662e-02 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 14 | 76 | 3.662e-02 |
| GO:BP | GO:0032347 | regulation of aldosterone biosynthetic process | 4 | 8 | 3.674e-02 |
| GO:BP | GO:0032344 | regulation of aldosterone metabolic process | 4 | 8 | 3.674e-02 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 66 | 594 | 3.755e-02 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 19 | 119 | 3.755e-02 |
| GO:BP | GO:0019082 | viral protein processing | 8 | 31 | 3.769e-02 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 9 | 38 | 3.843e-02 |
| GO:BP | GO:0038203 | TORC2 signaling | 5 | 13 | 3.901e-02 |
| GO:BP | GO:0040034 | regulation of development, heterochronic | 5 | 13 | 3.901e-02 |
| GO:BP | GO:0010944 | negative regulation of transcription by competitive promoter binding | 5 | 13 | 3.901e-02 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 50 | 425 | 3.901e-02 |
| GO:BP | GO:0090158 | endoplasmic reticulum membrane organization | 5 | 13 | 3.901e-02 |
| GO:BP | GO:0033365 | protein localization to organelle | 121 | 1209 | 3.913e-02 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 40 | 322 | 3.924e-02 |
| GO:BP | GO:0051098 | regulation of binding | 29 | 213 | 3.981e-02 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 21 | 138 | 4.030e-02 |
| GO:BP | GO:0141188 | nucleic acid catabolic process | 42 | 343 | 4.030e-02 |
| GO:BP | GO:0007498 | mesoderm development | 21 | 138 | 4.030e-02 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 20 | 129 | 4.040e-02 |
| GO:BP | GO:0016126 | sterol biosynthetic process | 13 | 69 | 4.078e-02 |
| GO:BP | GO:0048666 | neuron development | 118 | 1177 | 4.084e-02 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 96 | 928 | 4.084e-02 |
| GO:BP | GO:0140352 | export from cell | 96 | 928 | 4.084e-02 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 32 | 243 | 4.104e-02 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 7 | 25 | 4.126e-02 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 7 | 25 | 4.126e-02 |
| GO:BP | GO:0038202 | TORC1 signaling | 17 | 103 | 4.205e-02 |
| GO:BP | GO:0009725 | response to hormone | 94 | 907 | 4.227e-02 |
| GO:BP | GO:1903830 | magnesium ion transmembrane transport | 6 | 19 | 4.297e-02 |
| GO:BP | GO:0006491 | N-glycan processing | 6 | 19 | 4.297e-02 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 20 | 130 | 4.316e-02 |
| GO:BP | GO:0051028 | mRNA transport | 20 | 130 | 4.316e-02 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 66 | 599 | 4.336e-02 |
| GO:BP | GO:0031647 | regulation of protein stability | 41 | 335 | 4.382e-02 |
| GO:BP | GO:0008202 | steroid metabolic process | 41 | 335 | 4.382e-02 |
| GO:BP | GO:0060323 | head morphogenesis | 9 | 39 | 4.382e-02 |
| GO:BP | GO:0035751 | regulation of lysosomal lumen pH | 8 | 32 | 4.393e-02 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 38 | 305 | 4.487e-02 |
| GO:BP | GO:0009267 | cellular response to starvation | 25 | 177 | 4.487e-02 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 13 | 70 | 4.487e-02 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 22 | 149 | 4.509e-02 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 28 | 206 | 4.549e-02 |
| GO:BP | GO:0035050 | embryonic heart tube development | 15 | 87 | 4.643e-02 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 51 | 441 | 4.744e-02 |
| GO:BP | GO:0042113 | B cell activation | 36 | 286 | 4.771e-02 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 10 | 47 | 4.804e-02 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 16 | 96 | 4.830e-02 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 22 | 150 | 4.838e-02 |
| KEGG | KEGG:04218 | Cellular senescence | 30 | 155 | 6.753e-03 |
| KEGG | KEGG:01522 | Endocrine resistance | 22 | 95 | 6.753e-03 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 13 | 42 | 6.753e-03 |
| KEGG | KEGG:00310 | Lysine degradation | 16 | 63 | 1.069e-02 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 20 | 93 | 1.299e-02 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 17 | 72 | 1.299e-02 |
| KEGG | KEGG:00071 | Fatty acid degradation | 12 | 43 | 1.441e-02 |
| KEGG | KEGG:05224 | Breast cancer | 27 | 147 | 1.441e-02 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 23 | 121 | 1.845e-02 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 36 | 223 | 1.845e-02 |
| KEGG | KEGG:04934 | Cushing syndrome | 27 | 153 | 1.973e-02 |
| KEGG | KEGG:04915 | Estrogen signaling pathway | 24 | 136 | 2.661e-02 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 19 | 97 | 2.661e-02 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 25 | 143 | 2.661e-02 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 28 | 166 | 2.661e-02 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 19 | 98 | 2.661e-02 |
| KEGG | KEGG:04929 | GnRH secretion | 14 | 64 | 3.239e-02 |
| KEGG | KEGG:05226 | Gastric cancer | 25 | 148 | 3.564e-02 |
| KEGG | KEGG:05200 | Pathways in cancer | 68 | 527 | 3.697e-02 |
| KEGG | KEGG:04540 | Gap junction | 17 | 88 | 3.913e-02 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 12 | 53 | 4.066e-02 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 21 | 120 | 4.204e-02 |
| KEGG | KEGG:05214 | Glioma | 15 | 75 | 4.244e-02 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 11 | 48 | 4.687e-02 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 19 | 107 | 4.687e-02 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 16 | 84 | 4.687e-02 |
#write.csv(table_DOX24_up_genes, "output/table_DOX24_upreg_genes.csv")
#GO:BP
table_DOX24_up_genes_GOBP <- table_DOX24_up_genes %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
#saveRDS(table_motif1_GOBP_d, "data/table_motif1_GOBP_d.RDS")
table_DOX24_up_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Specific Up DEGs Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))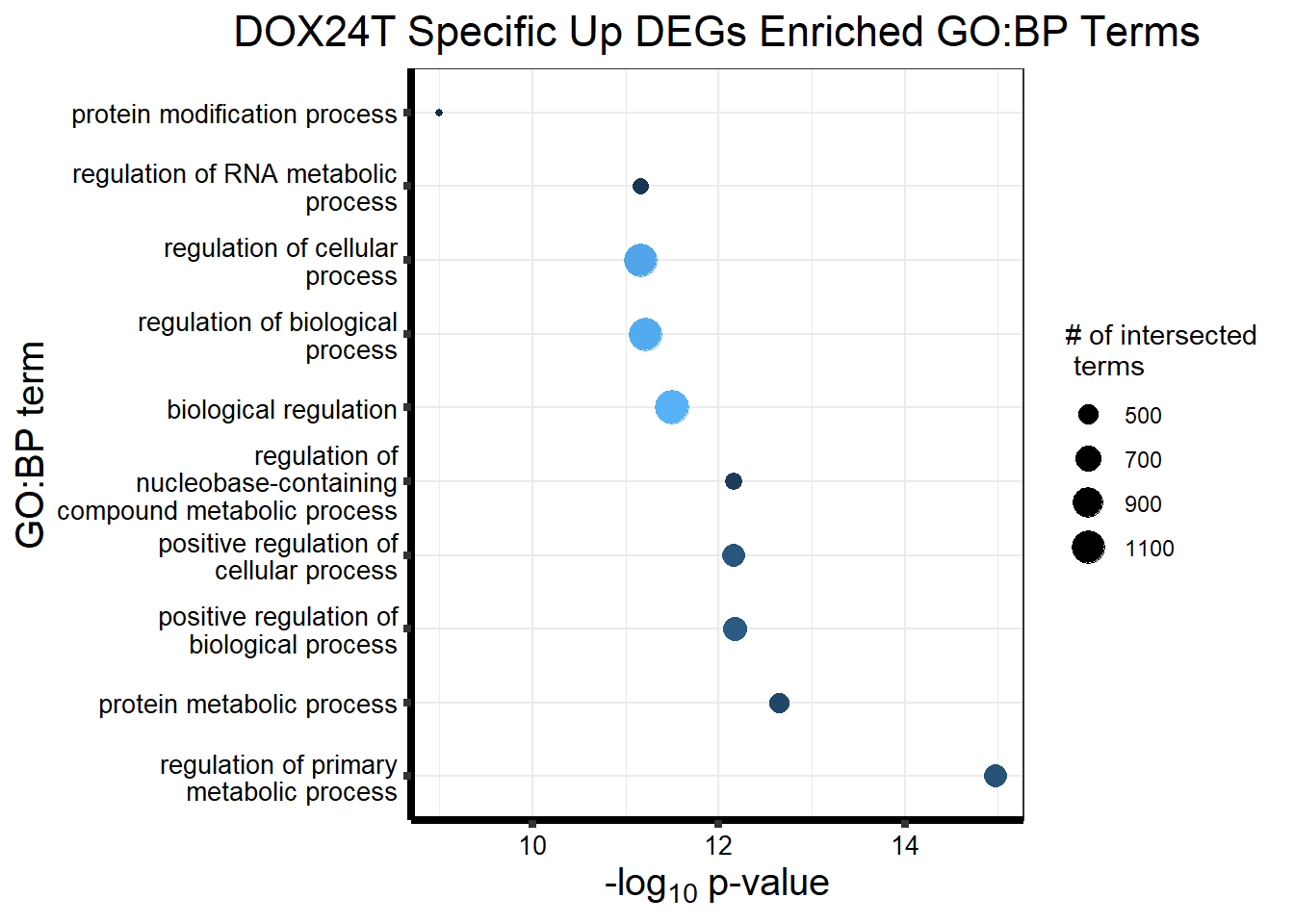
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_DOX24_up_genes_KEGG <- table_DOX24_up_genes %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24_up_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Specific Up DEGs Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))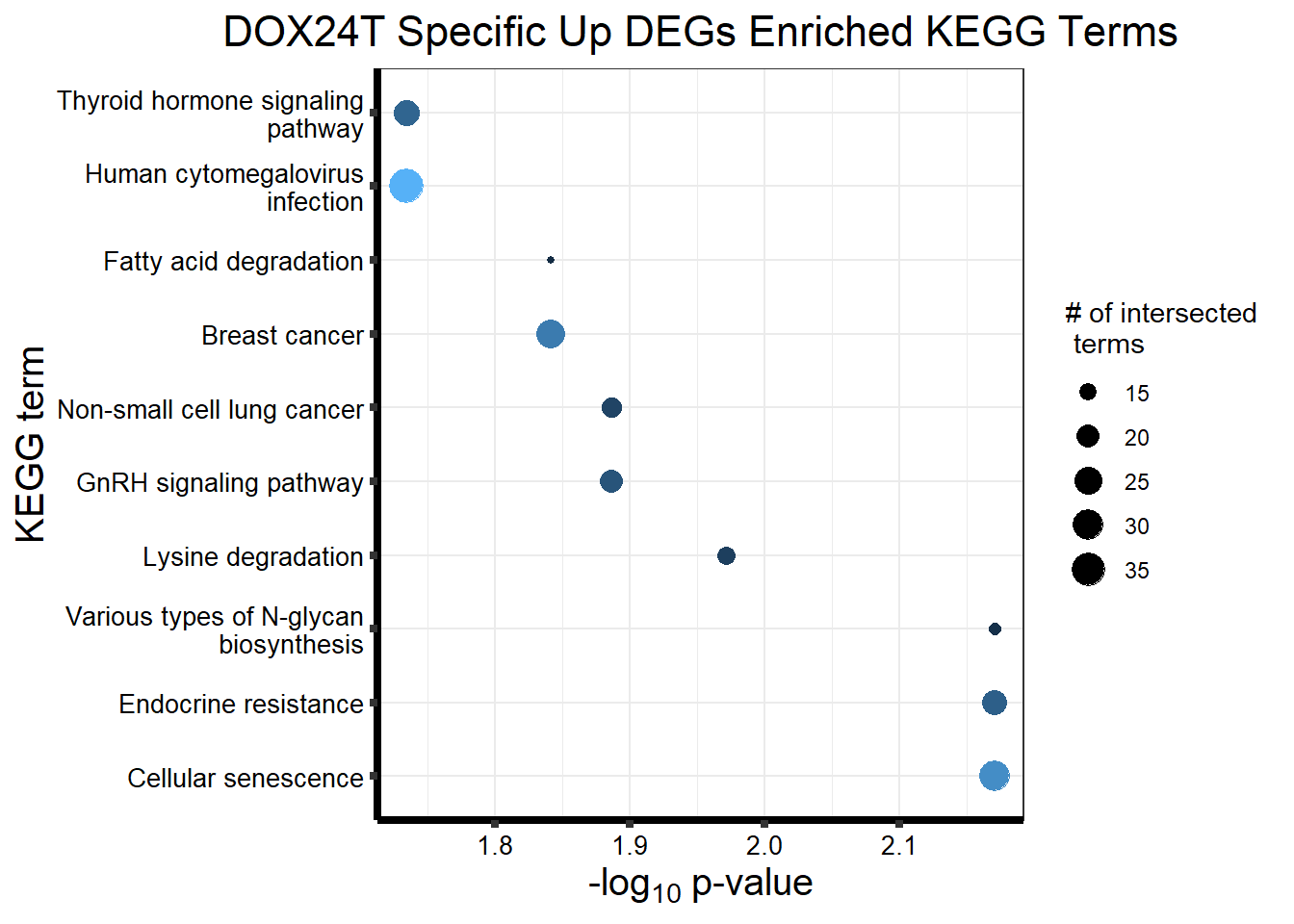
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####DOX24 Downregulated Genes#####
D24_DEGs_down_mat <- as.matrix(DOX24T_DEGs_GO_down)
DOX_24_down_dxr_gene <- gost(query = D24_DEGs_down_mat,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
DOX_24_down_gost_genes <- gostplot(DOX_24_down_dxr_gene, capped = FALSE, interactive = TRUE)
DOX_24_down_gost_genestable_DOX24_down_genes <- DOX_24_down_dxr_gene$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX24_down_genes %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0080090 | regulation of primary metabolic process | 779 | 5390 | 2.491e-36 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 608 | 3990 | 1.326e-32 |
| GO:BP | GO:0006351 | DNA-templated transcription | 554 | 3549 | 6.014e-32 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 560 | 3687 | 4.144e-29 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 523 | 3428 | 3.183e-27 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 520 | 3409 | 4.438e-27 |
| GO:BP | GO:0006996 | organelle organization | 533 | 3594 | 7.833e-25 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 68 | 168 | 2.418e-23 |
| GO:BP | GO:0044238 | primary metabolic process | 1413 | 12342 | 3.051e-23 |
| GO:BP | GO:0019538 | protein metabolic process | 647 | 4721 | 5.580e-22 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 417 | 2711 | 1.821e-21 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 823 | 6492 | 2.193e-19 |
| GO:BP | GO:0019222 | regulation of metabolic process | 877 | 7035 | 5.043e-19 |
| GO:BP | GO:0006412 | translation | 155 | 727 | 5.342e-19 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 390 | 2578 | 1.463e-18 |
| GO:BP | GO:0043412 | macromolecule modification | 442 | 3030 | 1.955e-18 |
| GO:BP | GO:0032543 | mitochondrial translation | 53 | 130 | 2.466e-18 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 459 | 3288 | 1.407e-15 |
| GO:BP | GO:0006399 | tRNA metabolic process | 65 | 210 | 2.240e-15 |
| GO:BP | GO:0008152 | metabolic process | 1534 | 14136 | 6.307e-15 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 311 | 2036 | 7.340e-15 |
| GO:BP | GO:0042254 | ribosome biogenesis | 83 | 323 | 1.691e-14 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 946 | 7992 | 4.027e-14 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 1336 | 12048 | 9.159e-14 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 703 | 5644 | 1.266e-13 |
| GO:BP | GO:0046907 | intracellular transport | 226 | 1381 | 1.531e-13 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 281 | 1835 | 1.986e-13 |
| GO:BP | GO:0010468 | regulation of gene expression | 690 | 5536 | 2.275e-13 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 481 | 3597 | 3.676e-13 |
| GO:BP | GO:0016072 | rRNA metabolic process | 70 | 266 | 9.680e-13 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 714 | 5809 | 1.238e-12 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 225 | 1417 | 4.106e-12 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 259 | 1697 | 4.106e-12 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 259 | 1699 | 4.513e-12 |
| GO:BP | GO:0090304 | nucleic acid metabolic process | 876 | 7434 | 5.303e-12 |
| GO:BP | GO:0051649 | establishment of localization in cell | 295 | 2010 | 6.845e-12 |
| GO:BP | GO:0036211 | protein modification process | 389 | 2846 | 1.566e-11 |
| GO:BP | GO:0009056 | catabolic process | 363 | 2639 | 5.516e-11 |
| GO:BP | GO:0006259 | DNA metabolic process | 169 | 1005 | 6.692e-11 |
| GO:BP | GO:0033043 | regulation of organelle organization | 187 | 1148 | 6.702e-11 |
| GO:BP | GO:0008033 | tRNA processing | 43 | 139 | 4.309e-10 |
| GO:BP | GO:0009451 | RNA modification | 49 | 174 | 6.303e-10 |
| GO:BP | GO:0015031 | protein transport | 219 | 1444 | 8.098e-10 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 706 | 5920 | 1.175e-09 |
| GO:BP | GO:0006364 | rRNA processing | 57 | 225 | 1.246e-09 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 350 | 2593 | 1.657e-09 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 136 | 786 | 1.657e-09 |
| GO:BP | GO:0033554 | cellular response to stress | 263 | 1830 | 1.680e-09 |
| GO:BP | GO:0006281 | DNA repair | 114 | 621 | 1.864e-09 |
| GO:BP | GO:0034654 | nucleobase-containing compound biosynthetic process | 822 | 7076 | 1.864e-09 |
| GO:BP | GO:0033036 | macromolecule localization | 419 | 3228 | 2.494e-09 |
| GO:BP | GO:0051641 | cellular localization | 465 | 3661 | 3.594e-09 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 116 | 643 | 3.600e-09 |
| GO:BP | GO:0006974 | DNA damage response | 150 | 908 | 4.395e-09 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 115 | 638 | 4.477e-09 |
| GO:BP | GO:0030163 | protein catabolic process | 165 | 1030 | 5.069e-09 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 113 | 628 | 7.193e-09 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 367 | 2782 | 7.625e-09 |
| GO:BP | GO:0016070 | RNA metabolic process | 796 | 6873 | 9.372e-09 |
| GO:BP | GO:0008104 | protein localization | 365 | 2770 | 9.621e-09 |
| GO:BP | GO:0048518 | positive regulation of biological process | 734 | 6264 | 9.621e-09 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 356 | 2696 | 1.334e-08 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 57 | 242 | 1.965e-08 |
| GO:BP | GO:0006886 | intracellular protein transport | 119 | 686 | 2.130e-08 |
| GO:BP | GO:0007005 | mitochondrion organization | 85 | 435 | 2.583e-08 |
| GO:BP | GO:0141187 | nucleic acid biosynthetic process | 783 | 6782 | 2.770e-08 |
| GO:BP | GO:0071705 | nitrogen compound transport | 267 | 1923 | 3.215e-08 |
| GO:BP | GO:0043687 | post-translational protein modification | 161 | 1027 | 3.856e-08 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 324 | 2433 | 3.908e-08 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 99 | 543 | 4.473e-08 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 93 | 500 | 5.161e-08 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 81 | 414 | 5.820e-08 |
| GO:BP | GO:0006325 | chromatin organization | 142 | 884 | 8.053e-08 |
| GO:BP | GO:1903008 | organelle disassembly | 26 | 71 | 8.857e-08 |
| GO:BP | GO:0009894 | regulation of catabolic process | 161 | 1040 | 8.954e-08 |
| GO:BP | GO:0016073 | snRNA metabolic process | 23 | 58 | 1.303e-07 |
| GO:BP | GO:0032774 | RNA biosynthetic process | 766 | 6671 | 1.381e-07 |
| GO:BP | GO:0032790 | ribosome disassembly | 19 | 41 | 1.479e-07 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 137 | 853 | 1.515e-07 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 152 | 975 | 1.561e-07 |
| GO:BP | GO:0043414 | macromolecule methylation | 36 | 127 | 1.930e-07 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 46 | 187 | 1.976e-07 |
| GO:BP | GO:0009987 | cellular process | 2002 | 20247 | 2.072e-07 |
| GO:BP | GO:0006403 | RNA localization | 48 | 200 | 2.089e-07 |
| GO:BP | GO:0006400 | tRNA modification | 30 | 95 | 2.456e-07 |
| GO:BP | GO:0050789 | regulation of biological process | 1316 | 12336 | 2.502e-07 |
| GO:BP | GO:0009059 | macromolecule biosynthetic process | 1058 | 9643 | 2.537e-07 |
| GO:BP | GO:0050794 | regulation of cellular process | 1278 | 11946 | 3.407e-07 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 103 | 597 | 3.517e-07 |
| GO:BP | GO:0006914 | autophagy | 103 | 597 | 3.517e-07 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 933 | 8393 | 4.831e-07 |
| GO:BP | GO:0051726 | regulation of cell cycle | 163 | 1087 | 5.831e-07 |
| GO:BP | GO:0009058 | biosynthetic process | 1152 | 10664 | 8.796e-07 |
| GO:BP | GO:0032259 | methylation | 52 | 236 | 9.568e-07 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 32 | 112 | 1.016e-06 |
| GO:BP | GO:0045184 | establishment of protein localization | 260 | 1937 | 1.016e-06 |
| GO:BP | GO:0006302 | double-strand break repair | 64 | 319 | 1.016e-06 |
| GO:BP | GO:0070925 | organelle assembly | 157 | 1046 | 1.016e-06 |
| GO:BP | GO:0065007 | biological regulation | 1348 | 12743 | 1.285e-06 |
| GO:BP | GO:0007049 | cell cycle | 228 | 1663 | 1.540e-06 |
| GO:BP | GO:0072344 | rescue of stalled ribosome | 16 | 34 | 1.728e-06 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 34 | 127 | 2.074e-06 |
| GO:BP | GO:0051276 | chromosome organization | 97 | 574 | 2.512e-06 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 248 | 1855 | 3.098e-06 |
| GO:BP | GO:0010467 | gene expression | 1007 | 9224 | 3.214e-06 |
| GO:BP | GO:0006282 | regulation of DNA repair | 49 | 225 | 3.285e-06 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 91 | 535 | 4.717e-06 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 34 | 134 | 8.176e-06 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 91 | 546 | 1.203e-05 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 60 | 312 | 1.214e-05 |
| GO:BP | GO:0006338 | chromatin remodeling | 113 | 725 | 1.507e-05 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 176 | 1256 | 1.543e-05 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 27 | 96 | 1.662e-05 |
| GO:BP | GO:0016567 | protein ubiquitination | 118 | 768 | 1.706e-05 |
| GO:BP | GO:0000723 | telomere maintenance | 38 | 164 | 1.827e-05 |
| GO:BP | GO:0038202 | TORC1 signaling | 28 | 103 | 2.122e-05 |
| GO:BP | GO:0006402 | mRNA catabolic process | 52 | 261 | 2.386e-05 |
| GO:BP | GO:0001510 | RNA methylation | 24 | 81 | 2.551e-05 |
| GO:BP | GO:0000725 | recombinational repair | 42 | 193 | 2.672e-05 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 41 | 188 | 3.412e-05 |
| GO:BP | GO:0051168 | nuclear export | 38 | 169 | 3.897e-05 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 24 | 83 | 4.055e-05 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 113 | 745 | 5.453e-05 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 85 | 519 | 5.670e-05 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 55 | 291 | 6.076e-05 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 15 | 38 | 6.378e-05 |
| GO:BP | GO:0000278 | mitotic cell cycle | 130 | 892 | 7.232e-05 |
| GO:BP | GO:0031929 | TOR signaling | 37 | 167 | 7.533e-05 |
| GO:BP | GO:0022411 | cellular component disassembly | 75 | 446 | 8.634e-05 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 33 | 143 | 1.060e-04 |
| GO:BP | GO:0051236 | establishment of RNA localization | 36 | 163 | 1.102e-04 |
| GO:BP | GO:0016180 | snRNA processing | 14 | 35 | 1.147e-04 |
| GO:BP | GO:0006417 | regulation of translation | 66 | 380 | 1.147e-04 |
| GO:BP | GO:0009303 | rRNA transcription | 15 | 40 | 1.293e-04 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 16 | 45 | 1.349e-04 |
| GO:BP | GO:0006401 | RNA catabolic process | 57 | 315 | 1.502e-04 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 33 | 146 | 1.635e-04 |
| GO:BP | GO:0000209 | protein polyubiquitination | 52 | 280 | 1.839e-04 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 31 | 134 | 1.906e-04 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 19 | 62 | 1.971e-04 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 59 | 334 | 2.134e-04 |
| GO:BP | GO:0051169 | nuclear transport | 59 | 334 | 2.134e-04 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 20 | 68 | 2.186e-04 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 56 | 312 | 2.235e-04 |
| GO:BP | GO:0022402 | cell cycle process | 172 | 1280 | 2.291e-04 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 17 | 52 | 2.291e-04 |
| GO:BP | GO:0016197 | endosomal transport | 53 | 290 | 2.291e-04 |
| GO:BP | GO:0006414 | translational elongation | 22 | 80 | 2.421e-04 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 29 | 123 | 2.439e-04 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 36 | 170 | 2.685e-04 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 31 | 137 | 2.859e-04 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 16 | 48 | 3.154e-04 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 106 | 720 | 3.729e-04 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 13 | 34 | 4.167e-04 |
| GO:BP | GO:0050657 | nucleic acid transport | 34 | 160 | 4.283e-04 |
| GO:BP | GO:0050658 | RNA transport | 34 | 160 | 4.283e-04 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 78 | 492 | 4.283e-04 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 22 | 83 | 4.321e-04 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 18 | 60 | 4.321e-04 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 31 | 140 | 4.321e-04 |
| GO:BP | GO:0141188 | nucleic acid catabolic process | 59 | 343 | 4.394e-04 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 41 | 210 | 4.795e-04 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 24 | 96 | 5.006e-04 |
| GO:BP | GO:0000154 | rRNA modification | 13 | 35 | 5.675e-04 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 20 | 73 | 6.181e-04 |
| GO:BP | GO:0032200 | telomere organization | 38 | 191 | 6.225e-04 |
| GO:BP | GO:0016043 | cellular component organization | 880 | 8184 | 6.783e-04 |
| GO:BP | GO:0006390 | mitochondrial transcription | 10 | 22 | 7.049e-04 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 44 | 236 | 7.446e-04 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 15 | 46 | 7.446e-04 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 27 | 118 | 8.004e-04 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 44 | 237 | 8.205e-04 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 18 | 63 | 8.434e-04 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 196 | 1529 | 8.494e-04 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 141 | 1039 | 8.774e-04 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 29 | 132 | 8.774e-04 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 100 | 686 | 8.774e-04 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 20 | 75 | 8.864e-04 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 61 | 369 | 9.852e-04 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 62 | 377 | 9.906e-04 |
| GO:BP | GO:0017004 | cytochrome complex assembly | 14 | 42 | 1.002e-03 |
| GO:BP | GO:0016236 | macroautophagy | 60 | 362 | 1.040e-03 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 19 | 70 | 1.051e-03 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 76 | 492 | 1.185e-03 |
| GO:BP | GO:0061024 | membrane organization | 115 | 821 | 1.257e-03 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 26 | 116 | 1.528e-03 |
| GO:BP | GO:0051028 | mRNA transport | 28 | 130 | 1.674e-03 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 16 | 55 | 1.740e-03 |
| GO:BP | GO:0006405 | RNA export from nucleus | 22 | 91 | 1.740e-03 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 59 | 361 | 1.758e-03 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 24 | 104 | 1.765e-03 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 24 | 104 | 1.765e-03 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 13 | 39 | 1.824e-03 |
| GO:BP | GO:0070979 | protein K11-linked ubiquitination | 13 | 39 | 1.824e-03 |
| GO:BP | GO:0060341 | regulation of cellular localization | 136 | 1013 | 1.856e-03 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 8 | 16 | 1.895e-03 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 25 | 112 | 2.172e-03 |
| GO:BP | GO:0007030 | Golgi organization | 31 | 153 | 2.187e-03 |
| GO:BP | GO:0035556 | intracellular signal transduction | 346 | 2965 | 2.331e-03 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 67 | 430 | 2.428e-03 |
| GO:BP | GO:0000819 | sister chromatid segregation | 42 | 234 | 2.429e-03 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 12 | 35 | 2.503e-03 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 54 | 327 | 2.569e-03 |
| GO:BP | GO:0010506 | regulation of autophagy | 59 | 367 | 2.642e-03 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 31 | 155 | 2.731e-03 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 14 | 46 | 2.759e-03 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 7 | 13 | 2.990e-03 |
| GO:BP | GO:0031503 | protein-containing complex localization | 39 | 215 | 3.334e-03 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 26 | 122 | 3.350e-03 |
| GO:BP | GO:0043038 | amino acid activation | 14 | 47 | 3.514e-03 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 15 | 53 | 3.785e-03 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 15 | 53 | 3.785e-03 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 27 | 130 | 3.859e-03 |
| GO:BP | GO:0010256 | endomembrane system organization | 88 | 613 | 3.863e-03 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 18 | 71 | 3.863e-03 |
| GO:BP | GO:0044782 | cilium organization | 66 | 431 | 4.289e-03 |
| GO:BP | GO:0006513 | protein monoubiquitination | 14 | 48 | 4.377e-03 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 28 | 139 | 4.928e-03 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 56 | 352 | 4.928e-03 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 36 | 197 | 4.949e-03 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 13 | 43 | 4.990e-03 |
| GO:BP | GO:0043144 | sno(s)RNA processing | 7 | 14 | 5.110e-03 |
| GO:BP | GO:0006370 | 7-methylguanosine mRNA capping | 5 | 7 | 5.202e-03 |
| GO:BP | GO:0040031 | snRNA modification | 5 | 7 | 5.202e-03 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 22 | 99 | 5.564e-03 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 21 | 93 | 6.100e-03 |
| GO:BP | GO:0007017 | microtubule-based process | 131 | 999 | 6.222e-03 |
| GO:BP | GO:0071806 | protein transmembrane transport | 18 | 74 | 6.405e-03 |
| GO:BP | GO:0030518 | nuclear receptor-mediated steroid hormone signaling pathway | 25 | 121 | 6.898e-03 |
| GO:BP | GO:1990928 | response to amino acid starvation | 15 | 56 | 6.898e-03 |
| GO:BP | GO:0090685 | RNA localization to nucleus | 8 | 19 | 7.117e-03 |
| GO:BP | GO:0090672 | telomerase RNA localization | 8 | 19 | 7.117e-03 |
| GO:BP | GO:0090671 | telomerase RNA localization to Cajal body | 8 | 19 | 7.117e-03 |
| GO:BP | GO:0090670 | RNA localization to Cajal body | 8 | 19 | 7.117e-03 |
| GO:BP | GO:0071025 | RNA surveillance | 8 | 19 | 7.117e-03 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 85 | 600 | 7.156e-03 |
| GO:BP | GO:0062125 | regulation of mitochondrial gene expression | 11 | 34 | 7.730e-03 |
| GO:BP | GO:0071028 | nuclear mRNA surveillance | 6 | 11 | 7.914e-03 |
| GO:BP | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process | 9 | 24 | 7.914e-03 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 9 | 24 | 7.914e-03 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 234 | 1953 | 7.914e-03 |
| GO:BP | GO:0062028 | regulation of stress granule assembly | 6 | 11 | 7.914e-03 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 9 | 24 | 7.914e-03 |
| GO:BP | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 10 | 29 | 7.924e-03 |
| GO:BP | GO:0043248 | proteasome assembly | 7 | 15 | 7.924e-03 |
| GO:BP | GO:0071027 | nuclear RNA surveillance | 7 | 15 | 7.924e-03 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 28 | 144 | 8.123e-03 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 195 | 1592 | 8.301e-03 |
| GO:BP | GO:0051179 | localization | 604 | 5555 | 8.322e-03 |
| GO:BP | GO:0030488 | tRNA methylation | 12 | 40 | 8.571e-03 |
| GO:BP | GO:0006457 | protein folding | 38 | 219 | 8.647e-03 |
| GO:BP | GO:0007010 | cytoskeleton organization | 188 | 1529 | 8.670e-03 |
| GO:BP | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway | 32 | 174 | 8.723e-03 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 17 | 70 | 8.764e-03 |
| GO:BP | GO:0140014 | mitotic nuclear division | 46 | 282 | 9.062e-03 |
| GO:BP | GO:0141084 | inflammasome-mediated signaling pathway | 13 | 46 | 9.062e-03 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 13 | 46 | 9.062e-03 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 11 | 35 | 9.457e-03 |
| GO:BP | GO:0006310 | DNA recombination | 54 | 347 | 9.531e-03 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 73 | 505 | 9.706e-03 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 21 | 97 | 9.752e-03 |
| GO:BP | GO:0030150 | protein import into mitochondrial matrix | 8 | 20 | 9.752e-03 |
| GO:BP | GO:0008535 | respiratory chain complex IV assembly | 10 | 30 | 1.015e-02 |
| GO:BP | GO:0141085 | regulation of inflammasome-mediated signaling pathway | 12 | 41 | 1.047e-02 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 92 | 670 | 1.060e-02 |
| GO:BP | GO:0009452 | 7-methylguanosine RNA capping | 5 | 8 | 1.075e-02 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 13 | 47 | 1.100e-02 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 22 | 105 | 1.127e-02 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 22 | 105 | 1.127e-02 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 74 | 517 | 1.147e-02 |
| GO:BP | GO:0032978 | protein insertion into membrane from inner side | 4 | 5 | 1.152e-02 |
| GO:BP | GO:0032979 | protein insertion into mitochondrial inner membrane from matrix | 4 | 5 | 1.152e-02 |
| GO:BP | GO:0016074 | sno(s)RNA metabolic process | 7 | 16 | 1.159e-02 |
| GO:BP | GO:0036260 | RNA capping | 7 | 16 | 1.159e-02 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 11 | 36 | 1.167e-02 |
| GO:BP | GO:0010212 | response to ionizing radiation | 27 | 141 | 1.197e-02 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 34 | 193 | 1.201e-02 |
| GO:BP | GO:0030031 | cell projection assembly | 85 | 613 | 1.221e-02 |
| GO:BP | GO:0006413 | translational initiation | 25 | 127 | 1.237e-02 |
| GO:BP | GO:0031126 | sno(s)RNA 3’-end processing | 6 | 12 | 1.248e-02 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 36 | 209 | 1.270e-02 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 13 | 48 | 1.303e-02 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 9 | 26 | 1.359e-02 |
| GO:BP | GO:0060271 | cilium assembly | 60 | 403 | 1.370e-02 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 31 | 172 | 1.371e-02 |
| GO:BP | GO:0032880 | regulation of protein localization | 118 | 907 | 1.380e-02 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 42 | 257 | 1.385e-02 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 46 | 289 | 1.397e-02 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 16 | 67 | 1.408e-02 |
| GO:BP | GO:0048284 | organelle fusion | 30 | 165 | 1.415e-02 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 39 | 234 | 1.435e-02 |
| GO:BP | GO:0033365 | protein localization to organelle | 151 | 1209 | 1.520e-02 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 17 | 74 | 1.520e-02 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 12 | 43 | 1.520e-02 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 14 | 55 | 1.520e-02 |
| GO:BP | GO:0006508 | proteolysis | 181 | 1486 | 1.520e-02 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 607 | 5629 | 1.591e-02 |
| GO:BP | GO:0051640 | organelle localization | 85 | 620 | 1.629e-02 |
| GO:BP | GO:0048285 | organelle fission | 71 | 500 | 1.682e-02 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 42 | 260 | 1.689e-02 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 9 | 27 | 1.753e-02 |
| GO:BP | GO:0009966 | regulation of signal transduction | 343 | 3034 | 1.783e-02 |
| GO:BP | GO:0043401 | steroid hormone receptor signaling pathway | 26 | 138 | 1.783e-02 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 8 | 22 | 1.786e-02 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 41 | 253 | 1.787e-02 |
| GO:BP | GO:0141137 | positive regulation of gene expression, epigenetic | 14 | 56 | 1.787e-02 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 171 | 1400 | 1.825e-02 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 16 | 69 | 1.868e-02 |
| GO:BP | GO:0016050 | vesicle organization | 56 | 376 | 1.884e-02 |
| GO:BP | GO:0009304 | tRNA transcription | 6 | 13 | 1.922e-02 |
| GO:BP | GO:0007059 | chromosome segregation | 62 | 427 | 1.957e-02 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 62 | 427 | 1.957e-02 |
| GO:BP | GO:0051865 | protein autoubiquitination | 17 | 76 | 1.975e-02 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 25 | 132 | 1.993e-02 |
| GO:BP | GO:0009218 | pyrimidine ribonucleotide metabolic process | 10 | 33 | 1.995e-02 |
| GO:BP | GO:0006997 | nucleus organization | 27 | 147 | 2.048e-02 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 29 | 162 | 2.050e-02 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 29 | 162 | 2.050e-02 |
| GO:BP | GO:0080134 | regulation of response to stress | 169 | 1387 | 2.104e-02 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 89 | 662 | 2.107e-02 |
| GO:BP | GO:0032205 | negative regulation of telomere maintenance | 11 | 39 | 2.107e-02 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 128 | 1011 | 2.107e-02 |
| GO:BP | GO:0007031 | peroxisome organization | 11 | 39 | 2.107e-02 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 11 | 39 | 2.107e-02 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 11 | 39 | 2.107e-02 |
| GO:BP | GO:0140285 | endosome fission | 3 | 3 | 2.112e-02 |
| GO:BP | GO:0009195 | pyrimidine ribonucleoside diphosphate catabolic process | 3 | 3 | 2.112e-02 |
| GO:BP | GO:0070478 | nuclear-transcribed mRNA catabolic process, 3’-5’ exonucleolytic nonsense-mediated decay | 3 | 3 | 2.112e-02 |
| GO:BP | GO:0009140 | pyrimidine nucleoside diphosphate catabolic process | 3 | 3 | 2.112e-02 |
| GO:BP | GO:0016094 | polyprenol biosynthetic process | 3 | 3 | 2.112e-02 |
| GO:BP | GO:0031120 | snRNA pseudouridine synthesis | 3 | 3 | 2.112e-02 |
| GO:BP | GO:0006256 | UDP catabolic process | 3 | 3 | 2.112e-02 |
| GO:BP | GO:0002128 | tRNA nucleoside ribose methylation | 3 | 3 | 2.112e-02 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 9 | 28 | 2.117e-02 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 9 | 28 | 2.117e-02 |
| GO:BP | GO:0006839 | mitochondrial transport | 31 | 178 | 2.117e-02 |
| GO:BP | GO:0141086 | negative regulation of inflammasome-mediated signaling pathway | 7 | 18 | 2.204e-02 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 138 | 1105 | 2.250e-02 |
| GO:BP | GO:0090042 | tubulin deacetylation | 8 | 23 | 2.250e-02 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 55 | 372 | 2.259e-02 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 21 | 105 | 2.261e-02 |
| GO:BP | GO:0098732 | macromolecule deacylation | 14 | 58 | 2.325e-02 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 16 | 71 | 2.333e-02 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 42 | 266 | 2.341e-02 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 49 | 323 | 2.341e-02 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 93 | 701 | 2.343e-02 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 30 | 172 | 2.387e-02 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 28 | 157 | 2.407e-02 |
| GO:BP | GO:0030520 | estrogen receptor signaling pathway | 13 | 52 | 2.407e-02 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 439 | 3993 | 2.407e-02 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 276 | 2407 | 2.419e-02 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 22 | 113 | 2.437e-02 |
| GO:BP | GO:0044546 | NLRP3 inflammasome complex assembly | 11 | 40 | 2.437e-02 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 12 | 46 | 2.438e-02 |
| GO:BP | GO:1990116 | ribosome-associated ubiquitin-dependent protein catabolic process | 4 | 6 | 2.438e-02 |
| GO:BP | GO:0000389 | mRNA 3’-splice site recognition | 4 | 6 | 2.438e-02 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 19 | 92 | 2.467e-02 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 20 | 99 | 2.467e-02 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 276 | 2410 | 2.545e-02 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 41 | 260 | 2.621e-02 |
| GO:BP | GO:0006998 | nuclear envelope organization | 14 | 59 | 2.635e-02 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 25 | 136 | 2.698e-02 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 52 | 351 | 2.732e-02 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 19 | 93 | 2.766e-02 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 19 | 93 | 2.766e-02 |
| GO:BP | GO:0051604 | protein maturation | 74 | 539 | 2.772e-02 |
| GO:BP | GO:0031647 | regulation of protein stability | 50 | 335 | 2.817e-02 |
| GO:BP | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 8 | 24 | 2.847e-02 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 12 | 47 | 2.871e-02 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 61 | 428 | 2.871e-02 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 11 | 41 | 2.895e-02 |
| GO:BP | GO:0045945 | positive regulation of transcription by RNA polymerase III | 7 | 19 | 2.897e-02 |
| GO:BP | GO:0016559 | peroxisome fission | 5 | 10 | 2.897e-02 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 5 | 10 | 2.897e-02 |
| GO:BP | GO:0000492 | box C/D snoRNP assembly | 5 | 10 | 2.897e-02 |
| GO:BP | GO:0018216 | peptidyl-arginine methylation | 5 | 10 | 2.897e-02 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 16 | 73 | 2.905e-02 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 18 | 87 | 3.043e-02 |
| GO:BP | GO:0045727 | positive regulation of translation | 25 | 138 | 3.187e-02 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 13 | 54 | 3.211e-02 |
| GO:BP | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening | 9 | 30 | 3.239e-02 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 55 | 380 | 3.288e-02 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 21 | 109 | 3.302e-02 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 29 | 169 | 3.334e-02 |
| GO:BP | GO:0006479 | protein methylation | 12 | 48 | 3.334e-02 |
| GO:BP | GO:0071763 | nuclear membrane organization | 12 | 48 | 3.334e-02 |
| GO:BP | GO:0008213 | protein alkylation | 12 | 48 | 3.334e-02 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 11 | 42 | 3.418e-02 |
| GO:BP | GO:0140632 | canonical inflammasome complex assembly | 11 | 42 | 3.418e-02 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 25 | 139 | 3.432e-02 |
| GO:BP | GO:0010646 | regulation of cell communication | 385 | 3486 | 3.432e-02 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 31 | 185 | 3.435e-02 |
| GO:BP | GO:0043487 | regulation of RNA stability | 32 | 193 | 3.479e-02 |
| GO:BP | GO:0033123 | positive regulation of purine nucleotide catabolic process | 8 | 25 | 3.529e-02 |
| GO:BP | GO:1903241 | U2-type prespliceosome assembly | 8 | 25 | 3.529e-02 |
| GO:BP | GO:0030813 | positive regulation of nucleotide catabolic process | 8 | 25 | 3.529e-02 |
| GO:BP | GO:0045821 | positive regulation of glycolytic process | 8 | 25 | 3.529e-02 |
| GO:BP | GO:0000470 | maturation of LSU-rRNA | 8 | 25 | 3.529e-02 |
| GO:BP | GO:0023051 | regulation of signaling | 384 | 3478 | 3.529e-02 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 53 | 365 | 3.529e-02 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 6 | 15 | 3.603e-02 |
| GO:BP | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | 13 | 55 | 3.603e-02 |
| GO:BP | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly | 6 | 15 | 3.603e-02 |
| GO:BP | GO:0009191 | ribonucleoside diphosphate catabolic process | 20 | 103 | 3.603e-02 |
| GO:BP | GO:0150011 | regulation of neuron projection arborization | 6 | 15 | 3.603e-02 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 54 | 374 | 3.603e-02 |
| GO:BP | GO:0006220 | pyrimidine nucleotide metabolic process | 13 | 55 | 3.603e-02 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 45 | 299 | 3.617e-02 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 16 | 75 | 3.617e-02 |
| GO:BP | GO:0006986 | response to unfolded protein | 25 | 140 | 3.622e-02 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 19 | 96 | 3.628e-02 |
| GO:BP | GO:0043543 | protein acylation | 18 | 89 | 3.654e-02 |
| GO:BP | GO:0018200 | peptidyl-glutamic acid modification | 7 | 20 | 3.676e-02 |
| GO:BP | GO:0009301 | snRNA transcription | 7 | 20 | 3.676e-02 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 22 | 118 | 3.712e-02 |
| GO:BP | GO:0042073 | intraciliary transport | 12 | 49 | 3.752e-02 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 24 | 133 | 3.754e-02 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 9 | 31 | 3.780e-02 |
| GO:BP | GO:0045010 | actin nucleation | 14 | 62 | 3.780e-02 |
| GO:BP | GO:0016226 | iron-sulfur cluster assembly | 9 | 31 | 3.780e-02 |
| GO:BP | GO:0031163 | metallo-sulfur cluster assembly | 9 | 31 | 3.780e-02 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 11 | 43 | 3.866e-02 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 54 | 376 | 3.866e-02 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 23 | 126 | 3.879e-02 |
| GO:BP | GO:1900225 | regulation of NLRP3 inflammasome complex assembly | 10 | 37 | 3.880e-02 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 85 | 646 | 3.880e-02 |
| GO:BP | GO:0016925 | protein sumoylation | 15 | 69 | 3.887e-02 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 42 | 276 | 3.887e-02 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 88 | 673 | 3.894e-02 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 19 | 97 | 3.929e-02 |
| GO:BP | GO:0097581 | lamellipodium organization | 18 | 90 | 3.979e-02 |
| GO:BP | GO:0019827 | stem cell population maintenance | 30 | 180 | 3.985e-02 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 14 | 63 | 4.209e-02 |
| GO:BP | GO:0071044 | histone mRNA catabolic process | 5 | 11 | 4.209e-02 |
| GO:BP | GO:0018195 | peptidyl-arginine modification | 5 | 11 | 4.209e-02 |
| GO:BP | GO:0042255 | ribosome assembly | 14 | 63 | 4.209e-02 |
| GO:BP | GO:0046048 | UDP metabolic process | 4 | 7 | 4.209e-02 |
| GO:BP | GO:0031167 | rRNA methylation | 8 | 26 | 4.209e-02 |
| GO:BP | GO:0150093 | amyloid-beta clearance by transcytosis | 4 | 7 | 4.209e-02 |
| GO:BP | GO:0170036 | import into the mitochondrion | 12 | 50 | 4.209e-02 |
| GO:BP | GO:0035601 | protein deacylation | 12 | 50 | 4.209e-02 |
| GO:BP | GO:0035617 | stress granule disassembly | 4 | 7 | 4.209e-02 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 30 | 181 | 4.209e-02 |
| GO:BP | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process | 4 | 7 | 4.209e-02 |
| GO:BP | GO:0051204 | protein insertion into mitochondrial membrane | 4 | 7 | 4.209e-02 |
| GO:BP | GO:0033617 | mitochondrial cytochrome c oxidase assembly | 8 | 26 | 4.209e-02 |
| GO:BP | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 10 | 38 | 4.590e-02 |
| GO:BP | GO:0090043 | regulation of tubulin deacetylation | 7 | 21 | 4.664e-02 |
| GO:BP | GO:0099188 | postsynaptic cytoskeleton organization | 7 | 21 | 4.664e-02 |
| GO:BP | GO:0030162 | regulation of proteolysis | 57 | 406 | 4.664e-02 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 24 | 136 | 4.690e-02 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 24 | 136 | 4.690e-02 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 24 | 136 | 4.690e-02 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 67 | 493 | 4.717e-02 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 6 | 16 | 4.717e-02 |
| GO:BP | GO:0046036 | CTP metabolic process | 6 | 16 | 4.717e-02 |
| GO:BP | GO:0098974 | postsynaptic actin cytoskeleton organization | 6 | 16 | 4.717e-02 |
| GO:BP | GO:0002218 | activation of innate immune response | 45 | 305 | 4.776e-02 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 37 | 239 | 4.778e-02 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 25 | 144 | 4.793e-02 |
| GO:BP | GO:0030032 | lamellipodium assembly | 15 | 71 | 4.838e-02 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 155 | 1296 | 4.962e-02 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 105 | 506 | 2.196e-09 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 35 | 140 | 1.825e-04 |
| KEGG | KEGG:03082 | ATP-dependent chromatin remodeling | 30 | 116 | 3.337e-04 |
| KEGG | KEGG:03040 | Spliceosome | 39 | 174 | 4.202e-04 |
| KEGG | KEGG:03018 | RNA degradation | 22 | 79 | 1.166e-03 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 15 | 44 | 1.572e-03 |
| KEGG | KEGG:03008 | Ribosome biogenesis in eukaryotes | 29 | 139 | 1.422e-02 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 58 | 363 | 4.086e-02 |
| KEGG | KEGG:04137 | Mitophagy - animal | 22 | 103 | 4.086e-02 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 10 | 32 | 4.368e-02 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 31 | 168 | 4.703e-02 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 12 | 44 | 4.703e-02 |
#write.csv(table_DOX24_down_genes, "output/table_DOX24_downreg_genes.csv")
#GO:BP
table_DOX24_down_genes_GOBP <- table_DOX24_down_genes %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
#saveRDS(table_motif1_GOBP_d, "data/table_motif1_GOBP_d.RDS")
table_DOX24_down_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Specific Down DEGs Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))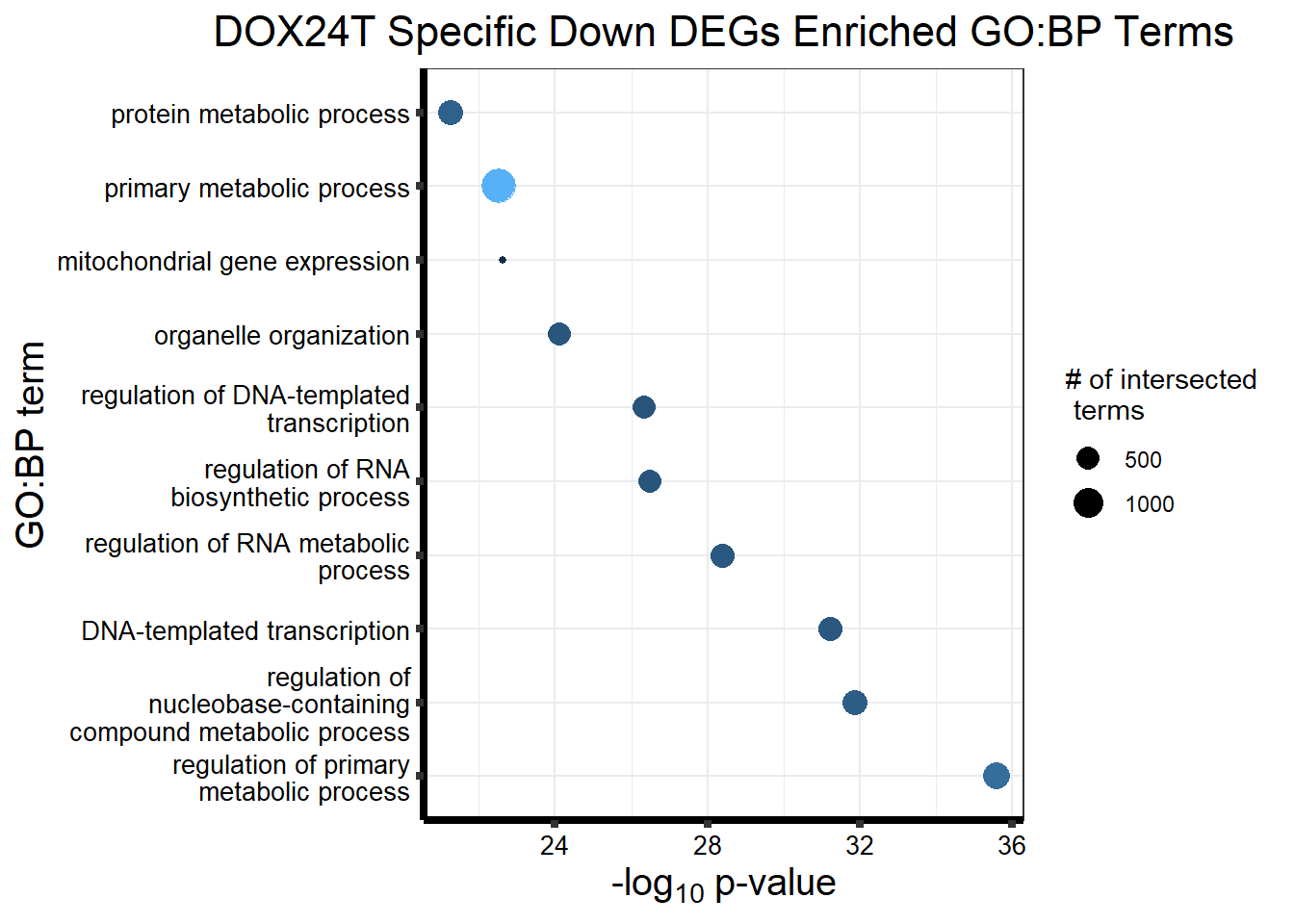
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_DOX24_down_genes_KEGG <- table_DOX24_down_genes %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24_down_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Specific Down DEGs Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))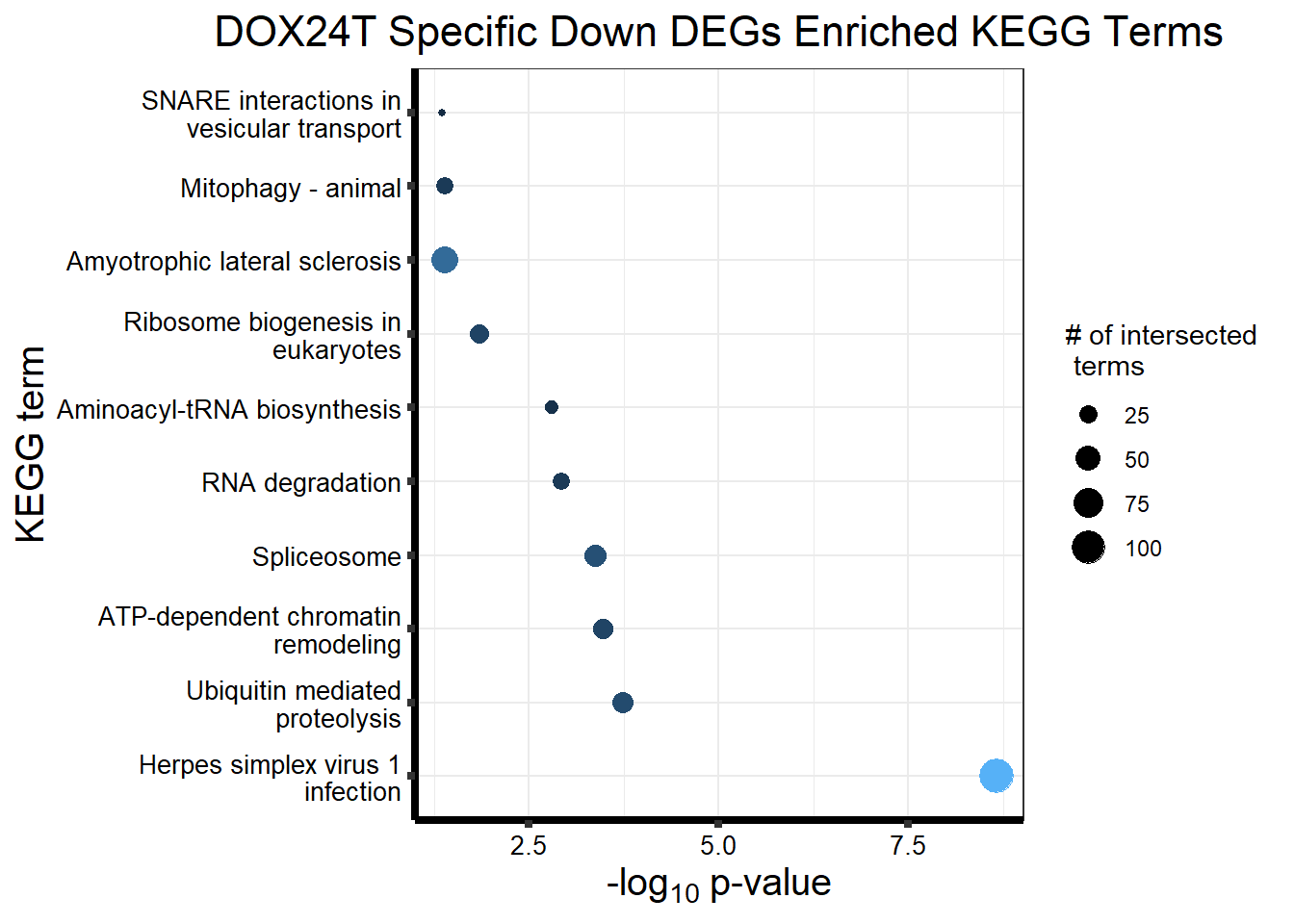
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####DOX24T share Upregulated DEGs GO KEGG#####
D24Tshare_DEGs_up_mat <- as.matrix(DOX24Tshare_DEGs_GO_up)
DOX_24Tshare_up_dxr_gene <- gost(query = D24Tshare_DEGs_up_mat,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
DOX_24Tshare_up_gost_genes <- gostplot(DOX_24Tshare_up_dxr_gene, capped = FALSE, interactive = TRUE)
DOX_24Tshare_up_gost_genestable_DOX24Tshare_up_genes <- DOX_24Tshare_up_dxr_gene$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX24Tshare_up_genes %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0080090 | regulation of primary metabolic process | 1318 | 5390 | 8.687e-45 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1482 | 6264 | 1.034e-43 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1410 | 5920 | 1.664e-42 |
| GO:BP | GO:0065007 | biological regulation | 2616 | 12743 | 2.240e-38 |
| GO:BP | GO:0050789 | regulation of biological process | 2540 | 12336 | 8.310e-37 |
| GO:BP | GO:0050794 | regulation of cellular process | 2470 | 11946 | 4.513e-36 |
| GO:BP | GO:0035556 | intracellular signal transduction | 773 | 2965 | 5.621e-32 |
| GO:BP | GO:0019538 | protein metabolic process | 1125 | 4721 | 3.115e-31 |
| GO:BP | GO:0007275 | multicellular organism development | 1124 | 4727 | 8.277e-31 |
| GO:BP | GO:0032502 | developmental process | 1474 | 6553 | 1.312e-30 |
| GO:BP | GO:0009056 | catabolic process | 695 | 2639 | 1.032e-29 |
| GO:BP | GO:0023051 | regulation of signaling | 865 | 3478 | 7.776e-29 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 967 | 3990 | 1.383e-28 |
| GO:BP | GO:0019222 | regulation of metabolic process | 1553 | 7035 | 1.500e-28 |
| GO:BP | GO:0048856 | anatomical structure development | 1356 | 5997 | 2.836e-28 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 904 | 3687 | 3.383e-28 |
| GO:BP | GO:0010646 | regulation of cell communication | 863 | 3486 | 3.767e-28 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1608 | 7376 | 4.173e-27 |
| GO:BP | GO:0050793 | regulation of developmental process | 643 | 2457 | 1.815e-26 |
| GO:BP | GO:0009966 | regulation of signal transduction | 762 | 3034 | 3.203e-26 |
| GO:BP | GO:0006351 | DNA-templated transcription | 863 | 3549 | 2.115e-25 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1308 | 5834 | 3.223e-25 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1269 | 5629 | 3.223e-25 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 950 | 3993 | 4.200e-25 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 871 | 3597 | 4.200e-25 |
| GO:BP | GO:0051179 | localization | 1250 | 5555 | 2.251e-24 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 829 | 3409 | 2.918e-24 |
| GO:BP | GO:0048731 | system development | 957 | 4053 | 3.759e-24 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 830 | 3428 | 1.093e-23 |
| GO:BP | GO:0030154 | cell differentiation | 1030 | 4437 | 1.112e-23 |
| GO:BP | GO:0048869 | cellular developmental process | 1030 | 4438 | 1.174e-23 |
| GO:BP | GO:0033554 | cellular response to stress | 497 | 1830 | 1.403e-23 |
| GO:BP | GO:0006810 | transport | 1023 | 4407 | 1.677e-23 |
| GO:BP | GO:0048468 | cell development | 714 | 2876 | 6.597e-23 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 398 | 1400 | 2.307e-22 |
| GO:BP | GO:0036211 | protein modification process | 704 | 2846 | 4.431e-22 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 726 | 2956 | 5.427e-22 |
| GO:BP | GO:0050896 | response to stimulus | 1872 | 8999 | 6.008e-22 |
| GO:BP | GO:0043412 | macromolecule modification | 739 | 3030 | 1.410e-21 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 606 | 2407 | 1.928e-20 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 606 | 2410 | 2.597e-20 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 1402 | 6492 | 2.871e-20 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 418 | 1529 | 4.384e-20 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 363 | 1280 | 4.396e-20 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 664 | 2711 | 1.662e-19 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 395 | 1436 | 2.116e-19 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 364 | 1296 | 2.213e-19 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 447 | 1680 | 3.613e-19 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 635 | 2578 | 3.684e-19 |
| GO:BP | GO:0023057 | negative regulation of signaling | 393 | 1435 | 5.869e-19 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 777 | 3288 | 7.089e-19 |
| GO:BP | GO:0006950 | response to stress | 907 | 3948 | 8.496e-19 |
| GO:BP | GO:0023052 | signaling | 1396 | 6515 | 9.974e-19 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 368 | 1326 | 1.047e-18 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 422 | 1576 | 1.435e-18 |
| GO:BP | GO:0032501 | multicellular organismal process | 1542 | 7322 | 3.081e-18 |
| GO:BP | GO:0051641 | cellular localization | 847 | 3661 | 3.518e-18 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 600 | 2433 | 4.079e-18 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 657 | 2713 | 4.330e-18 |
| GO:BP | GO:0007154 | cell communication | 1396 | 6540 | 4.632e-18 |
| GO:BP | GO:0051234 | establishment of localization | 1091 | 4928 | 6.715e-18 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 400 | 1489 | 8.301e-18 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 384 | 1418 | 1.159e-17 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 341 | 1223 | 1.290e-17 |
| GO:BP | GO:0009894 | regulation of catabolic process | 300 | 1040 | 1.322e-17 |
| GO:BP | GO:0008219 | cell death | 504 | 1991 | 3.372e-17 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 487 | 1912 | 4.219e-17 |
| GO:BP | GO:0033036 | macromolecule localization | 755 | 3228 | 4.249e-17 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 495 | 1953 | 5.560e-17 |
| GO:BP | GO:0012501 | programmed cell death | 502 | 1987 | 5.784e-17 |
| GO:BP | GO:0007165 | signal transduction | 1284 | 6002 | 2.228e-16 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 1247 | 5809 | 2.917e-16 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 378 | 1417 | 2.992e-16 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 279 | 970 | 3.861e-16 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 508 | 2036 | 4.464e-16 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 361 | 1343 | 5.083e-16 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 271 | 938 | 6.198e-16 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 213 | 689 | 8.698e-16 |
| GO:BP | GO:0070848 | response to growth factor | 220 | 721 | 1.280e-15 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 468 | 1855 | 1.340e-15 |
| GO:BP | GO:0006915 | apoptotic process | 482 | 1923 | 1.448e-15 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 638 | 2696 | 3.428e-15 |
| GO:BP | GO:0022008 | neurogenesis | 448 | 1771 | 4.430e-15 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 654 | 2782 | 5.708e-15 |
| GO:BP | GO:0051649 | establishment of localization in cell | 497 | 2010 | 6.104e-15 |
| GO:BP | GO:0007399 | nervous system development | 618 | 2604 | 6.237e-15 |
| GO:BP | GO:0008104 | protein localization | 651 | 2770 | 7.098e-15 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 460 | 1835 | 8.302e-15 |
| GO:BP | GO:0071705 | nitrogen compound transport | 478 | 1923 | 9.447e-15 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 614 | 2593 | 1.289e-14 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1203 | 5644 | 1.864e-14 |
| GO:BP | GO:0006996 | organelle organization | 811 | 3594 | 2.029e-14 |
| GO:BP | GO:0032879 | regulation of localization | 498 | 2029 | 2.233e-14 |
| GO:BP | GO:0046907 | intracellular transport | 362 | 1381 | 2.432e-14 |
| GO:BP | GO:0042592 | homeostatic process | 436 | 1736 | 4.068e-14 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 656 | 2819 | 4.707e-14 |
| GO:BP | GO:0030030 | cell projection organization | 413 | 1631 | 6.661e-14 |
| GO:BP | GO:0048513 | animal organ development | 707 | 3085 | 9.252e-14 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 425 | 1697 | 1.606e-13 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 425 | 1699 | 1.950e-13 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 167 | 528 | 2.858e-13 |
| GO:BP | GO:0035295 | tube development | 297 | 1101 | 2.896e-13 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 400 | 1588 | 4.631e-13 |
| GO:BP | GO:0048699 | generation of neurons | 389 | 1536 | 4.809e-13 |
| GO:BP | GO:0016310 | phosphorylation | 343 | 1320 | 5.530e-13 |
| GO:BP | GO:0015031 | protein transport | 369 | 1444 | 6.474e-13 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 170 | 546 | 7.479e-13 |
| GO:BP | GO:0030182 | neuron differentiation | 370 | 1451 | 8.118e-13 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 381 | 1506 | 1.077e-12 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 414 | 1667 | 1.450e-12 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 371 | 1462 | 1.528e-12 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 181 | 597 | 1.528e-12 |
| GO:BP | GO:0006914 | autophagy | 181 | 597 | 1.528e-12 |
| GO:BP | GO:0035239 | tube morphogenesis | 245 | 879 | 1.710e-12 |
| GO:BP | GO:0010468 | regulation of gene expression | 1166 | 5536 | 2.953e-12 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 202 | 692 | 2.953e-12 |
| GO:BP | GO:0009790 | embryo development | 300 | 1135 | 3.040e-12 |
| GO:BP | GO:0006468 | protein phosphorylation | 319 | 1227 | 4.517e-12 |
| GO:BP | GO:0009888 | tissue development | 486 | 2032 | 5.160e-12 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 518 | 2192 | 6.070e-12 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 253 | 925 | 6.103e-12 |
| GO:BP | GO:0008283 | cell population proliferation | 482 | 2015 | 6.315e-12 |
| GO:BP | GO:0043009 | chordate embryonic development | 196 | 671 | 6.315e-12 |
| GO:BP | GO:0007178 | cell surface receptor protein serine/threonine kinase signaling pathway | 123 | 364 | 6.506e-12 |
| GO:BP | GO:0065008 | regulation of biological quality | 668 | 2947 | 6.566e-12 |
| GO:BP | GO:0031175 | neuron projection development | 274 | 1023 | 7.560e-12 |
| GO:BP | GO:0048666 | neuron development | 307 | 1177 | 8.455e-12 |
| GO:BP | GO:0060284 | regulation of cell development | 235 | 847 | 9.156e-12 |
| GO:BP | GO:0006325 | chromatin organization | 243 | 884 | 1.021e-11 |
| GO:BP | GO:0065009 | regulation of molecular function | 372 | 1496 | 2.644e-11 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 114 | 336 | 3.707e-11 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 746 | 3375 | 4.505e-11 |
| GO:BP | GO:0030163 | protein catabolic process | 272 | 1030 | 5.150e-11 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 390 | 1592 | 6.240e-11 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 143 | 458 | 6.649e-11 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 190 | 662 | 8.171e-11 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 319 | 1256 | 9.391e-11 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 226 | 825 | 1.046e-10 |
| GO:BP | GO:0048732 | gland development | 141 | 454 | 1.455e-10 |
| GO:BP | GO:0030097 | hemopoiesis | 258 | 976 | 1.777e-10 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 537 | 2331 | 1.787e-10 |
| GO:BP | GO:0032880 | regulation of protein localization | 243 | 907 | 1.787e-10 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 114 | 344 | 1.960e-10 |
| GO:BP | GO:0051049 | regulation of transport | 390 | 1607 | 2.335e-10 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 281 | 1087 | 2.669e-10 |
| GO:BP | GO:0016236 | macroautophagy | 118 | 362 | 2.748e-10 |
| GO:BP | GO:0043687 | post-translational protein modification | 268 | 1027 | 2.876e-10 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 105 | 311 | 3.854e-10 |
| GO:BP | GO:0060429 | epithelium development | 311 | 1238 | 6.887e-10 |
| GO:BP | GO:0001701 | in utero embryonic development | 129 | 413 | 7.672e-10 |
| GO:BP | GO:0060341 | regulation of cellular localization | 263 | 1013 | 8.091e-10 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 150 | 503 | 8.447e-10 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 234 | 879 | 8.566e-10 |
| GO:BP | GO:0044238 | primary metabolic process | 2366 | 12342 | 9.418e-10 |
| GO:BP | GO:0006338 | chromatin remodeling | 200 | 725 | 9.700e-10 |
| GO:BP | GO:0032940 | secretion by cell | 228 | 854 | 1.155e-09 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 254 | 975 | 1.227e-09 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 398 | 1669 | 1.607e-09 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 692 | 3157 | 1.883e-09 |
| GO:BP | GO:0023056 | positive regulation of signaling | 423 | 1796 | 2.179e-09 |
| GO:BP | GO:0001503 | ossification | 131 | 430 | 3.322e-09 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 377 | 1578 | 4.395e-09 |
| GO:BP | GO:0046903 | secretion | 255 | 992 | 4.660e-09 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 185 | 671 | 5.733e-09 |
| GO:BP | GO:0040007 | growth | 241 | 929 | 6.130e-09 |
| GO:BP | GO:0044281 | small molecule metabolic process | 416 | 1776 | 6.879e-09 |
| GO:BP | GO:0000902 | cell morphogenesis | 255 | 996 | 7.015e-09 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 224 | 853 | 8.623e-09 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 178 | 643 | 8.956e-09 |
| GO:BP | GO:0140352 | export from cell | 240 | 928 | 8.965e-09 |
| GO:BP | GO:0080134 | regulation of response to stress | 336 | 1387 | 9.239e-09 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 419 | 1795 | 9.239e-09 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 106 | 331 | 9.257e-09 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 620 | 2813 | 9.533e-09 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 285 | 1144 | 1.196e-08 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 396 | 1685 | 1.238e-08 |
| GO:BP | GO:0006629 | lipid metabolic process | 336 | 1391 | 1.284e-08 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 176 | 638 | 1.459e-08 |
| GO:BP | GO:0010506 | regulation of autophagy | 114 | 367 | 1.475e-08 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 153 | 537 | 1.832e-08 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 270 | 1077 | 1.832e-08 |
| GO:BP | GO:0006508 | proteolysis | 354 | 1486 | 2.300e-08 |
| GO:BP | GO:0001775 | cell activation | 281 | 1132 | 2.302e-08 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 168 | 606 | 2.424e-08 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 217 | 831 | 2.630e-08 |
| GO:BP | GO:0007033 | vacuole organization | 82 | 239 | 2.647e-08 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 207 | 786 | 3.059e-08 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 85 | 252 | 3.299e-08 |
| GO:BP | GO:0006974 | DNA damage response | 233 | 908 | 3.299e-08 |
| GO:BP | GO:0048729 | tissue morphogenesis | 169 | 614 | 3.756e-08 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 178 | 658 | 5.652e-08 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 182 | 677 | 6.170e-08 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 85 | 255 | 6.200e-08 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 153 | 546 | 6.200e-08 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 115 | 380 | 6.383e-08 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 289 | 1182 | 6.408e-08 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 174 | 642 | 7.261e-08 |
| GO:BP | GO:0031399 | regulation of protein modification process | 261 | 1049 | 7.343e-08 |
| GO:BP | GO:0045184 | establishment of protein localization | 441 | 1937 | 8.777e-08 |
| GO:BP | GO:0048589 | developmental growth | 178 | 663 | 1.010e-07 |
| GO:BP | GO:0060485 | mesenchyme development | 100 | 320 | 1.150e-07 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 170 | 628 | 1.209e-07 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 598 | 2745 | 1.338e-07 |
| GO:BP | GO:0046649 | lymphocyte activation | 210 | 814 | 1.377e-07 |
| GO:BP | GO:0016567 | protein ubiquitination | 200 | 768 | 1.479e-07 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 271 | 1105 | 1.516e-07 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 87 | 268 | 1.612e-07 |
| GO:BP | GO:0001568 | blood vessel development | 192 | 732 | 1.665e-07 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 290 | 1199 | 1.807e-07 |
| GO:BP | GO:0045321 | leukocyte activation | 244 | 980 | 2.290e-07 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 99 | 320 | 2.367e-07 |
| GO:BP | GO:0040008 | regulation of growth | 163 | 604 | 3.123e-07 |
| GO:BP | GO:1901698 | response to nitrogen compound | 264 | 1080 | 3.296e-07 |
| GO:BP | GO:0001944 | vasculature development | 197 | 762 | 3.459e-07 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 232 | 928 | 3.746e-07 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 58 | 157 | 3.748e-07 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 65 | 184 | 3.813e-07 |
| GO:BP | GO:0001525 | angiogenesis | 150 | 547 | 3.934e-07 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 115 | 392 | 4.100e-07 |
| GO:BP | GO:0048863 | stem cell differentiation | 80 | 245 | 4.803e-07 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 127 | 446 | 5.013e-07 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 171 | 645 | 5.068e-07 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 81 | 250 | 5.704e-07 |
| GO:BP | GO:0001649 | osteoblast differentiation | 77 | 234 | 6.082e-07 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 248 | 1011 | 6.605e-07 |
| GO:BP | GO:0048608 | reproductive structure development | 96 | 314 | 7.284e-07 |
| GO:BP | GO:0009725 | response to hormone | 226 | 907 | 7.507e-07 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 138 | 499 | 8.455e-07 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 100 | 332 | 8.473e-07 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 85 | 269 | 8.984e-07 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 70 | 208 | 9.607e-07 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 115 | 398 | 9.812e-07 |
| GO:BP | GO:0045444 | fat cell differentiation | 78 | 241 | 1.066e-06 |
| GO:BP | GO:0009267 | cellular response to starvation | 62 | 177 | 1.119e-06 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 164 | 620 | 1.134e-06 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 65 | 189 | 1.160e-06 |
| GO:BP | GO:0016477 | cell migration | 353 | 1534 | 1.161e-06 |
| GO:BP | GO:0051960 | regulation of nervous system development | 128 | 457 | 1.188e-06 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 94 | 309 | 1.259e-06 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 110 | 378 | 1.259e-06 |
| GO:BP | GO:0072359 | circulatory system development | 274 | 1145 | 1.259e-06 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 167 | 635 | 1.259e-06 |
| GO:BP | GO:0006979 | response to oxidative stress | 115 | 400 | 1.267e-06 |
| GO:BP | GO:0061458 | reproductive system development | 96 | 318 | 1.348e-06 |
| GO:BP | GO:0001501 | skeletal system development | 146 | 540 | 1.472e-06 |
| GO:BP | GO:0007169 | cell surface receptor protein tyrosine kinase signaling pathway | 165 | 629 | 1.806e-06 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 181 | 706 | 2.352e-06 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 82 | 262 | 2.359e-06 |
| GO:BP | GO:0000165 | MAPK cascade | 189 | 744 | 2.426e-06 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 62 | 181 | 2.693e-06 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 49 | 131 | 2.919e-06 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 68 | 206 | 3.272e-06 |
| GO:BP | GO:0006886 | intracellular protein transport | 176 | 686 | 3.314e-06 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 94 | 315 | 3.314e-06 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 123 | 443 | 3.515e-06 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 95 | 320 | 3.671e-06 |
| GO:BP | GO:0042594 | response to starvation | 71 | 219 | 3.760e-06 |
| GO:BP | GO:0051338 | regulation of transferase activity | 135 | 498 | 3.857e-06 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 250 | 1042 | 3.857e-06 |
| GO:BP | GO:0007049 | cell cycle | 375 | 1663 | 3.932e-06 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 93 | 312 | 3.977e-06 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 59 | 171 | 3.977e-06 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 80 | 257 | 4.162e-06 |
| GO:BP | GO:0010720 | positive regulation of cell development | 126 | 458 | 4.162e-06 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 249 | 1039 | 4.423e-06 |
| GO:BP | GO:0016032 | viral process | 118 | 423 | 4.674e-06 |
| GO:BP | GO:0000045 | autophagosome assembly | 45 | 118 | 4.809e-06 |
| GO:BP | GO:0051223 | regulation of protein transport | 122 | 442 | 5.360e-06 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 184 | 729 | 5.464e-06 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 205 | 830 | 6.019e-06 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 231 | 957 | 6.970e-06 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 56 | 162 | 7.476e-06 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 92 | 312 | 7.693e-06 |
| GO:BP | GO:0001889 | liver development | 52 | 147 | 8.720e-06 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 52 | 147 | 8.720e-06 |
| GO:BP | GO:0032868 | response to insulin | 82 | 270 | 8.923e-06 |
| GO:BP | GO:0002682 | regulation of immune system process | 356 | 1581 | 9.394e-06 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 230 | 956 | 9.505e-06 |
| GO:BP | GO:0061448 | connective tissue development | 87 | 292 | 9.751e-06 |
| GO:BP | GO:0002521 | leukocyte differentiation | 163 | 636 | 9.756e-06 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 215 | 884 | 9.978e-06 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 100 | 349 | 1.024e-05 |
| GO:BP | GO:0051604 | protein maturation | 142 | 539 | 1.049e-05 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 117 | 425 | 1.054e-05 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 45 | 121 | 1.054e-05 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 32 | 74 | 1.083e-05 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 47 | 129 | 1.165e-05 |
| GO:BP | GO:0016049 | cell growth | 130 | 485 | 1.177e-05 |
| GO:BP | GO:0048870 | cell motility | 397 | 1793 | 1.177e-05 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 172 | 681 | 1.239e-05 |
| GO:BP | GO:0043434 | response to peptide hormone | 120 | 440 | 1.246e-05 |
| GO:BP | GO:0009987 | cellular process | 3634 | 20247 | 1.256e-05 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 38 | 96 | 1.365e-05 |
| GO:BP | GO:0006259 | DNA metabolic process | 239 | 1005 | 1.387e-05 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 281 | 1212 | 1.390e-05 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 67 | 210 | 1.447e-05 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 102 | 361 | 1.541e-05 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 92 | 317 | 1.541e-05 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 64 | 198 | 1.541e-05 |
| GO:BP | GO:0007417 | central nervous system development | 250 | 1061 | 1.609e-05 |
| GO:BP | GO:1903131 | mononuclear cell differentiation | 140 | 534 | 1.614e-05 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 44 | 119 | 1.624e-05 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 52 | 150 | 1.626e-05 |
| GO:BP | GO:2000145 | regulation of cell motility | 242 | 1022 | 1.626e-05 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 52 | 150 | 1.626e-05 |
| GO:BP | GO:0035282 | segmentation | 41 | 108 | 1.737e-05 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 66 | 207 | 1.743e-05 |
| GO:BP | GO:0034504 | protein localization to nucleus | 92 | 318 | 1.744e-05 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 146 | 563 | 1.788e-05 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 170 | 676 | 1.824e-05 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 175 | 701 | 2.062e-05 |
| GO:BP | GO:0061564 | axon development | 138 | 528 | 2.263e-05 |
| GO:BP | GO:0006887 | exocytosis | 101 | 360 | 2.354e-05 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 145 | 561 | 2.354e-05 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 48 | 136 | 2.354e-05 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 191 | 779 | 2.354e-05 |
| GO:BP | GO:0072175 | epithelial tube formation | 48 | 136 | 2.354e-05 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 94 | 329 | 2.355e-05 |
| GO:BP | GO:0030855 | epithelial cell differentiation | 184 | 746 | 2.446e-05 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 153 | 599 | 2.461e-05 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 52 | 152 | 2.465e-05 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 39 | 102 | 2.506e-05 |
| GO:BP | GO:0072164 | mesonephric tubule development | 39 | 102 | 2.506e-05 |
| GO:BP | GO:0051169 | nuclear transport | 95 | 334 | 2.511e-05 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 95 | 334 | 2.511e-05 |
| GO:BP | GO:0001558 | regulation of cell growth | 111 | 406 | 2.662e-05 |
| GO:BP | GO:1905037 | autophagosome organization | 45 | 125 | 2.671e-05 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 157 | 619 | 2.716e-05 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 161 | 638 | 2.740e-05 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 139 | 535 | 2.793e-05 |
| GO:BP | GO:0001822 | kidney development | 92 | 322 | 2.959e-05 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 55 | 165 | 2.972e-05 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 27 | 60 | 2.999e-05 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 33 | 81 | 3.195e-05 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 29 | 67 | 3.227e-05 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 216 | 905 | 3.396e-05 |
| GO:BP | GO:0060674 | placenta blood vessel development | 18 | 32 | 3.417e-05 |
| GO:BP | GO:0051726 | regulation of cell cycle | 253 | 1087 | 3.424e-05 |
| GO:BP | GO:0022612 | gland morphogenesis | 46 | 130 | 3.424e-05 |
| GO:BP | GO:0090183 | regulation of kidney development | 19 | 35 | 3.546e-05 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 165 | 660 | 3.586e-05 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 49 | 142 | 3.587e-05 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 63 | 199 | 3.690e-05 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 258 | 1113 | 3.702e-05 |
| GO:BP | GO:0006401 | RNA catabolic process | 90 | 315 | 3.714e-05 |
| GO:BP | GO:0098657 | import into cell | 219 | 922 | 4.009e-05 |
| GO:BP | GO:0030278 | regulation of ossification | 44 | 123 | 4.052e-05 |
| GO:BP | GO:0046777 | protein autophosphorylation | 58 | 179 | 4.133e-05 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1614 | 8393 | 4.147e-05 |
| GO:BP | GO:0042221 | response to chemical | 795 | 3909 | 4.147e-05 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 209 | 874 | 4.216e-05 |
| GO:BP | GO:1901652 | response to peptide | 227 | 962 | 4.216e-05 |
| GO:BP | GO:0030879 | mammary gland development | 48 | 139 | 4.346e-05 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 74 | 247 | 4.683e-05 |
| GO:BP | GO:0040012 | regulation of locomotion | 248 | 1067 | 4.685e-05 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 73 | 243 | 4.887e-05 |
| GO:BP | GO:0001657 | ureteric bud development | 38 | 101 | 4.985e-05 |
| GO:BP | GO:0035265 | organ growth | 54 | 164 | 5.291e-05 |
| GO:BP | GO:0035148 | tube formation | 50 | 148 | 5.432e-05 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 74 | 248 | 5.439e-05 |
| GO:BP | GO:0006402 | mRNA catabolic process | 77 | 261 | 5.473e-05 |
| GO:BP | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 19 | 36 | 5.882e-05 |
| GO:BP | GO:0040011 | locomotion | 285 | 1255 | 5.960e-05 |
| GO:BP | GO:0072001 | renal system development | 93 | 332 | 5.992e-05 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 139 | 543 | 6.057e-05 |
| GO:BP | GO:0048638 | regulation of developmental growth | 88 | 310 | 6.137e-05 |
| GO:BP | GO:0043603 | amide metabolic process | 120 | 455 | 6.285e-05 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 54 | 165 | 6.330e-05 |
| GO:BP | GO:0071772 | response to BMP | 54 | 165 | 6.330e-05 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 169 | 686 | 6.433e-05 |
| GO:BP | GO:0007423 | sensory organ development | 156 | 624 | 6.433e-05 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 46 | 133 | 6.451e-05 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 49 | 145 | 6.544e-05 |
| GO:BP | GO:0001823 | mesonephros development | 39 | 106 | 6.671e-05 |
| GO:BP | GO:0051640 | organelle localization | 155 | 620 | 6.855e-05 |
| GO:BP | GO:0060348 | bone development | 67 | 220 | 7.239e-05 |
| GO:BP | GO:0030334 | regulation of cell migration | 225 | 960 | 7.316e-05 |
| GO:BP | GO:0007548 | sex differentiation | 84 | 294 | 7.585e-05 |
| GO:BP | GO:0141188 | nucleic acid catabolic process | 95 | 343 | 7.638e-05 |
| GO:BP | GO:0000278 | mitotic cell cycle | 211 | 892 | 7.720e-05 |
| GO:BP | GO:0030509 | BMP signaling pathway | 51 | 154 | 7.883e-05 |
| GO:BP | GO:0051046 | regulation of secretion | 154 | 617 | 7.998e-05 |
| GO:BP | GO:0008152 | metabolic process | 2620 | 14136 | 8.156e-05 |
| GO:BP | GO:0034097 | response to cytokine | 222 | 947 | 8.231e-05 |
| GO:BP | GO:0008406 | gonad development | 71 | 238 | 8.231e-05 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 133 | 519 | 8.905e-05 |
| GO:BP | GO:0051216 | cartilage development | 65 | 213 | 9.142e-05 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 229 | 983 | 9.154e-05 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 36 | 96 | 9.311e-05 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 240 | 1038 | 9.432e-05 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 64 | 209 | 9.432e-05 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 43 | 123 | 9.432e-05 |
| GO:BP | GO:0048878 | chemical homeostasis | 241 | 1043 | 9.434e-05 |
| GO:BP | GO:0033043 | regulation of organelle organization | 262 | 1148 | 9.684e-05 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 58 | 184 | 9.710e-05 |
| GO:BP | GO:0098727 | maintenance of cell number | 58 | 184 | 9.710e-05 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 116 | 441 | 9.783e-05 |
| GO:BP | GO:0043583 | ear development | 70 | 235 | 9.879e-05 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 110 | 414 | 1.048e-04 |
| GO:BP | GO:0030098 | lymphocyte differentiation | 116 | 442 | 1.090e-04 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 45 | 132 | 1.192e-04 |
| GO:BP | GO:0045926 | negative regulation of growth | 68 | 228 | 1.276e-04 |
| GO:BP | GO:0050821 | protein stabilization | 67 | 224 | 1.338e-04 |
| GO:BP | GO:0072210 | metanephric nephron development | 20 | 41 | 1.392e-04 |
| GO:BP | GO:0048568 | embryonic organ development | 120 | 463 | 1.421e-04 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 49 | 149 | 1.430e-04 |
| GO:BP | GO:0016197 | endosomal transport | 82 | 290 | 1.430e-04 |
| GO:BP | GO:0007041 | lysosomal transport | 46 | 137 | 1.467e-04 |
| GO:BP | GO:0051050 | positive regulation of transport | 200 | 847 | 1.474e-04 |
| GO:BP | GO:0007034 | vacuolar transport | 55 | 174 | 1.507e-04 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 55 | 174 | 1.507e-04 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 139 | 553 | 1.514e-04 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 85 | 304 | 1.540e-04 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 85 | 304 | 1.540e-04 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 126 | 492 | 1.542e-04 |
| GO:BP | GO:0007267 | cell-cell signaling | 299 | 1342 | 1.546e-04 |
| GO:BP | GO:0072073 | kidney epithelium development | 50 | 154 | 1.689e-04 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 68 | 230 | 1.704e-04 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 64 | 213 | 1.761e-04 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 147 | 593 | 1.803e-04 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 99 | 369 | 1.912e-04 |
| GO:BP | GO:0045639 | positive regulation of myeloid cell differentiation | 37 | 103 | 2.019e-04 |
| GO:BP | GO:0038202 | TORC1 signaling | 37 | 103 | 2.019e-04 |
| GO:BP | GO:0019827 | stem cell population maintenance | 56 | 180 | 2.057e-04 |
| GO:BP | GO:0009306 | protein secretion | 101 | 379 | 2.102e-04 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 20 | 42 | 2.102e-04 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 48 | 147 | 2.104e-04 |
| GO:BP | GO:0043487 | regulation of RNA stability | 59 | 193 | 2.165e-04 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 207 | 887 | 2.179e-04 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 42 | 123 | 2.182e-04 |
| GO:BP | GO:0031667 | response to nutrient levels | 129 | 510 | 2.234e-04 |
| GO:BP | GO:0002064 | epithelial cell development | 65 | 219 | 2.267e-04 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 68 | 232 | 2.267e-04 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 52 | 164 | 2.297e-04 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 56 | 181 | 2.408e-04 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 200 | 854 | 2.408e-04 |
| GO:BP | GO:0072273 | metanephric nephron morphogenesis | 14 | 24 | 2.412e-04 |
| GO:BP | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib | 11 | 16 | 2.424e-04 |
| GO:BP | GO:0001890 | placenta development | 49 | 152 | 2.448e-04 |
| GO:BP | GO:0070482 | response to oxygen levels | 93 | 344 | 2.448e-04 |
| GO:BP | GO:1905332 | positive regulation of morphogenesis of an epithelium | 18 | 36 | 2.448e-04 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 55 | 177 | 2.460e-04 |
| GO:BP | GO:0031647 | regulation of protein stability | 91 | 335 | 2.460e-04 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 71 | 246 | 2.534e-04 |
| GO:BP | GO:0016054 | organic acid catabolic process | 71 | 246 | 2.534e-04 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 24 | 56 | 2.534e-04 |
| GO:BP | GO:0061053 | somite development | 32 | 85 | 2.538e-04 |
| GO:BP | GO:0002063 | chondrocyte development | 17 | 33 | 2.539e-04 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 44 | 132 | 2.605e-04 |
| GO:BP | GO:0007219 | Notch signaling pathway | 57 | 186 | 2.676e-04 |
| GO:BP | GO:0022414 | reproductive process | 349 | 1608 | 2.733e-04 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 101 | 382 | 2.832e-04 |
| GO:BP | GO:0010970 | transport along microtubule | 55 | 178 | 2.884e-04 |
| GO:BP | GO:0042886 | amide transport | 90 | 332 | 2.923e-04 |
| GO:BP | GO:0043542 | endothelial cell migration | 65 | 221 | 2.977e-04 |
| GO:BP | GO:0006260 | DNA replication | 79 | 283 | 3.127e-04 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 105 | 402 | 3.345e-04 |
| GO:BP | GO:0051656 | establishment of organelle localization | 124 | 491 | 3.370e-04 |
| GO:BP | GO:0007409 | axonogenesis | 118 | 463 | 3.454e-04 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 210 | 909 | 3.487e-04 |
| GO:BP | GO:0009605 | response to external stimulus | 503 | 2415 | 3.567e-04 |
| GO:BP | GO:0060322 | head development | 189 | 806 | 3.671e-04 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 35 | 98 | 3.714e-04 |
| GO:BP | GO:0043549 | regulation of kinase activity | 111 | 431 | 3.719e-04 |
| GO:BP | GO:0006082 | organic acid metabolic process | 211 | 915 | 3.723e-04 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 86 | 316 | 3.748e-04 |
| GO:BP | GO:0055088 | lipid homeostasis | 52 | 167 | 3.767e-04 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 36 | 102 | 3.768e-04 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 45 | 138 | 3.768e-04 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 63 | 214 | 3.768e-04 |
| GO:BP | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I | 13 | 22 | 3.881e-04 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 17 | 34 | 4.025e-04 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 28 | 72 | 4.173e-04 |
| GO:BP | GO:0030900 | forebrain development | 109 | 423 | 4.214e-04 |
| GO:BP | GO:0016043 | cellular component organization | 1561 | 8184 | 4.253e-04 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 53 | 172 | 4.270e-04 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 186 | 794 | 4.412e-04 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 20 | 44 | 4.466e-04 |
| GO:BP | GO:0060349 | bone morphogenesis | 36 | 103 | 4.715e-04 |
| GO:BP | GO:0050807 | regulation of synapse organization | 84 | 309 | 4.748e-04 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 37 | 107 | 4.764e-04 |
| GO:BP | GO:0030856 | regulation of epithelial cell differentiation | 49 | 156 | 4.891e-04 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 66 | 229 | 4.948e-04 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 126 | 505 | 5.023e-04 |
| GO:BP | GO:1905456 | regulation of lymphoid progenitor cell differentiation | 9 | 12 | 5.073e-04 |
| GO:BP | GO:0042110 | T cell activation | 142 | 582 | 5.234e-04 |
| GO:BP | GO:0030217 | T cell differentiation | 86 | 319 | 5.253e-04 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 101 | 388 | 5.253e-04 |
| GO:BP | GO:0001892 | embryonic placenta development | 32 | 88 | 5.272e-04 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 46 | 144 | 5.272e-04 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 55 | 182 | 5.397e-04 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 61 | 208 | 5.524e-04 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 50 | 161 | 5.562e-04 |
| GO:BP | GO:0060993 | kidney morphogenesis | 35 | 100 | 5.724e-04 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 41 | 124 | 5.771e-04 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 30 | 81 | 6.175e-04 |
| GO:BP | GO:0043276 | anoikis | 17 | 35 | 6.210e-04 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 83 | 307 | 6.332e-04 |
| GO:BP | GO:0000209 | protein polyubiquitination | 77 | 280 | 6.443e-04 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 217 | 954 | 6.626e-04 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 32 | 89 | 6.696e-04 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 33 | 93 | 6.893e-04 |
| GO:BP | GO:0002475 | antigen processing and presentation via MHC class Ib | 13 | 23 | 6.987e-04 |
| GO:BP | GO:0060444 | branching involved in mammary gland duct morphogenesis | 13 | 23 | 6.987e-04 |
| GO:BP | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 81 | 299 | 7.171e-04 |
| GO:BP | GO:0061061 | muscle structure development | 164 | 693 | 7.227e-04 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 25 | 63 | 7.227e-04 |
| GO:BP | GO:0031929 | TOR signaling | 51 | 167 | 7.436e-04 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 21 | 49 | 7.825e-04 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 44 | 138 | 7.936e-04 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 69 | 246 | 8.088e-04 |
| GO:BP | GO:0002484 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway | 10 | 15 | 8.121e-04 |
| GO:BP | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib | 10 | 15 | 8.121e-04 |
| GO:BP | GO:0048864 | stem cell development | 31 | 86 | 8.155e-04 |
| GO:BP | GO:0048511 | rhythmic process | 82 | 305 | 8.446e-04 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 122 | 492 | 8.482e-04 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 80 | 296 | 8.526e-04 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 72 | 260 | 8.641e-04 |
| GO:BP | GO:0099111 | microtubule-based transport | 63 | 220 | 8.641e-04 |
| GO:BP | GO:0050708 | regulation of protein secretion | 74 | 269 | 8.641e-04 |
| GO:BP | GO:0007032 | endosome organization | 34 | 98 | 8.654e-04 |
| GO:BP | GO:0061326 | renal tubule development | 36 | 106 | 8.708e-04 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 36 | 106 | 8.708e-04 |
| GO:BP | GO:0010586 | miRNA metabolic process | 36 | 106 | 8.708e-04 |
| GO:BP | GO:0072080 | nephron tubule development | 35 | 102 | 8.708e-04 |
| GO:BP | GO:0060541 | respiratory system development | 64 | 225 | 9.315e-04 |
| GO:BP | GO:0072006 | nephron development | 49 | 160 | 9.323e-04 |
| GO:BP | GO:0021700 | developmental maturation | 92 | 352 | 9.323e-04 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 29 | 79 | 9.474e-04 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 48 | 156 | 9.673e-04 |
| GO:BP | GO:0044282 | small molecule catabolic process | 96 | 371 | 9.724e-04 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 55 | 186 | 9.734e-04 |
| GO:BP | GO:0072028 | nephron morphogenesis | 30 | 83 | 9.807e-04 |
| GO:BP | GO:0097009 | energy homeostasis | 30 | 83 | 9.807e-04 |
| GO:BP | GO:0016050 | vesicle organization | 97 | 376 | 1.000e-03 |
| GO:BP | GO:0051168 | nuclear export | 51 | 169 | 1.000e-03 |
| GO:BP | GO:0072087 | renal vesicle development | 11 | 18 | 1.015e-03 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 84 | 316 | 1.015e-03 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 23 | 57 | 1.017e-03 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 32 | 91 | 1.029e-03 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 19 | 43 | 1.032e-03 |
| GO:BP | GO:0051259 | protein complex oligomerization | 74 | 271 | 1.080e-03 |
| GO:BP | GO:0043010 | camera-type eye development | 94 | 363 | 1.100e-03 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 52 | 174 | 1.111e-03 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 124 | 505 | 1.113e-03 |
| GO:BP | GO:0051898 | negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 24 | 61 | 1.129e-03 |
| GO:BP | GO:0007420 | brain development | 175 | 754 | 1.148e-03 |
| GO:BP | GO:0007042 | lysosomal lumen acidification | 13 | 24 | 1.171e-03 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 13 | 24 | 1.171e-03 |
| GO:BP | GO:0002483 | antigen processing and presentation of endogenous peptide antigen | 13 | 24 | 1.171e-03 |
| GO:BP | GO:0014032 | neural crest cell development | 29 | 80 | 1.188e-03 |
| GO:BP | GO:0034330 | cell junction organization | 191 | 834 | 1.190e-03 |
| GO:BP | GO:0032259 | methylation | 66 | 236 | 1.201e-03 |
| GO:BP | GO:0006606 | protein import into nucleus | 50 | 166 | 1.203e-03 |
| GO:BP | GO:0031348 | negative regulation of defense response | 70 | 254 | 1.210e-03 |
| GO:BP | GO:0061795 | Golgi lumen acidification | 9 | 13 | 1.211e-03 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 151 | 637 | 1.217e-03 |
| GO:BP | GO:0001843 | neural tube closure | 31 | 88 | 1.247e-03 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 99 | 388 | 1.274e-03 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 52 | 175 | 1.274e-03 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 69 | 250 | 1.280e-03 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 44 | 141 | 1.280e-03 |
| GO:BP | GO:0048255 | mRNA stabilization | 26 | 69 | 1.301e-03 |
| GO:BP | GO:0035329 | hippo signaling | 23 | 58 | 1.343e-03 |
| GO:BP | GO:0051402 | neuron apoptotic process | 79 | 296 | 1.378e-03 |
| GO:BP | GO:0061614 | miRNA transcription | 28 | 77 | 1.444e-03 |
| GO:BP | GO:0034205 | amyloid-beta formation | 19 | 44 | 1.447e-03 |
| GO:BP | GO:0006417 | regulation of translation | 97 | 380 | 1.455e-03 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 45 | 146 | 1.460e-03 |
| GO:BP | GO:0042063 | gliogenesis | 91 | 352 | 1.460e-03 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 92 | 357 | 1.511e-03 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 44 | 142 | 1.511e-03 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 44 | 142 | 1.511e-03 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 51 | 172 | 1.540e-03 |
| GO:BP | GO:0019080 | viral gene expression | 36 | 109 | 1.540e-03 |
| GO:BP | GO:0060606 | tube closure | 31 | 89 | 1.540e-03 |
| GO:BP | GO:0046620 | regulation of organ growth | 31 | 89 | 1.540e-03 |
| GO:BP | GO:0006066 | alcohol metabolic process | 93 | 362 | 1.547e-03 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 107 | 428 | 1.551e-03 |
| GO:BP | GO:0090398 | cellular senescence | 34 | 101 | 1.555e-03 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 25 | 66 | 1.565e-03 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 88 | 339 | 1.587e-03 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 88 | 339 | 1.587e-03 |
| GO:BP | GO:0031347 | regulation of defense response | 185 | 809 | 1.589e-03 |
| GO:BP | GO:0038066 | p38MAPK cascade | 22 | 55 | 1.589e-03 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 42 | 134 | 1.591e-03 |
| GO:BP | GO:0090596 | sensory organ morphogenesis | 78 | 293 | 1.606e-03 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 78 | 293 | 1.606e-03 |
| GO:BP | GO:0002520 | immune system development | 58 | 203 | 1.614e-03 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 26 | 70 | 1.642e-03 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 26 | 70 | 1.642e-03 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 20 | 48 | 1.652e-03 |
| GO:BP | GO:0033365 | protein localization to organelle | 264 | 1209 | 1.652e-03 |
| GO:BP | GO:0033993 | response to lipid | 209 | 930 | 1.652e-03 |
| GO:BP | GO:0046466 | membrane lipid catabolic process | 20 | 48 | 1.652e-03 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 20 | 48 | 1.652e-03 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 72 | 266 | 1.659e-03 |
| GO:BP | GO:0050865 | regulation of cell activation | 149 | 632 | 1.670e-03 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 133 | 554 | 1.670e-03 |
| GO:BP | GO:0001654 | eye development | 104 | 415 | 1.692e-03 |
| GO:BP | GO:0021915 | neural tube development | 48 | 160 | 1.717e-03 |
| GO:BP | GO:0048839 | inner ear development | 59 | 208 | 1.721e-03 |
| GO:BP | GO:0010256 | endomembrane system organization | 145 | 613 | 1.736e-03 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 38 | 118 | 1.740e-03 |
| GO:BP | GO:0043489 | RNA stabilization | 28 | 78 | 1.755e-03 |
| GO:BP | GO:0030162 | regulation of proteolysis | 102 | 406 | 1.756e-03 |
| GO:BP | GO:0046579 | positive regulation of Ras protein signal transduction | 11 | 19 | 1.790e-03 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 58 | 204 | 1.813e-03 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 35 | 106 | 1.828e-03 |
| GO:BP | GO:0019218 | regulation of steroid metabolic process | 34 | 102 | 1.839e-03 |
| GO:BP | GO:0044090 | positive regulation of vacuole organization | 13 | 25 | 1.839e-03 |
| GO:BP | GO:0009065 | glutamine family amino acid catabolic process | 13 | 25 | 1.839e-03 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 31 | 90 | 1.840e-03 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 46 | 152 | 1.846e-03 |
| GO:BP | GO:0009299 | mRNA transcription | 21 | 52 | 1.846e-03 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 70 | 258 | 1.846e-03 |
| GO:BP | GO:0006403 | RNA localization | 57 | 200 | 1.901e-03 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 19 | 45 | 1.917e-03 |
| GO:BP | GO:0010817 | regulation of hormone levels | 133 | 556 | 1.917e-03 |
| GO:BP | GO:0001756 | somitogenesis | 25 | 67 | 1.950e-03 |
| GO:BP | GO:0007610 | behavior | 157 | 674 | 1.983e-03 |
| GO:BP | GO:0046660 | female sex differentiation | 39 | 123 | 2.004e-03 |
| GO:BP | GO:0043062 | extracellular structure organization | 85 | 328 | 2.068e-03 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 28 | 79 | 2.195e-03 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 171 | 745 | 2.231e-03 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 29 | 83 | 2.234e-03 |
| GO:BP | GO:0009615 | response to virus | 103 | 414 | 2.346e-03 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 83 | 320 | 2.374e-03 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 24 | 64 | 2.390e-03 |
| GO:BP | GO:0150063 | visual system development | 104 | 419 | 2.390e-03 |
| GO:BP | GO:0072009 | nephron epithelium development | 39 | 124 | 2.393e-03 |
| GO:BP | GO:0061024 | membrane organization | 186 | 821 | 2.393e-03 |
| GO:BP | GO:0051170 | import into nucleus | 50 | 171 | 2.400e-03 |
| GO:BP | GO:0140673 | transcription elongation-coupled chromatin remodeling | 7 | 9 | 2.442e-03 |
| GO:BP | GO:0023061 | signal release | 121 | 501 | 2.459e-03 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 57 | 202 | 2.470e-03 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 122 | 506 | 2.486e-03 |
| GO:BP | GO:0002486 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent | 9 | 14 | 2.487e-03 |
| GO:BP | GO:0001666 | response to hypoxia | 79 | 302 | 2.487e-03 |
| GO:BP | GO:0048002 | antigen processing and presentation of peptide antigen | 25 | 68 | 2.490e-03 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 42 | 137 | 2.528e-03 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 26 | 72 | 2.591e-03 |
| GO:BP | GO:0046661 | male sex differentiation | 51 | 176 | 2.611e-03 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 78 | 298 | 2.653e-03 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 27 | 76 | 2.662e-03 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 22 | 57 | 2.676e-03 |
| GO:BP | GO:0051452 | intracellular pH reduction | 22 | 57 | 2.676e-03 |
| GO:BP | GO:0060537 | muscle tissue development | 106 | 430 | 2.676e-03 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 44 | 146 | 2.717e-03 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 32 | 96 | 2.732e-03 |
| GO:BP | GO:0060173 | limb development | 55 | 194 | 2.733e-03 |
| GO:BP | GO:0048736 | appendage development | 55 | 194 | 2.733e-03 |
| GO:BP | GO:0070925 | organelle assembly | 230 | 1046 | 2.770e-03 |
| GO:BP | GO:0044770 | cell cycle phase transition | 127 | 532 | 2.807e-03 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 63 | 230 | 2.807e-03 |
| GO:BP | GO:0034329 | cell junction assembly | 121 | 503 | 2.834e-03 |
| GO:BP | GO:0048880 | sensory system development | 105 | 426 | 2.837e-03 |
| GO:BP | GO:0034349 | glial cell apoptotic process | 10 | 17 | 2.895e-03 |
| GO:BP | GO:0072077 | renal vesicle morphogenesis | 10 | 17 | 2.895e-03 |
| GO:BP | GO:0030198 | extracellular matrix organization | 84 | 327 | 2.926e-03 |
| GO:BP | GO:0048705 | skeletal system morphogenesis | 64 | 235 | 2.963e-03 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 45 | 151 | 2.972e-03 |
| GO:BP | GO:0019318 | hexose metabolic process | 62 | 226 | 2.972e-03 |
| GO:BP | GO:0010721 | negative regulation of cell development | 71 | 267 | 2.972e-03 |
| GO:BP | GO:0045682 | regulation of epidermis development | 24 | 65 | 2.972e-03 |
| GO:BP | GO:0007035 | vacuolar acidification | 16 | 36 | 2.972e-03 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 24 | 65 | 2.972e-03 |
| GO:BP | GO:0044804 | nucleophagy | 12 | 23 | 2.972e-03 |
| GO:BP | GO:1902003 | regulation of amyloid-beta formation | 16 | 36 | 2.972e-03 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 12 | 23 | 2.972e-03 |
| GO:BP | GO:0016237 | microautophagy | 12 | 23 | 2.972e-03 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 16 | 36 | 2.972e-03 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 24 | 65 | 2.972e-03 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 12 | 23 | 2.972e-03 |
| GO:BP | GO:0007040 | lysosome organization | 36 | 113 | 2.985e-03 |
| GO:BP | GO:0080171 | lytic vacuole organization | 36 | 113 | 2.985e-03 |
| GO:BP | GO:2000786 | positive regulation of autophagosome assembly | 11 | 20 | 2.996e-03 |
| GO:BP | GO:0098739 | import across plasma membrane | 55 | 195 | 3.038e-03 |
| GO:BP | GO:0014706 | striated muscle tissue development | 101 | 408 | 3.050e-03 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 123 | 514 | 3.056e-03 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 25 | 69 | 3.058e-03 |
| GO:BP | GO:0006997 | nucleus organization | 44 | 147 | 3.066e-03 |
| GO:BP | GO:0051896 | regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 70 | 263 | 3.116e-03 |
| GO:BP | GO:0043112 | receptor metabolic process | 26 | 73 | 3.148e-03 |
| GO:BP | GO:0001841 | neural tube formation | 33 | 101 | 3.162e-03 |
| GO:BP | GO:0060996 | dendritic spine development | 33 | 101 | 3.162e-03 |
| GO:BP | GO:0071695 | anatomical structure maturation | 73 | 277 | 3.176e-03 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 66 | 245 | 3.198e-03 |
| GO:BP | GO:0009880 | embryonic pattern specification | 27 | 77 | 3.198e-03 |
| GO:BP | GO:0001656 | metanephros development | 31 | 93 | 3.225e-03 |
| GO:BP | GO:0019915 | lipid storage | 31 | 93 | 3.225e-03 |
| GO:BP | GO:0170062 | nutrient storage | 31 | 93 | 3.225e-03 |
| GO:BP | GO:0044419 | biological process involved in interspecies interaction between organisms | 367 | 1759 | 3.280e-03 |
| GO:BP | GO:0006575 | modified amino acid metabolic process | 51 | 178 | 3.280e-03 |
| GO:BP | GO:0006839 | mitochondrial transport | 51 | 178 | 3.280e-03 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 84 | 329 | 3.443e-03 |
| GO:BP | GO:0001658 | branching involved in ureteric bud morphogenesis | 23 | 62 | 3.564e-03 |
| GO:BP | GO:0006457 | protein folding | 60 | 219 | 3.690e-03 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 24 | 66 | 3.730e-03 |
| GO:BP | GO:0006897 | endocytosis | 162 | 709 | 3.730e-03 |
| GO:BP | GO:0002376 | immune system process | 576 | 2871 | 3.741e-03 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 20 | 51 | 3.776e-03 |
| GO:BP | GO:0008063 | Toll signaling pathway | 8 | 12 | 3.788e-03 |
| GO:BP | GO:0015833 | peptide transport | 75 | 288 | 3.798e-03 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 14 | 30 | 3.820e-03 |
| GO:BP | GO:0019725 | cellular homeostasis | 187 | 835 | 3.890e-03 |
| GO:BP | GO:0072089 | stem cell proliferation | 38 | 123 | 3.891e-03 |
| GO:BP | GO:0014020 | primary neural tube formation | 31 | 94 | 3.915e-03 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 122 | 513 | 4.059e-03 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 91 | 364 | 4.139e-03 |
| GO:BP | GO:0031099 | regeneration | 56 | 202 | 4.143e-03 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 36 | 115 | 4.171e-03 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 63 | 234 | 4.329e-03 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 82 | 322 | 4.329e-03 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 22 | 59 | 4.331e-03 |
| GO:BP | GO:0045604 | regulation of epidermal cell differentiation | 22 | 59 | 4.331e-03 |
| GO:BP | GO:0030282 | bone mineralization | 39 | 128 | 4.401e-03 |
| GO:BP | GO:0006672 | ceramide metabolic process | 34 | 107 | 4.421e-03 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 48 | 167 | 4.421e-03 |
| GO:BP | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis | 9 | 15 | 4.639e-03 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 9 | 15 | 4.639e-03 |
| GO:BP | GO:0009798 | axis specification | 32 | 99 | 4.642e-03 |
| GO:BP | GO:1902993 | positive regulation of amyloid precursor protein catabolic process | 12 | 24 | 4.650e-03 |
| GO:BP | GO:0042254 | ribosome biogenesis | 82 | 323 | 4.744e-03 |
| GO:BP | GO:0048592 | eye morphogenesis | 49 | 172 | 4.788e-03 |
| GO:BP | GO:0014015 | positive regulation of gliogenesis | 25 | 71 | 4.788e-03 |
| GO:BP | GO:0006536 | glutamate metabolic process | 15 | 34 | 4.815e-03 |
| GO:BP | GO:0019882 | antigen processing and presentation | 36 | 116 | 4.914e-03 |
| GO:BP | GO:0090189 | regulation of branching involved in ureteric bud morphogenesis | 11 | 21 | 4.914e-03 |
| GO:BP | GO:0042113 | B cell activation | 74 | 286 | 4.927e-03 |
| GO:BP | GO:0034727 | piecemeal microautophagy of the nucleus | 10 | 18 | 4.927e-03 |
| GO:BP | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis | 10 | 18 | 4.927e-03 |
| GO:BP | GO:0010544 | negative regulation of platelet activation | 10 | 18 | 4.927e-03 |
| GO:BP | GO:0022402 | cell cycle process | 273 | 1280 | 5.132e-03 |
| GO:BP | GO:0046879 | hormone secretion | 82 | 324 | 5.189e-03 |
| GO:BP | GO:0051098 | regulation of binding | 58 | 213 | 5.226e-03 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 21 | 56 | 5.243e-03 |
| GO:BP | GO:0051260 | protein homooligomerization | 56 | 204 | 5.243e-03 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 106 | 439 | 5.336e-03 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 165 | 730 | 5.347e-03 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 33 | 104 | 5.376e-03 |
| GO:BP | GO:0043124 | negative regulation of canonical NF-kappaB signal transduction | 22 | 60 | 5.506e-03 |
| GO:BP | GO:0019883 | antigen processing and presentation of endogenous antigen | 14 | 31 | 5.523e-03 |
| GO:BP | GO:0003338 | metanephros morphogenesis | 14 | 31 | 5.523e-03 |
| GO:BP | GO:0097421 | liver regeneration | 14 | 31 | 5.523e-03 |
| GO:BP | GO:0010001 | glial cell differentiation | 69 | 264 | 5.552e-03 |
| GO:BP | GO:0050658 | RNA transport | 46 | 160 | 5.582e-03 |
| GO:BP | GO:0050657 | nucleic acid transport | 46 | 160 | 5.582e-03 |
| GO:BP | GO:0007005 | mitochondrion organization | 105 | 435 | 5.667e-03 |
| GO:BP | GO:0072524 | pyridine-containing compound metabolic process | 48 | 169 | 5.712e-03 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 48 | 169 | 5.712e-03 |
| GO:BP | GO:0042632 | cholesterol homeostasis | 30 | 92 | 5.719e-03 |
| GO:BP | GO:0045995 | regulation of embryonic development | 30 | 92 | 5.719e-03 |
| GO:BP | GO:0060052 | neurofilament cytoskeleton organization | 7 | 10 | 5.746e-03 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 24 | 68 | 5.746e-03 |
| GO:BP | GO:2000739 | regulation of mesenchymal stem cell differentiation | 7 | 10 | 5.746e-03 |
| GO:BP | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin | 7 | 10 | 5.746e-03 |
| GO:BP | GO:0001655 | urogenital system development | 24 | 68 | 5.746e-03 |
| GO:BP | GO:0090184 | positive regulation of kidney development | 7 | 10 | 5.746e-03 |
| GO:BP | GO:1902105 | regulation of leukocyte differentiation | 83 | 330 | 5.746e-03 |
| GO:BP | GO:0099536 | synaptic signaling | 180 | 807 | 5.758e-03 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 29 | 88 | 5.758e-03 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 234 | 1082 | 5.770e-03 |
| GO:BP | GO:0022408 | negative regulation of cell-cell adhesion | 54 | 196 | 5.786e-03 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 25 | 72 | 5.786e-03 |
| GO:BP | GO:0061035 | regulation of cartilage development | 25 | 72 | 5.786e-03 |
| GO:BP | GO:0051101 | regulation of DNA binding | 25 | 72 | 5.786e-03 |
| GO:BP | GO:0009914 | hormone transport | 84 | 335 | 5.807e-03 |
| GO:BP | GO:0051028 | mRNA transport | 39 | 130 | 5.807e-03 |
| GO:BP | GO:0030857 | negative regulation of epithelial cell differentiation | 19 | 49 | 5.807e-03 |
| GO:BP | GO:0009063 | amino acid catabolic process | 35 | 113 | 5.817e-03 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 42 | 143 | 5.831e-03 |
| GO:BP | GO:0008584 | male gonad development | 44 | 152 | 6.085e-03 |
| GO:BP | GO:0032330 | regulation of chondrocyte differentiation | 20 | 53 | 6.226e-03 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 20 | 53 | 6.226e-03 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 118 | 500 | 6.341e-03 |
| GO:BP | GO:1903034 | regulation of response to wounding | 48 | 170 | 6.392e-03 |
| GO:BP | GO:0002790 | peptide secretion | 68 | 261 | 6.426e-03 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 21 | 57 | 6.565e-03 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 36 | 118 | 6.607e-03 |
| GO:BP | GO:0055092 | sterol homeostasis | 30 | 93 | 6.770e-03 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 30 | 93 | 6.770e-03 |
| GO:BP | GO:0030323 | respiratory tube development | 55 | 202 | 6.892e-03 |
| GO:BP | GO:0006006 | glucose metabolic process | 51 | 184 | 6.901e-03 |
| GO:BP | GO:0072665 | protein localization to vacuole | 29 | 89 | 6.901e-03 |
| GO:BP | GO:0038095 | Fc-epsilon receptor signaling pathway | 12 | 25 | 6.906e-03 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 44 | 153 | 6.973e-03 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 18 | 46 | 7.015e-03 |
| GO:BP | GO:0016485 | protein processing | 60 | 225 | 7.028e-03 |
| GO:BP | GO:0009952 | anterior/posterior pattern specification | 60 | 225 | 7.028e-03 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 24 | 69 | 7.059e-03 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 26 | 77 | 7.105e-03 |
| GO:BP | GO:0050890 | cognition | 82 | 328 | 7.235e-03 |
| GO:BP | GO:0038203 | TORC2 signaling | 8 | 13 | 7.346e-03 |
| GO:BP | GO:0034111 | negative regulation of homotypic cell-cell adhesion | 8 | 13 | 7.346e-03 |
| GO:BP | GO:0014013 | regulation of gliogenesis | 33 | 106 | 7.346e-03 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 11 | 22 | 7.530e-03 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 19 | 50 | 7.530e-03 |
| GO:BP | GO:0061709 | reticulophagy | 11 | 22 | 7.530e-03 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 70 | 272 | 7.548e-03 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 16 | 39 | 7.548e-03 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 62 | 235 | 7.625e-03 |
| GO:BP | GO:0060966 | regulation of gene silencing by regulatory ncRNA | 14 | 32 | 7.625e-03 |
| GO:BP | GO:0035751 | regulation of lysosomal lumen pH | 14 | 32 | 7.625e-03 |
| GO:BP | GO:0001659 | temperature homeostasis | 51 | 185 | 7.717e-03 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 236 | 1099 | 7.847e-03 |
| GO:BP | GO:0009084 | glutamine family amino acid biosynthetic process | 10 | 19 | 7.912e-03 |
| GO:BP | GO:0006491 | N-glycan processing | 10 | 19 | 7.912e-03 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 20 | 54 | 7.912e-03 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 20 | 54 | 7.912e-03 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 63 | 240 | 7.912e-03 |
| GO:BP | GO:0035627 | ceramide transport | 9 | 16 | 7.926e-03 |
| GO:BP | GO:0002065 | columnar/cuboidal epithelial cell differentiation | 35 | 115 | 7.926e-03 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 30 | 94 | 7.956e-03 |
| GO:BP | GO:0055085 | transmembrane transport | 328 | 1578 | 7.976e-03 |
| GO:BP | GO:0051236 | establishment of RNA localization | 46 | 163 | 7.999e-03 |
| GO:BP | GO:0009611 | response to wounding | 131 | 568 | 8.189e-03 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 43 | 150 | 8.302e-03 |
| GO:BP | GO:0032456 | endocytic recycling | 28 | 86 | 8.321e-03 |
| GO:BP | GO:0030149 | sphingolipid catabolic process | 17 | 43 | 8.336e-03 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 6 | 8 | 8.348e-03 |
| GO:BP | GO:0046984 | regulation of hemoglobin biosynthetic process | 6 | 8 | 8.348e-03 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 22 | 62 | 8.385e-03 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 23 | 66 | 8.530e-03 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 47 | 168 | 8.560e-03 |
| GO:BP | GO:0016042 | lipid catabolic process | 87 | 354 | 8.576e-03 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 13 | 29 | 8.739e-03 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 13 | 29 | 8.739e-03 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 13 | 29 | 8.739e-03 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 13 | 29 | 8.739e-03 |
| GO:BP | GO:0030072 | peptide hormone secretion | 66 | 255 | 8.744e-03 |
| GO:BP | GO:0006275 | regulation of DNA replication | 36 | 120 | 8.783e-03 |
| GO:BP | GO:0009411 | response to UV | 44 | 155 | 8.896e-03 |
| GO:BP | GO:0045055 | regulated exocytosis | 61 | 232 | 8.896e-03 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 48 | 173 | 9.105e-03 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 31 | 99 | 9.105e-03 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 30 | 95 | 9.409e-03 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 30 | 95 | 9.409e-03 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 173 | 781 | 9.422e-03 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 45 | 160 | 9.579e-03 |
| GO:BP | GO:0051047 | positive regulation of secretion | 80 | 322 | 9.647e-03 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 51 | 187 | 9.647e-03 |
| GO:BP | GO:0006405 | RNA export from nucleus | 29 | 91 | 9.657e-03 |
| GO:BP | GO:0002694 | regulation of leukocyte activation | 132 | 576 | 9.892e-03 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 20 | 55 | 9.946e-03 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 16 | 40 | 9.970e-03 |
| GO:BP | GO:0070227 | lymphocyte apoptotic process | 27 | 83 | 1.013e-02 |
| GO:BP | GO:0002181 | cytoplasmic translation | 46 | 165 | 1.023e-02 |
| GO:BP | GO:0061462 | protein localization to lysosome | 21 | 59 | 1.023e-02 |
| GO:BP | GO:1901342 | regulation of vasculature development | 75 | 299 | 1.028e-02 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 24 | 71 | 1.049e-02 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 23 | 67 | 1.049e-02 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 113 | 483 | 1.051e-02 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 144 | 637 | 1.058e-02 |
| GO:BP | GO:0006658 | phosphatidylserine metabolic process | 11 | 23 | 1.133e-02 |
| GO:BP | GO:0072202 | cell differentiation involved in metanephros development | 11 | 23 | 1.133e-02 |
| GO:BP | GO:0046985 | positive regulation of hemoglobin biosynthetic process | 5 | 6 | 1.141e-02 |
| GO:BP | GO:1904760 | regulation of myofibroblast differentiation | 5 | 6 | 1.141e-02 |
| GO:BP | GO:2000354 | regulation of ovarian follicle development | 5 | 6 | 1.141e-02 |
| GO:BP | GO:0022407 | regulation of cell-cell adhesion | 115 | 494 | 1.141e-02 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 52 | 193 | 1.141e-02 |
| GO:BP | GO:0042270 | protection from natural killer cell mediated cytotoxicity | 5 | 6 | 1.141e-02 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 52 | 193 | 1.141e-02 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 91 | 377 | 1.155e-02 |
| GO:BP | GO:0007254 | JNK cascade | 48 | 175 | 1.155e-02 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 18 | 48 | 1.156e-02 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 18 | 48 | 1.156e-02 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 171 | 775 | 1.167e-02 |
| GO:BP | GO:1904238 | pericyte cell differentiation | 7 | 11 | 1.167e-02 |
| GO:BP | GO:0035871 | protein K11-linked deubiquitination | 7 | 11 | 1.167e-02 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 171 | 775 | 1.167e-02 |
| GO:BP | GO:0090331 | negative regulation of platelet aggregation | 7 | 11 | 1.167e-02 |
| GO:BP | GO:0072497 | mesenchymal stem cell differentiation | 7 | 11 | 1.167e-02 |
| GO:BP | GO:0030324 | lung development | 53 | 198 | 1.186e-02 |
| GO:BP | GO:0002067 | glandular epithelial cell differentiation | 15 | 37 | 1.186e-02 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 19 | 52 | 1.202e-02 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 32 | 105 | 1.202e-02 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 32 | 105 | 1.202e-02 |
| GO:BP | GO:0050866 | negative regulation of cell activation | 56 | 212 | 1.215e-02 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by regulatory ncRNA | 13 | 30 | 1.219e-02 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 13 | 30 | 1.219e-02 |
| GO:BP | GO:0006537 | glutamate biosynthetic process | 4 | 4 | 1.224e-02 |
| GO:BP | GO:0035914 | skeletal muscle cell differentiation | 26 | 80 | 1.224e-02 |
| GO:BP | GO:1990170 | stress response to cadmium ion | 4 | 4 | 1.224e-02 |
| GO:BP | GO:1904761 | negative regulation of myofibroblast differentiation | 4 | 4 | 1.224e-02 |
| GO:BP | GO:0002477 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib | 4 | 4 | 1.224e-02 |
| GO:BP | GO:0071585 | detoxification of cadmium ion | 4 | 4 | 1.224e-02 |
| GO:BP | GO:0010868 | negative regulation of triglyceride biosynthetic process | 4 | 4 | 1.224e-02 |
| GO:BP | GO:0015693 | magnesium ion transport | 10 | 20 | 1.224e-02 |
| GO:BP | GO:0051051 | negative regulation of transport | 98 | 412 | 1.226e-02 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 20 | 56 | 1.226e-02 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 92 | 383 | 1.240e-02 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 21 | 60 | 1.249e-02 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 24 | 72 | 1.251e-02 |
| GO:BP | GO:0043414 | macromolecule methylation | 37 | 127 | 1.268e-02 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 44 | 158 | 1.275e-02 |
| GO:BP | GO:0044819 | mitotic G1/S transition checkpoint signaling | 8 | 14 | 1.279e-02 |
| GO:BP | GO:1901653 | cellular response to peptide | 8 | 14 | 1.279e-02 |
| GO:BP | GO:0007000 | nucleolus organization | 9 | 17 | 1.279e-02 |
| GO:BP | GO:0048388 | endosomal lumen acidification | 8 | 14 | 1.279e-02 |
| GO:BP | GO:0043117 | positive regulation of vascular permeability | 9 | 17 | 1.279e-02 |
| GO:BP | GO:0009749 | response to glucose | 55 | 208 | 1.279e-02 |
| GO:BP | GO:0140718 | facultative heterochromatin formation | 8 | 14 | 1.279e-02 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 16 | 41 | 1.280e-02 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 16 | 41 | 1.280e-02 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 58 | 222 | 1.289e-02 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 122 | 531 | 1.292e-02 |
| GO:BP | GO:0006281 | DNA repair | 140 | 621 | 1.307e-02 |
| GO:BP | GO:0070555 | response to interleukin-1 | 33 | 110 | 1.318e-02 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 29 | 93 | 1.318e-02 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 41 | 145 | 1.323e-02 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 73 | 293 | 1.361e-02 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 38 | 132 | 1.370e-02 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 17 | 45 | 1.372e-02 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 63 | 246 | 1.381e-02 |
| GO:BP | GO:0099173 | postsynapse organization | 63 | 246 | 1.381e-02 |
| GO:BP | GO:0034405 | response to fluid shear stress | 14 | 34 | 1.388e-02 |
| GO:BP | GO:0035270 | endocrine system development | 40 | 141 | 1.399e-02 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 55 | 209 | 1.408e-02 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 31 | 102 | 1.436e-02 |
| GO:BP | GO:0042551 | neuron maturation | 18 | 49 | 1.439e-02 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 53 | 200 | 1.440e-02 |
| GO:BP | GO:0032200 | telomere organization | 51 | 191 | 1.470e-02 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 62 | 242 | 1.475e-02 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 74 | 299 | 1.515e-02 |
| GO:BP | GO:0051648 | vesicle localization | 57 | 219 | 1.523e-02 |
| GO:BP | GO:0006879 | intracellular iron ion homeostasis | 23 | 69 | 1.523e-02 |
| GO:BP | GO:1905330 | regulation of morphogenesis of an epithelium | 22 | 65 | 1.525e-02 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 33 | 111 | 1.525e-02 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 22 | 65 | 1.525e-02 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 21 | 61 | 1.525e-02 |
| GO:BP | GO:0060350 | endochondral bone morphogenesis | 21 | 61 | 1.525e-02 |
| GO:BP | GO:0030258 | lipid modification | 52 | 196 | 1.530e-02 |
| GO:BP | GO:0016072 | rRNA metabolic process | 67 | 266 | 1.536e-02 |
| GO:BP | GO:0030513 | positive regulation of BMP signaling pathway | 15 | 38 | 1.537e-02 |
| GO:BP | GO:0051249 | regulation of lymphocyte activation | 117 | 509 | 1.537e-02 |
| GO:BP | GO:0030641 | regulation of cellular pH | 29 | 94 | 1.543e-02 |
| GO:BP | GO:0001817 | regulation of cytokine production | 171 | 781 | 1.553e-02 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 32 | 107 | 1.591e-02 |
| GO:BP | GO:0015695 | organic cation transport | 35 | 120 | 1.591e-02 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 32 | 107 | 1.591e-02 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 46 | 169 | 1.591e-02 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 65 | 257 | 1.591e-02 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 65 | 257 | 1.591e-02 |
| GO:BP | GO:0051453 | regulation of intracellular pH | 28 | 90 | 1.593e-02 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 28 | 90 | 1.593e-02 |
| GO:BP | GO:0001816 | cytokine production | 172 | 787 | 1.622e-02 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 13 | 31 | 1.636e-02 |
| GO:BP | GO:0098868 | bone growth | 13 | 31 | 1.636e-02 |
| GO:BP | GO:0000578 | embryonic axis specification | 16 | 42 | 1.644e-02 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 127 | 560 | 1.644e-02 |
| GO:BP | GO:0019953 | sexual reproduction | 237 | 1122 | 1.644e-02 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 127 | 560 | 1.644e-02 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 31 | 103 | 1.649e-02 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 31 | 103 | 1.649e-02 |
| GO:BP | GO:0048562 | embryonic organ morphogenesis | 75 | 305 | 1.649e-02 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 39 | 138 | 1.659e-02 |
| GO:BP | GO:0050728 | negative regulation of inflammatory response | 45 | 165 | 1.678e-02 |
| GO:BP | GO:0030073 | insulin secretion | 55 | 211 | 1.703e-02 |
| GO:BP | GO:0048469 | cell maturation | 55 | 211 | 1.703e-02 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 144 | 646 | 1.703e-02 |
| GO:BP | GO:0006364 | rRNA processing | 58 | 225 | 1.709e-02 |
| GO:BP | GO:0051100 | negative regulation of binding | 30 | 99 | 1.718e-02 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 138 | 616 | 1.720e-02 |
| GO:BP | GO:0032526 | response to retinoic acid | 33 | 112 | 1.736e-02 |
| GO:BP | GO:1901655 | cellular response to ketone | 33 | 112 | 1.736e-02 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 62 | 244 | 1.750e-02 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 46 | 170 | 1.765e-02 |
| GO:BP | GO:0042698 | ovulation cycle | 24 | 74 | 1.770e-02 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 24 | 74 | 1.770e-02 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 42 | 152 | 1.779e-02 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 18 | 50 | 1.780e-02 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 18 | 50 | 1.780e-02 |
| GO:BP | GO:0019377 | glycolipid catabolic process | 10 | 21 | 1.782e-02 |
| GO:BP | GO:0046514 | ceramide catabolic process | 10 | 21 | 1.782e-02 |
| GO:BP | GO:0045586 | regulation of gamma-delta T cell differentiation | 6 | 9 | 1.785e-02 |
| GO:BP | GO:0000183 | rDNA heterochromatin formation | 6 | 9 | 1.785e-02 |
| GO:BP | GO:0060363 | cranial suture morphogenesis | 6 | 9 | 1.785e-02 |
| GO:BP | GO:0097192 | extrinsic apoptotic signaling pathway in absence of ligand | 23 | 70 | 1.785e-02 |
| GO:BP | GO:0048318 | axial mesoderm development | 6 | 9 | 1.785e-02 |
| GO:BP | GO:0003418 | growth plate cartilage chondrocyte differentiation | 6 | 9 | 1.785e-02 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 23 | 70 | 1.785e-02 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 23 | 70 | 1.785e-02 |
| GO:BP | GO:0003279 | cardiac septum development | 32 | 108 | 1.799e-02 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 14 | 35 | 1.799e-02 |
| GO:BP | GO:0046545 | development of primary female sexual characteristics | 32 | 108 | 1.799e-02 |
| GO:BP | GO:0110110 | positive regulation of animal organ morphogenesis | 14 | 35 | 1.799e-02 |
| GO:BP | GO:0006024 | glycosaminoglycan biosynthetic process | 14 | 35 | 1.799e-02 |
| GO:BP | GO:0016486 | peptide hormone processing | 14 | 35 | 1.799e-02 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 72 | 292 | 1.812e-02 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 21 | 62 | 1.812e-02 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 37 | 130 | 1.819e-02 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 149 | 673 | 1.835e-02 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 61 | 240 | 1.835e-02 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 45 | 166 | 1.843e-02 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 39 | 139 | 1.843e-02 |
| GO:BP | GO:0008585 | female gonad development | 31 | 104 | 1.875e-02 |
| GO:BP | GO:0032801 | receptor catabolic process | 12 | 28 | 1.886e-02 |
| GO:BP | GO:0030031 | cell projection assembly | 137 | 613 | 1.894e-02 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 87 | 365 | 1.898e-02 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 48 | 180 | 1.906e-02 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 36 | 126 | 1.916e-02 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 36 | 126 | 1.916e-02 |
| GO:BP | GO:0006952 | defense response | 375 | 1853 | 1.923e-02 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 30 | 100 | 1.933e-02 |
| GO:BP | GO:2000027 | regulation of animal organ morphogenesis | 26 | 83 | 1.933e-02 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 15 | 39 | 1.933e-02 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 9 | 18 | 1.933e-02 |
| GO:BP | GO:1901606 | alpha-amino acid catabolic process | 30 | 100 | 1.933e-02 |
| GO:BP | GO:0019362 | pyridine nucleotide metabolic process | 44 | 162 | 1.933e-02 |
| GO:BP | GO:1902430 | negative regulation of amyloid-beta formation | 9 | 18 | 1.933e-02 |
| GO:BP | GO:0140448 | signaling receptor ligand precursor processing | 15 | 39 | 1.933e-02 |
| GO:BP | GO:0030279 | negative regulation of ossification | 15 | 39 | 1.933e-02 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 26 | 83 | 1.933e-02 |
| GO:BP | GO:0046496 | nicotinamide nucleotide metabolic process | 44 | 162 | 1.933e-02 |
| GO:BP | GO:0043691 | reverse cholesterol transport | 9 | 18 | 1.933e-02 |
| GO:BP | GO:0048565 | digestive tract development | 38 | 135 | 1.933e-02 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 51 | 194 | 1.933e-02 |
| GO:BP | GO:0060323 | head morphogenesis | 15 | 39 | 1.933e-02 |
| GO:BP | GO:0001776 | leukocyte homeostasis | 33 | 113 | 1.934e-02 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 42 | 153 | 1.940e-02 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 79 | 327 | 1.990e-02 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 133 | 594 | 1.992e-02 |
| GO:BP | GO:0061723 | glycophagy | 7 | 12 | 1.999e-02 |
| GO:BP | GO:0048505 | regulation of timing of cell differentiation | 7 | 12 | 1.999e-02 |
| GO:BP | GO:0060379 | cardiac muscle cell myoblast differentiation | 7 | 12 | 1.999e-02 |
| GO:BP | GO:0009746 | response to hexose | 55 | 213 | 1.999e-02 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 7 | 12 | 1.999e-02 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 7 | 12 | 1.999e-02 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 7 | 12 | 1.999e-02 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 7 | 12 | 1.999e-02 |
| GO:BP | GO:0048635 | negative regulation of muscle organ development | 7 | 12 | 1.999e-02 |
| GO:BP | GO:1905671 | regulation of lysosome organization | 7 | 12 | 1.999e-02 |
| GO:BP | GO:0048711 | positive regulation of astrocyte differentiation | 7 | 12 | 1.999e-02 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 29 | 96 | 2.002e-02 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 75 | 308 | 2.008e-02 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 32 | 109 | 2.008e-02 |
| GO:BP | GO:0016322 | neuron remodeling | 8 | 15 | 2.008e-02 |
| GO:BP | GO:0021537 | telencephalon development | 70 | 284 | 2.008e-02 |
| GO:BP | GO:0070243 | regulation of thymocyte apoptotic process | 8 | 15 | 2.008e-02 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 16 | 43 | 2.008e-02 |
| GO:BP | GO:0071496 | cellular response to external stimulus | 24 | 75 | 2.023e-02 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 43 | 158 | 2.023e-02 |
| GO:BP | GO:0030183 | B cell differentiation | 43 | 158 | 2.023e-02 |
| GO:BP | GO:0017157 | regulation of exocytosis | 48 | 181 | 2.066e-02 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 23 | 71 | 2.071e-02 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 17 | 47 | 2.077e-02 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 28 | 92 | 2.077e-02 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 17 | 47 | 2.077e-02 |
| GO:BP | GO:0060711 | labyrinthine layer development | 17 | 47 | 2.077e-02 |
| GO:BP | GO:0006509 | membrane protein ectodomain proteolysis | 17 | 47 | 2.077e-02 |
| GO:BP | GO:1903955 | positive regulation of protein targeting to mitochondrion | 13 | 32 | 2.081e-02 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 13 | 32 | 2.081e-02 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 13 | 32 | 2.081e-02 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 13 | 32 | 2.081e-02 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 122 | 540 | 2.088e-02 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 117 | 515 | 2.088e-02 |
| GO:BP | GO:0045927 | positive regulation of growth | 64 | 256 | 2.088e-02 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 31 | 105 | 2.088e-02 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 18 | 51 | 2.107e-02 |
| GO:BP | GO:0089718 | amino acid import across plasma membrane | 18 | 51 | 2.107e-02 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 18 | 51 | 2.107e-02 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 18 | 51 | 2.107e-02 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 61 | 242 | 2.116e-02 |
| GO:BP | GO:0032608 | interferon-beta production | 20 | 59 | 2.123e-02 |
| GO:BP | GO:0048286 | lung alveolus development | 19 | 55 | 2.123e-02 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 19 | 55 | 2.123e-02 |
| GO:BP | GO:0072170 | metanephric tubule development | 11 | 25 | 2.123e-02 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 20 | 59 | 2.123e-02 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 27 | 88 | 2.125e-02 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 40 | 145 | 2.125e-02 |
| GO:BP | GO:0010232 | vascular transport | 27 | 88 | 2.125e-02 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 185 | 861 | 2.164e-02 |
| GO:BP | GO:0007155 | cell adhesion | 313 | 1530 | 2.230e-02 |
| GO:BP | GO:0031960 | response to corticosteroid | 43 | 159 | 2.240e-02 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 14 | 36 | 2.265e-02 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 14 | 36 | 2.265e-02 |
| GO:BP | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion | 14 | 36 | 2.265e-02 |
| GO:BP | GO:0019395 | fatty acid oxidation | 32 | 110 | 2.275e-02 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 25 | 80 | 2.288e-02 |
| GO:BP | GO:0030500 | regulation of bone mineralization | 25 | 80 | 2.288e-02 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 25 | 80 | 2.288e-02 |
| GO:BP | GO:0002260 | lymphocyte homeostasis | 25 | 80 | 2.288e-02 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 131 | 587 | 2.294e-02 |
| GO:BP | GO:0071774 | response to fibroblast growth factor | 34 | 119 | 2.339e-02 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 24 | 76 | 2.366e-02 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 24 | 76 | 2.366e-02 |
| GO:BP | GO:0035108 | limb morphogenesis | 42 | 155 | 2.375e-02 |
| GO:BP | GO:0035107 | appendage morphogenesis | 42 | 155 | 2.375e-02 |
| GO:BP | GO:0006664 | glycolipid metabolic process | 31 | 106 | 2.389e-02 |
| GO:BP | GO:0055123 | digestive system development | 40 | 146 | 2.390e-02 |
| GO:BP | GO:0048753 | pigment granule organization | 15 | 40 | 2.400e-02 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 15 | 40 | 2.400e-02 |
| GO:BP | GO:0070266 | necroptotic process | 15 | 40 | 2.400e-02 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 23 | 72 | 2.426e-02 |
| GO:BP | GO:0038034 | signal transduction in absence of ligand | 23 | 72 | 2.426e-02 |
| GO:BP | GO:0072207 | metanephric epithelium development | 12 | 29 | 2.445e-02 |
| GO:BP | GO:0035493 | SNARE complex assembly | 10 | 22 | 2.445e-02 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 12 | 29 | 2.445e-02 |
| GO:BP | GO:0002066 | columnar/cuboidal epithelial cell development | 12 | 29 | 2.445e-02 |
| GO:BP | GO:0044342 | type B pancreatic cell proliferation | 12 | 29 | 2.445e-02 |
| GO:BP | GO:0070935 | 3’-UTR-mediated mRNA stabilization | 12 | 29 | 2.445e-02 |
| GO:BP | GO:0060384 | innervation | 12 | 29 | 2.445e-02 |
| GO:BP | GO:0007350 | blastoderm segmentation | 12 | 29 | 2.445e-02 |
| GO:BP | GO:0007611 | learning or memory | 69 | 282 | 2.459e-02 |
| GO:BP | GO:2000106 | regulation of leukocyte apoptotic process | 27 | 89 | 2.462e-02 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 27 | 89 | 2.462e-02 |
| GO:BP | GO:0033077 | T cell differentiation in thymus | 27 | 89 | 2.462e-02 |
| GO:BP | GO:0031100 | animal organ regeneration | 22 | 68 | 2.467e-02 |
| GO:BP | GO:0016125 | sterol metabolic process | 43 | 160 | 2.468e-02 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 30 | 102 | 2.481e-02 |
| GO:BP | GO:0015914 | phospholipid transport | 30 | 102 | 2.481e-02 |
| GO:BP | GO:0007369 | gastrulation | 51 | 197 | 2.489e-02 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 51 | 197 | 2.489e-02 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 41 | 151 | 2.490e-02 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 37 | 133 | 2.502e-02 |
| GO:BP | GO:0046883 | regulation of hormone secretion | 66 | 268 | 2.520e-02 |
| GO:BP | GO:0007292 | female gamete generation | 46 | 174 | 2.526e-02 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 17 | 48 | 2.530e-02 |
| GO:BP | GO:0070228 | regulation of lymphocyte apoptotic process | 20 | 60 | 2.541e-02 |
| GO:BP | GO:0007517 | muscle organ development | 86 | 365 | 2.542e-02 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 78 | 326 | 2.543e-02 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 118 | 524 | 2.588e-02 |
| GO:BP | GO:0033353 | S-adenosylmethionine cycle | 5 | 7 | 2.608e-02 |
| GO:BP | GO:0072197 | ureter morphogenesis | 5 | 7 | 2.608e-02 |
| GO:BP | GO:1905806 | regulation of synapse pruning | 5 | 7 | 2.608e-02 |
| GO:BP | GO:2000638 | regulation of SREBP signaling pathway | 5 | 7 | 2.608e-02 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 42 | 156 | 2.610e-02 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 34 | 120 | 2.612e-02 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 56 | 221 | 2.615e-02 |
| GO:BP | GO:0007507 | heart development | 134 | 605 | 2.635e-02 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 133 | 600 | 2.642e-02 |
| GO:BP | GO:0007498 | mesoderm development | 38 | 138 | 2.645e-02 |
| GO:BP | GO:0042692 | muscle cell differentiation | 94 | 405 | 2.649e-02 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 45 | 170 | 2.678e-02 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 13 | 33 | 2.678e-02 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 31 | 107 | 2.683e-02 |
| GO:BP | GO:1903509 | liposaccharide metabolic process | 31 | 107 | 2.683e-02 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 31 | 107 | 2.683e-02 |
| GO:BP | GO:0060221 | retinal rod cell differentiation | 9 | 19 | 2.726e-02 |
| GO:BP | GO:0097401 | synaptic vesicle lumen acidification | 9 | 19 | 2.726e-02 |
| GO:BP | GO:0060231 | mesenchymal to epithelial transition | 9 | 19 | 2.726e-02 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 87 | 371 | 2.726e-02 |
| GO:BP | GO:1902992 | negative regulation of amyloid precursor protein catabolic process | 9 | 19 | 2.726e-02 |
| GO:BP | GO:1903830 | magnesium ion transmembrane transport | 9 | 19 | 2.726e-02 |
| GO:BP | GO:1902004 | positive regulation of amyloid-beta formation | 9 | 19 | 2.726e-02 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 37 | 134 | 2.798e-02 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 37 | 134 | 2.798e-02 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 59 | 236 | 2.799e-02 |
| GO:BP | GO:0007249 | canonical NF-kappaB signal transduction | 74 | 308 | 2.804e-02 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 27 | 90 | 2.824e-02 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 83 | 352 | 2.839e-02 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 97 | 421 | 2.845e-02 |
| GO:BP | GO:0044782 | cilium organization | 99 | 431 | 2.855e-02 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis | 14 | 37 | 2.855e-02 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 11 | 26 | 2.855e-02 |
| GO:BP | GO:0033002 | muscle cell proliferation | 53 | 208 | 2.877e-02 |
| GO:BP | GO:0030199 | collagen fibril organization | 22 | 69 | 2.898e-02 |
| GO:BP | GO:0003013 | circulatory system process | 133 | 602 | 2.922e-02 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 40 | 148 | 2.931e-02 |
| GO:BP | GO:0043648 | dicarboxylic acid metabolic process | 29 | 99 | 2.962e-02 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 36 | 130 | 2.962e-02 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 21 | 65 | 2.968e-02 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 21 | 65 | 2.968e-02 |
| GO:BP | GO:0002931 | response to ischemia | 21 | 65 | 2.968e-02 |
| GO:BP | GO:0010958 | regulation of amino acid import across plasma membrane | 8 | 16 | 3.023e-02 |
| GO:BP | GO:1903789 | regulation of amino acid transmembrane transport | 8 | 16 | 3.023e-02 |
| GO:BP | GO:0045651 | positive regulation of macrophage differentiation | 8 | 16 | 3.023e-02 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 98 | 427 | 3.025e-02 |
| GO:BP | GO:0070231 | T cell apoptotic process | 20 | 61 | 3.025e-02 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 20 | 61 | 3.025e-02 |
| GO:BP | GO:0009314 | response to radiation | 100 | 437 | 3.034e-02 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 59 | 237 | 3.034e-02 |
| GO:BP | GO:0097306 | cellular response to alcohol | 31 | 108 | 3.043e-02 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 16 | 45 | 3.044e-02 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 16 | 45 | 3.044e-02 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 46 | 176 | 3.046e-02 |
| GO:BP | GO:0007389 | pattern specification process | 108 | 477 | 3.046e-02 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 25 | 82 | 3.048e-02 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 133 | 603 | 3.051e-02 |
| GO:BP | GO:0051607 | defense response to virus | 76 | 319 | 3.053e-02 |
| GO:BP | GO:0060688 | regulation of morphogenesis of a branching structure | 18 | 53 | 3.062e-02 |
| GO:BP | GO:0043114 | regulation of vascular permeability | 17 | 49 | 3.062e-02 |
| GO:BP | GO:0019083 | viral transcription | 18 | 53 | 3.062e-02 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 17 | 49 | 3.062e-02 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 18 | 53 | 3.062e-02 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 17 | 49 | 3.062e-02 |
| GO:BP | GO:0016358 | dendrite development | 60 | 242 | 3.067e-02 |
| GO:BP | GO:1990700 | nucleolar chromatin organization | 6 | 10 | 3.119e-02 |
| GO:BP | GO:0046643 | regulation of gamma-delta T cell activation | 6 | 10 | 3.119e-02 |
| GO:BP | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus | 6 | 10 | 3.119e-02 |
| GO:BP | GO:0036446 | myofibroblast differentiation | 6 | 10 | 3.119e-02 |
| GO:BP | GO:0031214 | biomineral tissue development | 47 | 181 | 3.136e-02 |
| GO:BP | GO:0032688 | negative regulation of interferon-beta production | 7 | 13 | 3.149e-02 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 12 | 30 | 3.149e-02 |
| GO:BP | GO:0031571 | mitotic G1 DNA damage checkpoint signaling | 7 | 13 | 3.149e-02 |
| GO:BP | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway | 7 | 13 | 3.149e-02 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 210 | 1000 | 3.149e-02 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 7 | 13 | 3.149e-02 |
| GO:BP | GO:0040034 | regulation of development, heterochronic | 7 | 13 | 3.149e-02 |
| GO:BP | GO:0019076 | viral release from host cell | 12 | 30 | 3.149e-02 |
| GO:BP | GO:0035891 | exit from host cell | 12 | 30 | 3.149e-02 |
| GO:BP | GO:0006596 | polyamine biosynthetic process | 7 | 13 | 3.149e-02 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 12 | 30 | 3.149e-02 |
| GO:BP | GO:0070914 | UV-damage excision repair | 7 | 13 | 3.149e-02 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 119 | 533 | 3.149e-02 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 54 | 214 | 3.165e-02 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 101 | 443 | 3.171e-02 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 27 | 91 | 3.190e-02 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 40 | 149 | 3.197e-02 |
| GO:BP | GO:0001707 | mesoderm formation | 23 | 74 | 3.243e-02 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 23 | 74 | 3.243e-02 |
| GO:BP | GO:0038061 | non-canonical NF-kappaB signal transduction | 34 | 122 | 3.254e-02 |
| GO:BP | GO:0072234 | metanephric nephron tubule development | 10 | 23 | 3.296e-02 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 46 | 177 | 3.319e-02 |
| GO:BP | GO:0045833 | negative regulation of lipid metabolic process | 29 | 100 | 3.325e-02 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 26 | 87 | 3.347e-02 |
| GO:BP | GO:0098609 | cell-cell adhesion | 205 | 976 | 3.392e-02 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 13 | 34 | 3.392e-02 |
| GO:BP | GO:0001569 | branching involved in blood vessel morphogenesis | 13 | 34 | 3.392e-02 |
| GO:BP | GO:0042476 | odontogenesis | 37 | 136 | 3.452e-02 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 54 | 215 | 3.465e-02 |
| GO:BP | GO:0010565 | regulation of ketone metabolic process | 35 | 127 | 3.465e-02 |
| GO:BP | GO:0045216 | cell-cell junction organization | 54 | 215 | 3.465e-02 |
| GO:BP | GO:0008088 | axo-dendritic transport | 25 | 83 | 3.502e-02 |
| GO:BP | GO:0008543 | fibroblast growth factor receptor signaling pathway | 25 | 83 | 3.502e-02 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 59 | 239 | 3.534e-02 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 64 | 263 | 3.538e-02 |
| GO:BP | GO:0034284 | response to monosaccharide | 55 | 220 | 3.544e-02 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 14 | 38 | 3.548e-02 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 14 | 38 | 3.548e-02 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 14 | 38 | 3.548e-02 |
| GO:BP | GO:0002763 | positive regulation of myeloid leukocyte differentiation | 20 | 62 | 3.548e-02 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 14 | 38 | 3.548e-02 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 20 | 62 | 3.548e-02 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 40 | 150 | 3.550e-02 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 40 | 150 | 3.550e-02 |
| GO:BP | GO:0006885 | regulation of pH | 30 | 105 | 3.585e-02 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 56 | 225 | 3.605e-02 |
| GO:BP | GO:0014009 | glial cell proliferation | 19 | 58 | 3.616e-02 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 46 | 178 | 3.645e-02 |
| GO:BP | GO:0070232 | regulation of T cell apoptotic process | 15 | 42 | 3.645e-02 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 15 | 42 | 3.645e-02 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 34 | 123 | 3.653e-02 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 18 | 54 | 3.661e-02 |
| GO:BP | GO:0060324 | face development | 18 | 54 | 3.661e-02 |
| GO:BP | GO:0010761 | fibroblast migration | 18 | 54 | 3.661e-02 |
| GO:BP | GO:0051301 | cell division | 143 | 658 | 3.674e-02 |
| GO:BP | GO:0045687 | positive regulation of glial cell differentiation | 16 | 46 | 3.681e-02 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 16 | 46 | 3.681e-02 |
| GO:BP | GO:0009957 | epidermal cell fate specification | 4 | 5 | 3.700e-02 |
| GO:BP | GO:0071072 | negative regulation of phospholipid biosynthetic process | 4 | 5 | 3.700e-02 |
| GO:BP | GO:0006113 | fermentation | 4 | 5 | 3.700e-02 |
| GO:BP | GO:0003431 | growth plate cartilage chondrocyte development | 4 | 5 | 3.700e-02 |
| GO:BP | GO:0070345 | negative regulation of fat cell proliferation | 4 | 5 | 3.700e-02 |
| GO:BP | GO:0097500 | receptor localization to non-motile cilium | 4 | 5 | 3.700e-02 |
| GO:BP | GO:0015818 | isoleucine transport | 4 | 5 | 3.700e-02 |
| GO:BP | GO:1905908 | positive regulation of amyloid fibril formation | 4 | 5 | 3.700e-02 |
| GO:BP | GO:1903801 | L-leucine import across plasma membrane | 4 | 5 | 3.700e-02 |
| GO:BP | GO:0007386 | compartment pattern specification | 4 | 5 | 3.700e-02 |
| GO:BP | GO:0051176 | positive regulation of sulfur metabolic process | 4 | 5 | 3.700e-02 |
| GO:BP | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway | 4 | 5 | 3.700e-02 |
| GO:BP | GO:0030325 | adrenal gland development | 11 | 27 | 3.700e-02 |
| GO:BP | GO:0007416 | synapse assembly | 63 | 259 | 3.700e-02 |
| GO:BP | GO:0060740 | prostate gland epithelium morphogenesis | 11 | 27 | 3.700e-02 |
| GO:BP | GO:0016188 | synaptic vesicle maturation | 11 | 27 | 3.700e-02 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 11 | 27 | 3.700e-02 |
| GO:BP | GO:0009408 | response to heat | 29 | 101 | 3.728e-02 |
| GO:BP | GO:0022411 | cellular component disassembly | 101 | 446 | 3.728e-02 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 70 | 293 | 3.733e-02 |
| GO:BP | GO:0060716 | labyrinthine layer blood vessel development | 9 | 20 | 3.756e-02 |
| GO:BP | GO:0007351 | tripartite regional subdivision | 9 | 20 | 3.756e-02 |
| GO:BP | GO:0006684 | sphingomyelin metabolic process | 9 | 20 | 3.756e-02 |
| GO:BP | GO:0048339 | paraxial mesoderm development | 9 | 20 | 3.756e-02 |
| GO:BP | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway | 9 | 20 | 3.756e-02 |
| GO:BP | GO:0008595 | anterior/posterior axis specification, embryo | 9 | 20 | 3.756e-02 |
| GO:BP | GO:0032332 | positive regulation of chondrocyte differentiation | 9 | 20 | 3.756e-02 |
| GO:BP | GO:0051384 | response to glucocorticoid | 37 | 137 | 3.763e-02 |
| GO:BP | GO:0071396 | cellular response to lipid | 134 | 613 | 3.781e-02 |
| GO:BP | GO:0001824 | blastocyst development | 33 | 119 | 3.804e-02 |
| GO:BP | GO:0009743 | response to carbohydrate | 60 | 245 | 3.804e-02 |
| GO:BP | GO:0060041 | retina development in camera-type eye | 45 | 174 | 3.805e-02 |
| GO:BP | GO:0009607 | response to biotic stimulus | 329 | 1634 | 3.860e-02 |
| GO:BP | GO:0032835 | glomerulus development | 22 | 71 | 3.860e-02 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 87 | 377 | 3.862e-02 |
| GO:BP | GO:0050808 | synapse organization | 124 | 563 | 3.954e-02 |
| GO:BP | GO:0051952 | regulation of amine transport | 25 | 84 | 3.960e-02 |
| GO:BP | GO:0006094 | gluconeogenesis | 25 | 84 | 3.960e-02 |
| GO:BP | GO:0071347 | cellular response to interleukin-1 | 25 | 84 | 3.960e-02 |
| GO:BP | GO:0019082 | viral protein processing | 12 | 31 | 4.021e-02 |
| GO:BP | GO:0006506 | GPI anchor biosynthetic process | 12 | 31 | 4.021e-02 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 34 | 124 | 4.039e-02 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 110 | 493 | 4.135e-02 |
| GO:BP | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway | 20 | 63 | 4.142e-02 |
| GO:BP | GO:0045580 | regulation of T cell differentiation | 48 | 189 | 4.185e-02 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 123 | 559 | 4.189e-02 |
| GO:BP | GO:0042060 | wound healing | 98 | 433 | 4.191e-02 |
| GO:BP | GO:0170033 | L-amino acid metabolic process | 45 | 175 | 4.210e-02 |
| GO:BP | GO:0061157 | mRNA destabilization | 29 | 102 | 4.245e-02 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 13 | 35 | 4.252e-02 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 13 | 35 | 4.252e-02 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 19 | 59 | 4.252e-02 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 35 | 129 | 4.262e-02 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 35 | 129 | 4.262e-02 |
| GO:BP | GO:0055091 | phospholipid homeostasis | 8 | 17 | 4.321e-02 |
| GO:BP | GO:2000696 | regulation of epithelial cell differentiation involved in kidney development | 8 | 17 | 4.321e-02 |
| GO:BP | GO:0045778 | positive regulation of ossification | 18 | 55 | 4.348e-02 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 14 | 39 | 4.385e-02 |
| GO:BP | GO:0032438 | melanosome organization | 14 | 39 | 4.385e-02 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 54 | 218 | 4.388e-02 |
| GO:BP | GO:0030850 | prostate gland development | 17 | 51 | 4.401e-02 |
| GO:BP | GO:0019058 | viral life cycle | 73 | 310 | 4.401e-02 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 17 | 51 | 4.401e-02 |
| GO:BP | GO:0097066 | response to thyroid hormone | 10 | 24 | 4.401e-02 |
| GO:BP | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 17 | 51 | 4.401e-02 |
| GO:BP | GO:0098719 | sodium ion import across plasma membrane | 10 | 24 | 4.401e-02 |
| GO:BP | GO:0045088 | regulation of innate immune response | 101 | 449 | 4.411e-02 |
| GO:BP | GO:0006023 | aminoglycan biosynthetic process | 15 | 43 | 4.419e-02 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 15 | 43 | 4.419e-02 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 15 | 43 | 4.419e-02 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 15 | 43 | 4.419e-02 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 16 | 47 | 4.424e-02 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 44 | 171 | 4.456e-02 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 22 | 72 | 4.505e-02 |
| GO:BP | GO:0032606 | type I interferon production | 34 | 125 | 4.505e-02 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 34 | 125 | 4.505e-02 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 30 | 107 | 4.509e-02 |
| GO:BP | GO:0034440 | lipid oxidation | 32 | 116 | 4.525e-02 |
| GO:BP | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0019661 | glucose catabolic process to lactate via pyruvate | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0002051 | osteoblast fate commitment | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0035621 | ER to Golgi ceramide transport | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0033693 | neurofilament bundle assembly | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0071403 | cellular response to high density lipoprotein particle stimulus | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0035494 | SNARE complex disassembly | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 45 | 176 | 4.548e-02 |
| GO:BP | GO:0019660 | glycolytic fermentation | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0140961 | cellular detoxification of metal ion | 3 | 3 | 4.548e-02 |
| GO:BP | GO:1904023 | regulation of glucose catabolic process to lactate via pyruvate | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0098849 | cellular detoxification of cadmium ion | 3 | 3 | 4.548e-02 |
| GO:BP | GO:1905532 | regulation of L-leucine import across plasma membrane | 3 | 3 | 4.548e-02 |
| GO:BP | GO:1905952 | regulation of lipid localization | 42 | 162 | 4.548e-02 |
| GO:BP | GO:1990637 | response to prolactin | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0099185 | postsynaptic intermediate filament cytoskeleton organization | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0042335 | cuticle development | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0007113 | endomitotic cell cycle | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0072034 | renal vesicle induction | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0019659 | glucose catabolic process to lactate | 3 | 3 | 4.548e-02 |
| GO:BP | GO:2000367 | regulation of acrosomal vesicle exocytosis | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0009826 | unidimensional cell growth | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0097355 | protein localization to heterochromatin | 3 | 3 | 4.548e-02 |
| GO:BP | GO:0042593 | glucose homeostasis | 63 | 262 | 4.548e-02 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 97 | 430 | 4.610e-02 |
| GO:BP | GO:0009994 | oocyte differentiation | 21 | 68 | 4.611e-02 |
| GO:BP | GO:0060021 | roof of mouth development | 27 | 94 | 4.627e-02 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 29 | 103 | 4.718e-02 |
| GO:BP | GO:0072283 | metanephric renal vesicle morphogenesis | 7 | 14 | 4.723e-02 |
| GO:BP | GO:0035970 | peptidyl-threonine dephosphorylation | 7 | 14 | 4.723e-02 |
| GO:BP | GO:0090153 | regulation of sphingolipid biosynthetic process | 7 | 14 | 4.723e-02 |
| GO:BP | GO:0098814 | spontaneous synaptic transmission | 7 | 14 | 4.723e-02 |
| GO:BP | GO:0036151 | phosphatidylcholine acyl-chain remodeling | 7 | 14 | 4.723e-02 |
| GO:BP | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation | 7 | 14 | 4.723e-02 |
| GO:BP | GO:0015803 | branched-chain amino acid transport | 7 | 14 | 4.723e-02 |
| GO:BP | GO:0003416 | endochondral bone growth | 11 | 28 | 4.761e-02 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 11 | 28 | 4.761e-02 |
| GO:BP | GO:0033059 | cellular pigmentation | 20 | 64 | 4.764e-02 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 38 | 144 | 4.764e-02 |
| GO:BP | GO:0048599 | oocyte development | 20 | 64 | 4.764e-02 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 38 | 144 | 4.764e-02 |
| GO:BP | GO:0043604 | amide biosynthetic process | 44 | 172 | 4.809e-02 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 102 | 456 | 4.831e-02 |
| GO:BP | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis | 5 | 8 | 4.839e-02 |
| GO:BP | GO:1902746 | regulation of lens fiber cell differentiation | 5 | 8 | 4.839e-02 |
| GO:BP | GO:0006538 | glutamate catabolic process | 5 | 8 | 4.839e-02 |
| GO:BP | GO:0045647 | negative regulation of erythrocyte differentiation | 5 | 8 | 4.839e-02 |
| GO:BP | GO:0032344 | regulation of aldosterone metabolic process | 5 | 8 | 4.839e-02 |
| GO:BP | GO:0072033 | renal vesicle formation | 5 | 8 | 4.839e-02 |
| GO:BP | GO:0032347 | regulation of aldosterone biosynthetic process | 5 | 8 | 4.839e-02 |
| GO:BP | GO:0003223 | ventricular compact myocardium morphogenesis | 5 | 8 | 4.839e-02 |
| GO:BP | GO:1904026 | regulation of collagen fibril organization | 5 | 8 | 4.839e-02 |
| GO:BP | GO:0006290 | pyrimidine dimer repair | 5 | 8 | 4.839e-02 |
| GO:BP | GO:0007422 | peripheral nervous system development | 26 | 90 | 4.839e-02 |
| GO:BP | GO:0007623 | circadian rhythm | 52 | 210 | 4.839e-02 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 63 | 263 | 4.855e-02 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 23 | 77 | 4.881e-02 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 19 | 60 | 4.883e-02 |
| GO:BP | GO:0002448 | mast cell mediated immunity | 19 | 60 | 4.883e-02 |
| GO:BP | GO:0071168 | protein localization to chromatin | 19 | 60 | 4.883e-02 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 34 | 126 | 4.905e-02 |
| GO:BP | GO:0002444 | myeloid leukocyte mediated immunity | 32 | 117 | 4.955e-02 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 123 | 563 | 4.955e-02 |
| GO:BP | GO:0036075 | replacement ossification | 12 | 32 | 4.960e-02 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 12 | 32 | 4.960e-02 |
| GO:BP | GO:0001958 | endochondral ossification | 12 | 32 | 4.960e-02 |
| GO:BP | GO:1900038 | negative regulation of cellular response to hypoxia | 6 | 11 | 4.960e-02 |
| GO:BP | GO:0180046 | GPI anchored protein biosynthesis | 12 | 32 | 4.960e-02 |
| GO:BP | GO:0045684 | positive regulation of epidermis development | 12 | 32 | 4.960e-02 |
| GO:BP | GO:0060767 | epithelial cell proliferation involved in prostate gland development | 6 | 11 | 4.960e-02 |
| GO:BP | GO:0045843 | negative regulation of striated muscle tissue development | 6 | 11 | 4.960e-02 |
| GO:BP | GO:0006505 | GPI anchor metabolic process | 12 | 32 | 4.960e-02 |
| GO:BP | GO:2000644 | regulation of receptor catabolic process | 6 | 11 | 4.960e-02 |
| GO:BP | GO:0071108 | protein K48-linked deubiquitination | 12 | 32 | 4.960e-02 |
| GO:BP | GO:0006954 | inflammatory response | 181 | 862 | 4.960e-02 |
| GO:BP | GO:0060397 | growth hormone receptor signaling pathway via JAK-STAT | 6 | 11 | 4.960e-02 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 12 | 32 | 4.960e-02 |
| GO:BP | GO:0031943 | regulation of glucocorticoid metabolic process | 6 | 11 | 4.960e-02 |
| GO:BP | GO:0003337 | mesenchymal to epithelial transition involved in metanephros morphogenesis | 6 | 11 | 4.960e-02 |
| GO:BP | GO:0010807 | regulation of synaptic vesicle priming | 6 | 11 | 4.960e-02 |
| GO:BP | GO:1990928 | response to amino acid starvation | 18 | 56 | 4.976e-02 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 37 | 140 | 4.986e-02 |
| KEGG | KEGG:04140 | Autophagy - animal | 65 | 165 | 3.285e-05 |
| KEGG | KEGG:05169 | Epstein-Barr virus infection | 72 | 198 | 1.376e-04 |
| KEGG | KEGG:05161 | Hepatitis B | 61 | 162 | 1.678e-04 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 39 | 97 | 1.723e-03 |
| KEGG | KEGG:05210 | Colorectal cancer | 35 | 86 | 2.681e-03 |
| KEGG | KEGG:04218 | Cellular senescence | 54 | 155 | 4.155e-03 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 40 | 107 | 4.155e-03 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 50 | 143 | 4.155e-03 |
| KEGG | KEGG:05131 | Shigellosis | 78 | 246 | 4.155e-03 |
| KEGG | KEGG:05212 | Pancreatic cancer | 31 | 76 | 4.155e-03 |
| KEGG | KEGG:04115 | p53 signaling pathway | 30 | 74 | 4.653e-03 |
| KEGG | KEGG:01522 | Endocrine resistance | 36 | 95 | 5.025e-03 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 64 | 201 | 8.161e-03 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 28 | 70 | 8.161e-03 |
| KEGG | KEGG:04142 | Lysosome | 45 | 131 | 9.339e-03 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 96 | 330 | 1.101e-02 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 28 | 72 | 1.116e-02 |
| KEGG | KEGG:05226 | Gastric cancer | 49 | 148 | 1.160e-02 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 28 | 73 | 1.253e-02 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 88 | 301 | 1.253e-02 |
| KEGG | KEGG:05200 | Pathways in cancer | 143 | 527 | 1.262e-02 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 44 | 131 | 1.262e-02 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 50 | 155 | 1.555e-02 |
| KEGG | KEGG:05167 | Kaposi sarcoma-associated herpesvirus infection | 60 | 194 | 1.633e-02 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 66 | 218 | 1.651e-02 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 46 | 141 | 1.651e-02 |
| KEGG | KEGG:04750 | Inflammatory mediator regulation of TRP channels | 34 | 98 | 2.094e-02 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 30 | 84 | 2.220e-02 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 40 | 121 | 2.274e-02 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 25 | 67 | 2.439e-02 |
| KEGG | KEGG:05162 | Measles | 44 | 138 | 2.664e-02 |
| KEGG | KEGG:04934 | Cushing syndrome | 48 | 153 | 2.664e-02 |
| KEGG | KEGG:04710 | Circadian rhythm | 15 | 34 | 2.664e-02 |
| KEGG | KEGG:00600 | Sphingolipid metabolism | 21 | 54 | 2.664e-02 |
| KEGG | KEGG:05215 | Prostate cancer | 33 | 97 | 2.702e-02 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 39 | 120 | 2.842e-02 |
| KEGG | KEGG:05213 | Endometrial cancer | 22 | 58 | 2.842e-02 |
| KEGG | KEGG:05224 | Breast cancer | 46 | 147 | 2.885e-02 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 19 | 48 | 2.894e-02 |
| KEGG | KEGG:04137 | Mitophagy - animal | 34 | 103 | 3.573e-02 |
| KEGG | KEGG:05217 | Basal cell carcinoma | 23 | 63 | 3.585e-02 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 48 | 157 | 3.585e-02 |
| KEGG | KEGG:04360 | Axon guidance | 54 | 181 | 3.694e-02 |
| KEGG | KEGG:04330 | Notch signaling pathway | 22 | 60 | 3.843e-02 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 50 | 166 | 3.847e-02 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 31 | 93 | 3.847e-02 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 19 | 50 | 4.078e-02 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 27 | 79 | 4.290e-02 |
| KEGG | KEGG:04668 | TNF signaling pathway | 36 | 113 | 4.355e-02 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 10 | 21 | 4.750e-02 |
#write.csv(table_DOX24R_up_genes, "output/table_DOX24R_upreg_genes.csv")
#GO:BP
table_DOX24Tshare_up_genes_GOBP <- table_DOX24Tshare_up_genes %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
#saveRDS(table_motif1_GOBP_d, "data/table_motif1_GOBP_d.RDS")
table_DOX24Tshare_up_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Shared Up DEGs Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))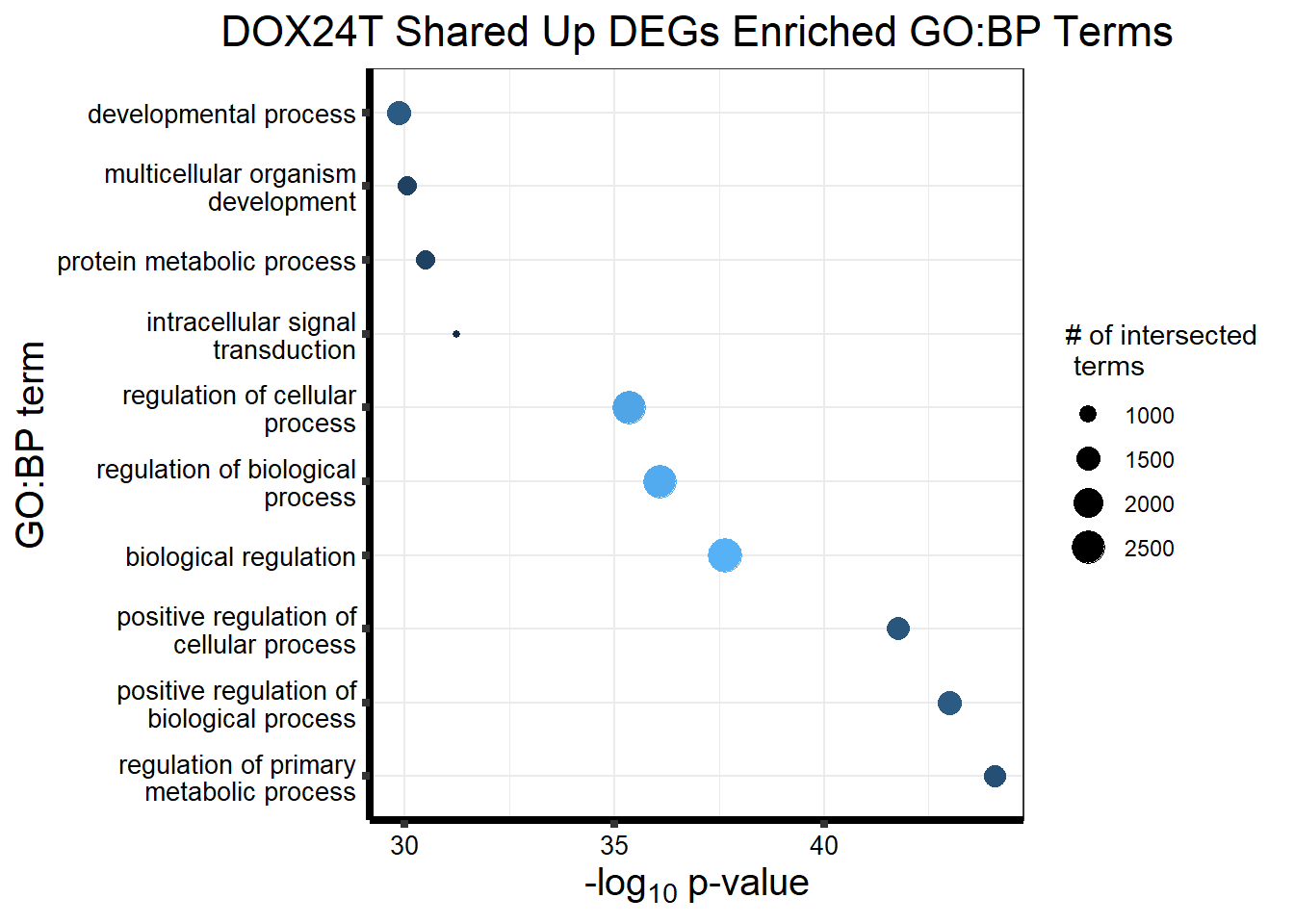
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_DOX24Tshare_up_genes_KEGG <- table_DOX24Tshare_up_genes %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24Tshare_up_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Shared Up DEGs Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))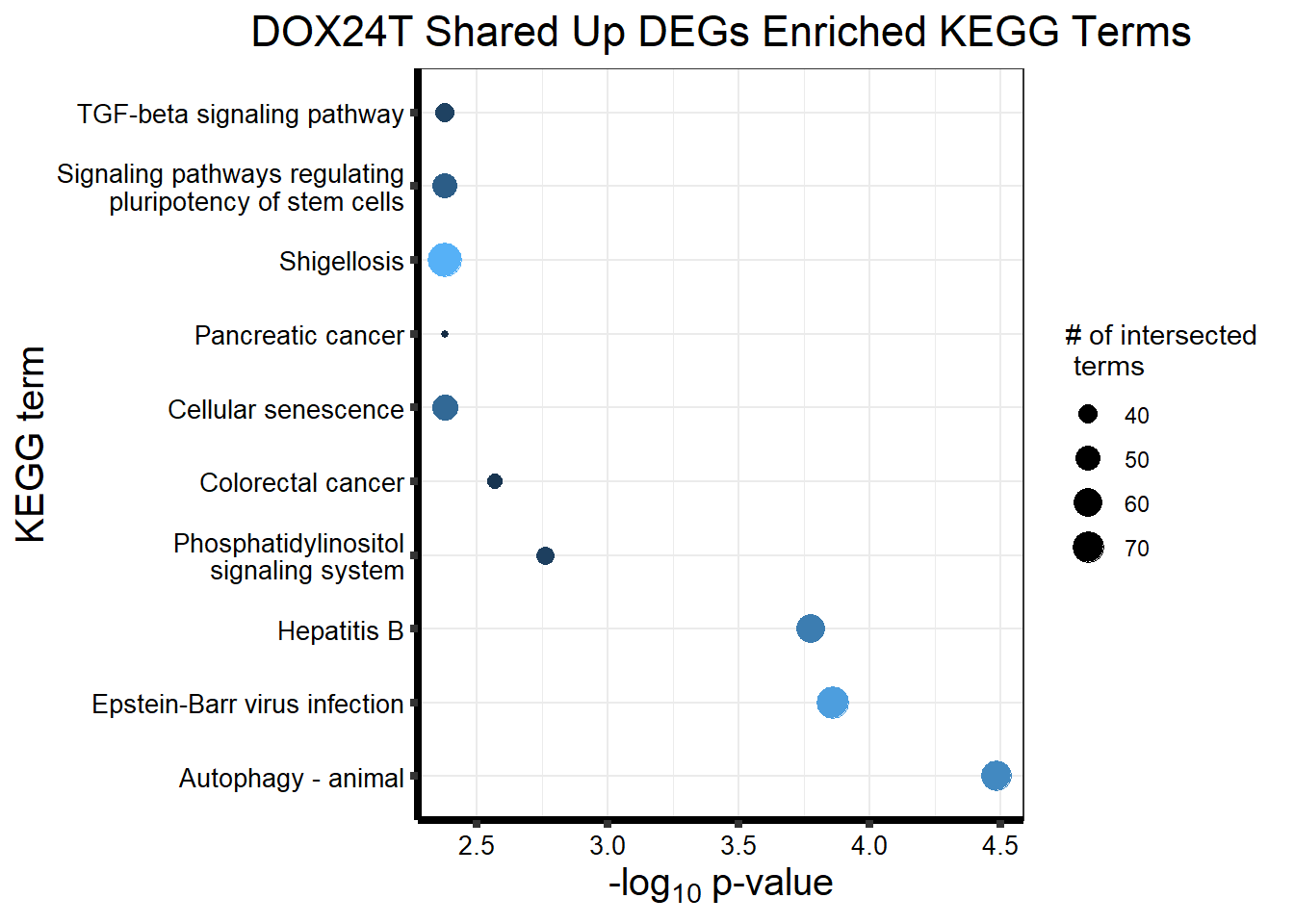
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####DOX24R Downregulated DEGs GO KEGG#####
D24Tshare_DEGs_down_mat <- as.matrix(DOX24Tshare_DEGs_GO_down)
DOX_24Tshare_down_dxr_gene <- gost(query = D24Tshare_DEGs_down_mat,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
DOX_24Tshare_down_gost_genes <- gostplot(DOX_24Tshare_down_dxr_gene, capped = FALSE, interactive = TRUE)
DOX_24Tshare_down_gost_genestable_DOX24Tshare_down_genes <- DOX_24Tshare_down_dxr_gene$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX24Tshare_down_genes %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006996 | organelle organization | 1110 | 3594 | 7.044e-70 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 1421 | 5390 | 3.141e-44 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 1082 | 3990 | 2.874e-36 |
| GO:BP | GO:0033043 | regulation of organelle organization | 404 | 1148 | 9.181e-35 |
| GO:BP | GO:0019538 | protein metabolic process | 1226 | 4721 | 6.559e-33 |
| GO:BP | GO:0043412 | macromolecule modification | 847 | 3030 | 9.423e-32 |
| GO:BP | GO:0007049 | cell cycle | 522 | 1663 | 8.045e-31 |
| GO:BP | GO:0000278 | mitotic cell cycle | 325 | 892 | 8.045e-31 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 104 | 168 | 1.292e-30 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 284 | 745 | 1.456e-30 |
| GO:BP | GO:0051641 | cellular localization | 982 | 3661 | 1.755e-30 |
| GO:BP | GO:0022402 | cell cycle process | 423 | 1280 | 7.396e-30 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 984 | 3687 | 1.039e-29 |
| GO:BP | GO:0006351 | DNA-templated transcription | 948 | 3549 | 1.616e-28 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 694 | 2433 | 4.282e-28 |
| GO:BP | GO:0051649 | establishment of localization in cell | 594 | 2010 | 7.886e-28 |
| GO:BP | GO:0032543 | mitochondrial translation | 84 | 130 | 1.298e-26 |
| GO:BP | GO:0070925 | organelle assembly | 352 | 1046 | 3.555e-26 |
| GO:BP | GO:0051726 | regulation of cell cycle | 361 | 1087 | 1.084e-25 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 905 | 3428 | 3.848e-25 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 900 | 3409 | 5.437e-25 |
| GO:BP | GO:0065007 | biological regulation | 2779 | 12743 | 6.710e-25 |
| GO:BP | GO:0050789 | regulation of biological process | 2700 | 12336 | 1.833e-24 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1431 | 5920 | 3.987e-24 |
| GO:BP | GO:0006399 | tRNA metabolic process | 110 | 210 | 4.400e-24 |
| GO:BP | GO:0046907 | intracellular transport | 428 | 1381 | 7.648e-24 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 259 | 720 | 1.351e-23 |
| GO:BP | GO:0006281 | DNA repair | 232 | 621 | 1.491e-23 |
| GO:BP | GO:0006974 | DNA damage response | 307 | 908 | 4.118e-23 |
| GO:BP | GO:0033554 | cellular response to stress | 532 | 1830 | 6.259e-23 |
| GO:BP | GO:0033036 | macromolecule localization | 848 | 3228 | 1.217e-22 |
| GO:BP | GO:0006259 | DNA metabolic process | 330 | 1005 | 1.519e-22 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1491 | 6264 | 2.280e-22 |
| GO:BP | GO:0050794 | regulation of cellular process | 2612 | 11946 | 2.808e-22 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 859 | 3288 | 3.054e-22 |
| GO:BP | GO:0036211 | protein modification process | 758 | 2846 | 1.330e-21 |
| GO:BP | GO:0051276 | chromosome organization | 214 | 574 | 1.373e-21 |
| GO:BP | GO:0007010 | cytoskeleton organization | 455 | 1529 | 1.528e-21 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 743 | 2782 | 1.743e-21 |
| GO:BP | GO:0140014 | mitotic nuclear division | 128 | 282 | 5.779e-21 |
| GO:BP | GO:0009056 | catabolic process | 708 | 2639 | 7.216e-21 |
| GO:BP | GO:0008104 | protein localization | 737 | 2770 | 8.093e-21 |
| GO:BP | GO:0000819 | sister chromatid segregation | 111 | 234 | 5.847e-20 |
| GO:BP | GO:0006412 | translation | 250 | 727 | 8.368e-20 |
| GO:BP | GO:0051179 | localization | 1328 | 5555 | 8.368e-20 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 913 | 3597 | 9.276e-20 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 566 | 2036 | 9.379e-20 |
| GO:BP | GO:0019222 | regulation of metabolic process | 1624 | 7035 | 1.083e-18 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 111 | 242 | 1.440e-18 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 231 | 670 | 2.119e-18 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 1510 | 6492 | 3.485e-18 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 94 | 193 | 7.917e-18 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 413 | 1417 | 1.132e-17 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 705 | 2711 | 3.933e-17 |
| GO:BP | GO:0007059 | chromosome segregation | 161 | 427 | 1.209e-16 |
| GO:BP | GO:0006302 | double-strand break repair | 130 | 319 | 1.767e-16 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 161 | 430 | 2.556e-16 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 503 | 1835 | 4.817e-16 |
| GO:BP | GO:0044238 | primary metabolic process | 2643 | 12342 | 5.402e-16 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 130 | 323 | 5.667e-16 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 175 | 492 | 3.036e-15 |
| GO:BP | GO:0048285 | organelle fission | 177 | 500 | 3.356e-15 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 665 | 2578 | 3.489e-15 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 1869 | 8393 | 4.898e-15 |
| GO:BP | GO:0051301 | cell division | 217 | 658 | 8.236e-15 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 213 | 643 | 9.039e-15 |
| GO:BP | GO:0008033 | tRNA processing | 71 | 139 | 9.039e-15 |
| GO:BP | GO:0007017 | microtubule-based process | 301 | 999 | 1.189e-14 |
| GO:BP | GO:0035556 | intracellular signal transduction | 746 | 2965 | 1.408e-14 |
| GO:BP | GO:0015031 | protein transport | 406 | 1444 | 1.431e-14 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 248 | 786 | 1.857e-14 |
| GO:BP | GO:0009894 | regulation of catabolic process | 310 | 1040 | 1.945e-14 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 208 | 628 | 1.981e-14 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 210 | 638 | 2.966e-14 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 462 | 1699 | 5.133e-14 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 461 | 1697 | 6.597e-14 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 184 | 543 | 9.242e-14 |
| GO:BP | GO:0000280 | nuclear division | 160 | 452 | 9.522e-14 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 79 | 169 | 9.742e-14 |
| GO:BP | GO:0044770 | cell cycle phase transition | 181 | 532 | 9.919e-14 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 153 | 428 | 1.505e-13 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 114 | 289 | 2.705e-13 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 152 | 427 | 2.761e-13 |
| GO:BP | GO:0006914 | autophagy | 196 | 597 | 3.558e-13 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 196 | 597 | 3.558e-13 |
| GO:BP | GO:0009987 | cellular process | 4016 | 20247 | 3.952e-13 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 148 | 414 | 4.105e-13 |
| GO:BP | GO:0009451 | RNA modification | 79 | 174 | 6.642e-13 |
| GO:BP | GO:0030163 | protein catabolic process | 301 | 1030 | 7.821e-13 |
| GO:BP | GO:0016072 | rRNA metabolic process | 106 | 266 | 1.061e-12 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 123 | 327 | 1.262e-12 |
| GO:BP | GO:0071705 | nitrogen compound transport | 505 | 1923 | 1.364e-12 |
| GO:BP | GO:0007005 | mitochondrion organization | 152 | 435 | 1.489e-12 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 169 | 500 | 1.521e-12 |
| GO:BP | GO:0007051 | spindle organization | 88 | 206 | 1.580e-12 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 129 | 352 | 2.667e-12 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 81 | 185 | 3.326e-12 |
| GO:BP | GO:0043687 | post-translational protein modification | 297 | 1027 | 4.604e-12 |
| GO:BP | GO:0042254 | ribosome biogenesis | 120 | 323 | 6.808e-12 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 485 | 1855 | 1.017e-11 |
| GO:BP | GO:0006400 | tRNA modification | 51 | 95 | 1.096e-11 |
| GO:BP | GO:0007052 | mitotic spindle organization | 64 | 134 | 1.183e-11 |
| GO:BP | GO:0051640 | organelle localization | 196 | 620 | 1.812e-11 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 64 | 136 | 2.743e-11 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 604 | 2407 | 2.826e-11 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 604 | 2410 | 3.587e-11 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 643 | 2593 | 4.638e-11 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 176 | 546 | 4.638e-11 |
| GO:BP | GO:0016043 | cellular component organization | 1792 | 8184 | 5.521e-11 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 162 | 492 | 5.521e-11 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 251 | 853 | 5.521e-11 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 132 | 377 | 5.757e-11 |
| GO:BP | GO:0061024 | membrane organization | 243 | 821 | 6.555e-11 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 288 | 1011 | 6.989e-11 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 279 | 975 | 9.220e-11 |
| GO:BP | GO:0051234 | establishment of localization | 1132 | 4928 | 9.845e-11 |
| GO:BP | GO:0006810 | transport | 1024 | 4407 | 1.044e-10 |
| GO:BP | GO:0030029 | actin filament-based process | 243 | 825 | 1.102e-10 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 96 | 248 | 1.120e-10 |
| GO:BP | GO:0045184 | establishment of protein localization | 497 | 1937 | 1.136e-10 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 167 | 517 | 1.355e-10 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 89 | 225 | 1.751e-10 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 167 | 519 | 1.908e-10 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 660 | 2696 | 2.582e-10 |
| GO:BP | GO:0000725 | recombinational repair | 79 | 193 | 3.787e-10 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 77 | 187 | 4.898e-10 |
| GO:BP | GO:0032259 | methylation | 91 | 236 | 5.168e-10 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 49 | 97 | 5.179e-10 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 77 | 188 | 6.573e-10 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 60 | 132 | 7.581e-10 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 72 | 172 | 8.942e-10 |
| GO:BP | GO:0007507 | heart development | 186 | 605 | 9.038e-10 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 62 | 139 | 9.179e-10 |
| GO:BP | GO:0022411 | cellular component disassembly | 146 | 446 | 1.038e-09 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 209 | 701 | 1.125e-09 |
| GO:BP | GO:0006282 | regulation of DNA repair | 87 | 225 | 1.163e-09 |
| GO:BP | GO:0006364 | rRNA processing | 87 | 225 | 1.163e-09 |
| GO:BP | GO:0043414 | macromolecule methylation | 58 | 127 | 1.241e-09 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 47 | 93 | 1.244e-09 |
| GO:BP | GO:0016236 | macroautophagy | 124 | 362 | 1.244e-09 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1268 | 5644 | 1.452e-09 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 48 | 97 | 1.975e-09 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 110 | 312 | 2.318e-09 |
| GO:BP | GO:0006310 | DNA recombination | 119 | 347 | 2.833e-09 |
| GO:BP | GO:0016073 | snRNA metabolic process | 34 | 58 | 3.758e-09 |
| GO:BP | GO:0006325 | chromatin organization | 250 | 884 | 4.247e-09 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 1297 | 5809 | 4.425e-09 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 47 | 96 | 4.887e-09 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 109 | 312 | 5.122e-09 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 48 | 100 | 7.326e-09 |
| GO:BP | GO:0010468 | regulation of gene expression | 1239 | 5536 | 8.159e-09 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 192 | 646 | 9.148e-09 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 65 | 156 | 9.574e-09 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 50 | 107 | 9.829e-09 |
| GO:BP | GO:0016567 | protein ubiquitination | 221 | 768 | 9.829e-09 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 198 | 673 | 1.191e-08 |
| GO:BP | GO:0006886 | intracellular protein transport | 201 | 686 | 1.265e-08 |
| GO:BP | GO:0000723 | telomere maintenance | 67 | 164 | 1.356e-08 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 54 | 121 | 1.555e-08 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 102 | 291 | 1.555e-08 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 164 | 535 | 1.577e-08 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 270 | 983 | 1.752e-08 |
| GO:BP | GO:0006403 | RNA localization | 77 | 200 | 1.793e-08 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 51 | 112 | 1.951e-08 |
| GO:BP | GO:0016197 | endosomal transport | 101 | 290 | 2.769e-08 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 52 | 117 | 3.821e-08 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 484 | 1953 | 4.271e-08 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 152 | 493 | 4.374e-08 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 178 | 600 | 4.568e-08 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 48 | 105 | 5.266e-08 |
| GO:BP | GO:0051656 | establishment of organelle localization | 151 | 491 | 5.948e-08 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 85 | 234 | 6.166e-08 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 85 | 234 | 6.166e-08 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 91 | 257 | 7.270e-08 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 36 | 69 | 7.377e-08 |
| GO:BP | GO:0009966 | regulation of signal transduction | 713 | 3034 | 7.584e-08 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 40 | 81 | 7.584e-08 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 51 | 116 | 8.266e-08 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 31 | 55 | 8.301e-08 |
| GO:BP | GO:0051310 | metaphase chromosome alignment | 47 | 104 | 1.186e-07 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 54 | 127 | 1.189e-07 |
| GO:BP | GO:0010646 | regulation of cell communication | 806 | 3486 | 1.201e-07 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 120 | 372 | 1.341e-07 |
| GO:BP | GO:0030031 | cell projection assembly | 179 | 613 | 1.428e-07 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 119 | 369 | 1.573e-07 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 32 | 59 | 1.609e-07 |
| GO:BP | GO:0016180 | snRNA processing | 23 | 35 | 1.662e-07 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 47 | 105 | 1.662e-07 |
| GO:BP | GO:0023051 | regulation of signaling | 803 | 3478 | 1.662e-07 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 401 | 1592 | 1.674e-07 |
| GO:BP | GO:0033365 | protein localization to organelle | 316 | 1209 | 1.676e-07 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 40 | 83 | 1.738e-07 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 39 | 80 | 1.788e-07 |
| GO:BP | GO:0051783 | regulation of nuclear division | 60 | 149 | 1.837e-07 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 36 | 71 | 1.837e-07 |
| GO:BP | GO:0044782 | cilium organization | 134 | 431 | 2.122e-07 |
| GO:BP | GO:0060271 | cilium assembly | 127 | 403 | 2.158e-07 |
| GO:BP | GO:0051225 | spindle assembly | 56 | 136 | 2.282e-07 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 152 | 505 | 2.322e-07 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 58 | 143 | 2.364e-07 |
| GO:BP | GO:0050000 | chromosome localization | 53 | 126 | 2.377e-07 |
| GO:BP | GO:0006417 | regulation of translation | 121 | 380 | 2.473e-07 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 285 | 1077 | 2.708e-07 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 205 | 729 | 2.784e-07 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 291 | 1105 | 3.064e-07 |
| GO:BP | GO:0032790 | ribosome disassembly | 25 | 41 | 3.064e-07 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 60 | 151 | 3.154e-07 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 55 | 134 | 3.411e-07 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 50 | 118 | 4.610e-07 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 120 | 380 | 4.710e-07 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 116 | 365 | 5.523e-07 |
| GO:BP | GO:0055006 | cardiac cell development | 40 | 86 | 5.700e-07 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 63 | 164 | 6.113e-07 |
| GO:BP | GO:0010256 | endomembrane system organization | 176 | 613 | 6.478e-07 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 37 | 77 | 6.611e-07 |
| GO:BP | GO:1903008 | organelle disassembly | 35 | 71 | 6.994e-07 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 51 | 123 | 7.618e-07 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 55 | 137 | 8.248e-07 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 118 | 376 | 8.853e-07 |
| GO:BP | GO:0051304 | chromosome separation | 39 | 84 | 8.871e-07 |
| GO:BP | GO:0009303 | rRNA transcription | 24 | 40 | 9.021e-07 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 382 | 1529 | 9.523e-07 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 82 | 236 | 9.767e-07 |
| GO:BP | GO:0030010 | establishment of cell polarity | 61 | 159 | 1.040e-06 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 273 | 1039 | 1.059e-06 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 131 | 430 | 1.067e-06 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 48 | 114 | 1.067e-06 |
| GO:BP | GO:0060341 | regulation of cellular localization | 267 | 1013 | 1.107e-06 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 163 | 563 | 1.122e-06 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 49 | 118 | 1.288e-06 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 111 | 351 | 1.394e-06 |
| GO:BP | GO:0006402 | mRNA catabolic process | 88 | 261 | 1.458e-06 |
| GO:BP | GO:0007399 | nervous system development | 611 | 2604 | 1.501e-06 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 36 | 76 | 1.515e-06 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 71 | 197 | 1.515e-06 |
| GO:BP | GO:0043038 | amino acid activation | 26 | 47 | 2.243e-06 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 64 | 173 | 2.269e-06 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 81 | 237 | 2.535e-06 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 32 | 65 | 2.700e-06 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 32 | 65 | 2.700e-06 |
| GO:BP | GO:0030030 | cell projection organization | 401 | 1631 | 2.702e-06 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 31 | 62 | 2.717e-06 |
| GO:BP | GO:0051236 | establishment of RNA localization | 61 | 163 | 2.778e-06 |
| GO:BP | GO:0031503 | protein-containing complex localization | 75 | 215 | 2.915e-06 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 37 | 81 | 3.098e-06 |
| GO:BP | GO:0072344 | rescue of stalled ribosome | 21 | 34 | 3.098e-06 |
| GO:BP | GO:0061337 | cardiac conduction | 41 | 94 | 3.100e-06 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 55 | 142 | 3.105e-06 |
| GO:BP | GO:0055002 | striated muscle cell development | 63 | 171 | 3.263e-06 |
| GO:BP | GO:0007030 | Golgi organization | 58 | 153 | 3.342e-06 |
| GO:BP | GO:0031032 | actomyosin structure organization | 74 | 212 | 3.355e-06 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 25 | 45 | 3.388e-06 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 54 | 139 | 3.542e-06 |
| GO:BP | GO:0003007 | heart morphogenesis | 88 | 266 | 3.554e-06 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 896 | 3993 | 3.742e-06 |
| GO:BP | GO:0007275 | multicellular organism development | 1046 | 4727 | 3.826e-06 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 28 | 54 | 4.078e-06 |
| GO:BP | GO:0055001 | muscle cell development | 70 | 198 | 4.101e-06 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 46 | 112 | 4.454e-06 |
| GO:BP | GO:0044281 | small molecule metabolic process | 430 | 1776 | 5.388e-06 |
| GO:BP | GO:0003015 | heart process | 84 | 253 | 5.646e-06 |
| GO:BP | GO:0032879 | regulation of localization | 484 | 2029 | 5.717e-06 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 167 | 594 | 5.731e-06 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 25 | 46 | 5.775e-06 |
| GO:BP | GO:0010212 | response to ionizing radiation | 54 | 141 | 5.886e-06 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 62 | 170 | 5.889e-06 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 389 | 1588 | 5.971e-06 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 52 | 134 | 5.998e-06 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 26 | 49 | 6.031e-06 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 26 | 49 | 6.031e-06 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 26 | 49 | 6.031e-06 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 27 | 52 | 6.269e-06 |
| GO:BP | GO:0006839 | mitochondrial transport | 64 | 178 | 6.645e-06 |
| GO:BP | GO:0006338 | chromatin remodeling | 197 | 725 | 6.645e-06 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 87 | 266 | 6.861e-06 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1225 | 5629 | 7.694e-06 |
| GO:BP | GO:0014706 | striated muscle tissue development | 122 | 408 | 8.089e-06 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 72 | 209 | 8.121e-06 |
| GO:BP | GO:0060047 | heart contraction | 81 | 244 | 8.813e-06 |
| GO:BP | GO:0042692 | muscle cell differentiation | 121 | 405 | 9.417e-06 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 85 | 260 | 9.436e-06 |
| GO:BP | GO:0001510 | RNA methylation | 36 | 81 | 9.436e-06 |
| GO:BP | GO:0060537 | muscle tissue development | 127 | 430 | 9.556e-06 |
| GO:BP | GO:0032200 | telomere organization | 67 | 191 | 9.663e-06 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 26 | 50 | 9.738e-06 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 89 | 276 | 9.747e-06 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 28 | 56 | 9.875e-06 |
| GO:BP | GO:0032880 | regulation of protein localization | 237 | 907 | 1.147e-05 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 78 | 234 | 1.169e-05 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 95 | 301 | 1.177e-05 |
| GO:BP | GO:0061061 | muscle structure development | 188 | 693 | 1.337e-05 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 87 | 270 | 1.337e-05 |
| GO:BP | GO:0008016 | regulation of heart contraction | 70 | 204 | 1.337e-05 |
| GO:BP | GO:0007080 | mitotic metaphase chromosome alignment | 30 | 63 | 1.430e-05 |
| GO:BP | GO:0031399 | regulation of protein modification process | 268 | 1049 | 1.499e-05 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 28 | 57 | 1.530e-05 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 45 | 113 | 1.530e-05 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 26 | 51 | 1.533e-05 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 26 | 51 | 1.533e-05 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 26 | 51 | 1.533e-05 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 26 | 51 | 1.533e-05 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 52 | 138 | 1.579e-05 |
| GO:BP | GO:0050658 | RNA transport | 58 | 160 | 1.616e-05 |
| GO:BP | GO:0050657 | nucleic acid transport | 58 | 160 | 1.616e-05 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 33 | 73 | 1.686e-05 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 36 | 83 | 1.812e-05 |
| GO:BP | GO:0000209 | protein polyubiquitination | 89 | 280 | 1.836e-05 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 21 | 37 | 1.867e-05 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 57 | 157 | 1.876e-05 |
| GO:BP | GO:0032502 | developmental process | 1405 | 6553 | 1.928e-05 |
| GO:BP | GO:0048856 | anatomical structure development | 1293 | 5997 | 2.212e-05 |
| GO:BP | GO:0008152 | metabolic process | 2879 | 14136 | 2.212e-05 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 23 | 43 | 2.242e-05 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 114 | 383 | 2.268e-05 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 28 | 58 | 2.270e-05 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 51 | 136 | 2.292e-05 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 51 | 136 | 2.292e-05 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 33 | 74 | 2.375e-05 |
| GO:BP | GO:0010506 | regulation of autophagy | 110 | 367 | 2.397e-05 |
| GO:BP | GO:0006986 | response to unfolded protein | 52 | 140 | 2.501e-05 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 32 | 71 | 2.580e-05 |
| GO:BP | GO:0065009 | regulation of molecular function | 364 | 1496 | 2.611e-05 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 95 | 307 | 2.842e-05 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 91 | 291 | 2.888e-05 |
| GO:BP | GO:0006915 | apoptotic process | 455 | 1923 | 3.029e-05 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 49 | 130 | 3.066e-05 |
| GO:BP | GO:0006950 | response to stress | 876 | 3948 | 3.267e-05 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 83 | 260 | 3.344e-05 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 33 | 75 | 3.344e-05 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 311 | 1256 | 3.403e-05 |
| GO:BP | GO:0048731 | system development | 897 | 4053 | 3.569e-05 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 26 | 53 | 3.606e-05 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 26 | 53 | 3.606e-05 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 26 | 53 | 3.606e-05 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 24 | 47 | 3.663e-05 |
| GO:BP | GO:0051169 | nuclear transport | 101 | 334 | 4.072e-05 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 101 | 334 | 4.072e-05 |
| GO:BP | GO:0072359 | circulatory system development | 286 | 1145 | 4.245e-05 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 39 | 96 | 4.257e-05 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 105 | 351 | 4.393e-05 |
| GO:BP | GO:0141188 | nucleic acid catabolic process | 103 | 343 | 4.530e-05 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 83 | 262 | 4.547e-05 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 228 | 884 | 4.566e-05 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1563 | 7376 | 4.907e-05 |
| GO:BP | GO:0006401 | RNA catabolic process | 96 | 315 | 4.941e-05 |
| GO:BP | GO:0006260 | DNA replication | 88 | 283 | 5.271e-05 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 13 | 18 | 5.306e-05 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 13 | 18 | 5.306e-05 |
| GO:BP | GO:0070979 | protein K11-linked ubiquitination | 21 | 39 | 5.306e-05 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 13 | 18 | 5.306e-05 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 31 | 70 | 5.557e-05 |
| GO:BP | GO:0006941 | striated muscle contraction | 64 | 189 | 5.766e-05 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 80 | 253 | 7.316e-05 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 19 | 34 | 7.773e-05 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 78 | 246 | 8.599e-05 |
| GO:BP | GO:0034330 | cell junction organization | 215 | 834 | 9.060e-05 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1252 | 5834 | 9.076e-05 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 43 | 113 | 9.423e-05 |
| GO:BP | GO:0012501 | programmed cell death | 464 | 1987 | 9.770e-05 |
| GO:BP | GO:0051168 | nuclear export | 58 | 169 | 1.038e-04 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 28 | 62 | 1.049e-04 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 28 | 62 | 1.049e-04 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 181 | 686 | 1.194e-04 |
| GO:BP | GO:0008219 | cell death | 464 | 1991 | 1.208e-04 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 18 | 32 | 1.220e-04 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 15 | 24 | 1.295e-04 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 37 | 93 | 1.303e-04 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 220 | 861 | 1.303e-04 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 19 | 35 | 1.330e-04 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 19 | 35 | 1.330e-04 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 51 | 144 | 1.373e-04 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 51 | 144 | 1.373e-04 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 23 | 47 | 1.398e-04 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 20 | 38 | 1.398e-04 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 45 | 122 | 1.423e-04 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 175 | 662 | 1.466e-04 |
| GO:BP | GO:0016482 | cytosolic transport | 50 | 141 | 1.604e-04 |
| GO:BP | GO:0051028 | mRNA transport | 47 | 130 | 1.656e-04 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 30 | 70 | 1.699e-04 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 45 | 123 | 1.801e-04 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 53 | 153 | 1.858e-04 |
| GO:BP | GO:0000910 | cytokinesis | 63 | 192 | 2.045e-04 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 41 | 109 | 2.062e-04 |
| GO:BP | GO:0048284 | organelle fusion | 56 | 165 | 2.077e-04 |
| GO:BP | GO:0090148 | membrane fission | 22 | 45 | 2.218e-04 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 37 | 95 | 2.219e-04 |
| GO:BP | GO:0032205 | negative regulation of telomere maintenance | 20 | 39 | 2.255e-04 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 20 | 39 | 2.255e-04 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 164 | 619 | 2.421e-04 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 11 | 15 | 2.477e-04 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 15 | 25 | 2.484e-04 |
| GO:BP | GO:0046034 | ATP metabolic process | 72 | 229 | 2.547e-04 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 29 | 68 | 2.608e-04 |
| GO:BP | GO:0051653 | spindle localization | 29 | 68 | 2.608e-04 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 25 | 55 | 2.741e-04 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 31 | 75 | 2.810e-04 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 49 | 140 | 2.824e-04 |
| GO:BP | GO:0007098 | centrosome cycle | 49 | 140 | 2.824e-04 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 67 | 210 | 2.941e-04 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 41 | 111 | 3.304e-04 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 32 | 79 | 3.320e-04 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 443 | 1912 | 3.411e-04 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 54 | 160 | 3.421e-04 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 21 | 43 | 3.429e-04 |
| GO:BP | GO:0062125 | regulation of mitochondrial gene expression | 18 | 34 | 3.429e-04 |
| GO:BP | GO:0140058 | neuron projection arborization | 18 | 34 | 3.429e-04 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 18 | 34 | 3.429e-04 |
| GO:BP | GO:0016050 | vesicle organization | 107 | 376 | 3.466e-04 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 39 | 104 | 3.466e-04 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 26 | 59 | 3.493e-04 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 19 | 37 | 3.493e-04 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 97 | 334 | 3.493e-04 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 26 | 59 | 3.493e-04 |
| GO:BP | GO:0051865 | protein autoubiquitination | 31 | 76 | 3.699e-04 |
| GO:BP | GO:0046602 | regulation of mitotic centrosome separation | 8 | 9 | 3.793e-04 |
| GO:BP | GO:0031647 | regulation of protein stability | 97 | 335 | 3.948e-04 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 36 | 94 | 4.193e-04 |
| GO:BP | GO:0086009 | membrane repolarization | 24 | 53 | 4.193e-04 |
| GO:BP | GO:0006414 | translational elongation | 32 | 80 | 4.315e-04 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 39 | 105 | 4.365e-04 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 27 | 63 | 4.371e-04 |
| GO:BP | GO:0051293 | establishment of spindle localization | 27 | 63 | 4.371e-04 |
| GO:BP | GO:0097352 | autophagosome maturation | 27 | 63 | 4.371e-04 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 46 | 131 | 4.414e-04 |
| GO:BP | GO:0099643 | signal release from synapse | 51 | 150 | 4.414e-04 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 51 | 150 | 4.414e-04 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 60 | 185 | 4.436e-04 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 231 | 928 | 4.483e-04 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 333 | 1400 | 4.648e-04 |
| GO:BP | GO:0036503 | ERAD pathway | 40 | 109 | 4.655e-04 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 54 | 162 | 4.770e-04 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 26 | 60 | 4.808e-04 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 21 | 44 | 5.040e-04 |
| GO:BP | GO:0016358 | dendrite development | 74 | 242 | 5.109e-04 |
| GO:BP | GO:0006139 | nucleobase-containing compound metabolic process | 1669 | 7992 | 5.164e-04 |
| GO:BP | GO:0071478 | cellular response to radiation | 59 | 182 | 5.164e-04 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 28 | 67 | 5.252e-04 |
| GO:BP | GO:1901875 | positive regulation of post-translational protein modification | 46 | 132 | 5.324e-04 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 46 | 132 | 5.324e-04 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 19 | 38 | 5.328e-04 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 52 | 155 | 5.393e-04 |
| GO:BP | GO:0071806 | protein transmembrane transport | 30 | 74 | 5.452e-04 |
| GO:BP | GO:0051383 | kinetochore organization | 13 | 21 | 5.499e-04 |
| GO:BP | GO:0050808 | synapse organization | 149 | 563 | 5.688e-04 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 37 | 99 | 5.707e-04 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 73 | 239 | 5.871e-04 |
| GO:BP | GO:0031297 | replication fork processing | 23 | 51 | 6.347e-04 |
| GO:BP | GO:1903522 | regulation of blood circulation | 77 | 256 | 6.435e-04 |
| GO:BP | GO:0043009 | chordate embryonic development | 173 | 671 | 6.435e-04 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 42 | 118 | 6.535e-04 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 74 | 244 | 6.690e-04 |
| GO:BP | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 14 | 24 | 6.720e-04 |
| GO:BP | GO:0006479 | protein methylation | 22 | 48 | 6.807e-04 |
| GO:BP | GO:0008213 | protein alkylation | 22 | 48 | 6.807e-04 |
| GO:BP | GO:0090224 | regulation of spindle organization | 22 | 48 | 6.807e-04 |
| GO:BP | GO:0001508 | action potential | 56 | 172 | 6.860e-04 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 57 | 176 | 6.930e-04 |
| GO:BP | GO:0016310 | phosphorylation | 314 | 1320 | 7.512e-04 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 33 | 86 | 8.040e-04 |
| GO:BP | GO:0003205 | cardiac chamber development | 55 | 169 | 8.074e-04 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 47 | 138 | 8.096e-04 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 47 | 138 | 8.096e-04 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 29 | 72 | 8.178e-04 |
| GO:BP | GO:0071025 | RNA surveillance | 12 | 19 | 8.201e-04 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 146 | 554 | 8.201e-04 |
| GO:BP | GO:0110154 | RNA decapping | 12 | 19 | 8.201e-04 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 17 | 33 | 8.238e-04 |
| GO:BP | GO:0008053 | mitochondrial fusion | 17 | 33 | 8.238e-04 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 605 | 2713 | 8.955e-04 |
| GO:BP | GO:0009314 | response to radiation | 119 | 437 | 9.174e-04 |
| GO:BP | GO:0030239 | myofibril assembly | 30 | 76 | 9.365e-04 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 63 | 202 | 9.675e-04 |
| GO:BP | GO:0003012 | muscle system process | 119 | 438 | 1.015e-03 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 43 | 124 | 1.026e-03 |
| GO:BP | GO:0006390 | mitochondrial transcription | 13 | 22 | 1.030e-03 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 36 | 98 | 1.045e-03 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 115 | 421 | 1.045e-03 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 27 | 66 | 1.052e-03 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 101 | 361 | 1.069e-03 |
| GO:BP | GO:0051648 | vesicle localization | 67 | 219 | 1.085e-03 |
| GO:BP | GO:0007015 | actin filament organization | 124 | 461 | 1.137e-03 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 176 | 692 | 1.146e-03 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 134 | 505 | 1.151e-03 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 26 | 63 | 1.192e-03 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 19 | 40 | 1.199e-03 |
| GO:BP | GO:0030488 | tRNA methylation | 19 | 40 | 1.199e-03 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 11 | 17 | 1.219e-03 |
| GO:BP | GO:1905037 | autophagosome organization | 43 | 125 | 1.236e-03 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 76 | 257 | 1.243e-03 |
| GO:BP | GO:0071539 | protein localization to centrosome | 18 | 37 | 1.250e-03 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 15 | 28 | 1.250e-03 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 17 | 34 | 1.284e-03 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 62 | 200 | 1.292e-03 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 55 | 172 | 1.294e-03 |
| GO:BP | GO:0065008 | regulation of biological quality | 651 | 2947 | 1.314e-03 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 8 | 10 | 1.314e-03 |
| GO:BP | GO:0032506 | cytokinetic process | 22 | 50 | 1.349e-03 |
| GO:BP | GO:0000045 | autophagosome assembly | 41 | 118 | 1.376e-03 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 48 | 145 | 1.408e-03 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 49 | 149 | 1.433e-03 |
| GO:BP | GO:0034475 | U4 snRNA 3’-end processing | 7 | 8 | 1.440e-03 |
| GO:BP | GO:0150012 | positive regulation of neuron projection arborization | 7 | 8 | 1.440e-03 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 21 | 47 | 1.479e-03 |
| GO:BP | GO:0051668 | localization within membrane | 226 | 924 | 1.526e-03 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 12 | 20 | 1.556e-03 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 89 | 314 | 1.665e-03 |
| GO:BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | 41 | 119 | 1.679e-03 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 58 | 186 | 1.767e-03 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 10 | 15 | 1.767e-03 |
| GO:BP | GO:0071027 | nuclear RNA surveillance | 10 | 15 | 1.767e-03 |
| GO:BP | GO:0010970 | transport along microtubule | 56 | 178 | 1.767e-03 |
| GO:BP | GO:0001325 | formation of extrachromosomal circular DNA | 10 | 15 | 1.767e-03 |
| GO:BP | GO:0150011 | regulation of neuron projection arborization | 10 | 15 | 1.767e-03 |
| GO:BP | GO:0090656 | t-circle formation | 10 | 15 | 1.767e-03 |
| GO:BP | GO:0090737 | telomere maintenance via telomere trimming | 10 | 15 | 1.767e-03 |
| GO:BP | GO:0009790 | embryo development | 271 | 1135 | 1.772e-03 |
| GO:BP | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 18 | 38 | 1.845e-03 |
| GO:BP | GO:0045214 | sarcomere organization | 22 | 51 | 1.860e-03 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 45 | 135 | 1.870e-03 |
| GO:BP | GO:0000154 | rRNA modification | 17 | 35 | 1.936e-03 |
| GO:BP | GO:0002027 | regulation of heart rate | 36 | 101 | 1.972e-03 |
| GO:BP | GO:0099625 | ventricular cardiac muscle cell membrane repolarization | 15 | 29 | 1.998e-03 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 57 | 183 | 2.042e-03 |
| GO:BP | GO:0006513 | protein monoubiquitination | 21 | 48 | 2.060e-03 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 21 | 48 | 2.060e-03 |
| GO:BP | GO:0006508 | proteolysis | 345 | 1486 | 2.110e-03 |
| GO:BP | GO:0030048 | actin filament-based movement | 45 | 136 | 2.240e-03 |
| GO:BP | GO:0006220 | pyrimidine nucleotide metabolic process | 23 | 55 | 2.271e-03 |
| GO:BP | GO:0009208 | pyrimidine ribonucleoside triphosphate metabolic process | 11 | 18 | 2.378e-03 |
| GO:BP | GO:0017004 | cytochrome complex assembly | 19 | 42 | 2.508e-03 |
| GO:BP | GO:0099173 | postsynapse organization | 72 | 246 | 2.512e-03 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 30 | 80 | 2.519e-03 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 9 | 13 | 2.561e-03 |
| GO:BP | GO:0051258 | protein polymerization | 81 | 284 | 2.568e-03 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 63 | 209 | 2.573e-03 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 76 | 263 | 2.580e-03 |
| GO:BP | GO:0043487 | regulation of RNA stability | 59 | 193 | 2.692e-03 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 59 | 193 | 2.692e-03 |
| GO:BP | GO:0007031 | peroxisome organization | 18 | 39 | 2.692e-03 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 56 | 181 | 2.759e-03 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 12 | 21 | 2.789e-03 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 36 | 103 | 2.990e-03 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 16 | 33 | 3.035e-03 |
| GO:BP | GO:0051642 | centrosome localization | 16 | 33 | 3.035e-03 |
| GO:BP | GO:0009218 | pyrimidine ribonucleotide metabolic process | 16 | 33 | 3.035e-03 |
| GO:BP | GO:1990928 | response to amino acid starvation | 23 | 56 | 3.046e-03 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 60 | 198 | 3.058e-03 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 37 | 107 | 3.100e-03 |
| GO:BP | GO:0008535 | respiratory chain complex IV assembly | 15 | 30 | 3.113e-03 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 15 | 30 | 3.113e-03 |
| GO:BP | GO:0006284 | base-excision repair | 20 | 46 | 3.164e-03 |
| GO:BP | GO:0080134 | regulation of response to stress | 322 | 1387 | 3.430e-03 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 22 | 53 | 3.453e-03 |
| GO:BP | GO:0034394 | protein localization to cell surface | 26 | 67 | 3.504e-03 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 19 | 43 | 3.513e-03 |
| GO:BP | GO:0071028 | nuclear mRNA surveillance | 8 | 11 | 3.553e-03 |
| GO:BP | GO:0030953 | astral microtubule organization | 8 | 11 | 3.553e-03 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 10 | 16 | 3.576e-03 |
| GO:BP | GO:0016074 | sno(s)RNA metabolic process | 10 | 16 | 3.576e-03 |
| GO:BP | GO:0046036 | CTP metabolic process | 10 | 16 | 3.576e-03 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 362 | 1578 | 3.583e-03 |
| GO:BP | GO:0035418 | protein localization to synapse | 32 | 89 | 3.614e-03 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 76 | 266 | 3.629e-03 |
| GO:BP | GO:0003279 | cardiac septum development | 37 | 108 | 3.727e-03 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 37 | 108 | 3.727e-03 |
| GO:BP | GO:0031929 | TOR signaling | 52 | 167 | 3.737e-03 |
| GO:BP | GO:0001701 | in utero embryonic development | 110 | 413 | 3.744e-03 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 93 | 339 | 3.744e-03 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 93 | 339 | 3.744e-03 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 34 | 97 | 4.015e-03 |
| GO:BP | GO:0006275 | regulation of DNA replication | 40 | 120 | 4.048e-03 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 65 | 221 | 4.143e-03 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 28 | 75 | 4.222e-03 |
| GO:BP | GO:0060039 | pericardium development | 11 | 19 | 4.290e-03 |
| GO:BP | GO:0006936 | muscle contraction | 95 | 349 | 4.297e-03 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 26 | 68 | 4.456e-03 |
| GO:BP | GO:0097120 | receptor localization to synapse | 29 | 79 | 4.581e-03 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 59 | 197 | 4.615e-03 |
| GO:BP | GO:0043622 | cortical microtubule organization | 7 | 9 | 4.650e-03 |
| GO:BP | GO:0000467 | exonucleolytic trimming to generate mature 3’-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 7 | 9 | 4.650e-03 |
| GO:BP | GO:0003162 | atrioventricular node development | 7 | 9 | 4.650e-03 |
| GO:BP | GO:0006304 | DNA modification | 15 | 31 | 4.671e-03 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 33 | 94 | 4.671e-03 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 12 | 22 | 4.680e-03 |
| GO:BP | GO:0086013 | membrane repolarization during cardiac muscle cell action potential | 13 | 25 | 4.857e-03 |
| GO:BP | GO:0046785 | microtubule polymerization | 34 | 98 | 4.867e-03 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 30 | 83 | 4.867e-03 |
| GO:BP | GO:0003292 | cardiac septum cell differentiation | 5 | 5 | 4.897e-03 |
| GO:BP | GO:0060922 | atrioventricular node cell differentiation | 5 | 5 | 4.897e-03 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 43 | 133 | 4.985e-03 |
| GO:BP | GO:0009152 | purine ribonucleotide biosynthetic process | 50 | 161 | 5.026e-03 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 301 | 1296 | 5.026e-03 |
| GO:BP | GO:0061157 | mRNA destabilization | 35 | 102 | 5.027e-03 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 25 | 65 | 5.073e-03 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 31 | 87 | 5.129e-03 |
| GO:BP | GO:0050779 | RNA destabilization | 36 | 106 | 5.181e-03 |
| GO:BP | GO:0000902 | cell morphogenesis | 237 | 996 | 5.181e-03 |
| GO:BP | GO:0043144 | sno(s)RNA processing | 9 | 14 | 5.276e-03 |
| GO:BP | GO:0007100 | mitotic centrosome separation | 9 | 14 | 5.276e-03 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 159 | 637 | 5.382e-03 |
| GO:BP | GO:0071035 | nuclear polyadenylation-dependent rRNA catabolic process | 6 | 7 | 5.422e-03 |
| GO:BP | GO:0071038 | TRAMP-dependent tRNA surveillance pathway | 6 | 7 | 5.422e-03 |
| GO:BP | GO:0040031 | snRNA modification | 6 | 7 | 5.422e-03 |
| GO:BP | GO:0022008 | neurogenesis | 400 | 1771 | 5.749e-03 |
| GO:BP | GO:0098534 | centriole assembly | 20 | 48 | 5.752e-03 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 297 | 1280 | 5.752e-03 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 34 | 99 | 5.846e-03 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 201 | 831 | 5.865e-03 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 22 | 55 | 5.898e-03 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 96 | 357 | 5.975e-03 |
| GO:BP | GO:0038202 | TORC1 signaling | 35 | 103 | 6.025e-03 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 128 | 499 | 6.173e-03 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 31 | 88 | 6.271e-03 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 167 | 676 | 6.375e-03 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 28 | 77 | 6.524e-03 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 168 | 681 | 6.559e-03 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 48 | 155 | 6.712e-03 |
| GO:BP | GO:0045333 | cellular respiration | 69 | 242 | 6.712e-03 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 48 | 155 | 6.712e-03 |
| GO:BP | GO:0006413 | translational initiation | 41 | 127 | 6.712e-03 |
| GO:BP | GO:0009142 | nucleoside triphosphate biosynthetic process | 41 | 127 | 6.712e-03 |
| GO:BP | GO:0003231 | cardiac ventricle development | 41 | 127 | 6.712e-03 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 123 | 478 | 6.839e-03 |
| GO:BP | GO:0086010 | membrane depolarization during action potential | 14 | 29 | 7.184e-03 |
| GO:BP | GO:0034508 | centromere complex assembly | 14 | 29 | 7.184e-03 |
| GO:BP | GO:0042407 | cristae formation | 11 | 20 | 7.206e-03 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 18 | 42 | 7.222e-03 |
| GO:BP | GO:0001570 | vasculogenesis | 30 | 85 | 7.279e-03 |
| GO:BP | GO:0070129 | regulation of mitochondrial translation | 13 | 26 | 7.432e-03 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 12 | 23 | 7.457e-03 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 12 | 23 | 7.457e-03 |
| GO:BP | GO:1903578 | regulation of ATP metabolic process | 27 | 74 | 7.514e-03 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 267 | 1144 | 7.514e-03 |
| GO:BP | GO:1904779 | regulation of protein localization to centrosome | 8 | 12 | 7.745e-03 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 63 | 218 | 7.745e-03 |
| GO:BP | GO:0031126 | sno(s)RNA 3’-end processing | 8 | 12 | 7.745e-03 |
| GO:BP | GO:0017148 | negative regulation of translation | 42 | 132 | 7.864e-03 |
| GO:BP | GO:0072657 | protein localization to membrane | 199 | 827 | 7.922e-03 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 17 | 39 | 7.988e-03 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 28 | 78 | 7.988e-03 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 305 | 1326 | 8.329e-03 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 34 | 101 | 8.396e-03 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 19 | 46 | 8.648e-03 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 26 | 71 | 8.753e-03 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 21 | 53 | 8.761e-03 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 30 | 86 | 8.842e-03 |
| GO:BP | GO:0090630 | activation of GTPase activity | 16 | 36 | 8.842e-03 |
| GO:BP | GO:0006301 | postreplication repair | 16 | 36 | 8.842e-03 |
| GO:BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | 37 | 113 | 8.865e-03 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 166 | 677 | 9.046e-03 |
| GO:BP | GO:0030162 | regulation of proteolysis | 106 | 406 | 9.166e-03 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 73 | 262 | 9.215e-03 |
| GO:BP | GO:0070085 | glycosylation | 69 | 245 | 9.282e-03 |
| GO:BP | GO:0061512 | protein localization to cilium | 27 | 75 | 9.282e-03 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 226 | 956 | 9.450e-03 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 68 | 241 | 9.543e-03 |
| GO:BP | GO:0007099 | centriole replication | 18 | 43 | 9.719e-03 |
| GO:BP | GO:0099111 | microtubule-based transport | 63 | 220 | 9.736e-03 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 22 | 57 | 9.787e-03 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 162 | 660 | 9.814e-03 |
| GO:BP | GO:0106074 | aminoacyl-tRNA metabolism involved in translational fidelity | 9 | 15 | 9.842e-03 |
| GO:BP | GO:0043248 | proteasome assembly | 9 | 15 | 9.842e-03 |
| GO:BP | GO:0009209 | pyrimidine ribonucleoside triphosphate biosynthetic process | 9 | 15 | 9.842e-03 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 9 | 15 | 9.842e-03 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 226 | 957 | 9.881e-03 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 69 | 246 | 1.029e-02 |
| GO:BP | GO:0009165 | nucleotide biosynthetic process | 65 | 229 | 1.029e-02 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 46 | 150 | 1.032e-02 |
| GO:BP | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening | 14 | 30 | 1.035e-02 |
| GO:BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | 37 | 114 | 1.036e-02 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 43 | 138 | 1.051e-02 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 40 | 126 | 1.052e-02 |
| GO:BP | GO:0006468 | protein phosphorylation | 283 | 1227 | 1.052e-02 |
| GO:BP | GO:0035050 | embryonic heart tube development | 30 | 87 | 1.058e-02 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 30 | 87 | 1.058e-02 |
| GO:BP | GO:0006405 | RNA export from nucleus | 31 | 91 | 1.096e-02 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 13 | 27 | 1.096e-02 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 51 | 171 | 1.100e-02 |
| GO:BP | GO:0031125 | rRNA 3’-end processing | 7 | 10 | 1.102e-02 |
| GO:BP | GO:0009220 | pyrimidine ribonucleotide biosynthetic process | 12 | 24 | 1.138e-02 |
| GO:BP | GO:0009147 | pyrimidine nucleoside triphosphate metabolic process | 12 | 24 | 1.138e-02 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 161 | 658 | 1.143e-02 |
| GO:BP | GO:0006450 | regulation of translational fidelity | 11 | 21 | 1.144e-02 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 33 | 99 | 1.149e-02 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 34 | 103 | 1.172e-02 |
| GO:BP | GO:0007033 | vacuole organization | 67 | 239 | 1.203e-02 |
| GO:BP | GO:0003230 | cardiac atrium development | 16 | 37 | 1.205e-02 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 133 | 531 | 1.205e-02 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 37 | 115 | 1.211e-02 |
| GO:BP | GO:0090257 | regulation of muscle system process | 65 | 231 | 1.276e-02 |
| GO:BP | GO:0014896 | muscle hypertrophy | 30 | 88 | 1.280e-02 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 176 | 729 | 1.280e-02 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 20 | 51 | 1.280e-02 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 49 | 164 | 1.292e-02 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 71 | 257 | 1.315e-02 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 26 | 73 | 1.315e-02 |
| GO:BP | GO:0060914 | heart formation | 15 | 34 | 1.337e-02 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 58 | 202 | 1.371e-02 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 34 | 104 | 1.392e-02 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 91 | 345 | 1.422e-02 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 37 | 116 | 1.426e-02 |
| GO:BP | GO:0032509 | endosome transport via multivesicular body sorting pathway | 17 | 41 | 1.456e-02 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 14 | 31 | 1.474e-02 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 14 | 31 | 1.474e-02 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 48 | 161 | 1.502e-02 |
| GO:BP | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport | 8 | 13 | 1.503e-02 |
| GO:BP | GO:0120180 | cell-substrate junction disassembly | 6 | 8 | 1.534e-02 |
| GO:BP | GO:0120181 | focal adhesion disassembly | 6 | 8 | 1.534e-02 |
| GO:BP | GO:0106354 | tRNA surveillance | 6 | 8 | 1.534e-02 |
| GO:BP | GO:0071051 | poly(A)-dependent snoRNA 3’-end processing | 6 | 8 | 1.534e-02 |
| GO:BP | GO:1904354 | negative regulation of telomere capping | 6 | 8 | 1.534e-02 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 98 | 377 | 1.537e-02 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 197 | 830 | 1.546e-02 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 13 | 28 | 1.597e-02 |
| GO:BP | GO:0051298 | centrosome duplication | 26 | 74 | 1.613e-02 |
| GO:BP | GO:0006836 | neurotransmitter transport | 60 | 212 | 1.629e-02 |
| GO:BP | GO:0030518 | nuclear receptor-mediated steroid hormone signaling pathway | 38 | 121 | 1.658e-02 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 38 | 121 | 1.658e-02 |
| GO:BP | GO:0032981 | mitochondrial respiratory chain complex I assembly | 23 | 63 | 1.676e-02 |
| GO:BP | GO:0010257 | NADH dehydrogenase complex assembly | 23 | 63 | 1.676e-02 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 23 | 63 | 1.676e-02 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 18 | 45 | 1.677e-02 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 12 | 25 | 1.692e-02 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 12 | 25 | 1.692e-02 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 12 | 25 | 1.692e-02 |
| GO:BP | GO:1903241 | U2-type prespliceosome assembly | 12 | 25 | 1.692e-02 |
| GO:BP | GO:0110156 | mRNA methylguanosine-cap decapping | 9 | 16 | 1.694e-02 |
| GO:BP | GO:0051299 | centrosome separation | 9 | 16 | 1.694e-02 |
| GO:BP | GO:0043328 | protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 9 | 16 | 1.694e-02 |
| GO:BP | GO:1990166 | protein localization to site of double-strand break | 9 | 16 | 1.694e-02 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 11 | 22 | 1.761e-02 |
| GO:BP | GO:0000212 | meiotic spindle organization | 11 | 22 | 1.761e-02 |
| GO:BP | GO:0001947 | heart looping | 24 | 67 | 1.769e-02 |
| GO:BP | GO:0045945 | positive regulation of transcription by RNA polymerase III | 10 | 19 | 1.770e-02 |
| GO:BP | GO:1904505 | regulation of telomere maintenance in response to DNA damage | 10 | 19 | 1.770e-02 |
| GO:BP | GO:0048699 | generation of neurons | 345 | 1536 | 1.781e-02 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 44 | 146 | 1.790e-02 |
| GO:BP | GO:0034644 | cellular response to UV | 30 | 90 | 1.808e-02 |
| GO:BP | GO:0006486 | protein glycosylation | 63 | 226 | 1.827e-02 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 63 | 226 | 1.827e-02 |
| GO:BP | GO:0045727 | positive regulation of translation | 42 | 138 | 1.831e-02 |
| GO:BP | GO:0043401 | steroid hormone receptor signaling pathway | 42 | 138 | 1.831e-02 |
| GO:BP | GO:0106027 | neuron projection organization | 31 | 94 | 1.833e-02 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 19 | 49 | 1.865e-02 |
| GO:BP | GO:0042073 | intraciliary transport | 19 | 49 | 1.865e-02 |
| GO:BP | GO:0034502 | protein localization to chromosome | 39 | 126 | 1.878e-02 |
| GO:BP | GO:0003013 | circulatory system process | 147 | 602 | 1.883e-02 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 53 | 184 | 1.914e-02 |
| GO:BP | GO:0051661 | maintenance of centrosome location | 5 | 6 | 1.926e-02 |
| GO:BP | GO:0043954 | cellular component maintenance | 26 | 75 | 1.926e-02 |
| GO:BP | GO:0051096 | positive regulation of helicase activity | 5 | 6 | 1.926e-02 |
| GO:BP | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration | 5 | 6 | 1.926e-02 |
| GO:BP | GO:1990116 | ribosome-associated ubiquitin-dependent protein catabolic process | 5 | 6 | 1.926e-02 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 70 | 257 | 1.950e-02 |
| GO:BP | GO:0009411 | response to UV | 46 | 155 | 1.950e-02 |
| GO:BP | GO:0098910 | regulation of atrial cardiac muscle cell action potential | 4 | 4 | 1.950e-02 |
| GO:BP | GO:0048597 | post-embryonic camera-type eye morphogenesis | 4 | 4 | 1.950e-02 |
| GO:BP | GO:0035107 | appendage morphogenesis | 46 | 155 | 1.950e-02 |
| GO:BP | GO:0035108 | limb morphogenesis | 46 | 155 | 1.950e-02 |
| GO:BP | GO:1904506 | negative regulation of telomere maintenance in response to DNA damage | 4 | 4 | 1.950e-02 |
| GO:BP | GO:0035879 | plasma membrane lactate transport | 4 | 4 | 1.950e-02 |
| GO:BP | GO:1901698 | response to nitrogen compound | 249 | 1080 | 1.950e-02 |
| GO:BP | GO:0060928 | atrioventricular node cell development | 4 | 4 | 1.950e-02 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 51 | 176 | 1.978e-02 |
| GO:BP | GO:0036258 | multivesicular body assembly | 14 | 32 | 1.979e-02 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 14 | 32 | 1.979e-02 |
| GO:BP | GO:2000279 | negative regulation of DNA biosynthetic process | 14 | 32 | 1.979e-02 |
| GO:BP | GO:0006997 | nucleus organization | 44 | 147 | 1.998e-02 |
| GO:BP | GO:0006164 | purine nucleotide biosynthetic process | 55 | 193 | 2.004e-02 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 28 | 83 | 2.015e-02 |
| GO:BP | GO:0042594 | response to starvation | 61 | 219 | 2.109e-02 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 18 | 46 | 2.109e-02 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 18 | 46 | 2.109e-02 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 18 | 46 | 2.109e-02 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 16 | 39 | 2.110e-02 |
| GO:BP | GO:0090174 | organelle membrane fusion | 38 | 123 | 2.145e-02 |
| GO:BP | GO:0021915 | neural tube development | 47 | 160 | 2.146e-02 |
| GO:BP | GO:1902414 | protein localization to cell junction | 37 | 119 | 2.154e-02 |
| GO:BP | GO:0009134 | nucleoside diphosphate catabolic process | 34 | 107 | 2.166e-02 |
| GO:BP | GO:0086011 | membrane repolarization during action potential | 13 | 29 | 2.181e-02 |
| GO:BP | GO:0062028 | regulation of stress granule assembly | 7 | 11 | 2.187e-02 |
| GO:BP | GO:0018195 | peptidyl-arginine modification | 7 | 11 | 2.187e-02 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 7 | 11 | 2.187e-02 |
| GO:BP | GO:0009267 | cellular response to starvation | 51 | 177 | 2.204e-02 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 155 | 642 | 2.221e-02 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 26 | 76 | 2.282e-02 |
| GO:BP | GO:0007019 | microtubule depolymerization | 19 | 50 | 2.332e-02 |
| GO:BP | GO:0071985 | multivesicular body sorting pathway | 19 | 50 | 2.332e-02 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 328 | 1462 | 2.344e-02 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 48 | 165 | 2.369e-02 |
| GO:BP | GO:0002026 | regulation of the force of heart contraction | 12 | 26 | 2.369e-02 |
| GO:BP | GO:0033617 | mitochondrial cytochrome c oxidase assembly | 12 | 26 | 2.369e-02 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 12 | 26 | 2.369e-02 |
| GO:BP | GO:0031167 | rRNA methylation | 12 | 26 | 2.369e-02 |
| GO:BP | GO:1900542 | regulation of purine nucleotide metabolic process | 28 | 84 | 2.393e-02 |
| GO:BP | GO:0031667 | response to nutrient levels | 126 | 510 | 2.402e-02 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 56 | 199 | 2.412e-02 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 38 | 124 | 2.452e-02 |
| GO:BP | GO:0009135 | purine nucleoside diphosphate metabolic process | 36 | 116 | 2.491e-02 |
| GO:BP | GO:0009179 | purine ribonucleoside diphosphate metabolic process | 36 | 116 | 2.491e-02 |
| GO:BP | GO:0003283 | atrial septum development | 11 | 23 | 2.537e-02 |
| GO:BP | GO:0015919 | peroxisomal membrane transport | 11 | 23 | 2.537e-02 |
| GO:BP | GO:0046599 | regulation of centriole replication | 11 | 23 | 2.537e-02 |
| GO:BP | GO:0043574 | peroxisomal transport | 11 | 23 | 2.537e-02 |
| GO:BP | GO:0015780 | nucleotide-sugar transmembrane transport | 8 | 14 | 2.539e-02 |
| GO:BP | GO:1904353 | regulation of telomere capping | 8 | 14 | 2.539e-02 |
| GO:BP | GO:0006241 | CTP biosynthetic process | 8 | 14 | 2.539e-02 |
| GO:BP | GO:0016925 | protein sumoylation | 24 | 69 | 2.539e-02 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 24 | 69 | 2.539e-02 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 24 | 69 | 2.539e-02 |
| GO:BP | GO:0043170 | macromolecule metabolic process | 2425 | 12048 | 2.593e-02 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 25 | 73 | 2.639e-02 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 63 | 230 | 2.646e-02 |
| GO:BP | GO:0036257 | multivesicular body organization | 14 | 33 | 2.661e-02 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 14 | 33 | 2.661e-02 |
| GO:BP | GO:0051382 | kinetochore assembly | 9 | 17 | 2.663e-02 |
| GO:BP | GO:0085020 | protein K6-linked ubiquitination | 9 | 17 | 2.663e-02 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 21 | 58 | 2.675e-02 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 26 | 77 | 2.704e-02 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 16 | 40 | 2.730e-02 |
| GO:BP | GO:0034329 | cell junction assembly | 124 | 503 | 2.731e-02 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 67 | 248 | 2.809e-02 |
| GO:BP | GO:0051261 | protein depolymerization | 37 | 121 | 2.816e-02 |
| GO:BP | GO:0006906 | vesicle fusion | 37 | 121 | 2.816e-02 |
| GO:BP | GO:0006140 | regulation of nucleotide metabolic process | 28 | 85 | 2.816e-02 |
| GO:BP | GO:0042391 | regulation of membrane potential | 114 | 458 | 2.842e-02 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 29 | 89 | 2.845e-02 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 33 | 105 | 2.890e-02 |
| GO:BP | GO:0060996 | dendritic spine development | 32 | 101 | 2.890e-02 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 42 | 142 | 2.991e-02 |
| GO:BP | GO:0006520 | amino acid metabolic process | 78 | 297 | 2.991e-02 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 17 | 44 | 3.075e-02 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 24 | 70 | 3.075e-02 |
| GO:BP | GO:0099633 | protein localization to postsynaptic specialization membrane | 15 | 37 | 3.128e-02 |
| GO:BP | GO:0099645 | neurotransmitter receptor localization to postsynaptic specialization membrane | 15 | 37 | 3.128e-02 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 25 | 74 | 3.168e-02 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 57 | 206 | 3.186e-02 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 395 | 1795 | 3.192e-02 |
| GO:BP | GO:0006457 | protein folding | 60 | 219 | 3.203e-02 |
| GO:BP | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 6 | 9 | 3.249e-02 |
| GO:BP | GO:0036500 | ATF6-mediated unfolded protein response | 6 | 9 | 3.249e-02 |
| GO:BP | GO:0023056 | positive regulation of signaling | 395 | 1796 | 3.296e-02 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 12 | 27 | 3.296e-02 |
| GO:BP | GO:0016233 | telomere capping | 12 | 27 | 3.296e-02 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 12 | 27 | 3.296e-02 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 12 | 27 | 3.296e-02 |
| GO:BP | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway | 12 | 27 | 3.296e-02 |
| GO:BP | GO:0022406 | membrane docking | 28 | 86 | 3.307e-02 |
| GO:BP | GO:0051049 | regulation of transport | 356 | 1607 | 3.307e-02 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 28 | 86 | 3.307e-02 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 18 | 48 | 3.333e-02 |
| GO:BP | GO:0014020 | primary neural tube formation | 30 | 94 | 3.333e-02 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 67 | 250 | 3.333e-02 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 31 | 98 | 3.333e-02 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 18 | 48 | 3.333e-02 |
| GO:BP | GO:0007032 | endosome organization | 31 | 98 | 3.333e-02 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 32 | 102 | 3.333e-02 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 335 | 1506 | 3.348e-02 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 41 | 139 | 3.413e-02 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 50 | 177 | 3.475e-02 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 16 | 41 | 3.501e-02 |
| GO:BP | GO:0003203 | endocardial cushion morphogenesis | 16 | 41 | 3.501e-02 |
| GO:BP | GO:0042776 | proton motive force-driven mitochondrial ATP synthesis | 23 | 67 | 3.550e-02 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 11 | 24 | 3.591e-02 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 11 | 24 | 3.591e-02 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 11 | 24 | 3.591e-02 |
| GO:BP | GO:0000423 | mitophagy | 24 | 71 | 3.654e-02 |
| GO:BP | GO:0060322 | head development | 188 | 806 | 3.697e-02 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 78 | 300 | 3.806e-02 |
| GO:BP | GO:1901799 | negative regulation of proteasomal protein catabolic process | 20 | 56 | 3.808e-02 |
| GO:BP | GO:0141137 | positive regulation of gene expression, epigenetic | 20 | 56 | 3.808e-02 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 113 | 458 | 3.808e-02 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 20 | 56 | 3.808e-02 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 32 | 103 | 3.830e-02 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 17 | 45 | 3.830e-02 |
| GO:BP | GO:0003207 | cardiac chamber formation | 7 | 12 | 3.830e-02 |
| GO:BP | GO:0009191 | ribonucleoside diphosphate catabolic process | 32 | 103 | 3.830e-02 |
| GO:BP | GO:0086067 | AV node cell to bundle of His cell communication | 7 | 12 | 3.830e-02 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 10 | 21 | 3.830e-02 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 33 | 107 | 3.830e-02 |
| GO:BP | GO:1901661 | quinone metabolic process | 10 | 21 | 3.830e-02 |
| GO:BP | GO:0051657 | maintenance of organelle location | 7 | 12 | 3.830e-02 |
| GO:BP | GO:0072710 | response to hydroxyurea | 7 | 12 | 3.830e-02 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 41 | 140 | 3.830e-02 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 32 | 103 | 3.830e-02 |
| GO:BP | GO:0097061 | dendritic spine organization | 27 | 83 | 3.830e-02 |
| GO:BP | GO:0046051 | UTP metabolic process | 7 | 12 | 3.830e-02 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 27 | 83 | 3.830e-02 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 30 | 95 | 3.830e-02 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 10 | 21 | 3.830e-02 |
| GO:BP | GO:0006285 | base-excision repair, AP site formation | 7 | 12 | 3.830e-02 |
| GO:BP | GO:0099188 | postsynaptic cytoskeleton organization | 10 | 21 | 3.830e-02 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 7 | 12 | 3.830e-02 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 40 | 136 | 3.873e-02 |
| GO:BP | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway | 49 | 174 | 3.908e-02 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 13 | 31 | 3.910e-02 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 13 | 31 | 3.910e-02 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 21 | 60 | 3.910e-02 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 9 | 18 | 4.006e-02 |
| GO:BP | GO:0001522 | pseudouridine synthesis | 9 | 18 | 4.006e-02 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 38 | 128 | 4.016e-02 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 8 | 15 | 4.051e-02 |
| GO:BP | GO:0023057 | negative regulation of signaling | 319 | 1435 | 4.134e-02 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 23 | 68 | 4.185e-02 |
| GO:BP | GO:0035825 | homologous recombination | 23 | 68 | 4.185e-02 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 23 | 68 | 4.185e-02 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 34 | 112 | 4.287e-02 |
| GO:BP | GO:0043149 | stress fiber assembly | 33 | 108 | 4.336e-02 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 33 | 108 | 4.336e-02 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 16 | 42 | 4.403e-02 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 12 | 28 | 4.415e-02 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 12 | 28 | 4.415e-02 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 12 | 28 | 4.415e-02 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 29 | 92 | 4.446e-02 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 14 | 35 | 4.547e-02 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 14 | 35 | 4.547e-02 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 38 | 129 | 4.565e-02 |
| GO:BP | GO:0006370 | 7-methylguanosine mRNA capping | 5 | 7 | 4.565e-02 |
| GO:BP | GO:0051204 | protein insertion into mitochondrial membrane | 5 | 7 | 4.565e-02 |
| GO:BP | GO:1990048 | anterograde neuronal dense core vesicle transport | 5 | 7 | 4.565e-02 |
| GO:BP | GO:1903436 | regulation of mitotic cytokinetic process | 5 | 7 | 4.565e-02 |
| GO:BP | GO:0086028 | bundle of His cell to Purkinje myocyte signaling | 5 | 7 | 4.565e-02 |
| GO:BP | GO:0086043 | bundle of His cell action potential | 5 | 7 | 4.565e-02 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 57 | 210 | 4.565e-02 |
| GO:BP | GO:0032954 | regulation of cytokinetic process | 5 | 7 | 4.565e-02 |
| GO:BP | GO:0046048 | UDP metabolic process | 5 | 7 | 4.565e-02 |
| GO:BP | GO:0099545 | trans-synaptic signaling by trans-synaptic complex | 5 | 7 | 4.565e-02 |
| GO:BP | GO:0018023 | peptidyl-lysine trimethylation | 5 | 7 | 4.565e-02 |
| GO:BP | GO:1903438 | positive regulation of mitotic cytokinetic process | 5 | 7 | 4.565e-02 |
| GO:BP | GO:0017183 | protein histidyl modification to diphthamide | 5 | 7 | 4.565e-02 |
| GO:BP | GO:0009193 | pyrimidine ribonucleoside diphosphate metabolic process | 5 | 7 | 4.565e-02 |
| GO:BP | GO:0035735 | intraciliary transport involved in cilium assembly | 5 | 7 | 4.565e-02 |
| GO:BP | GO:0035617 | stress granule disassembly | 5 | 7 | 4.565e-02 |
| GO:BP | GO:0060972 | left/right pattern formation | 42 | 146 | 4.637e-02 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 42 | 146 | 4.637e-02 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 17 | 46 | 4.702e-02 |
| GO:BP | GO:0007020 | microtubule nucleation | 17 | 46 | 4.702e-02 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 181 | 779 | 4.773e-02 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 77 | 299 | 4.840e-02 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 11 | 25 | 4.845e-02 |
| GO:BP | GO:0035994 | response to muscle stretch | 11 | 25 | 4.845e-02 |
| GO:BP | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 11 | 25 | 4.845e-02 |
| GO:BP | GO:0030813 | positive regulation of nucleotide catabolic process | 11 | 25 | 4.845e-02 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 11 | 25 | 4.845e-02 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 11 | 25 | 4.845e-02 |
| GO:BP | GO:0033123 | positive regulation of purine nucleotide catabolic process | 11 | 25 | 4.845e-02 |
| GO:BP | GO:0045821 | positive regulation of glycolytic process | 11 | 25 | 4.845e-02 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 23 | 69 | 4.914e-02 |
| GO:BP | GO:0001578 | microtubule bundle formation | 39 | 134 | 4.958e-02 |
| GO:BP | GO:0001841 | neural tube formation | 31 | 101 | 4.970e-02 |
| GO:BP | GO:0170036 | import into the mitochondrion | 18 | 50 | 4.970e-02 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 24 | 73 | 4.970e-02 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 318 | 1436 | 4.970e-02 |
| GO:BP | GO:0003281 | ventricular septum development | 24 | 73 | 4.970e-02 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 18 | 50 | 4.970e-02 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 27 | 44 | 6.758e-06 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 162 | 506 | 8.870e-06 |
| KEGG | KEGG:03018 | RNA degradation | 38 | 79 | 2.840e-05 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 55 | 140 | 2.147e-04 |
| KEGG | KEGG:03082 | ATP-dependent chromatin remodeling | 46 | 116 | 8.407e-04 |
| KEGG | KEGG:04110 | Cell cycle | 55 | 157 | 5.361e-03 |
| KEGG | KEGG:03440 | Homologous recombination | 20 | 41 | 5.361e-03 |
| KEGG | KEGG:03410 | Base excision repair | 21 | 44 | 5.361e-03 |
| KEGG | KEGG:03040 | Spliceosome | 60 | 174 | 5.361e-03 |
| KEGG | KEGG:04137 | Mitophagy - animal | 38 | 103 | 1.192e-02 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 56 | 168 | 1.192e-02 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 107 | 363 | 1.192e-02 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 16 | 32 | 1.192e-02 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 39 | 108 | 1.345e-02 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 23 | 57 | 3.113e-02 |
#write.csv(table_DOX24R_down_genes, "output/table_DOX24R_downreg_genes.csv")
#GO:BP
table_DOX24Tshare_down_genes_GOBP <- table_DOX24Tshare_down_genes %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
#saveRDS(table_motif1_GOBP_d, "data/table_motif1_GOBP_d.RDS")
table_DOX24Tshare_down_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Shared Down DEGs Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))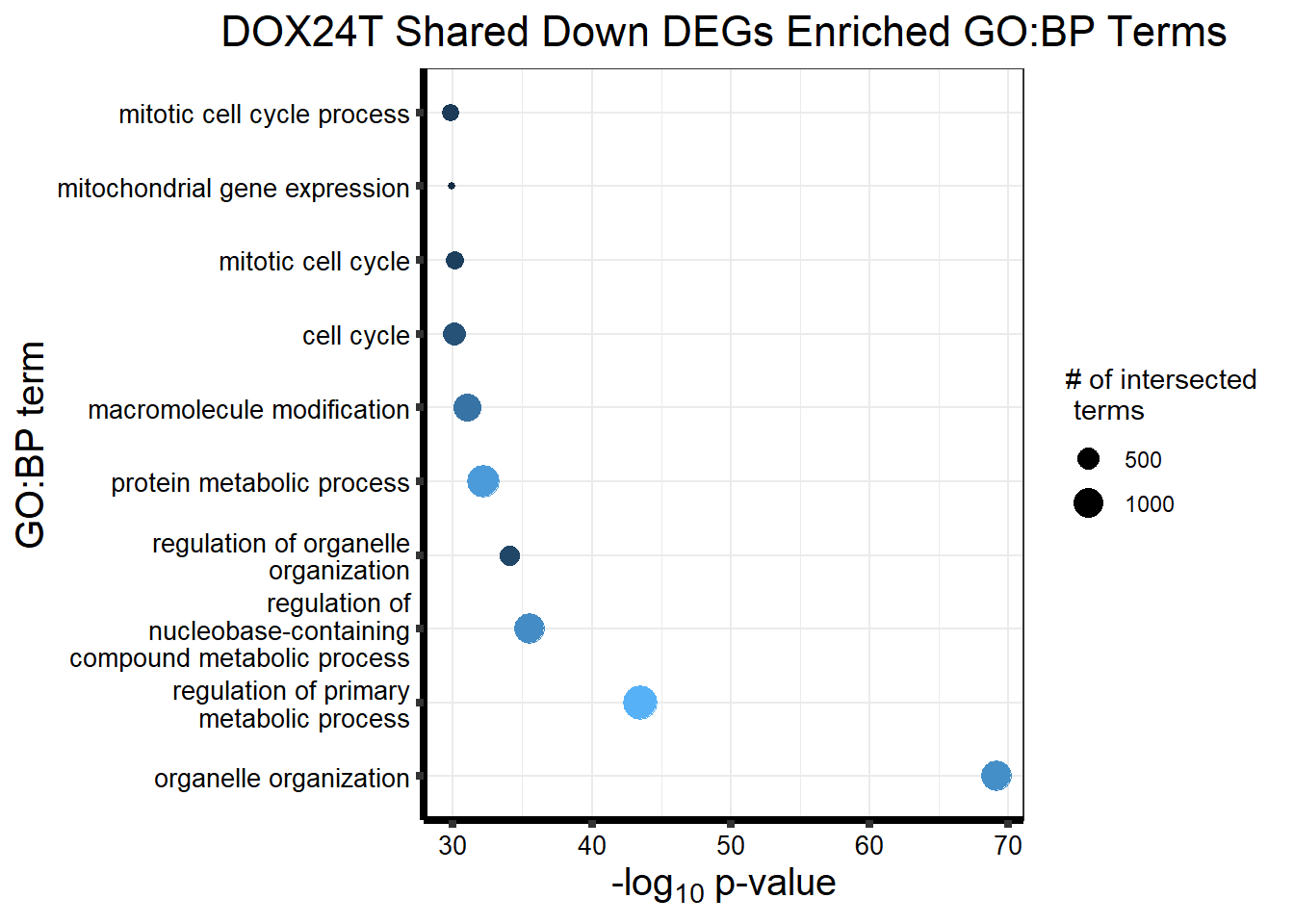
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_DOX24Tshare_down_genes_KEGG <- table_DOX24Tshare_down_genes %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24Tshare_down_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Shared Down DEGs Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))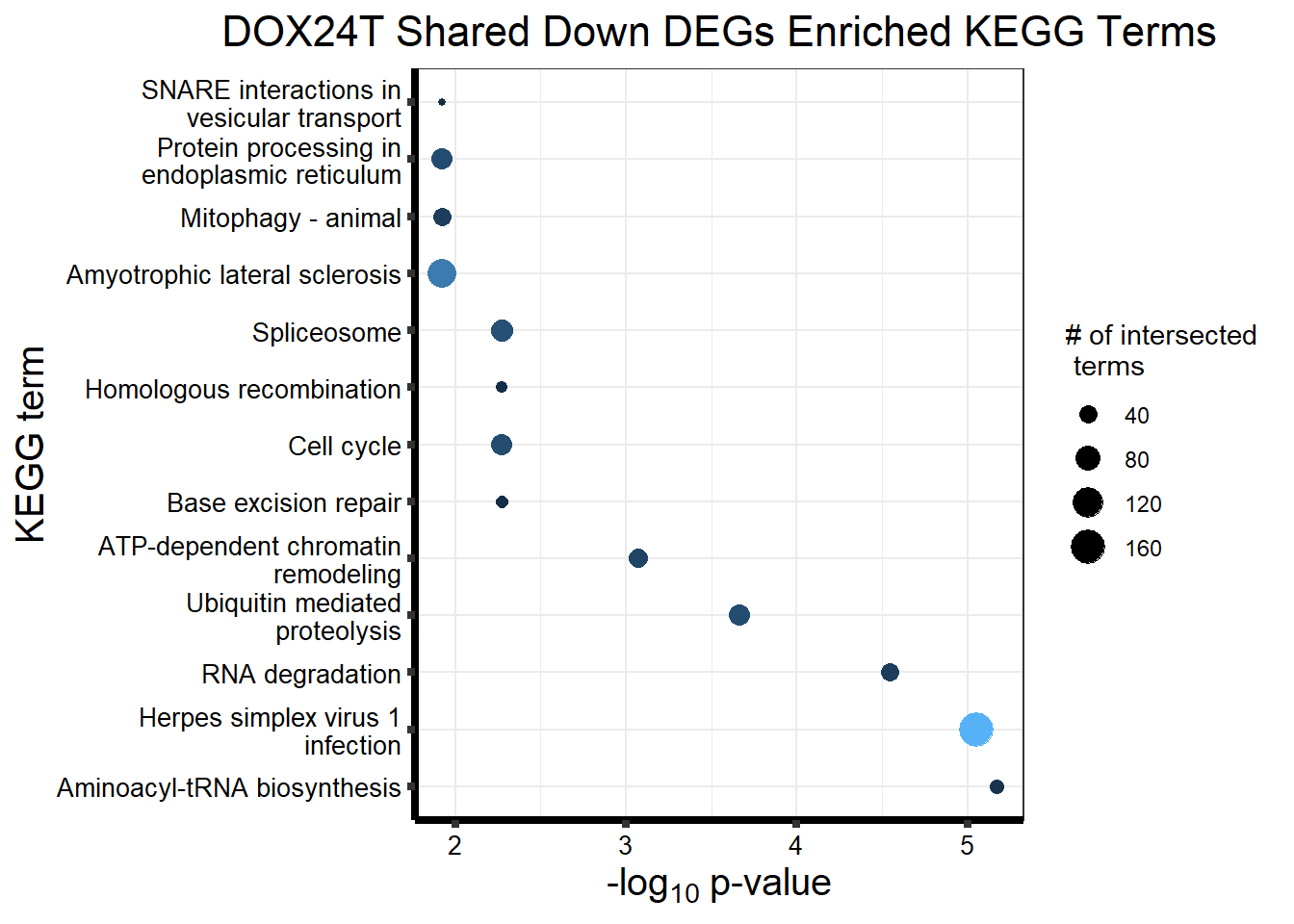
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
###RUVs GO/KEGG for All Conditions
#####DOX24 Genes Specific#####
D24_DEGs_mat_RUV <- as.matrix(DOX24T_DEGs_GO_RUV)
D24_DEGs_vec_RUV <- as.character(DOX24T_DEGs_GO_RUV$Entrez_ID)
#saveRDS(D24_DEGs_mat_RUV, "data/new/RUV/D24_DEGs_mat_RUV.RDS")
DOX_24_dxr_gene_RUV <- gost(query = D24_DEGs_vec_RUV,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
#check if there are any KEGG terms popping up
unique(DOX_24_dxr_gene_RUV$result$source)[1] "GO:BP"#only GO:BP appearing
DOX_24_gost_genes_RUV <- gostplot(DOX_24_dxr_gene_RUV, capped = FALSE, interactive = TRUE)
DOX_24_gost_genes_RUVtable_DOX24_genes_RUV <- DOX_24_dxr_gene_RUV$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX24_genes_RUV %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0048518 | positive regulation of biological process | 353 | 6264 | 1.727e-07 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 313 | 5390 | 1.727e-07 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 335 | 5920 | 3.353e-07 |
| GO:BP | GO:0065007 | biological regulation | 617 | 12743 | 1.482e-05 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 233 | 3990 | 3.463e-05 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 218 | 3687 | 3.463e-05 |
| GO:BP | GO:0050794 | regulation of cellular process | 581 | 11946 | 3.463e-05 |
| GO:BP | GO:0050789 | regulation of biological process | 597 | 12336 | 3.463e-05 |
| GO:BP | GO:0006351 | DNA-templated transcription | 209 | 3549 | 8.673e-05 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 201 | 3409 | 1.245e-04 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 153 | 2433 | 1.245e-04 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 201 | 3428 | 1.717e-04 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 108 | 1588 | 2.373e-04 |
| GO:BP | GO:0030030 | cell projection organization | 110 | 1631 | 2.565e-04 |
| GO:BP | GO:0019538 | protein metabolic process | 260 | 4721 | 3.103e-04 |
| GO:BP | GO:0007049 | cell cycle | 111 | 1663 | 3.283e-04 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 163 | 2711 | 4.684e-04 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 60 | 745 | 4.684e-04 |
| GO:BP | GO:0051726 | regulation of cell cycle | 79 | 1087 | 5.400e-04 |
| GO:BP | GO:0043412 | macromolecule modification | 178 | 3030 | 5.400e-04 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 153 | 2578 | 1.815e-03 |
| GO:BP | GO:0009056 | catabolic process | 156 | 2639 | 1.815e-03 |
| GO:BP | GO:0022402 | cell cycle process | 87 | 1280 | 1.966e-03 |
| GO:BP | GO:0033554 | cellular response to stress | 115 | 1830 | 2.408e-03 |
| GO:BP | GO:0000278 | mitotic cell cycle | 65 | 892 | 3.346e-03 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 93 | 1417 | 3.352e-03 |
| GO:BP | GO:0019222 | regulation of metabolic process | 355 | 7035 | 3.845e-03 |
| GO:BP | GO:0051716 | cellular response to stimulus | 370 | 7376 | 3.845e-03 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 331 | 6492 | 3.845e-03 |
| GO:BP | GO:0051641 | cellular localization | 202 | 3661 | 3.987e-03 |
| GO:BP | GO:0035556 | intracellular signal transduction | 169 | 2965 | 3.987e-03 |
| GO:BP | GO:0007275 | multicellular organism development | 250 | 4727 | 5.918e-03 |
| GO:BP | GO:0036211 | protein modification process | 162 | 2846 | 6.255e-03 |
| GO:BP | GO:0060021 | roof of mouth development | 14 | 94 | 7.333e-03 |
| GO:BP | GO:0048731 | system development | 218 | 4053 | 7.688e-03 |
| GO:BP | GO:0006402 | mRNA catabolic process | 26 | 261 | 8.420e-03 |
| GO:BP | GO:0141188 | nucleic acid catabolic process | 31 | 343 | 1.034e-02 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 38 | 458 | 1.034e-02 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 288 | 5629 | 1.133e-02 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 25 | 252 | 1.133e-02 |
| GO:BP | GO:0006401 | RNA catabolic process | 29 | 315 | 1.170e-02 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 8 | 35 | 1.248e-02 |
| GO:BP | GO:0001967 | suckling behavior | 5 | 12 | 1.248e-02 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 42 | 535 | 1.290e-02 |
| GO:BP | GO:0007507 | heart development | 46 | 605 | 1.290e-02 |
| GO:BP | GO:0140639 | positive regulation of pyroptotic inflammatory response | 4 | 7 | 1.310e-02 |
| GO:BP | GO:0060485 | mesenchyme development | 29 | 320 | 1.310e-02 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 110 | 1835 | 1.310e-02 |
| GO:BP | GO:0060039 | pericardium development | 6 | 19 | 1.310e-02 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 21 | 198 | 1.310e-02 |
| GO:BP | GO:0006996 | organelle organization | 194 | 3594 | 1.330e-02 |
| GO:BP | GO:0033043 | regulation of organelle organization | 75 | 1148 | 1.373e-02 |
| GO:BP | GO:0006974 | DNA damage response | 62 | 908 | 1.598e-02 |
| GO:BP | GO:0051649 | establishment of localization in cell | 118 | 2010 | 1.598e-02 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 110 | 1855 | 1.754e-02 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 65 | 970 | 1.754e-02 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 286 | 5644 | 1.754e-02 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 193 | 3597 | 1.754e-02 |
| GO:BP | GO:0044770 | cell cycle phase transition | 41 | 532 | 1.758e-02 |
| GO:BP | GO:0048519 | negative regulation of biological process | 294 | 5834 | 1.835e-02 |
| GO:BP | GO:0007399 | nervous system development | 146 | 2604 | 1.835e-02 |
| GO:BP | GO:0042475 | odontogenesis of dentin-containing tooth | 13 | 95 | 1.835e-02 |
| GO:BP | GO:0044238 | primary metabolic process | 572 | 12342 | 1.848e-02 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 151 | 2713 | 1.866e-02 |
| GO:BP | GO:0043487 | regulation of RNA stability | 20 | 193 | 1.996e-02 |
| GO:BP | GO:0010468 | regulation of gene expression | 280 | 5536 | 2.115e-02 |
| GO:BP | GO:0006950 | response to stress | 208 | 3948 | 2.181e-02 |
| GO:BP | GO:0048666 | neuron development | 75 | 1177 | 2.252e-02 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 155 | 2819 | 2.382e-02 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 177 | 3288 | 2.382e-02 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 101 | 1699 | 2.382e-02 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 101 | 1697 | 2.382e-02 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 28 | 323 | 2.382e-02 |
| GO:BP | GO:0061635 | regulation of protein complex stability | 5 | 15 | 2.433e-02 |
| GO:BP | GO:0072359 | circulatory system development | 73 | 1145 | 2.433e-02 |
| GO:BP | GO:0048863 | stem cell differentiation | 23 | 245 | 2.437e-02 |
| GO:BP | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis | 3 | 4 | 2.437e-02 |
| GO:BP | GO:0006574 | valine catabolic process | 3 | 4 | 2.437e-02 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 291 | 5809 | 2.437e-02 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 20 | 199 | 2.437e-02 |
| GO:BP | GO:0048856 | anatomical structure development | 299 | 5997 | 2.592e-02 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 31 | 377 | 2.592e-02 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 92 | 1529 | 2.668e-02 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 27 | 312 | 2.715e-02 |
| GO:BP | GO:0044782 | cilium organization | 34 | 431 | 2.832e-02 |
| GO:BP | GO:0009966 | regulation of signal transduction | 164 | 3034 | 2.890e-02 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 49 | 701 | 2.890e-02 |
| GO:BP | GO:0003214 | cardiac left ventricle morphogenesis | 5 | 16 | 2.890e-02 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 50 | 720 | 2.905e-02 |
| GO:BP | GO:0060271 | cilium assembly | 32 | 403 | 3.510e-02 |
| GO:BP | GO:0061564 | axon development | 39 | 528 | 3.778e-02 |
| GO:BP | GO:0030182 | neuron differentiation | 87 | 1451 | 3.906e-02 |
| GO:BP | GO:0065009 | regulation of molecular function | 89 | 1496 | 3.921e-02 |
| GO:BP | GO:0009987 | cellular process | 871 | 20247 | 3.921e-02 |
| GO:BP | GO:0001822 | kidney development | 27 | 322 | 3.921e-02 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 6 | 26 | 3.921e-02 |
| GO:BP | GO:0006399 | tRNA metabolic process | 20 | 210 | 3.921e-02 |
| GO:BP | GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 6 | 26 | 3.921e-02 |
| GO:BP | GO:0051179 | localization | 277 | 5555 | 3.921e-02 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 46 | 658 | 3.921e-02 |
| GO:BP | GO:0048699 | generation of neurons | 91 | 1536 | 3.921e-02 |
| GO:BP | GO:0048208 | COPII vesicle coating | 6 | 26 | 3.921e-02 |
| GO:BP | GO:0007059 | chromosome segregation | 33 | 427 | 3.954e-02 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 74 | 1199 | 3.954e-02 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 16 | 151 | 3.954e-02 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 33 | 427 | 3.954e-02 |
| GO:BP | GO:0031175 | neuron projection development | 65 | 1023 | 3.954e-02 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 10 | 70 | 3.954e-02 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 18 | 181 | 3.954e-02 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 55 | 831 | 3.954e-02 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 84 | 1400 | 3.954e-02 |
| GO:BP | GO:0060029 | convergent extension involved in organogenesis | 3 | 5 | 4.022e-02 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 8 | 47 | 4.022e-02 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 115 | 2036 | 4.022e-02 |
| GO:BP | GO:0010646 | regulation of cell communication | 183 | 3486 | 4.039e-02 |
| GO:BP | GO:0001503 | ossification | 33 | 430 | 4.053e-02 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 26 | 311 | 4.102e-02 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 16 | 153 | 4.102e-02 |
| GO:BP | GO:0006508 | proteolysis | 88 | 1486 | 4.102e-02 |
| GO:BP | GO:0022008 | neurogenesis | 102 | 1771 | 4.102e-02 |
| GO:BP | GO:0030031 | cell projection assembly | 43 | 613 | 4.354e-02 |
| GO:BP | GO:0023051 | regulation of signaling | 182 | 3478 | 4.581e-02 |
| GO:BP | GO:0035239 | tube morphogenesis | 57 | 879 | 4.581e-02 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 17 | 170 | 4.582e-02 |
| GO:BP | GO:0000902 | cell morphogenesis | 63 | 996 | 4.611e-02 |
| GO:BP | GO:0072001 | renal system development | 27 | 332 | 4.696e-02 |
| GO:BP | GO:0140014 | mitotic nuclear division | 24 | 282 | 4.711e-02 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 92 | 1578 | 4.745e-02 |
| GO:BP | GO:0003007 | heart morphogenesis | 23 | 266 | 4.745e-02 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 42 | 600 | 4.793e-02 |
| GO:BP | GO:0000819 | sister chromatid segregation | 21 | 234 | 4.793e-02 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 36 | 492 | 4.959e-02 |
#GO:BP
table_DOX24_genes_GOBP_RUV <- table_DOX24_genes_RUV %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24_genes_GOBP_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Specific DEGs Enriched GO:BP Terms RUVs")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))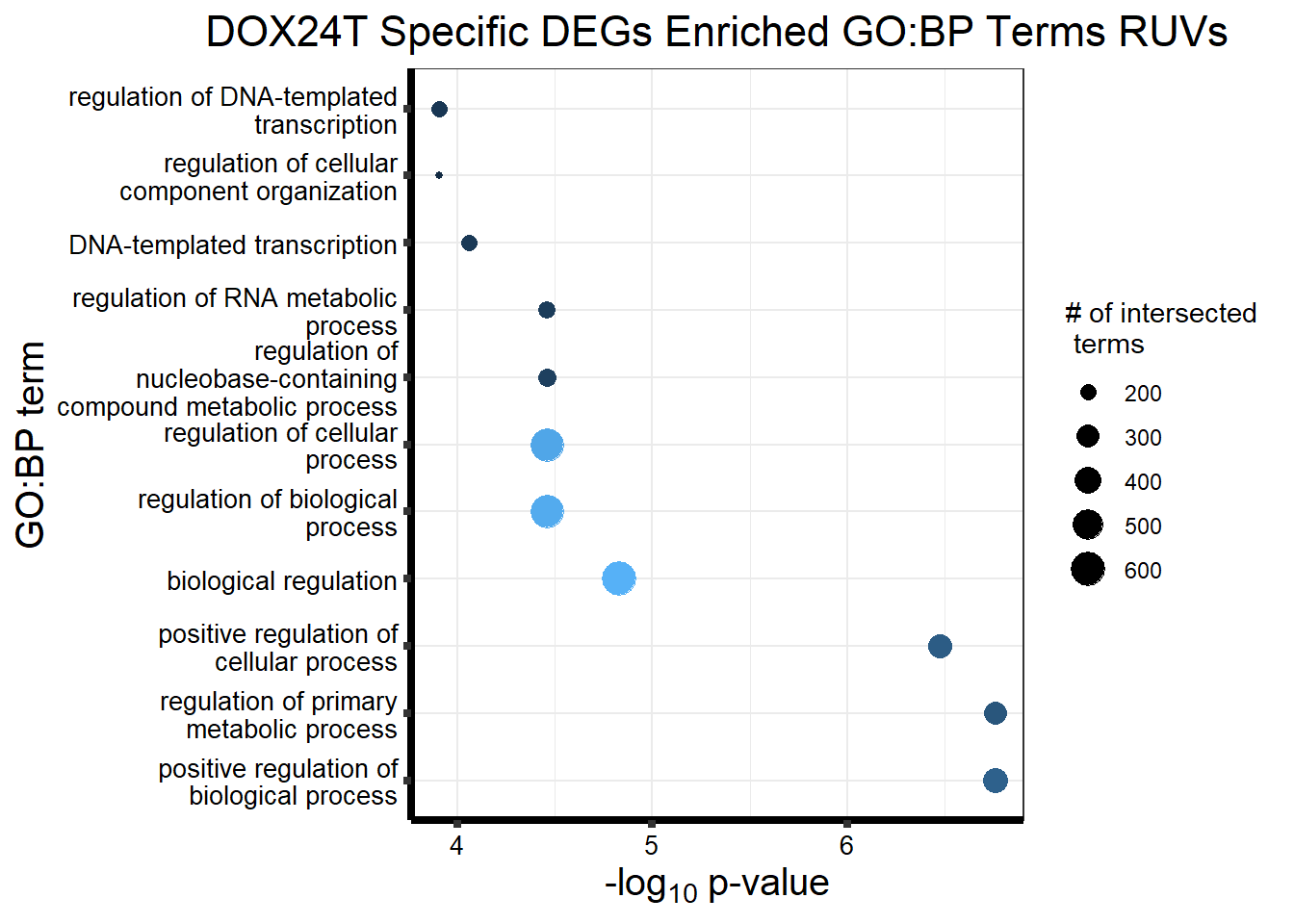
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
# #KEGG - no KEGG terms popped up for this one?
# table_DOX24_genes_KEGG_RUV <- table_DOX24_genes_RUV %>%
# dplyr::filter(source=="KEGG") %>%
# dplyr::select(p_value, term_name, intersection_size) %>%
# dplyr::slice_min(., n=10, order_by=p_value) %>%
# mutate(log_val = -log10(p_value))
#
# table_DOX24_genes_KEGG_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
# geom_point(aes(size = intersection_size)) +
# ggtitle("DOX24T Specific DEGs Enriched KEGG Terms RUVs")+
# xlab(expression("-log"[10]~"p-value"))+
# guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
# ylab("KEGG term")+
# scale_y_discrete(labels = scales::label_wrap(30))+
# theme_bw()+
# theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
# axis.title = element_text(size = 15, colour = "black"),
# axis.ticks = element_line(linewidth = 1.5),
# axis.line = element_line(linewidth = 1.5),
# axis.text = element_text(size = 10, colour = "black", angle = 0),
# strip.text = element_text(size = 15, colour = "black", face = "bold"))
#####DOX24T shared DEGs GO KEGG#####
D24Tshare_DEGs_mat_RUV <- as.matrix(DOX24T_share_DEGs_GO_plot_RUV)
DOX_24Tshare_dxr_gene_RUV <- gost(query = D24Tshare_DEGs_mat_RUV,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
#check if there are any KEGG terms popping up
unique(DOX_24Tshare_dxr_gene_RUV$result$source)[1] "GO:BP"#only GO:BP appearing
DOX_24Tshare_gost_genes_RUV <-
gostplot(DOX_24Tshare_dxr_gene_RUV, capped = FALSE, interactive = TRUE)
DOX_24Tshare_gost_genes_RUVtable_DOX24Tshare_genes_RUV <- DOX_24Tshare_dxr_gene_RUV$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX24Tshare_genes_RUV %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0006996 | organelle organization | 291 | 3594 | 4.937e-10 |
| GO:BP | GO:0035556 | intracellular signal transduction | 249 | 2965 | 6.072e-10 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 389 | 5390 | 5.622e-08 |
| GO:BP | GO:0051179 | localization | 398 | 5555 | 5.869e-08 |
| GO:BP | GO:0019538 | protein metabolic process | 346 | 4721 | 1.339e-07 |
| GO:BP | GO:0048518 | positive regulation of biological process | 434 | 6264 | 3.250e-07 |
| GO:BP | GO:0051649 | establishment of localization in cell | 173 | 2010 | 3.250e-07 |
| GO:BP | GO:0051641 | cellular localization | 278 | 3661 | 3.991e-07 |
| GO:BP | GO:0006810 | transport | 322 | 4407 | 7.021e-07 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 296 | 3990 | 7.212e-07 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 272 | 3597 | 7.212e-07 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 411 | 5920 | 7.212e-07 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 277 | 3687 | 8.117e-07 |
| GO:BP | GO:0023051 | regulation of signaling | 263 | 3478 | 1.246e-06 |
| GO:BP | GO:0009056 | catabolic process | 210 | 2639 | 1.330e-06 |
| GO:BP | GO:0046907 | intracellular transport | 126 | 1381 | 1.706e-06 |
| GO:BP | GO:0010646 | regulation of cell communication | 262 | 3486 | 2.013e-06 |
| GO:BP | GO:0032502 | developmental process | 443 | 6553 | 2.464e-06 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 249 | 3288 | 2.523e-06 |
| GO:BP | GO:0009966 | regulation of signal transduction | 232 | 3034 | 4.014e-06 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 160 | 1912 | 4.014e-06 |
| GO:BP | GO:0009894 | regulation of catabolic process | 100 | 1040 | 4.365e-06 |
| GO:BP | GO:0050789 | regulation of biological process | 755 | 12336 | 7.218e-06 |
| GO:BP | GO:0065007 | biological regulation | 776 | 12743 | 7.325e-06 |
| GO:BP | GO:0048856 | anatomical structure development | 407 | 5997 | 7.864e-06 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 192 | 2433 | 8.227e-06 |
| GO:BP | GO:0007275 | multicellular organism development | 332 | 4727 | 9.381e-06 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 166 | 2036 | 9.381e-06 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 61 | 546 | 1.612e-05 |
| GO:BP | GO:0050794 | regulation of cellular process | 731 | 11946 | 1.927e-05 |
| GO:BP | GO:0051234 | establishment of localization | 341 | 4928 | 2.406e-05 |
| GO:BP | GO:0033036 | macromolecule localization | 239 | 3228 | 2.406e-05 |
| GO:BP | GO:0033043 | regulation of organelle organization | 104 | 1148 | 3.044e-05 |
| GO:BP | GO:0048731 | system development | 288 | 4053 | 3.283e-05 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 150 | 1835 | 3.283e-05 |
| GO:BP | GO:0006351 | DNA-templated transcription | 257 | 3549 | 4.024e-05 |
| GO:BP | GO:0007032 | endosome organization | 20 | 98 | 4.695e-05 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 121 | 1417 | 5.892e-05 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 248 | 3428 | 7.138e-05 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 132 | 1592 | 7.990e-05 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 246 | 3409 | 9.463e-05 |
| GO:BP | GO:0032880 | regulation of protein localization | 84 | 907 | 1.636e-04 |
| GO:BP | GO:0033554 | cellular response to stress | 146 | 1830 | 1.636e-04 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 206 | 2782 | 1.660e-04 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 126 | 1529 | 1.839e-04 |
| GO:BP | GO:0051716 | cellular response to stimulus | 474 | 7376 | 1.935e-04 |
| GO:BP | GO:0060341 | regulation of cellular localization | 91 | 1013 | 1.935e-04 |
| GO:BP | GO:0019222 | regulation of metabolic process | 454 | 7035 | 2.394e-04 |
| GO:BP | GO:1902473 | regulation of protein localization to synapse | 10 | 29 | 2.416e-04 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 278 | 3993 | 2.468e-04 |
| GO:BP | GO:0042254 | ribosome biogenesis | 39 | 323 | 3.382e-04 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 116 | 1400 | 3.396e-04 |
| GO:BP | GO:0008104 | protein localization | 203 | 2770 | 3.433e-04 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 191 | 2578 | 3.562e-04 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 67 | 692 | 3.734e-04 |
| GO:BP | GO:0016072 | rRNA metabolic process | 34 | 266 | 3.844e-04 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 135 | 1699 | 4.088e-04 |
| GO:BP | GO:0043412 | macromolecule modification | 218 | 3030 | 4.486e-04 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 63 | 643 | 4.696e-04 |
| GO:BP | GO:0043009 | chordate embryonic development | 65 | 671 | 4.737e-04 |
| GO:BP | GO:0051640 | organelle localization | 61 | 620 | 5.398e-04 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 103 | 1223 | 5.398e-04 |
| GO:BP | GO:0006364 | rRNA processing | 30 | 225 | 5.398e-04 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 134 | 1697 | 5.398e-04 |
| GO:BP | GO:0023052 | signaling | 421 | 6515 | 5.398e-04 |
| GO:BP | GO:0032879 | regulation of localization | 155 | 2029 | 5.797e-04 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 150 | 1953 | 6.120e-04 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 190 | 2593 | 6.120e-04 |
| GO:BP | GO:0007399 | nervous system development | 190 | 2604 | 7.862e-04 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 31 | 242 | 7.940e-04 |
| GO:BP | GO:0070925 | organelle assembly | 90 | 1046 | 8.425e-04 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 72 | 786 | 8.643e-04 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 196 | 2711 | 9.123e-04 |
| GO:BP | GO:0071705 | nitrogen compound transport | 147 | 1923 | 9.123e-04 |
| GO:BP | GO:0007154 | cell communication | 420 | 6540 | 9.269e-04 |
| GO:BP | GO:0061024 | membrane organization | 74 | 821 | 1.077e-03 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 61 | 638 | 1.091e-03 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 58 | 597 | 1.091e-03 |
| GO:BP | GO:0006914 | autophagy | 58 | 597 | 1.091e-03 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 194 | 2696 | 1.254e-03 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 60 | 628 | 1.254e-03 |
| GO:BP | GO:0016050 | vesicle organization | 41 | 376 | 1.462e-03 |
| GO:BP | GO:0048513 | animal organ development | 217 | 3085 | 1.527e-03 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 40 | 365 | 1.605e-03 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 141 | 1855 | 1.629e-03 |
| GO:BP | GO:0016197 | endosomal transport | 34 | 290 | 1.635e-03 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 81 | 938 | 1.730e-03 |
| GO:BP | GO:1902414 | protein localization to cell junction | 19 | 119 | 1.738e-03 |
| GO:BP | GO:0009790 | embryo development | 94 | 1135 | 1.921e-03 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 520 | 8393 | 1.973e-03 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 53 | 543 | 1.973e-03 |
| GO:BP | GO:0015031 | protein transport | 114 | 1444 | 2.087e-03 |
| GO:BP | GO:0060996 | dendritic spine development | 17 | 101 | 2.174e-03 |
| GO:BP | GO:0051656 | establishment of organelle localization | 49 | 491 | 2.174e-03 |
| GO:BP | GO:0051276 | chromosome organization | 55 | 574 | 2.235e-03 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 14 | 73 | 2.403e-03 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 87 | 1039 | 2.404e-03 |
| GO:BP | GO:0006974 | DNA damage response | 78 | 908 | 2.613e-03 |
| GO:BP | GO:0006950 | response to stress | 266 | 3948 | 2.794e-03 |
| GO:BP | GO:0031503 | protein-containing complex localization | 27 | 215 | 2.950e-03 |
| GO:BP | GO:0097500 | receptor localization to non-motile cilium | 4 | 5 | 2.950e-03 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 38 | 352 | 2.950e-03 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 411 | 6492 | 3.822e-03 |
| GO:BP | GO:0051301 | cell division | 60 | 658 | 3.822e-03 |
| GO:BP | GO:0036211 | protein modification process | 199 | 2846 | 4.177e-03 |
| GO:BP | GO:0001701 | in utero embryonic development | 42 | 413 | 4.608e-03 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 172 | 2407 | 4.648e-03 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 42 | 414 | 4.777e-03 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 172 | 2410 | 4.829e-03 |
| GO:BP | GO:0140352 | export from cell | 78 | 928 | 4.829e-03 |
| GO:BP | GO:0050896 | response to stimulus | 549 | 8999 | 4.844e-03 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 12 | 60 | 4.883e-03 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 102 | 1296 | 5.102e-03 |
| GO:BP | GO:0042592 | homeostatic process | 130 | 1736 | 5.102e-03 |
| GO:BP | GO:0035418 | protein localization to synapse | 15 | 89 | 5.102e-03 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 101 | 1280 | 5.102e-03 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 25 | 200 | 5.208e-03 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 190 | 2713 | 5.208e-03 |
| GO:BP | GO:0048519 | negative regulation of biological process | 372 | 5834 | 5.279e-03 |
| GO:BP | GO:0048285 | organelle fission | 48 | 500 | 5.353e-03 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 114 | 1489 | 5.729e-03 |
| GO:BP | GO:0007165 | signal transduction | 381 | 6002 | 5.918e-03 |
| GO:BP | GO:0060322 | head development | 69 | 806 | 6.498e-03 |
| GO:BP | GO:0007417 | central nervous system development | 86 | 1061 | 6.507e-03 |
| GO:BP | GO:1902683 | regulation of receptor localization to synapse | 8 | 29 | 6.531e-03 |
| GO:BP | GO:1905870 | positive regulation of 3’-UTR-mediated mRNA stabilization | 4 | 6 | 6.855e-03 |
| GO:BP | GO:0065008 | regulation of biological quality | 203 | 2947 | 6.856e-03 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 359 | 5629 | 6.961e-03 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 12 | 63 | 6.961e-03 |
| GO:BP | GO:0001889 | liver development | 20 | 147 | 7.373e-03 |
| GO:BP | GO:0043112 | receptor metabolic process | 13 | 73 | 7.373e-03 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 195 | 2819 | 7.373e-03 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 57 | 637 | 7.540e-03 |
| GO:BP | GO:0050793 | regulation of developmental process | 173 | 2457 | 7.616e-03 |
| GO:BP | GO:0051648 | vesicle localization | 26 | 219 | 7.803e-03 |
| GO:BP | GO:0140014 | mitotic nuclear division | 31 | 282 | 7.890e-03 |
| GO:BP | GO:0007249 | canonical NF-kappaB signal transduction | 33 | 308 | 7.928e-03 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 98 | 1256 | 8.057e-03 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 16 | 105 | 8.511e-03 |
| GO:BP | GO:0043401 | steroid hormone receptor signaling pathway | 19 | 138 | 8.593e-03 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 18 | 127 | 8.672e-03 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 15 | 95 | 8.833e-03 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 20 | 150 | 8.929e-03 |
| GO:BP | GO:0030163 | protein catabolic process | 83 | 1030 | 8.929e-03 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 102 | 1326 | 9.474e-03 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 33 | 312 | 9.474e-03 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 57 | 646 | 9.833e-03 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 47 | 505 | 1.056e-02 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 367 | 5809 | 1.059e-02 |
| GO:BP | GO:0006401 | RNA catabolic process | 33 | 315 | 1.101e-02 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 49 | 535 | 1.119e-02 |
| GO:BP | GO:0033365 | protein localization to organelle | 94 | 1209 | 1.128e-02 |
| GO:BP | GO:0141193 | nuclear receptor-mediated signaling pathway | 22 | 177 | 1.135e-02 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 118 | 1588 | 1.137e-02 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 6 | 18 | 1.226e-02 |
| GO:BP | GO:0043084 | penile erection | 4 | 7 | 1.226e-02 |
| GO:BP | GO:0002328 | pro-B cell differentiation | 5 | 12 | 1.226e-02 |
| GO:BP | GO:1903441 | protein localization to ciliary membrane | 5 | 12 | 1.226e-02 |
| GO:BP | GO:1990456 | mitochondrion-endoplasmic reticulum membrane tethering | 4 | 7 | 1.226e-02 |
| GO:BP | GO:0000278 | mitotic cell cycle | 73 | 892 | 1.250e-02 |
| GO:BP | GO:0055088 | lipid homeostasis | 21 | 167 | 1.261e-02 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 85 | 1077 | 1.309e-02 |
| GO:BP | GO:0051338 | regulation of transferase activity | 46 | 498 | 1.319e-02 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 63 | 745 | 1.338e-02 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 60 | 701 | 1.346e-02 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 29 | 268 | 1.362e-02 |
| GO:BP | GO:0030030 | cell projection organization | 120 | 1631 | 1.371e-02 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 356 | 5644 | 1.412e-02 |
| GO:BP | GO:0032940 | secretion by cell | 70 | 854 | 1.505e-02 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 37 | 377 | 1.534e-02 |
| GO:BP | GO:0098696 | regulation of neurotransmitter receptor localization to postsynaptic specialization membrane | 6 | 19 | 1.573e-02 |
| GO:BP | GO:0140058 | neuron projection arborization | 8 | 34 | 1.578e-02 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 54 | 619 | 1.607e-02 |
| GO:BP | GO:0007507 | heart development | 53 | 605 | 1.632e-02 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 18 | 136 | 1.646e-02 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 12 | 71 | 1.646e-02 |
| GO:BP | GO:2000303 | regulation of ceramide biosynthetic process | 5 | 13 | 1.688e-02 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 21 | 172 | 1.688e-02 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 21 | 172 | 1.688e-02 |
| GO:BP | GO:0097499 | protein localization to non-motile cilium | 5 | 13 | 1.688e-02 |
| GO:BP | GO:0048869 | cellular developmental process | 286 | 4438 | 1.717e-02 |
| GO:BP | GO:0006402 | mRNA catabolic process | 28 | 261 | 1.798e-02 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 20 | 161 | 1.801e-02 |
| GO:BP | GO:0016233 | telomere capping | 7 | 27 | 1.812e-02 |
| GO:BP | GO:0030325 | adrenal gland development | 7 | 27 | 1.812e-02 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 15 | 104 | 1.842e-02 |
| GO:BP | GO:0000280 | nuclear division | 42 | 452 | 1.842e-02 |
| GO:BP | GO:0044238 | primary metabolic process | 721 | 12342 | 1.858e-02 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 199 | 2956 | 1.876e-02 |
| GO:BP | GO:1905868 | regulation of 3’-UTR-mediated mRNA stabilization | 4 | 8 | 1.933e-02 |
| GO:BP | GO:2000973 | regulation of pro-B cell differentiation | 4 | 8 | 1.933e-02 |
| GO:BP | GO:0141188 | nucleic acid catabolic process | 34 | 343 | 1.935e-02 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 15 | 105 | 1.970e-02 |
| GO:BP | GO:0001501 | skeletal system development | 48 | 540 | 1.970e-02 |
| GO:BP | GO:0071396 | cellular response to lipid | 53 | 613 | 1.975e-02 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 19 | 151 | 1.975e-02 |
| GO:BP | GO:0006403 | RNA localization | 23 | 200 | 1.991e-02 |
| GO:BP | GO:0030154 | cell differentiation | 285 | 4437 | 1.999e-02 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 57 | 673 | 2.021e-02 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 24 | 213 | 2.021e-02 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 13 | 84 | 2.027e-02 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 40 | 428 | 2.045e-02 |
| GO:BP | GO:0046903 | secretion | 78 | 992 | 2.057e-02 |
| GO:BP | GO:0022008 | neurogenesis | 127 | 1771 | 2.082e-02 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 55 | 645 | 2.082e-02 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 11 | 64 | 2.096e-02 |
| GO:BP | GO:0023057 | negative regulation of signaling | 106 | 1435 | 2.144e-02 |
| GO:BP | GO:0090153 | regulation of sphingolipid biosynthetic process | 5 | 14 | 2.144e-02 |
| GO:BP | GO:0031860 | telomeric 3’ overhang formation | 3 | 4 | 2.144e-02 |
| GO:BP | GO:0016482 | cytosolic transport | 18 | 141 | 2.144e-02 |
| GO:BP | GO:0000209 | protein polyubiquitination | 29 | 280 | 2.201e-02 |
| GO:BP | GO:0010468 | regulation of gene expression | 347 | 5536 | 2.232e-02 |
| GO:BP | GO:0032543 | mitochondrial translation | 17 | 130 | 2.243e-02 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 10 | 55 | 2.243e-02 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 100 | 1343 | 2.289e-02 |
| GO:BP | GO:0016358 | dendrite development | 26 | 242 | 2.309e-02 |
| GO:BP | GO:0007420 | brain development | 62 | 754 | 2.309e-02 |
| GO:BP | GO:0008219 | cell death | 140 | 1991 | 2.309e-02 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 25 | 230 | 2.437e-02 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 32 | 323 | 2.456e-02 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 10 | 56 | 2.518e-02 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 54 | 638 | 2.552e-02 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 51 | 594 | 2.564e-02 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 22 | 193 | 2.567e-02 |
| GO:BP | GO:0043487 | regulation of RNA stability | 22 | 193 | 2.567e-02 |
| GO:BP | GO:0021537 | telencephalon development | 29 | 284 | 2.579e-02 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 36 | 380 | 2.629e-02 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 16 | 121 | 2.631e-02 |
| GO:BP | GO:0030518 | nuclear receptor-mediated steroid hormone signaling pathway | 16 | 121 | 2.631e-02 |
| GO:BP | GO:0006259 | DNA metabolic process | 78 | 1005 | 2.642e-02 |
| GO:BP | GO:0030900 | forebrain development | 39 | 423 | 2.685e-02 |
| GO:BP | GO:0010506 | regulation of autophagy | 35 | 367 | 2.685e-02 |
| GO:BP | GO:0070837 | dehydroascorbic acid transport | 4 | 9 | 2.713e-02 |
| GO:BP | GO:0010994 | free ubiquitin chain polymerization | 4 | 9 | 2.713e-02 |
| GO:BP | GO:0006886 | intracellular protein transport | 57 | 686 | 2.713e-02 |
| GO:BP | GO:0012501 | programmed cell death | 139 | 1987 | 2.715e-02 |
| GO:BP | GO:0032402 | melanosome transport | 6 | 22 | 2.715e-02 |
| GO:BP | GO:0010256 | endomembrane system organization | 52 | 613 | 2.800e-02 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 7 | 30 | 2.800e-02 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 105 | 1436 | 2.814e-02 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 31 | 314 | 2.820e-02 |
| GO:BP | GO:0043122 | regulation of canonical NF-kappaB signal transduction | 29 | 287 | 2.820e-02 |
| GO:BP | GO:0000819 | sister chromatid segregation | 25 | 234 | 2.820e-02 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 24 | 221 | 2.820e-02 |
| GO:BP | GO:0016043 | cellular component organization | 493 | 8184 | 2.831e-02 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 120 | 1680 | 2.831e-02 |
| GO:BP | GO:0048732 | gland development | 41 | 454 | 2.831e-02 |
| GO:BP | GO:0001881 | receptor recycling | 9 | 48 | 2.836e-02 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 57 | 689 | 2.837e-02 |
| GO:BP | GO:0007059 | chromosome segregation | 39 | 427 | 2.962e-02 |
| GO:BP | GO:0022402 | cell cycle process | 95 | 1280 | 2.962e-02 |
| GO:BP | GO:0032259 | methylation | 25 | 236 | 3.071e-02 |
| GO:BP | GO:0097120 | receptor localization to synapse | 12 | 79 | 3.077e-02 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 76 | 983 | 3.089e-02 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 35 | 372 | 3.098e-02 |
| GO:BP | GO:0051904 | pigment granule transport | 6 | 23 | 3.234e-02 |
| GO:BP | GO:0032401 | establishment of melanosome localization | 6 | 23 | 3.234e-02 |
| GO:BP | GO:0031399 | regulation of protein modification process | 80 | 1049 | 3.338e-02 |
| GO:BP | GO:0051783 | regulation of nuclear division | 18 | 149 | 3.338e-02 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 24 | 225 | 3.374e-02 |
| GO:BP | GO:0043549 | regulation of kinase activity | 39 | 431 | 3.386e-02 |
| GO:BP | GO:0072359 | circulatory system development | 86 | 1145 | 3.436e-02 |
| GO:BP | GO:0006412 | translation | 59 | 727 | 3.485e-02 |
| GO:BP | GO:0034330 | cell junction organization | 66 | 834 | 3.496e-02 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 11 | 70 | 3.515e-02 |
| GO:BP | GO:0050808 | synapse organization | 48 | 563 | 3.515e-02 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 14 | 103 | 3.567e-02 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 12 | 81 | 3.634e-02 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 23 | 214 | 3.691e-02 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 17 | 139 | 3.732e-02 |
| GO:BP | GO:0033993 | response to lipid | 72 | 930 | 3.732e-02 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 17 | 139 | 3.732e-02 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 43 | 492 | 3.732e-02 |
| GO:BP | GO:0045444 | fat cell differentiation | 25 | 241 | 3.797e-02 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 43 | 493 | 3.810e-02 |
| GO:BP | GO:0051905 | establishment of pigment granule localization | 6 | 24 | 3.810e-02 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 46 | 537 | 3.810e-02 |
| GO:BP | GO:0002320 | lymphoid progenitor cell differentiation | 6 | 24 | 3.810e-02 |
| GO:BP | GO:0045900 | negative regulation of translational elongation | 3 | 5 | 3.834e-02 |
| GO:BP | GO:1904978 | regulation of endosome organization | 3 | 5 | 3.834e-02 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 33 | 351 | 3.892e-02 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 10 | 61 | 3.915e-02 |
| GO:BP | GO:0106027 | neuron projection organization | 13 | 94 | 4.213e-02 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 26 | 257 | 4.253e-02 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 50 | 600 | 4.253e-02 |
| GO:BP | GO:0070848 | response to growth factor | 58 | 721 | 4.296e-02 |
| GO:BP | GO:0016567 | protein ubiquitination | 61 | 768 | 4.441e-02 |
| GO:BP | GO:0006915 | apoptotic process | 133 | 1923 | 4.441e-02 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 15 | 118 | 4.477e-02 |
| GO:BP | GO:0051046 | regulation of secretion | 51 | 617 | 4.484e-02 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 31 | 327 | 4.502e-02 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 13 | 95 | 4.502e-02 |
| GO:BP | GO:0032967 | positive regulation of collagen biosynthetic process | 6 | 25 | 4.505e-02 |
| GO:BP | GO:0032400 | melanosome localization | 6 | 25 | 4.505e-02 |
| GO:BP | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 6 | 25 | 4.505e-02 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 21 | 193 | 4.549e-02 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 149 | 2192 | 4.549e-02 |
| GO:BP | GO:0007049 | cell cycle | 117 | 1663 | 4.596e-02 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 19 | 168 | 4.678e-02 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 8 | 43 | 4.694e-02 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 10 | 63 | 4.716e-02 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 20 | 181 | 4.725e-02 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 71 | 928 | 4.881e-02 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 117 | 1667 | 4.887e-02 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 17 | 144 | 4.912e-02 |
| GO:BP | GO:0007614 | short-term memory | 4 | 11 | 4.942e-02 |
| GO:BP | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane | 4 | 11 | 4.942e-02 |
| GO:BP | GO:0006784 | heme A biosynthetic process | 4 | 11 | 4.942e-02 |
| GO:BP | GO:0046160 | heme a metabolic process | 4 | 11 | 4.942e-02 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 117 | 1669 | 4.974e-02 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 38 | 430 | 4.974e-02 |
#GO:BP
table_DOX24Tshare_genes_GOBP_RUV <- table_DOX24Tshare_genes_RUV %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24Tshare_genes_GOBP_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24T Shared DEGs Enriched GO:BP Terms RUVs")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))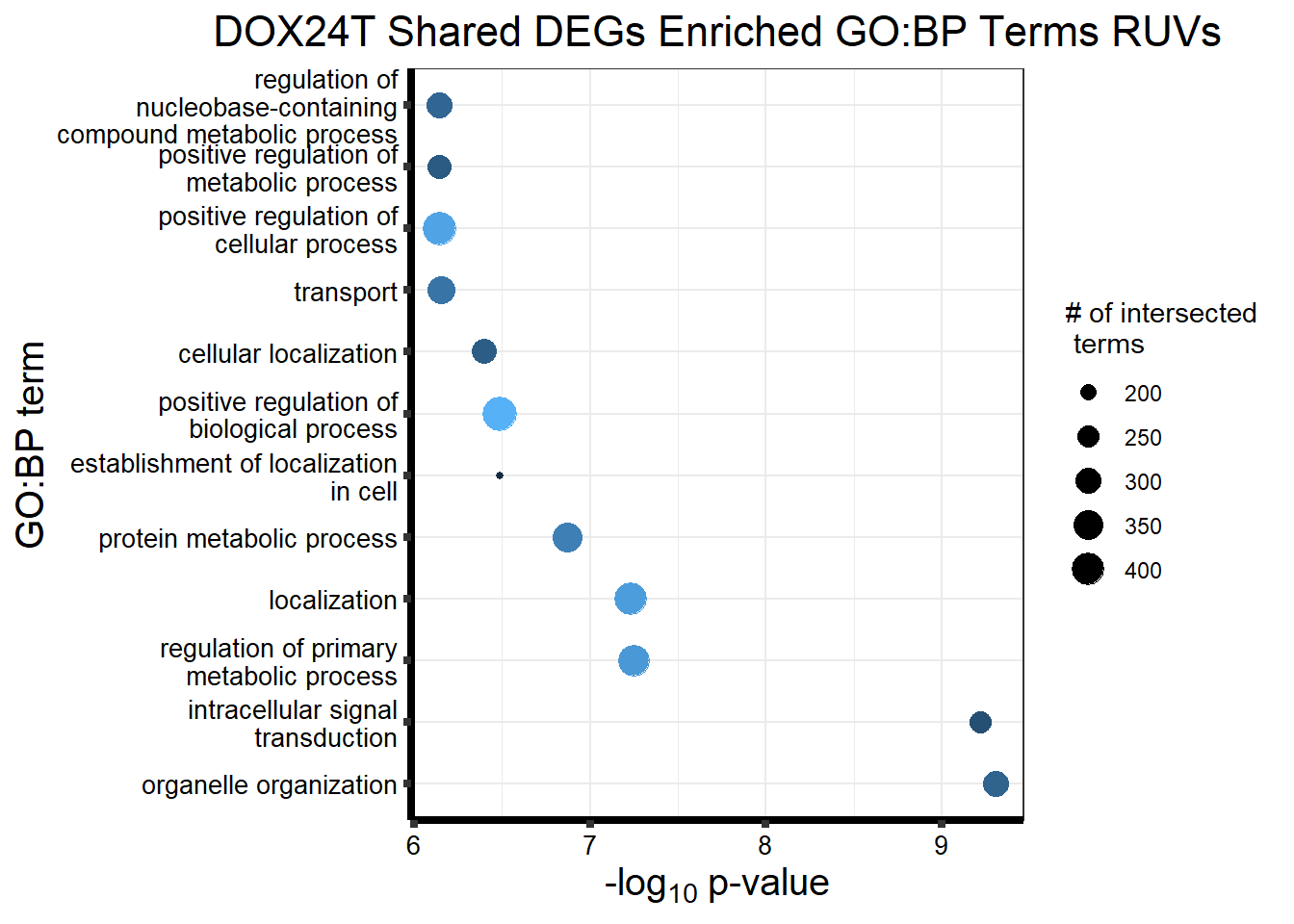
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
# #KEGG
# table_DOX24Tshare_genes_KEGG_RUV <- table_DOX24Tshare_genes_RUV %>%
# dplyr::filter(source=="KEGG") %>%
# dplyr::select(p_value, term_name, intersection_size) %>%
# dplyr::slice_min(., n=10, order_by=p_value) %>%
# mutate(log_val = -log10(p_value))
#
# table_DOX24Tshare_genes_KEGG_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
# geom_point(aes(size = intersection_size)) +
# ggtitle("DOX24T Shared DEGs Enriched KEGG Terms RUVs")+
# xlab(expression("-log"[10]~"p-value"))+
# guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
# ylab("KEGG term")+
# scale_y_discrete(labels = scales::label_wrap(30))+
# theme_bw()+
# theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
# axis.title = element_text(size = 15, colour = "black"),
# axis.ticks = element_line(linewidth = 1.5),
# axis.line = element_line(linewidth = 1.5),
# axis.text = element_text(size = 10, colour = "black", angle = 0),
# strip.text = element_text(size = 15, colour = "black", face = "bold"))
#####DOX24R DEGs GO KEGG#####
D24R_DEGs_mat_RUV <- as.matrix(DOX24R_DEGs_GO_RUV)
DOX_24R_dxr_gene_RUV <- gost(query = D24R_DEGs_mat_RUV,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
#check if there are any KEGG terms popping up
unique(DOX_24R_dxr_gene_RUV$result$source)[1] "GO:BP"#only GO:BP appearing
DOX_24R_gost_genes_RUV <-
gostplot(DOX_24R_dxr_gene_RUV, capped = FALSE, interactive = TRUE)
DOX_24R_gost_genes_RUVtable_DOX24R_genes_RUV <- DOX_24R_dxr_gene_RUV$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX24R_genes_RUV %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0019538 | protein metabolic process | 150 | 4721 | 7.286e-05 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 87 | 2407 | 4.484e-04 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 87 | 2410 | 4.484e-04 |
| GO:BP | GO:0048731 | system development | 124 | 4053 | 2.938e-03 |
| GO:BP | GO:0048518 | positive regulation of biological process | 176 | 6264 | 2.938e-03 |
| GO:BP | GO:0036211 | protein modification process | 94 | 2846 | 2.938e-03 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 168 | 5920 | 2.938e-03 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 36 | 779 | 4.837e-03 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 67 | 1855 | 4.837e-03 |
| GO:BP | GO:0006629 | lipid metabolic process | 54 | 1391 | 4.837e-03 |
| GO:BP | GO:0031399 | regulation of protein modification process | 44 | 1049 | 4.955e-03 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 153 | 5390 | 5.410e-03 |
| GO:BP | GO:0032879 | regulation of localization | 70 | 2029 | 8.866e-03 |
| GO:BP | GO:0006468 | protein phosphorylation | 48 | 1227 | 9.315e-03 |
| GO:BP | GO:0034311 | diol metabolic process | 6 | 31 | 9.494e-03 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 8 | 61 | 9.494e-03 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 44 | 1105 | 9.494e-03 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 22 | 398 | 9.494e-03 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 36 | 830 | 9.494e-03 |
| GO:BP | GO:0048519 | negative regulation of biological process | 161 | 5834 | 9.494e-03 |
| GO:BP | GO:0043412 | macromolecule modification | 95 | 3030 | 9.494e-03 |
| GO:BP | GO:0006950 | response to stress | 117 | 3948 | 9.494e-03 |
| GO:BP | GO:0007049 | cell cycle | 59 | 1663 | 1.114e-02 |
| GO:BP | GO:0007275 | multicellular organism development | 134 | 4727 | 1.238e-02 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 39 | 956 | 1.238e-02 |
| GO:BP | GO:0048513 | animal organ development | 95 | 3085 | 1.238e-02 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 39 | 957 | 1.238e-02 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 17 | 272 | 1.273e-02 |
| GO:BP | GO:0032502 | developmental process | 175 | 6553 | 1.356e-02 |
| GO:BP | GO:0034312 | diol biosynthetic process | 5 | 22 | 1.356e-02 |
| GO:BP | GO:0016310 | phosphorylation | 49 | 1320 | 1.356e-02 |
| GO:BP | GO:0042311 | vasodilation | 7 | 51 | 1.429e-02 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 29 | 637 | 1.429e-02 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 24 | 483 | 1.485e-02 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 30 | 677 | 1.608e-02 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 16 | 257 | 1.640e-02 |
| GO:BP | GO:0033554 | cellular response to stress | 62 | 1830 | 1.640e-02 |
| GO:BP | GO:0033036 | macromolecule localization | 97 | 3228 | 1.669e-02 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 58 | 1685 | 1.726e-02 |
| GO:BP | GO:0080134 | regulation of response to stress | 50 | 1387 | 1.726e-02 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 9 | 94 | 2.015e-02 |
| GO:BP | GO:0035556 | intracellular signal transduction | 90 | 2965 | 2.015e-02 |
| GO:BP | GO:0072359 | circulatory system development | 43 | 1145 | 2.031e-02 |
| GO:BP | GO:0006259 | DNA metabolic process | 39 | 1005 | 2.055e-02 |
| GO:BP | GO:0006996 | organelle organization | 105 | 3594 | 2.055e-02 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 15 | 242 | 2.199e-02 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 24 | 506 | 2.199e-02 |
| GO:BP | GO:0006974 | DNA damage response | 36 | 908 | 2.263e-02 |
| GO:BP | GO:0051726 | regulation of cell cycle | 41 | 1087 | 2.318e-02 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 76 | 2433 | 2.563e-02 |
| GO:BP | GO:0043010 | camera-type eye development | 19 | 363 | 2.733e-02 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 15 | 250 | 2.769e-02 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 8 | 80 | 2.769e-02 |
| GO:BP | GO:0008283 | cell population proliferation | 65 | 2015 | 2.908e-02 |
| GO:BP | GO:0006729 | tetrahydrobiopterin biosynthetic process | 3 | 7 | 2.908e-02 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 29 | 686 | 2.908e-02 |
| GO:BP | GO:0048856 | anatomical structure development | 159 | 5997 | 2.947e-02 |
| GO:BP | GO:0097009 | energy homeostasis | 8 | 83 | 3.155e-02 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 40 | 1077 | 3.155e-02 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 30 | 729 | 3.322e-02 |
| GO:BP | GO:0051049 | regulation of transport | 54 | 1607 | 3.475e-02 |
| GO:BP | GO:0032880 | regulation of protein localization | 35 | 907 | 3.532e-02 |
| GO:BP | GO:0055006 | cardiac cell development | 8 | 86 | 3.577e-02 |
| GO:BP | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway | 2 | 2 | 3.577e-02 |
| GO:BP | GO:0060661 | submandibular salivary gland formation | 2 | 2 | 3.577e-02 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 5 | 32 | 3.577e-02 |
| GO:BP | GO:0046146 | tetrahydrobiopterin metabolic process | 3 | 8 | 3.577e-02 |
| GO:BP | GO:0007399 | nervous system development | 79 | 2604 | 3.577e-02 |
| GO:BP | GO:0007417 | central nervous system development | 39 | 1061 | 3.577e-02 |
| GO:BP | GO:0010646 | regulation of cell communication | 100 | 3486 | 3.577e-02 |
| GO:BP | GO:0019751 | polyol metabolic process | 9 | 109 | 3.577e-02 |
| GO:BP | GO:0023051 | regulation of signaling | 100 | 3478 | 3.577e-02 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 5 | 32 | 3.577e-02 |
| GO:BP | GO:0022402 | cell cycle process | 45 | 1280 | 3.577e-02 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 7 | 67 | 3.577e-02 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 36 | 954 | 3.606e-02 |
| GO:BP | GO:0001654 | eye development | 20 | 415 | 3.638e-02 |
| GO:BP | GO:0030540 | female genitalia development | 4 | 19 | 3.921e-02 |
| GO:BP | GO:0150063 | visual system development | 20 | 419 | 3.985e-02 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 149 | 5629 | 3.985e-02 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 26 | 619 | 4.255e-02 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 40 | 1113 | 4.255e-02 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 19 | 392 | 4.255e-02 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 29 | 720 | 4.255e-02 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 14 | 244 | 4.255e-02 |
| GO:BP | GO:0006281 | DNA repair | 26 | 621 | 4.376e-02 |
| GO:BP | GO:0048880 | sensory system development | 20 | 426 | 4.449e-02 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 111 | 3993 | 4.449e-02 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 24 | 560 | 4.707e-02 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 24 | 560 | 4.707e-02 |
| GO:BP | GO:0030154 | cell differentiation | 121 | 4437 | 4.707e-02 |
| GO:BP | GO:0043542 | endothelial cell migration | 13 | 221 | 4.707e-02 |
| GO:BP | GO:0048869 | cellular developmental process | 121 | 4438 | 4.707e-02 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 9 | 117 | 4.707e-02 |
| GO:BP | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | 10 | 142 | 4.715e-02 |
| GO:BP | GO:0050789 | regulation of biological process | 291 | 12336 | 4.775e-02 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 16 | 309 | 4.824e-02 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 10 | 143 | 4.825e-02 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 19 | 402 | 4.874e-02 |
| GO:BP | GO:0065007 | biological regulation | 299 | 12743 | 4.967e-02 |
#GO:BP
table_DOX24R_genes_GOBP_RUV <- table_DOX24R_genes_RUV %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX24R_genes_GOBP_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX24R Specific DEGs Enriched GO:BP Terms RUVs")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))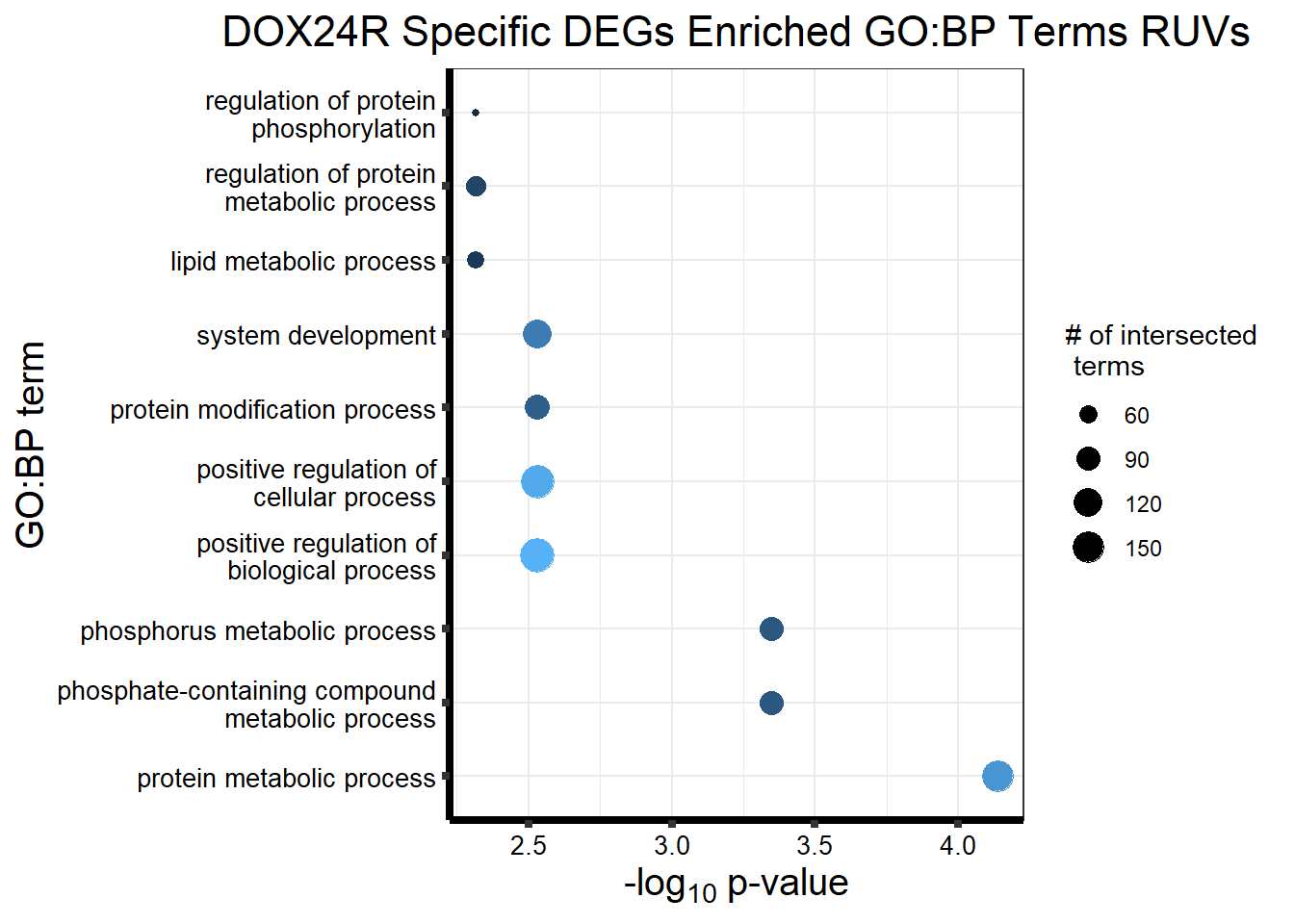
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
# #KEGG
# table_DOX24R_genes_KEGG_RUV <- table_DOX24R_genes_RUV %>%
# dplyr::filter(source=="KEGG") %>%
# dplyr::select(p_value, term_name, intersection_size) %>%
# dplyr::slice_min(., n=10, order_by=p_value) %>%
# mutate(log_val = -log10(p_value))
#
# table_DOX24R_genes_KEGG_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
# geom_point(aes(size = intersection_size)) +
# ggtitle("DOX24R DEGs Enriched KEGG Terms RUVs")+
# xlab(expression("-log"[10]~"p-value"))+
# guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
# ylab("KEGG term")+
# scale_y_discrete(labels = scales::label_wrap(30))+
# theme_bw()+
# theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
# axis.title = element_text(size = 15, colour = "black"),
# axis.ticks = element_line(linewidth = 1.5),
# axis.line = element_line(linewidth = 1.5),
# axis.text = element_text(size = 10, colour = "black", angle = 0),
# strip.text = element_text(size = 15, colour = "black", face = "bold"))
####DOX144R DEGs GO KEGG#####
D144R_DEGs_mat_RUV <- as.matrix(DOX144R_DEGs_GO_RUV)
DOX_144R_dxr_gene_RUV <- gost(query = D144R_DEGs_mat_RUV,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
#check if there are any KEGG terms popping up
unique(DOX_144R_dxr_gene_RUV$result$source)[1] "GO:BP" "KEGG" #GO:BP and KEGG appearing here
DOX_144R_gost_genes_RUV <-
gostplot(DOX_144R_dxr_gene_RUV, capped = FALSE, interactive = TRUE)
DOX_144R_gost_genes_RUVtable_DOX144R_genes_RUV <- DOX_144R_dxr_gene_RUV$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX144R_genes_RUV %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0033058 | directional locomotion | 1 | 3 | 1.726e-02 |
| GO:BP | GO:0007215 | glutamate receptor signaling pathway | 1 | 51 | 2.781e-02 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 1 | 33 | 2.781e-02 |
| GO:BP | GO:1990806 | ligand-gated ion channel signaling pathway | 1 | 37 | 2.781e-02 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 1 | 55 | 2.781e-02 |
| GO:BP | GO:0098976 | excitatory chemical synaptic transmission | 1 | 12 | 2.781e-02 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 1 | 50 | 2.781e-02 |
| GO:BP | GO:0051968 | positive regulation of synaptic transmission, glutamatergic | 1 | 36 | 2.781e-02 |
| GO:BP | GO:0050885 | neuromuscular process controlling balance | 1 | 56 | 2.781e-02 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 1 | 56 | 2.781e-02 |
| GO:BP | GO:0048168 | regulation of neuronal synaptic plasticity | 1 | 58 | 2.781e-02 |
| GO:BP | GO:0035235 | ionotropic glutamate receptor signaling pathway | 1 | 23 | 2.781e-02 |
| GO:BP | GO:1902414 | protein localization to cell junction | 1 | 119 | 3.261e-02 |
| GO:BP | GO:0051966 | regulation of synaptic transmission, glutamatergic | 1 | 77 | 3.261e-02 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 1 | 117 | 3.261e-02 |
| GO:BP | GO:0060079 | excitatory postsynaptic potential | 1 | 108 | 3.261e-02 |
| GO:BP | GO:0060291 | long-term synaptic potentiation | 1 | 110 | 3.261e-02 |
| GO:BP | GO:0035418 | protein localization to synapse | 1 | 89 | 3.261e-02 |
| GO:BP | GO:0035249 | synaptic transmission, glutamatergic | 1 | 109 | 3.261e-02 |
| GO:BP | GO:0099565 | chemical synaptic transmission, postsynaptic | 1 | 118 | 3.261e-02 |
| GO:BP | GO:0031644 | regulation of nervous system process | 1 | 118 | 3.261e-02 |
| GO:BP | GO:0060078 | regulation of postsynaptic membrane potential | 1 | 148 | 3.871e-02 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 1 | 180 | 4.166e-02 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 1 | 181 | 4.166e-02 |
| GO:BP | GO:0050905 | neuromuscular process | 1 | 175 | 4.166e-02 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 1 | 223 | 4.936e-02 |
| GO:BP | GO:0019722 | calcium-mediated signaling | 1 | 234 | 4.987e-02 |
| KEGG | KEGG:05031 | Amphetamine addiction | 1 | 69 | 3.253e-02 |
| KEGG | KEGG:05030 | Cocaine addiction | 1 | 49 | 3.253e-02 |
| KEGG | KEGG:05033 | Nicotine addiction | 1 | 40 | 3.253e-02 |
| KEGG | KEGG:04720 | Long-term potentiation | 1 | 67 | 3.253e-02 |
| KEGG | KEGG:04724 | Glutamatergic synapse | 1 | 115 | 3.615e-02 |
| KEGG | KEGG:04713 | Circadian entrainment | 1 | 97 | 3.615e-02 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 1 | 141 | 3.799e-02 |
| KEGG | KEGG:05034 | Alcoholism | 1 | 187 | 4.408e-02 |
| KEGG | KEGG:04020 | Calcium signaling pathway | 1 | 251 | 4.646e-02 |
| KEGG | KEGG:05020 | Prion disease | 1 | 271 | 4.646e-02 |
| KEGG | KEGG:04024 | cAMP signaling pathway | 1 | 223 | 4.646e-02 |
#GO:BP
table_DOX144R_genes_GOBP_RUV <- table_DOX144R_genes_RUV %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX144R_genes_GOBP_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX144R Specific DEGs Enriched GO:BP Terms RUVs")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))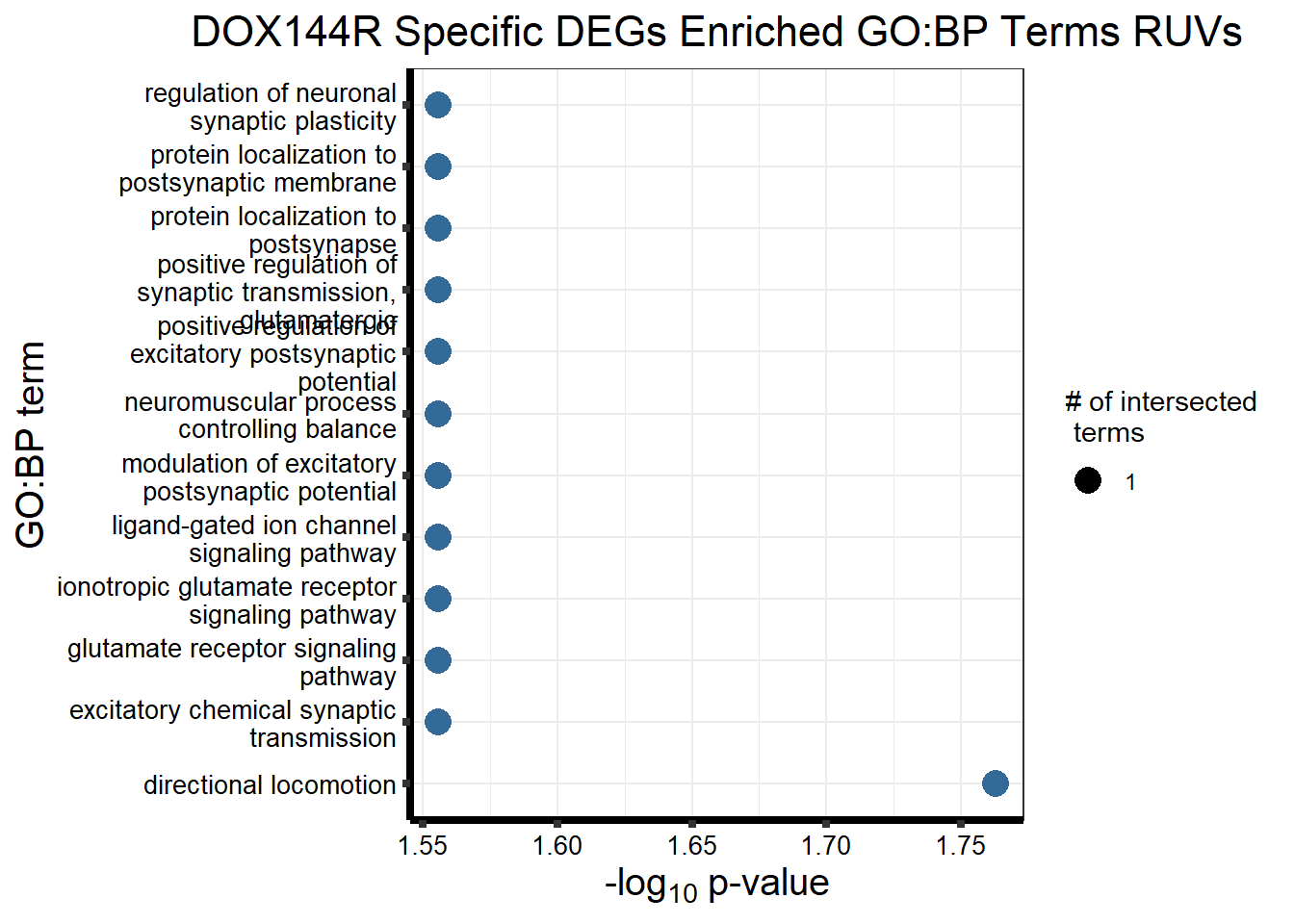
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_DOX144R_genes_KEGG_RUV <- table_DOX144R_genes_RUV %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX144R_genes_KEGG_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX144R DEGs Enriched KEGG Terms RUVs")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))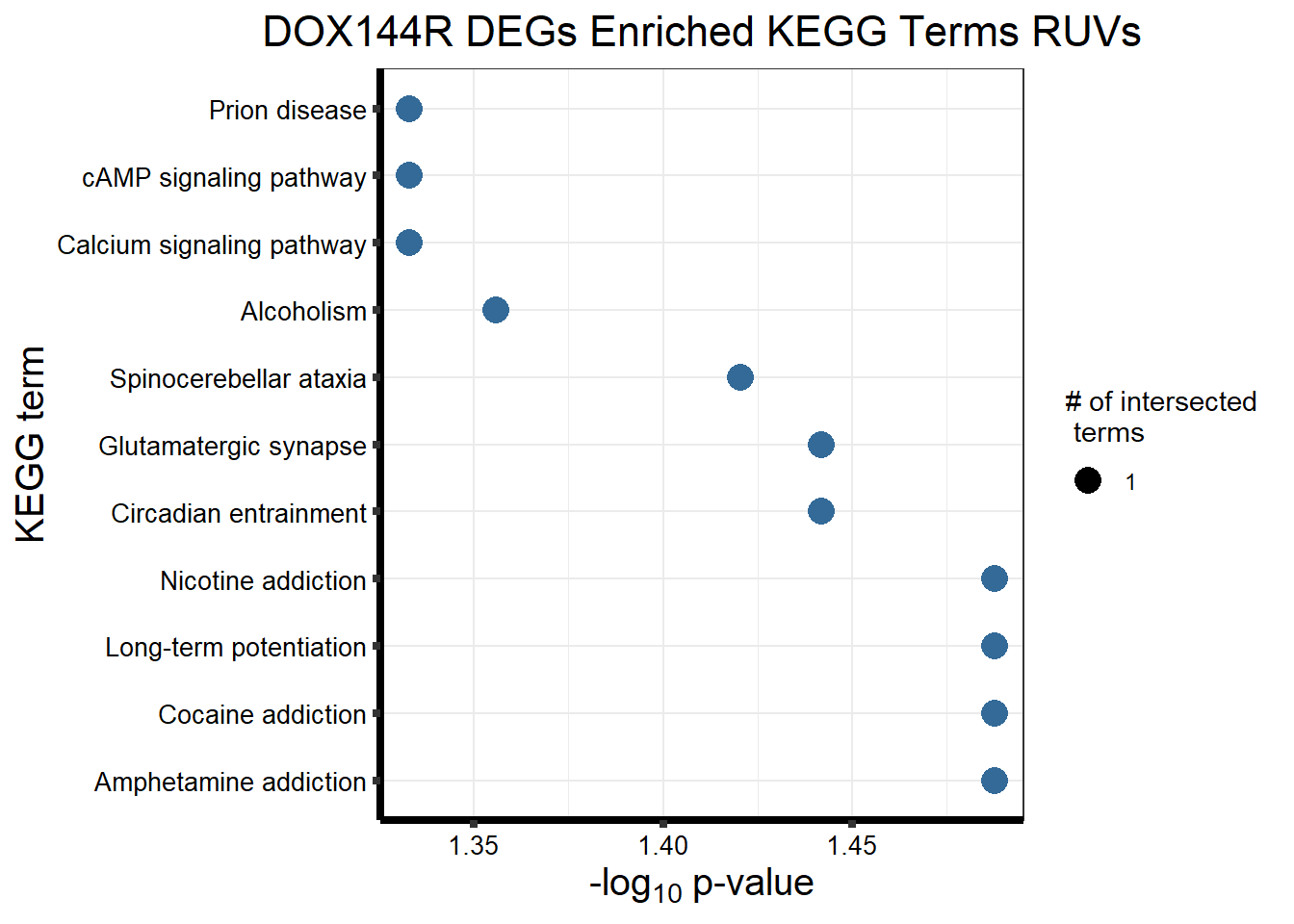
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####DOX144R shared DEGs GO KEGG#####
D144Rshare_DEGs_mat_RUV <- as.matrix(DOX144Rshare_DEGs_GO_plot_RUV)
DOX_144Rshare_dxr_gene_RUV <- gost(query = D144Rshare_DEGs_mat_RUV,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
#check if any KEGG terms are appearing
unique(DOX_144Rshare_dxr_gene_RUV$result$source)[1] "GO:BP"#only GO:BP appearing here
DOX_144Rshare_gost_genes_RUV <-
gostplot(DOX_144Rshare_dxr_gene_RUV, capped = FALSE, interactive = TRUE)
DOX_144Rshare_gost_genes_RUVtable_DOX144Rshare_genes_RUV <- DOX_144Rshare_dxr_gene_RUV$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_DOX144Rshare_genes_RUV %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 2 | 55 | 1.412e-02 |
| GO:BP | GO:0000278 | mitotic cell cycle | 4 | 892 | 1.412e-02 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 3 | 351 | 1.412e-02 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 4 | 745 | 1.412e-02 |
| GO:BP | GO:0051293 | establishment of spindle localization | 2 | 63 | 1.412e-02 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 2 | 47 | 1.412e-02 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 2 | 56 | 1.412e-02 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 3 | 262 | 1.412e-02 |
| GO:BP | GO:0051653 | spindle localization | 2 | 68 | 1.463e-02 |
| GO:BP | GO:1904860 | DNA synthesis involved in mitotic DNA replication | 1 | 1 | 1.598e-02 |
| GO:BP | GO:0090592 | DNA synthesis involved in DNA replication | 1 | 1 | 1.598e-02 |
| GO:BP | GO:0035418 | protein localization to synapse | 2 | 89 | 1.878e-02 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 2 | 93 | 1.893e-02 |
| GO:BP | GO:0051225 | spindle assembly | 2 | 136 | 2.197e-02 |
| GO:BP | GO:0046120 | deoxyribonucleoside biosynthetic process | 1 | 3 | 2.197e-02 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 2 | 114 | 2.197e-02 |
| GO:BP | GO:0046126 | pyrimidine deoxyribonucleoside biosynthetic process | 1 | 3 | 2.197e-02 |
| GO:BP | GO:0033058 | directional locomotion | 1 | 3 | 2.197e-02 |
| GO:BP | GO:1902414 | protein localization to cell junction | 2 | 119 | 2.197e-02 |
| GO:BP | GO:0110032 | positive regulation of G2/MI transition of meiotic cell cycle | 1 | 3 | 2.197e-02 |
| GO:BP | GO:0046105 | thymidine biosynthetic process | 1 | 3 | 2.197e-02 |
| GO:BP | GO:0022402 | cell cycle process | 4 | 1280 | 2.197e-02 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 2 | 124 | 2.197e-02 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 2 | 121 | 2.197e-02 |
| GO:BP | GO:0051296 | establishment of meiotic spindle orientation | 1 | 4 | 2.440e-02 |
| GO:BP | GO:0090233 | negative regulation of spindle checkpoint | 1 | 5 | 2.440e-02 |
| GO:BP | GO:0051783 | regulation of nuclear division | 2 | 149 | 2.440e-02 |
| GO:BP | GO:0140499 | negative regulation of mitotic spindle assembly checkpoint signaling | 1 | 5 | 2.440e-02 |
| GO:BP | GO:1901995 | positive regulation of meiotic cell cycle phase transition | 1 | 4 | 2.440e-02 |
| GO:BP | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process | 1 | 5 | 2.440e-02 |
| GO:BP | GO:0110030 | regulation of G2/MI transition of meiotic cell cycle | 1 | 5 | 2.440e-02 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 2 | 164 | 2.440e-02 |
| GO:BP | GO:0051301 | cell division | 3 | 658 | 2.440e-02 |
| GO:BP | GO:0046134 | pyrimidine nucleoside biosynthetic process | 1 | 5 | 2.440e-02 |
| GO:BP | GO:0046104 | thymidine metabolic process | 1 | 5 | 2.440e-02 |
| GO:BP | GO:0030010 | establishment of cell polarity | 2 | 159 | 2.440e-02 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 3 | 720 | 2.458e-02 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 2 | 186 | 2.494e-02 |
| GO:BP | GO:0007049 | cell cycle | 4 | 1663 | 2.494e-02 |
| GO:BP | GO:0051661 | maintenance of centrosome location | 1 | 6 | 2.570e-02 |
| GO:BP | GO:1904778 | positive regulation of protein localization to cell cortex | 1 | 6 | 2.570e-02 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 1 | 7 | 2.794e-02 |
| GO:BP | GO:1901977 | negative regulation of cell cycle checkpoint | 1 | 7 | 2.794e-02 |
| GO:BP | GO:0007051 | spindle organization | 2 | 206 | 2.794e-02 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 2 | 224 | 2.926e-02 |
| GO:BP | GO:0016311 | dephosphorylation | 2 | 218 | 2.926e-02 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 1 | 8 | 2.926e-02 |
| GO:BP | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process | 1 | 8 | 2.926e-02 |
| GO:BP | GO:1904776 | regulation of protein localization to cell cortex | 1 | 9 | 3.038e-02 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 2 | 234 | 3.038e-02 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 1 | 9 | 3.038e-02 |
| GO:BP | GO:0051295 | establishment of meiotic spindle localization | 1 | 9 | 3.038e-02 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 2 | 266 | 3.566e-02 |
| GO:BP | GO:0009120 | deoxyribonucleoside metabolic process | 1 | 11 | 3.566e-02 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 1 | 11 | 3.566e-02 |
| GO:BP | GO:0051657 | maintenance of organelle location | 1 | 12 | 3.569e-02 |
| GO:BP | GO:0007056 | spindle assembly involved in female meiosis | 1 | 12 | 3.569e-02 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 1 | 12 | 3.569e-02 |
| GO:BP | GO:0098976 | excitatory chemical synaptic transmission | 1 | 12 | 3.569e-02 |
| GO:BP | GO:0140014 | mitotic nuclear division | 2 | 282 | 3.664e-02 |
| GO:BP | GO:0051321 | meiotic cell cycle | 2 | 297 | 3.987e-02 |
| GO:BP | GO:0061469 | regulation of type B pancreatic cell proliferation | 1 | 15 | 4.110e-02 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 1 | 15 | 4.110e-02 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 1 | 15 | 4.110e-02 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 2 | 327 | 4.383e-02 |
| GO:BP | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition | 1 | 17 | 4.383e-02 |
| GO:BP | GO:0051726 | regulation of cell cycle | 3 | 1087 | 4.383e-02 |
| GO:BP | GO:1902969 | mitotic DNA replication | 1 | 17 | 4.383e-02 |
| GO:BP | GO:0006213 | pyrimidine nucleoside metabolic process | 1 | 18 | 4.507e-02 |
| GO:BP | GO:1902101 | positive regulation of metaphase/anaphase transition of cell cycle | 1 | 18 | 4.507e-02 |
| GO:BP | GO:0034404 | nucleobase-containing small molecule biosynthetic process | 1 | 19 | 4.561e-02 |
| GO:BP | GO:0006089 | lactate metabolic process | 1 | 19 | 4.561e-02 |
| GO:BP | GO:0009163 | nucleoside biosynthetic process | 1 | 19 | 4.561e-02 |
| GO:BP | GO:0033043 | regulation of organelle organization | 3 | 1148 | 4.614e-02 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 2 | 365 | 4.778e-02 |
| GO:BP | GO:0006670 | sphingosine metabolic process | 1 | 21 | 4.778e-02 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 1 | 21 | 4.778e-02 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 4 | 2407 | 4.814e-02 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 4 | 2410 | 4.814e-02 |
| GO:BP | GO:0000212 | meiotic spindle organization | 1 | 22 | 4.817e-02 |
| GO:BP | GO:0046519 | sphingoid metabolic process | 1 | 23 | 4.912e-02 |
| GO:BP | GO:0035235 | ionotropic glutamate receptor signaling pathway | 1 | 23 | 4.912e-02 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 1 | 24 | 4.944e-02 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 1 | 24 | 4.944e-02 |
| GO:BP | GO:0009162 | deoxyribonucleoside monophosphate metabolic process | 1 | 24 | 4.944e-02 |
| GO:BP | GO:1901659 | glycosyl compound biosynthetic process | 1 | 25 | 4.974e-02 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 1 | 25 | 4.974e-02 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 1 | 25 | 4.974e-02 |
#GO:BP
table_DOX144Rshare_genes_GOBP_RUV <- table_DOX144Rshare_genes_RUV %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_DOX144Rshare_genes_GOBP_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("DOX144R Shared DEGs Enriched GO:BP Terms RUVs")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))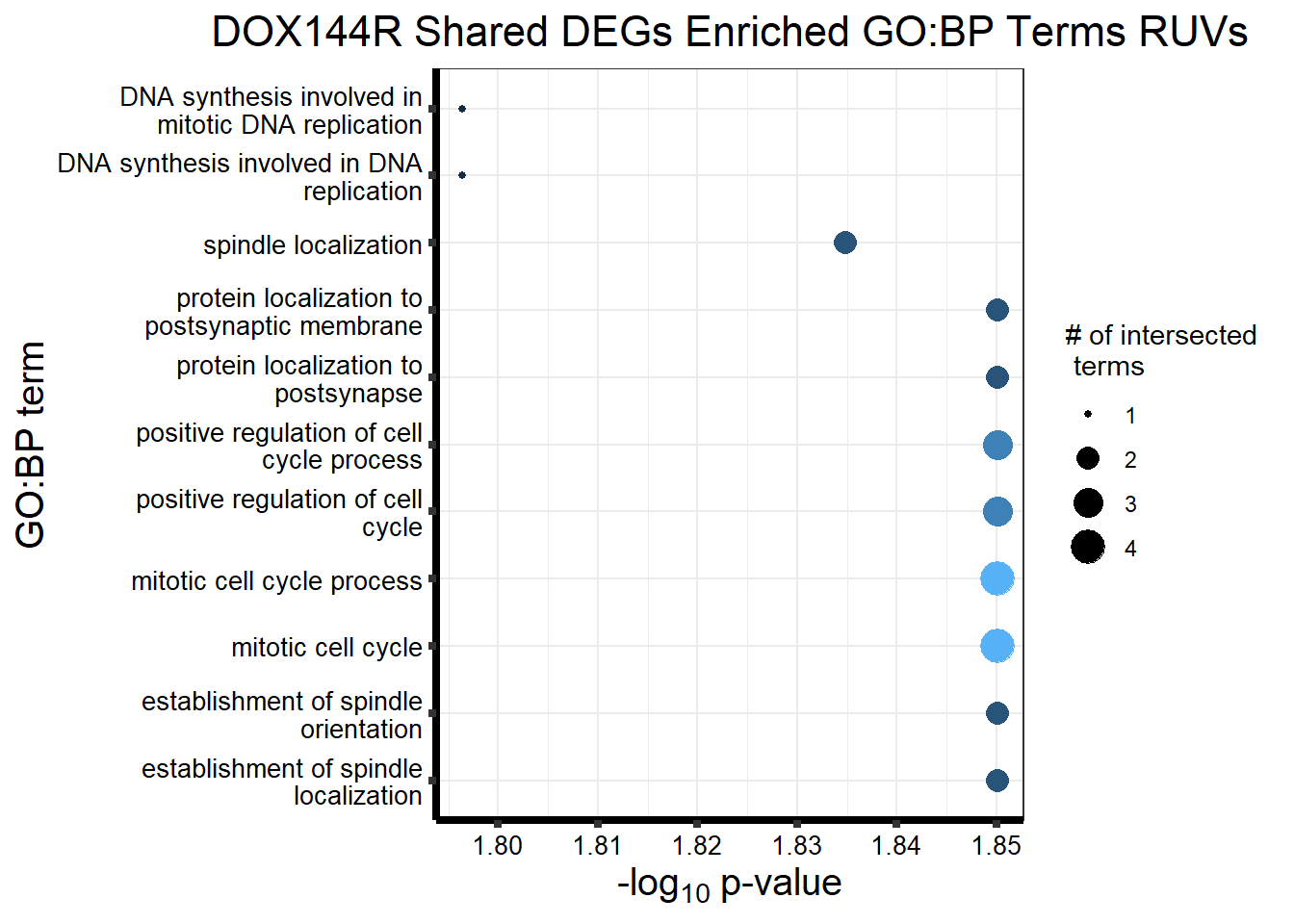
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
# #KEGG
# table_DOX144Rshare_genes_KEGG_RUV <- table_DOX144Rshare_genes_RUV %>%
# dplyr::filter(source=="KEGG") %>%
# dplyr::select(p_value, term_name, intersection_size) %>%
# dplyr::slice_min(., n=10, order_by=p_value) %>%
# mutate(log_val = -log10(p_value))
#
# table_DOX144Rshare_genes_KEGG_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
# geom_point(aes(size = intersection_size)) +
# ggtitle("DOX144R Shared DEGs Enriched KEGG Terms RUVs")+
# xlab(expression("-log"[10]~"p-value"))+
# guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
# ylab("KEGG term")+
# scale_y_discrete(labels = scales::label_wrap(30))+
# theme_bw()+
# theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
# axis.title = element_text(size = 15, colour = "black"),
# axis.ticks = element_line(linewidth = 1.5),
# axis.line = element_line(linewidth = 1.5),
# axis.text = element_text(size = 10, colour = "black", angle = 0),
# strip.text = element_text(size = 15, colour = "black", face = "bold"))#Plot DDR Genes logFC Heatmap RUVs k=1
#DDR Gene Expression Heatmap — DOX Over Recovery Time (68 genes, with categories)
#now add in the RUV corrected data and see if this makes a difference at all!
# Load libraries
# library(circlize)
# library(grid)
# library(reshape2)
# Load DEG files
load_deg <- function(path) read.csv(path)
DOX_24T_R <- load_deg("data/new/DEGs/Toptable_RUV_24T_final.csv")
DOX_24R_R <- load_deg("data/new/DEGs/Toptable_RUV_24R_final.csv")
DOX_144R_R <- load_deg("data/new/DEGs/Toptable_RUV_144R_final.csv")
# Final Entrez IDs and categories (68 genes)
#no need to change the name of this as it's the same
entrez_category_DDR <- tribble(
~ENTREZID, ~Category,
317, "Apoptosis", 355, "Apoptosis", 581, "Apoptosis", 637, "Apoptosis",
836, "Apoptosis", 841, "Apoptosis", 842, "Apoptosis", 27113, "Apoptosis",
5366, "Apoptosis", 54205, "Apoptosis", 55367, "Apoptosis", 8795, "Apoptosis",
1026, "Cell Cycle / Checkpoint", 1027, "Cell Cycle / Checkpoint", 595, "Cell Cycle / Checkpoint",
894, "Cell Cycle / Checkpoint", 896, "Cell Cycle / Checkpoint", 898, "Cell Cycle / Checkpoint",
9133, "Cell Cycle / Checkpoint", 9134, "Cell Cycle / Checkpoint", 891, "Cell Cycle / Checkpoint",
983, "Cell Cycle / Checkpoint", 1017, "Cell Cycle / Checkpoint", 1019, "Cell Cycle / Checkpoint",
1020, "Cell Cycle / Checkpoint", 1021, "Cell Cycle / Checkpoint", 993, "Cell Cycle / Checkpoint",
995, "Cell Cycle / Checkpoint", 1869, "Cell Cycle / Checkpoint", 4609, "Cell Cycle / Checkpoint",
5925, "Cell Cycle / Checkpoint", 9874, "Cell Cycle / Checkpoint", 11011, "Cell Cycle / Checkpoint",
1385, "Cell Cycle / Checkpoint",
472, "Damage Sensors / Signal Transducers", 545, "Damage Sensors / Signal Transducers",
5591, "Damage Sensors / Signal Transducers", 5810, "Damage Sensors / Signal Transducers",
5883, "Damage Sensors / Signal Transducers", 5884, "Damage Sensors / Signal Transducers",
6118, "Damage Sensors / Signal Transducers", 4361, "Damage Sensors / Signal Transducers",
10111, "Damage Sensors / Signal Transducers", 4683, "Damage Sensors / Signal Transducers",
84126, "Damage Sensors / Signal Transducers", 3014, "Damage Sensors / Signal Transducers",
672, "DNA Repair", 2177, "DNA Repair", 5888, "DNA Repair", 5893, "DNA Repair",
1647, "DNA Repair", 4616, "DNA Repair", 10912, "DNA Repair", 1111, "DNA Repair",
11200, "DNA Repair", 1643, "DNA Repair", 8243, "DNA Repair", 5981, "DNA Repair",
7157, "p53 Regulators / Targets", 4193, "p53 Regulators / Targets", 5371, "p53 Regulators / Targets",
27244, "p53 Regulators / Targets", 50484, "p53 Regulators / Targets",
207, "Miscellaneous / Broad", 25, "Miscellaneous / Broad"
)
entrez_ids_DDR_R <- entrez_category_DDR$ENTREZID
# saveRDS(entrez_ids_DDR_R, "data/new/RUV/DDR_genes_entrezid.RDS")
# Extract relevant DEG values
extract_data_DDR_R <- function(df, name) {
df %>%
filter(Entrez_ID %in% entrez_ids_DDR_R) %>%
mutate(
Gene = mapIds(org.Hs.eg.db, as.character(Entrez_ID),
column = "SYMBOL", keytype = "ENTREZID", multiVals = "first"),
Condition = name,
Signif = ifelse(adj.P.Val < 0.05, "*", "")
)
}
# DEG list
deg_list_RUV <- list("DOX_24T" = DOX_24T_R,
"DOX_24R" = DOX_24R_R,
"DOX_144R" = DOX_144R_R
)
# Combine all DEGs and annotate
all_data_DDR_RUV <- bind_rows(mapply(extract_data_DDR_R, deg_list_RUV, names(deg_list_RUV), SIMPLIFY = FALSE)) %>%
left_join(entrez_category_DDR, by = c("Entrez_ID" = "ENTREZID"))'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns# DDR_genesymbols_list_RUV <- all_data_DDR_RUV$Gene
# write.csv(DDR_genesymbols_list_RUV, "data/new/RUV/DDR_Genes_list_RUV.csv")
# Create matrices
logFC_matddr_R <- acast(all_data_DDR_RUV, Gene ~ Condition, value.var = "logFC")
signif_matddr_R <- acast(all_data_DDR_RUV, Gene ~ Condition, value.var = "Signif")
# Set desired order
desired_order <- c("DOX_24T",
"DOX_24R",
"DOX_144R")
logFC_mat_DDR_RUV <- logFC_matddr_R[, desired_order, drop = FALSE]
signif_mat_DDR_RUV <- signif_matddr_R[, desired_order, drop = FALSE]
# Column annotation
meta_DDR_R <- str_split_fixed(colnames(logFC_mat_DDR_RUV), "_", 2)
col_annot <- HeatmapAnnotation(
Drug = meta_DDR_R[, 1],
Time = meta_DDR_R[, 2],
col = list(
Drug = c("DOX" = "#499FBD"),
Time = c("24T" = "#238B45",
"24R" = "#74C476",
"144R" = "#C7E9C0")
),
annotation_height = unit(c(1, 1, 1), "cm")
)
# Row annotation
gene_order_df_DDR_RUV <- all_data_DDR_RUV %>%
distinct(Gene, Category) %>%
arrange(factor(Category, levels = sort(unique(entrez_category_DDR$Category))), Gene)
ordered_genes_DDR_RUV <- gene_order_df_DDR_RUV$Gene
logFC_mat_DDR_RUV <- logFC_mat_DDR_RUV[ordered_genes_DDR_RUV, ]
signif_mat_DDR_RUV <- signif_mat_DDR_RUV[ordered_genes_DDR_RUV, ]
category_colors_DDR <- structure(
c("darkorange", "steelblue", "darkgreen", "firebrick", "mediumpurple", "gray60"),
names = sort(unique(entrez_category_DDR$Category))
)
ha_left_DDR_RUV <- rowAnnotation(
Category = gene_order_df_DDR_RUV$Category,
col = list(Category = category_colors_DDR),
annotation_name_side = "top"
)
# Final Heatmap
Heatmap(logFC_mat_DDR_RUV,
name = "logFC",
top_annotation = col_annot,
left_annotation = ha_left_DDR_RUV,
cluster_columns = FALSE,
cluster_rows = FALSE,
show_row_names = TRUE,
show_column_names = FALSE,
row_names_gp = gpar(fontsize = 10),
column_title = "DDR Gene Expression Response (n = 68)\n DOX Recovery (RUVs Corrected)",
column_title_gp = gpar(fontsize = 14, fontface = "bold"),
cell_fun = function(j, i, x, y, width, height, fill) {
grid.text(signif_mat_DDR[i, j], x, y, gp = gpar(fontsize = 9))
}
)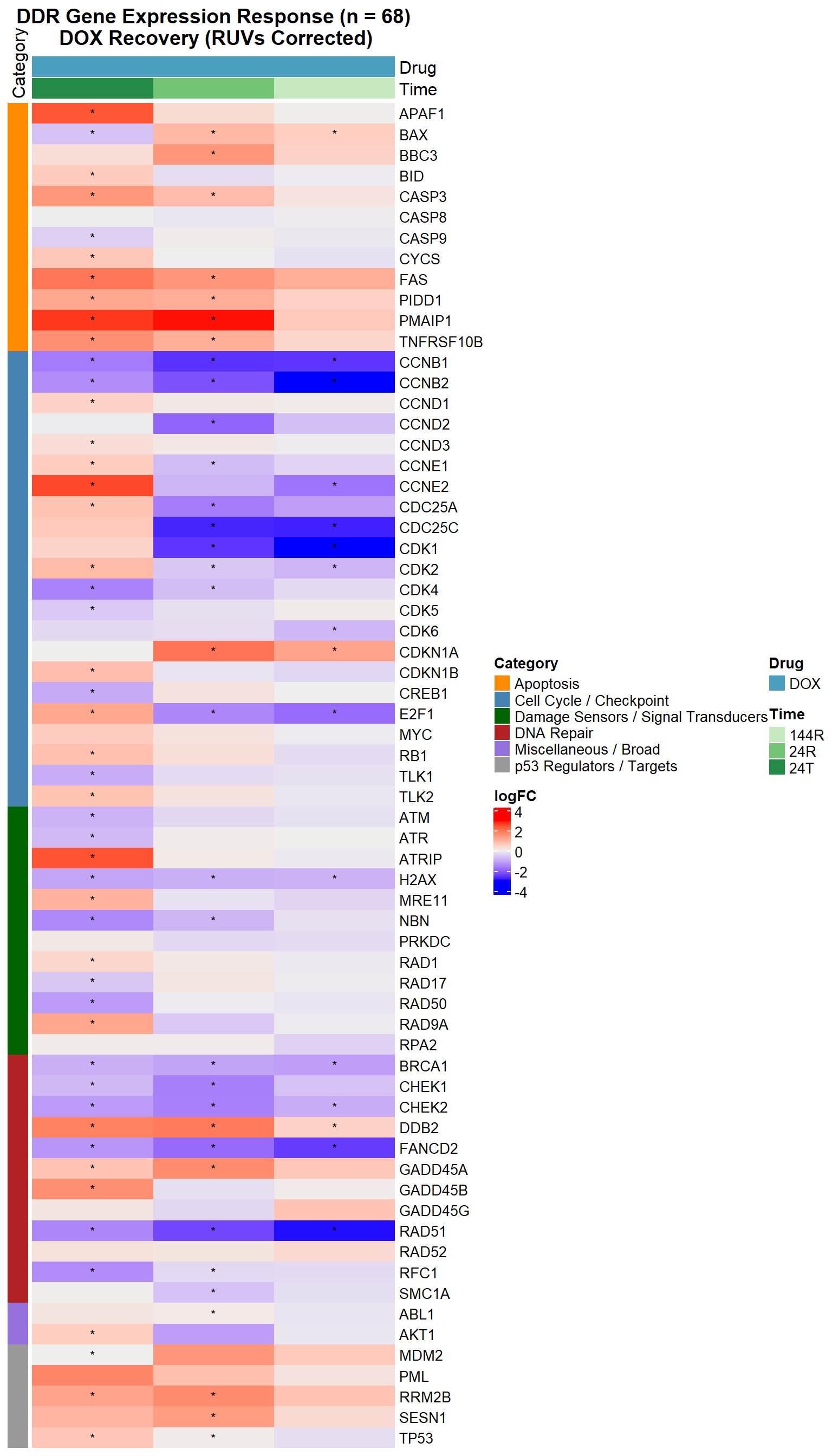
##LogFC of DDR Gene Set after RUVs Correction
library(car)Loading required package: carData
Attaching package: 'car'The following object is masked from 'package:VennDiagram':
ellipseThe following object is masked from 'package:dplyr':
recodeThe following object is masked from 'package:purrr':
some#get an overall idea of the response occurring in all genes with logFC across time
#start with the logFC and then do the abs logFC
####data setup####
#Read DNA Damage Response Gene List
#this had an extra column that was unncessesary
# DNA_damage <- read.csv("data/new/RUV/DDR_Genes_list_RUV.csv", stringsAsFactors = FALSE) %>%
# dplyr::select(-(X))
#
# names(DNA_damage)[names(DNA_damage) == "x"] <- "Symbol"
#
#saveRDS(DNA_damage, "data/new/RUV/DNA_damage_genesymbols.RDS")
#this is the final processed version with a single list of symbols with col name
DNA_damage <- readRDS("data/new/RUV/DNA_damage_genesymbols.RDS")
#Convert gene symbols to Entrez IDs
DNA_damage <- DNA_damage %>%
mutate(Entrez_ID = mapIds(org.Hs.eg.db,
keys = Symbol,
column = "ENTREZID",
keytype = "SYMBOL",
multiVals = "first"))'select()' returned 1:1 mapping between keys and columnsDNA_damage_genes <- na.omit(DNA_damage$Entrez_ID)
time_colors <- c("24T" = "#238B45",
"24R" = "#74C476",
"144R" = "#C7E9C0")
#load in my DEGs which I'll use to create a combined toptable
load_deg <- function(path) read.csv(path)
DOX_24T_R <- load_deg("data/new/DEGs/Toptable_RUV_24T_final.csv")
DOX_24R_R <- load_deg("data/new/DEGs/Toptable_RUV_24R_final.csv")
DOX_144R_R <- load_deg("data/new/DEGs/Toptable_RUV_144R_final.csv")
all_toptables_RUV <- bind_rows(
DOX_24T_R %>% mutate(Drug = "DOX", Timepoint = "24T"),
DOX_24R_R %>% mutate(Drug = "DOX", Timepoint = "24R"),
DOX_144R_R %>% mutate(Drug = "DOX", Timepoint = "144R"),
)
filtered_toptables_RUV <- all_toptables_RUV %>%
filter(Entrez_ID %in% DNA_damage_genes)
filtered_toptables_RUV <- filtered_toptables_RUV %>%
mutate(
Drug = factor(Drug, levels = c("DOX")),
Timepoint = factor(Timepoint, levels = c("24T", "24R", "144R"),
labels = c("Timepoint: 24T", "Timepoint: 24R", "Timepoint: 144R"))
)
levene_results_RUV <- filtered_toptables_RUV %>%
group_by(Timepoint) %>%
summarise(p_value = leveneTest(logFC ~ Timepoint, data = .)$`Pr(>F)`[1], .groups = "drop") %>%
mutate(significance = ifelse(p_value < 0.05, "*", "")) # Use p < 0.05 threshold for stars
# **🔹 Determine Y-axis position for Stars**
star_positions <- filtered_toptables_RUV %>%
group_by(Timepoint,Drug) %>%
summarise(y_pos = max(logFC, na.rm = TRUE) + 0.5, .groups = "drop") %>%
group_by(Timepoint) %>%
summarise(y_pos = max(y_pos), .groups = "drop")
# **🔹 Merge Levene test results with Y positions**
levene_results_plot_RUV <- levene_results_RUV %>%
left_join(star_positions, by = c("Timepoint")) %>%
mutate(x_position = 1.5)
ggplot(filtered_toptables_RUV, aes(x = Drug, y = logFC, fill = Timepoint)) +
geom_violin(trim = FALSE, alpha = 0.5) + # Violin plot for logFC
geom_boxplot(width = 0.1, outlier.shape = NA, color = "black", alpha = 0.5) + # Add boxplot inside violin
scale_fill_manual(values = c(
"Timepoint: 24T" = "#238B45",
"Timepoint: 24R" = "#74C476",
"Timepoint: 144R" = "#C7E9C0")) +
facet_grid(Drug ~ Timepoint) +
geom_text(
data = levene_results_plot_RUV %>% filter(significance == "*"), # Only plot significant comparisons
aes(x = x_position, y = y_pos, label = significance),
size = 6, fontface = "bold", color = "black", inherit.aes = FALSE
) +
theme_bw() +
xlab("") +
ylab("Log Fold Change") +
ggtitle("LogFC Distribution for DDR Genes") +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.line = element_line(linewidth = 1.5),
strip.background = element_rect(fill = "gray"),
strip.text = element_text(size = 12, color = "black", face = "bold"),
axis.text.x = element_text(size = 8, color = "black", angle = 0)
)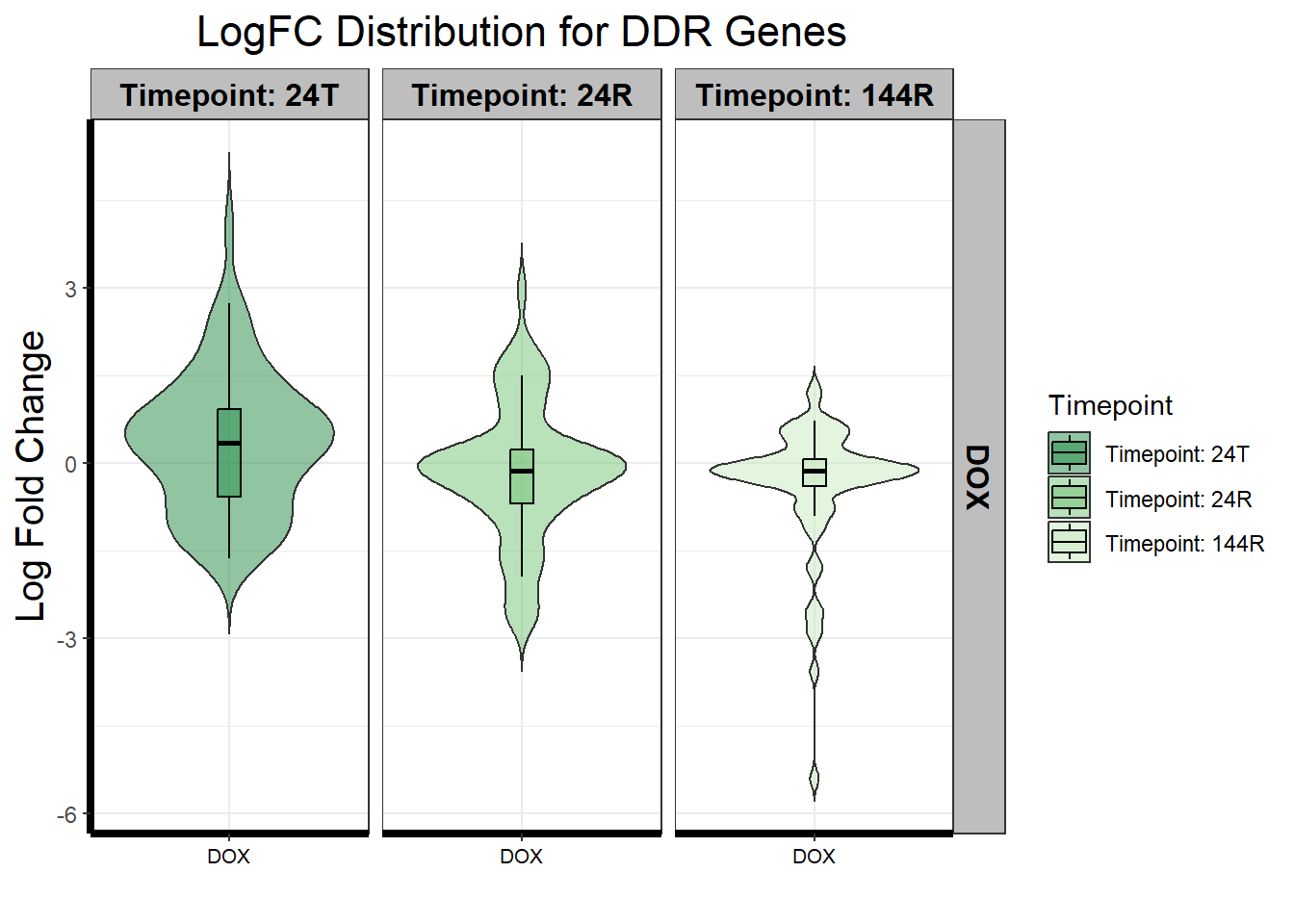
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
# Read DNA Damage Genes List
# DNA_damage
#
# # Convert gene symbols to Entrez IDs
# DNA_damage <- DNA_damage %>%
# mutate(Entrez_ID = mapIds(org.Hs.eg.db,
# keys = Symbol,
# column = "ENTREZID",
# keytype = "SYMBOL",
# multiVals = "first"))
#
# # Extract DNA damage gene Entrez IDs
# DNA_damage_genes <- na.omit(DNA_damage$Entrez_ID)
# total_DNA_damage_genes <- length(DNA_damage_genes) # Total number of DNA damage genes
#
# # Define DEG lists (with significant ones only)
# DOX_DEGs <- list(
# "DOX_24T" = DEG1_RUV, "DOX_24R" = DEG2_RUV, "DOX_144R" = DEG3_RUV
# )
#
# # Function to calculate the presence of DNA damage genes in DEGs
# calculate_proportion_DDR <- function(deg_list, drug_name) {
# data.frame(
# Sample = names(deg_list),
# Drug = drug_name,
# DNA_Damage_DEGs = sapply(deg_list, function(ids) sum(ids %in% DNA_damage_genes)), # DEGs present in DNA damage set
# Non_DNA_Damage_DEGs = sapply(deg_list, function(ids) total_DNA_damage_genes - sum(ids %in% DNA_damage_genes)) # Remaining DNA damage genes
# ) %>%
# mutate(
# Yes_Proportion = (DNA_Damage_DEGs / total_DNA_damage_genes) * 100, # Percentage of DEGs in DNA damage genes
# No_Proportion = (Non_DNA_Damage_DEGs / total_DNA_damage_genes) * 100 # Remaining DNA damage genes as No
# )
# }
#
# # Calculate proportions for CX-5461 and DOX
# DOX_proportion <- calculate_proportion_DDR(DOX_DEGs, "DOX")
#
# # Convert to long format for stacked bar plot
# proportion_long <- DOX_proportion %>%
# dplyr::select(Sample, Drug, Yes_Proportion, No_Proportion) %>%
# pivot_longer(cols = c(Yes_Proportion, No_Proportion), names_to = "Category", values_to = "Percentage") %>%
# mutate(Category = ifelse(Category == "Yes_Proportion", "Yes", "No"))
#
# # **Ensure correct order of samples on X-axis**
# sample_order <- c(
# "DOX_24T", "DOX_24R", "DOX_144R"
# )
# proportion_long$Sample <- factor(proportion_long$Sample, levels = sample_order, ordered = TRUE)
#
# # **Fix: Ensure "Yes" is on top and "No" is at the bottom in stacked bars**
# proportion_long$Category <- factor(proportion_long$Category, levels = c("Yes", "No")) # Ensures "Yes" on top, "No" at bottom
#
# # **Perform Chi-Square Test for DOX across timepoints**
# chi_square_results <- data.frame(Sample = character(), P_Value = numeric())
#
# for (i in seq(1, 6)) { #comparing across timepoints
# tx24_sample <- sample_order[i] #DOX24T
# rec24_sample <- sample_order[i + 6] #DOX24R
# rec144_sample <- sample_order[i + 12] #DOX144R
#
# tx24_data <- filter(DOX_proportion, Sample == tx24_sample)
# rec24_data <- filter(DOX_proportion, Sample == rec24_sample)
# rec144_data <- filter(DOX_proportion, Sample == rec144_sample)
#
# # Construct contingency table for Chi-Square test
# contingency_table <- matrix(
# c(tx24_data$DNA_Damage_DEGs, tx24_data$Non_DNA_Damage_DEGs,
# rec24_data$DNA_Damage_DEGs, rec24_data$Non_DNA_Damage_DEGs,
# rec144_data$DNA_Damage_DEGs, rec144_data$Non_DNA_Damage_DEGs),
# nrow = 3, byrow = TRUE
# )
#
# # Run Chi-Square Test
# test_result <- chisq.test(contingency_table)
# p_value <- test_result$p.value
#
# # Store results
# chi_square_results <- rbind(chi_square_results, data.frame(Sample = tx24_sample, P_Value = p_value))
# }
#
# # Add significance stars
# chi_square_results$Significant <- ifelse(chi_square_results$P_Value < 0.05, "*", "")
#
# # Merge Chi-Square results
# proportion_long <- left_join(proportion_long, chi_square_results, by = "Sample")
#
# # **Save output**
# write.csv(proportion_long, "C:/Work/Postdoc_UTMB/CX-5461 Project/Transcriptome literatures/lit2/Proportion_Stacked_DNA_Damage_DEGs_with_ChiSquare.csv", row.names = FALSE)
#
#
# # Define correct factor orders for samples
# sample_order <- c(
# "CX_0.1_3", "CX_0.1_24", "CX_0.1_48", "CX_0.5_3", "CX_0.5_24", "CX_0.5_48",
# "DOX_0.1_3", "DOX_0.1_24", "DOX_0.1_48", "DOX_0.5_3", "DOX_0.5_24", "DOX_0.5_48"
# )
#
# # Reapply factor levels for correct order in both proportion_data and proportion_long
# proportion_data$Sample <- factor(proportion_data$Sample, levels = sample_order, ordered = TRUE)
# proportion_long$Sample <- factor(proportion_long$Sample, levels = sample_order, ordered = TRUE)
#
# # **Fix: Ensure "Yes" is on top and "No" is at the bottom in stacked bars**
# proportion_long$Category <- factor(proportion_long$Category, levels = c("Yes", "No")##LogFC of DDR Genes Across Timepoints RUVs
library(rstatix)
Attaching package: 'rstatix'The following object is masked from 'package:AnnotationDbi':
selectThe following object is masked from 'package:IRanges':
descThe following object is masked from 'package:biomaRt':
selectThe following object is masked from 'package:stats':
filter# Read DNA Damage Response Gene List
# DNA_damage as above
# Convert gene symbols to Entrez IDs
# DNA_damage <- DNA_damage %>%
# mutate(Entrez_ID = mapIds(org.Hs.eg.db,
# keys = Symbol,
# column = "ENTREZID",
# keytype = "SYMBOL",
# multiVals = "first"))
# DNA_damage_genes <- na.omit(DNA_damage$Entrez_ID)
# saveRDS(DNA_damage_genes, "data/new/RUV/DNA_damage_genes_symbolentrez.RDS")
DNA_damage_genes <- readRDS("data/new/RUV/DNA_damage_genes_symbolentrez.RDS")
all_toptables_RUV <- bind_rows(
DOX_24T_R %>% mutate(Drug = "DOX", Timepoint = "24T"),
DOX_24R_R %>% mutate(Drug = "DOX", Timepoint = "24R"),
DOX_144R_R %>% mutate(Drug = "DOX", Timepoint = "144R"),
)
filtered_toptables_RUV <- all_toptables_RUV %>%
filter(Entrez_ID %in% DNA_damage_genes) %>%
mutate(abs_logFC = abs(logFC))
filtered_toptables_RUV <- filtered_toptables_RUV %>%
mutate(
Drug = factor(Drug, levels = c("DOX")),
Timepoint = factor(Timepoint, levels = c("24T", "24R", "144R"),
labels = c("Timepoint: 24T", "Timepoint: 24R", "Timepoint: 144R"))
)
wilcox_results <- filtered_toptables_RUV %>%
wilcox_test(abs_logFC ~ Timepoint) %>%
adjust_pvalue(method = "bonferroni") %>%
mutate(significance = ifelse(p < 0.05, "*", ""))
#make a little thing of your timepoint comparisons
pairs <- wilcox_results %>%
dplyr::select(group1, group2) %>%
distinct()
max_per_timepoint <- filtered_toptables_RUV %>%
group_by(Timepoint) %>%
summarise(max_logFC = max(abs_logFC, na.rm = TRUE), .groups = "drop")
#Join max values for group1
star_positions <- pairs %>%
left_join(max_per_timepoint, by = c("group1" = "Timepoint"))
names(star_positions)[names(star_positions) == "max_logFC"] <- "max1"
#Join max values for group2, with suffixes to avoid name conflict
star_positions <- star_positions %>%
left_join(max_per_timepoint, by = c("group2" = "Timepoint"), suffix = c(".group1", ".group2"))
names(star_positions)[names(star_positions) == "max_logFC"] <- "max2"
star_positions <- star_positions %>%
mutate(y_pos = pmax(max1, max2, na.rm = TRUE) + 0.2)
#now star positions has the same group1 and group2 as wilcoxon
wilcox_results_plot <- wilcox_results %>%
left_join(star_positions, by = c("group1", "group2")) %>%
mutate(x_position = 1.5)
wilcox_results_plot <- wilcox_results_plot %>%
mutate(group1 = gsub("Timepoint: ", "", group1),
group2 = gsub("Timepoint: ", "", group2))
wilcox_results_plot <- wilcox_results_plot %>%
mutate(x_position = case_when(
group1 == "24T" & group2 == "24R" ~ 1.5,
group1 == "24T" & group2 == "144R" ~ 2,
group1 == "24R" & group2 == "144R" ~ 2.5
))
####logFC####
ggplot(filtered_toptables_RUV, aes(x = Timepoint, y = logFC, fill = Timepoint)) +
geom_boxplot() +
scale_fill_manual(values = c(
"Timepoint: 24T" = "#238B45",
"Timepoint: 24R" = "#74C476",
"Timepoint: 144R" = "#C7E9C0")) +
facet_wrap(~ Drug) +
geom_text(
data = wilcox_results_plot,
aes(x = x_position, y = y_pos, label = significance),
size = 6, fontface = "bold", color = "black", inherit.aes = FALSE
) +
theme_bw() +
xlab("") +
ylab("Log Fold Change") +
ggtitle("Log Fold Change for DDR Genes") +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.line = element_line(linewidth = 1.5),
strip.background = element_rect(fill = "gray"),
strip.text = element_text(size = 12, color = "black", face = "bold"),
axis.text.x = element_text(size = 8, color = "black", angle = 0)
)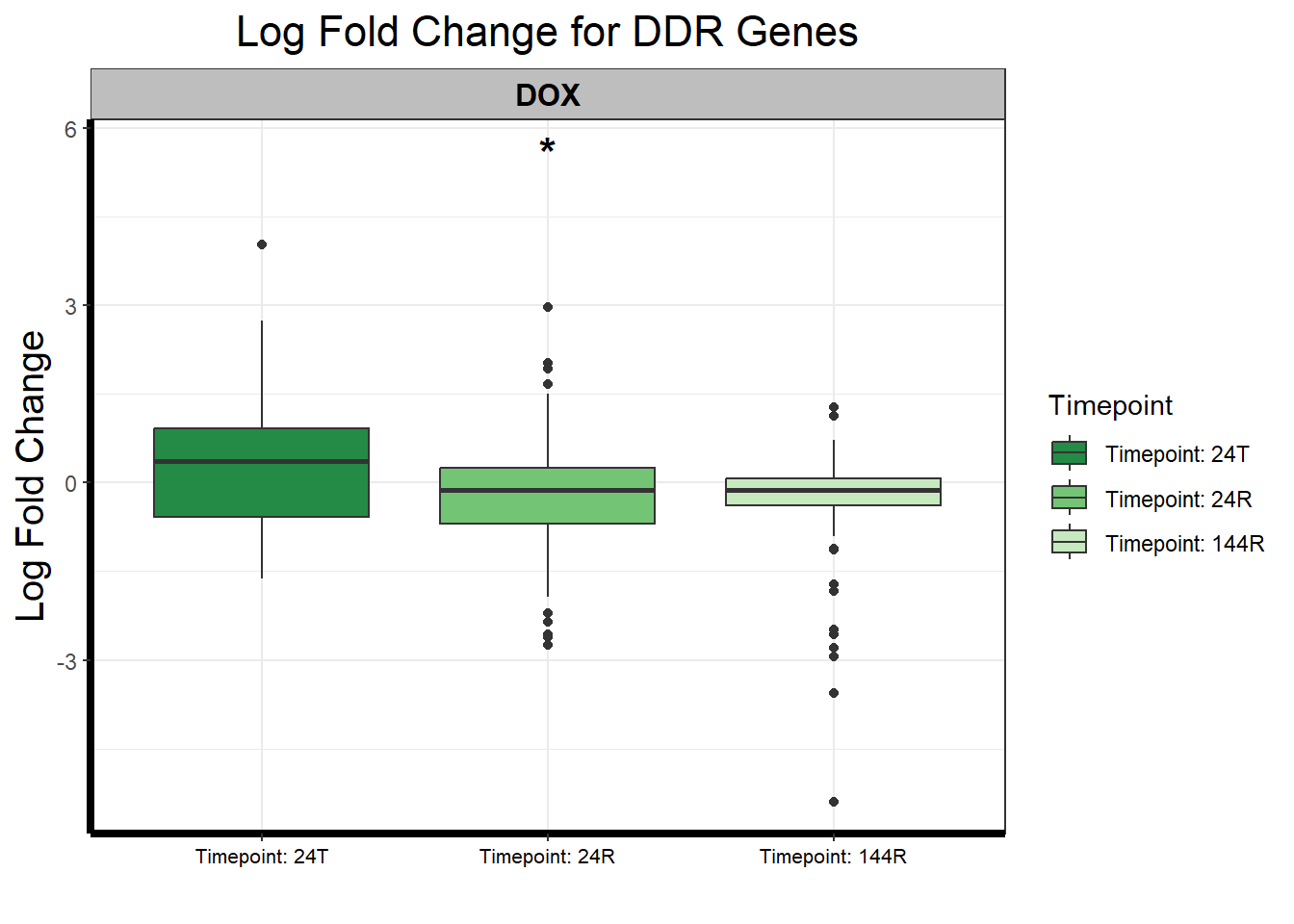
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
####abs logFC####
ggplot(filtered_toptables_RUV, aes(x = Timepoint, y = abs_logFC, fill = Timepoint)) +
geom_boxplot() +
scale_fill_manual(values = c(
"Timepoint: 24T" = "#238B45",
"Timepoint: 24R" = "#74C476",
"Timepoint: 144R" = "#C7E9C0")) +
facet_wrap(~ Drug) +
geom_text(
data = wilcox_results_plot,
aes(x = x_position, y = y_pos, label = significance),
size = 6, fontface = "bold", color = "black", inherit.aes = FALSE
) +
theme_bw() +
xlab("") +
ylab("|Log Fold Change|") +
ggtitle("|Log Fold Change| for DDR Genes") +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.line = element_line(linewidth = 1.5),
strip.background = element_rect(fill = "gray"),
strip.text = element_text(size = 12, color = "black", face = "bold"),
axis.text.x = element_text(size = 8, color = "black", angle = 0)
)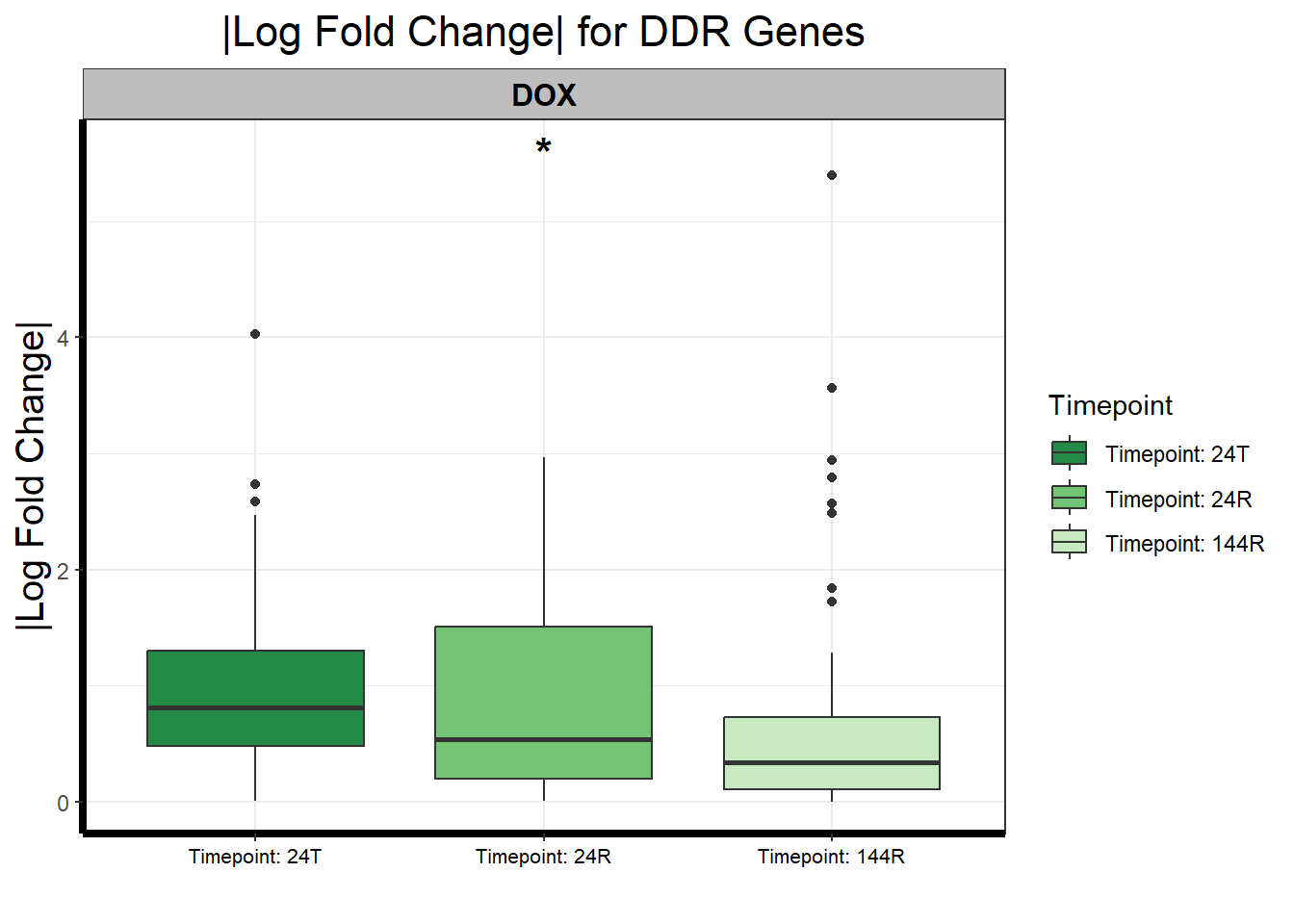
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#DIC Genes RUVs Corrected
#plot a list of 29 functionally validated DIC genes
# Load DEG files
load_deg <- function(path) read.csv(path)
DOX_24T_R <- load_deg("data/new/DEGs/Toptable_RUV_24T_final.csv")
DOX_24R_R <- load_deg("data/new/DEGs/Toptable_RUV_24R_final.csv")
DOX_144R_R <- load_deg("data/new/DEGs/Toptable_RUV_144R_final.csv")
#the data I have is in hgnc_symbols, I want to convert this to entrez_id like my df
DIC_genes <- tribble(
~SYMBOL, ~Category,
"CAT", "ROS Generation / Handling",
"CBR1", "ROS Generation / Handling",
"CBR3", "ROS Generation / Handling",
"ERBB2", "ROS Generation / Handling",
"GPX3", "ROS Generation / Handling",
"GSTM1", "ROS Generation / Handling",
"GSTP", "ROS Generation / Handling",
"HAS3", "ROS Generation / Handling",
"NOS3", "ROS Generation / Handling",
"PLCE1", "ROS Generation / Handling",
"RAC2", "ROS Generation / Handling",
"SPG7", "ROS Generation / Handling",
"PRDM2", "DNA Damage",
"MLH1", "DNA Damage",
"RARG", "DNA Damage",
"HFE", "Iron Uptake & Homeostasis",
"SLC22A17", "DOX Uptake",
"SLC28A1", "DOX Uptake",
"SLC28A3", "DOX Uptake",
"ABCB4", "DOX Efflux",
"ABCC2", "DOX Efflux",
"ABCC5", "DOX Efflux",
"ABCC9", "DOX Efflux",
"ABCC10", "DOX Efflux",
"CELF4", "Calcium Handling",
"MYH7", "Calcium Handling",
"CYP2J2", "Cardiac Electrical Activity",
"RIN3", "Cardiac Electrical Activity",
"ZFN521", "Cardiac Electrical Activity")
gene_df_DIC_RUV <- tibble(HGNC = DIC_genes)
gene_df_DIC_RUV <- gene_df_DIC_RUV %>%
mutate(Entrez_ID = mapIds(org.Hs.eg.db,
keys = DIC_genes$SYMBOL,
column = "ENTREZID",
keytype = "SYMBOL",
multiVals = "first")) %>%
unnest_wider(HGNC) %>%
mutate(Entrez_ID = as.character(Entrez_ID))'select()' returned 1:1 mapping between keys and columns# DIC_genes_symb <- gene_df_DIC_RUV %>%
# dplyr::select("SYMBOL", "Entrez_ID")
# saveRDS(DIC_genes_symb, "data/new/RUV/DIC_genelist_symbolentrez_RUV.RDS")
#now I've put together a dataframe with the HGNC, Category, and Entrez_ID
#plus I've ensured that Entrez_ID is a character for later joining
entrez_ids_DIC_RUV <- gene_df_DIC_RUV$Entrez_ID
# Extract relevant DEG values
extract_data_DIC_RUV <- function(df, name) {
df %>%
filter(Entrez_ID %in% entrez_ids_DIC_RUV) %>%
mutate(
Gene = mapIds(org.Hs.eg.db, as.character(Entrez_ID),
column = "SYMBOL", keytype = "ENTREZID", multiVals = "first"),
Condition = name,
Signif = ifelse(adj.P.Val < 0.05, "*", "")
)
}
# DEG list
deg_list_RUV <- list("DOX_24T" = DOX_24T_R,
"DOX_24R" = DOX_24R_R,
"DOX_144R" = DOX_144R_R
)
# Combine all DEGs and annotate
all_data_DIC_RUV <- bind_rows(mapply(extract_data_DIC_RUV,
deg_list_RUV,
names(deg_list_RUV),
SIMPLIFY = FALSE)) %>%
mutate(Entrez_ID = as.character(Entrez_ID)) %>%
left_join(gene_df_DIC_RUV,
by = "Entrez_ID")'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns#I've also made sure here that Entrez_ID is a character and not an integer
# Create matrices
logFC_matdic_RUV <- acast(all_data_DIC_RUV, Gene ~ Condition, value.var = "logFC")
signif_matdic_RUV <- acast(all_data_DIC_RUV, Gene ~ Condition, value.var = "Signif")
# Set desired order
desired_order <- c("DOX_24T",
"DOX_24R",
"DOX_144R")
logFC_mat_DIC_RUV <- logFC_matdic_RUV[, desired_order, drop = FALSE]
signif_mat_DIC_RUV <- signif_matdic_RUV[, desired_order, drop = FALSE]
# Column annotation
meta_DIC_R <- str_split_fixed(colnames(logFC_mat_DIC_RUV), "_", 3)
meta_DIC_R <- str_split_fixed(colnames(logFC_mat_DIC_RUV), "_", 2)
col_annot_RUV <- HeatmapAnnotation(
Drug = meta_DIC_R[, 1],
Time = meta_DIC_R[, 2],
col = list(
Drug = c("DOX" = "#499FBD"),
Time = c("24T" = "#238B45",
"24R" = "#74C476",
"144R" = "#C7E9C0")
),
annotation_height = unit(c(2, 2, 2), "cm")
)
#for this I can leave off the DMSO as it's already from a pairwise comparison
#make an extra object with my category order as well
category_order_DIC <- c(
"ROS Generation / Handling",
"DNA Damage",
"Calcium Handling",
"DOX Uptake",
"Iron Uptake & Homeostasis",
"DOX Efflux",
"Cardiac Electrical Activity"
)
# Row annotation
gene_order_df_DIC_RUV <- all_data_DIC_RUV %>%
distinct(Gene, Category) %>%
mutate(Category = factor(Category, levels = category_order_DIC)) %>%
arrange(Category, Gene)
ordered_genes_DIC_RUV <- gene_order_df_DIC_RUV$Gene
logFC_mat_DIC_RUV <- logFC_mat_DIC_RUV[ordered_genes_DIC_RUV, ]
signif_mat_DIC_RUV <- signif_mat_DIC_RUV[ordered_genes_DIC_RUV, ]
#add in your colors for each category
category_colors_DIC <- structure(
c("darkorange", "steelblue", "darkgreen", "firebrick", "gold", "mediumpurple", "gray60"),
names = category_order_DIC
)
ha_left_DIC_RUV <- rowAnnotation(
Category = gene_order_df_DIC_RUV$Category,
col = list(Category = category_colors_DIC),
annotation_name_side = "top"
)
# Draw heatmap
Heatmap(logFC_mat_DIC_RUV,
name = "logFC",
top_annotation = col_annot,
left_annotation = ha_left_DIC_RUV,
cluster_columns = FALSE,
cluster_rows = FALSE,
show_row_names = TRUE,
show_column_names = FALSE,
cell_fun = function(j, i, x, y, width, height, fill) {
grid.text(signif_mat_DIC[i, j], x, y, gp = gpar(fontsize = 9))
},
column_title = "DIC Genes Expression (n=29)\nDOX Recovery (RUVs Corrected)",
column_title_gp = gpar(fontsize = 14, fontface = "bold")
)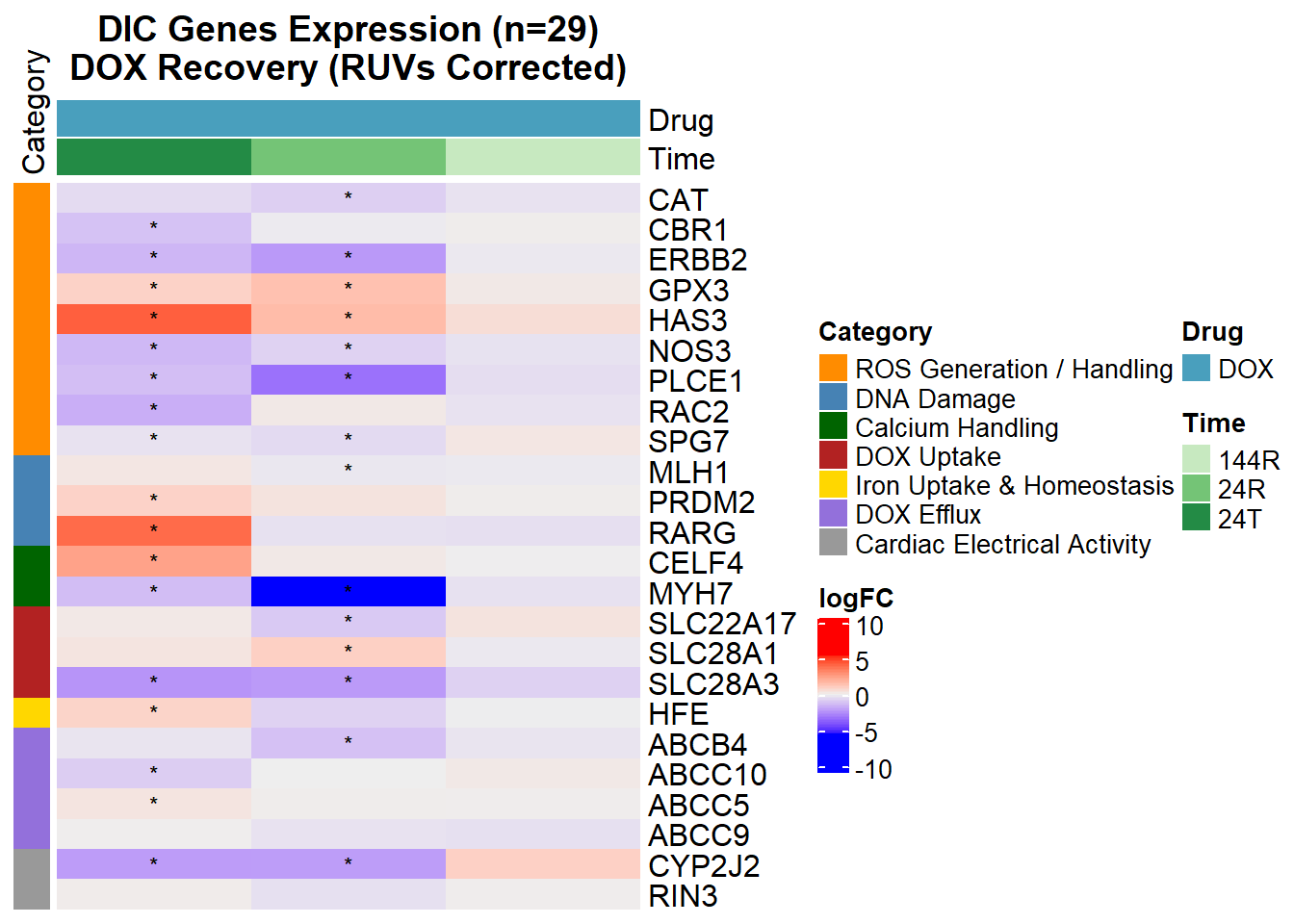
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#get an overall idea of the logFC of these genes over timepoints
library(rstatix)
# Read DIC gene list - named DOX_cardiotox
# DOX_cardiotox <- readRDS("data/new/RUV/DIC_genelist_symbolentrez_RUV.RDS")
#
# # Convert gene symbols to Entrez IDs
# DOX_cardiotox <- DOX_cardiotox %>%
# mutate(Entrez_ID = mapIds(org.Hs.eg.db,
# keys = SYMBOL,
# column = "ENTREZID",
# keytype = "SYMBOL",
# multiVals = "first"))
#
# DOX_cardiotox_genes <- na.omit(DOX_cardiotox$Entrez_ID)
# saveRDS(DOX_cardiotox_genes, "data/new/RUV/DIC_genes_symbolentrez.RDS")
#this already has both SYMBOL and Entrez_ID
DOX_cardiotox_genes <- readRDS("data/new/RUV/DIC_genes_symbolentrez.RDS")
all_toptables_RUV <- bind_rows(
DOX_24T_R %>% mutate(Drug = "DOX", Timepoint = "24T"),
DOX_24R_R %>% mutate(Drug = "DOX", Timepoint = "24R"),
DOX_144R_R %>% mutate(Drug = "DOX", Timepoint = "144R"),
)
filtered_toptables_RUV_DIC <- all_toptables_RUV %>%
filter(Entrez_ID %in% DOX_cardiotox_genes) %>%
mutate(abs_logFC = abs(logFC))
filtered_toptables_RUV_DIC <- filtered_toptables_RUV_DIC %>%
mutate(
Drug = factor(Drug, levels = c("DOX")),
Timepoint = factor(Timepoint, levels = c("24T", "24R", "144R"),
labels = c("Timepoint: 24T", "Timepoint: 24R", "Timepoint: 144R"))
)
wilcox_results_DIC <- filtered_toptables_RUV_DIC %>%
wilcox_test(abs_logFC ~ Timepoint) %>%
adjust_pvalue(method = "bonferroni") %>%
mutate(significance = ifelse(p < 0.05, "*", ""))
#make a little thing of your timepoint comparisons
pairs_DIC <- wilcox_results_DIC %>%
dplyr::select(group1, group2) %>%
distinct()
max_per_timepoint_DIC <- filtered_toptables_RUV_DIC %>%
group_by(Timepoint) %>%
summarise(max_logFC = max(abs_logFC, na.rm = TRUE), .groups = "drop")
#Join max values for group1
star_positions_DIC <- pairs_DIC %>%
left_join(max_per_timepoint_DIC, by = c("group1" = "Timepoint"))
names(star_positions_DIC)[names(star_positions_DIC) == "max_logFC"] <- "max1"
#Join max values for group2, with suffixes to avoid name conflict
star_positions_DIC <- star_positions_DIC %>%
left_join(max_per_timepoint_DIC, by = c("group2" = "Timepoint"), suffix = c(".group1", ".group2"))
names(star_positions_DIC)[names(star_positions_DIC) == "max_logFC"] <- "max2"
star_positions_DIC <- star_positions_DIC %>%
mutate(y_pos = pmax(max1, max2, na.rm = TRUE) + 0.2)
#now star positions has the same group1 and group2 as wilcoxon
wilcox_results_plot_DIC <- wilcox_results_DIC %>%
left_join(star_positions_DIC, by = c("group1", "group2")) %>%
mutate(x_position = 1.5)
wilcox_results_plot_DIC <- wilcox_results_plot_DIC %>%
mutate(group1 = gsub("Timepoint: ", "", group1),
group2 = gsub("Timepoint: ", "", group2))
wilcox_results_plot_DIC <- wilcox_results_plot_DIC %>%
mutate(x_position = case_when(
group1 == "24T" & group2 == "24R" ~ 1.5,
group1 == "24T" & group2 == "144R" ~ 2,
group1 == "24R" & group2 == "144R" ~ 2.5
))
####logFC####
ggplot(filtered_toptables_RUV_DIC, aes(x = Timepoint, y = logFC, fill = Timepoint)) +
geom_boxplot() +
scale_fill_manual(values = c(
"Timepoint: 24T" = "#238B45",
"Timepoint: 24R" = "#74C476",
"Timepoint: 144R" = "#C7E9C0")) +
facet_wrap(~ Drug) +
geom_text(
data = wilcox_results_plot_DIC,
aes(x = x_position, y = y_pos, label = significance),
size = 6, fontface = "bold", color = "black", inherit.aes = FALSE
) +
theme_bw() +
xlab("") +
ylab("Log Fold Change") +
ggtitle("Log Fold Change for DIC Genes") +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.line = element_line(linewidth = 1.5),
strip.background = element_rect(fill = "gray"),
strip.text = element_text(size = 12, color = "black", face = "bold"),
axis.text.x = element_text(size = 8, color = "black", angle = 0)
)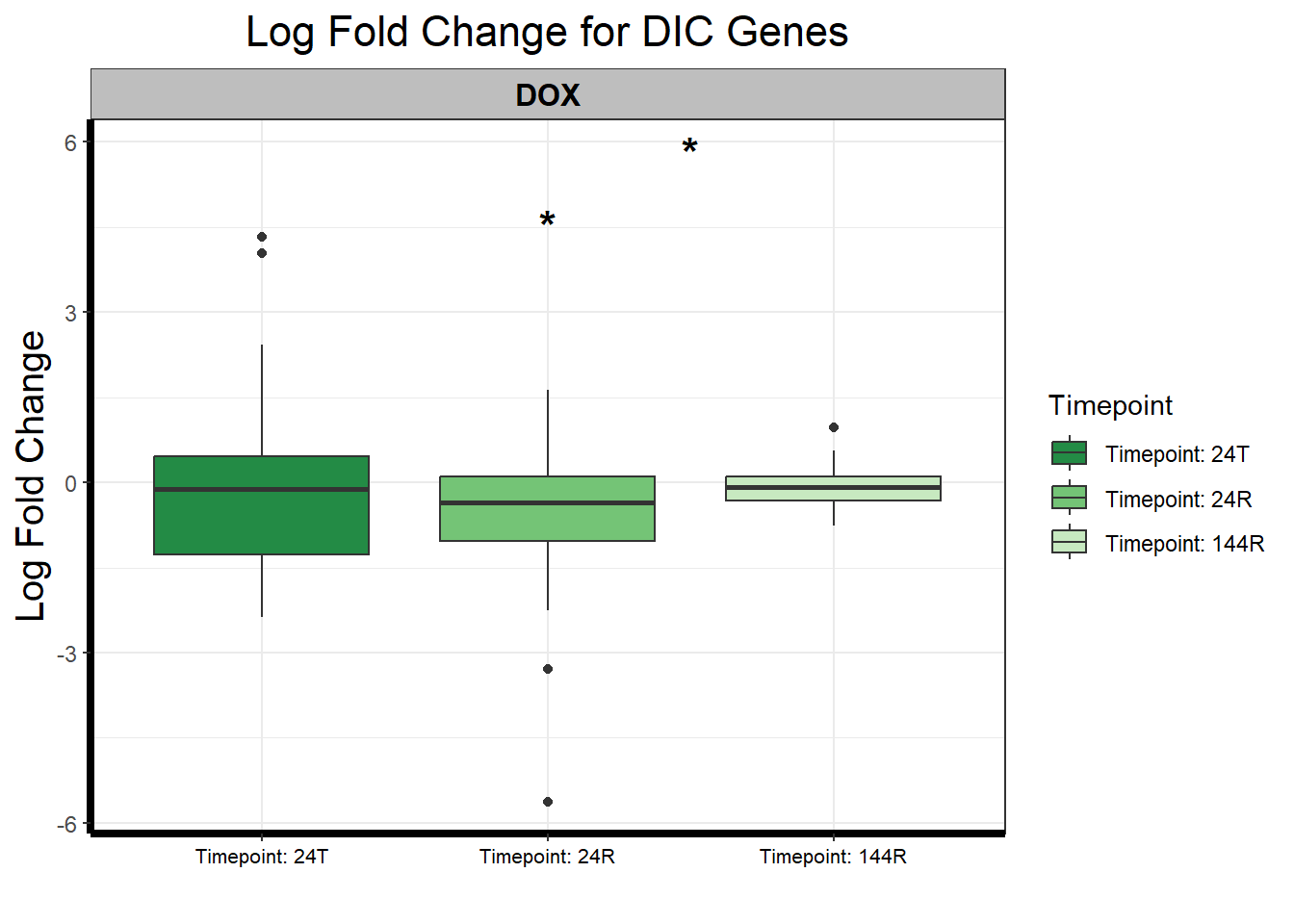
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
####abs logFC####
ggplot(filtered_toptables_RUV_DIC, aes(x = Timepoint, y = abs_logFC, fill = Timepoint)) +
geom_boxplot() +
scale_fill_manual(values = c(
"Timepoint: 24T" = "#238B45",
"Timepoint: 24R" = "#74C476",
"Timepoint: 144R" = "#C7E9C0")) +
facet_wrap(~ Drug) +
geom_text(
data = wilcox_results_plot_DIC,
aes(x = x_position, y = y_pos, label = significance),
size = 6, fontface = "bold", color = "black", inherit.aes = FALSE
) +
theme_bw() +
xlab("") +
ylab("|Log Fold Change|") +
ggtitle("|Log Fold Change| for DIC Genes") +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.line = element_line(linewidth = 1.5),
strip.background = element_rect(fill = "gray"),
strip.text = element_text(size = 12, color = "black", face = "bold"),
axis.text.x = element_text(size = 8, color = "black", angle = 0)
)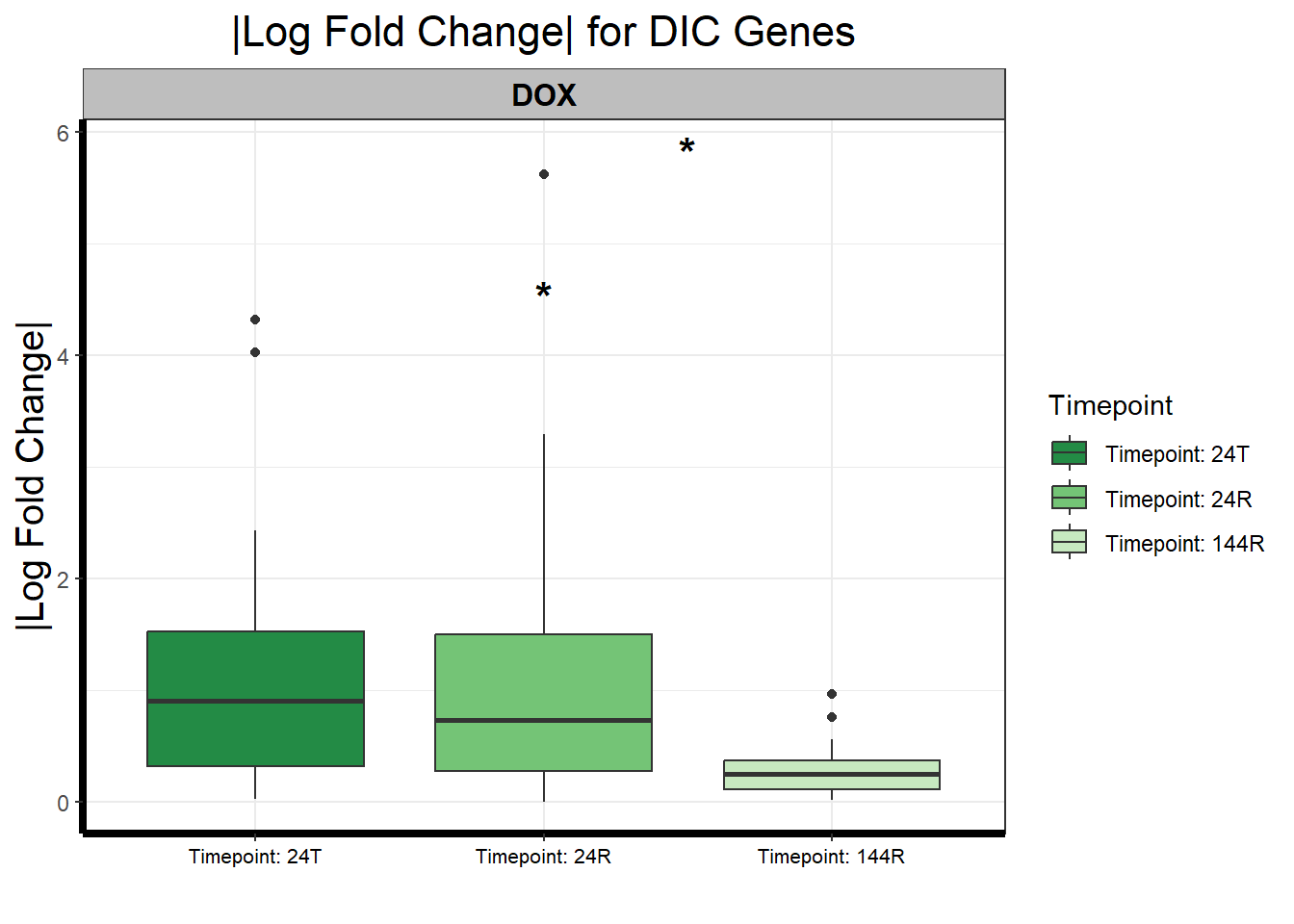
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#p53 Target Genes logFC Heatmap RUVs Corrected
# Load libraries
# Load DEG files
load_deg <- function(path) read.csv(path)
DOX_24T_R <- load_deg("data/new/DEGs/Toptable_RUV_24T_final.csv")
DOX_24R_R <- load_deg("data/new/DEGs/Toptable_RUV_24R_final.csv")
DOX_144R_R <- load_deg("data/new/DEGs/Toptable_RUV_144R_final.csv")
#P53 target Entrez_ID
entrez_ids_p53 <- c(1026,50484,4193,9766,9518,7832,1643,1647,1263,57103,51065,8795,51499,64393,581,
5228,5429,8493,55959,7508,64782,282991,355,53836,4814,10769,9050,27244,9540,94241,
26154,57763,900,26999,55332,26263,23479,23612,29950,9618,10346,8824,134147,55294,
22824,4254,6560,467,27113,60492,8444,60401,1969,220965,2232,3976,55191,84284,93129,
5564,7803,83667,7779,132671,7039,51768,137695,93134,7633,10973,340485,307,27350,
23245,3732,29965,1363,1435,196513,8507,8061,2517,51278,53354,54858,23228,5366,5912,
6236,51222,26152,59,1907,50650,91012,780,9249,11072,144455,64787,116151,27165,2876,
57822,55733,57722,121457,375449,85377,4851,5875,127544,29901,84958,8797,8793,441631,
220001,54541,5889,5054,25816,25987,5111,98,317,598,604,10904,1294,80315,53944,
1606,2770,3628,3675,3985,4035,4163,84552,29085,55367,5371,5791,54884,5980,8794,
1462,50808,220,583,694,1056,9076,10978,54677,1612,55040,114907,2274,127707,4000,
8079,4646,4747,27445,5143,80055,79156,5360,5364,23654,5565,5613,5625,10076,56963,
6004,390,255488,6326,6330,23513,7869,283130,204962,83959,6548,6774,9263,10228,
22954,10475,85363,494514,10142,79714,1006,8446,9648,79828,5507,55240,63874,25841,
9289,84883,154810,51321,421,8553,655,119032,84280,10950,824,839,57828,857,8812,
8837,94027,113189,22837,132864,10898,3300,81704,1847,1849,1947,9538,24139,5168,
147965,115548,9873,23768,2632,2817,3280,3265,23308,3490,51477,182,3856,8844,144811,
9404,4043,9848,2872,23041,740,343263,4638,26509,4792,22861,57523,55214,80025,164091,
57060,64065,51090,5453,8496,333926,55671,5900,55544,23179,8601,389,6223,55800,6385,
4088,6643,122809,257397,285343,7011,54790,374618,55362,51754,7157,9537,22906,7205,
80705,219699,55245,83719,7748,25946,118738)
# saveRDS(entrez_ids_p53, "data/new/RUV/p53_targetgenes_entrezid.RDS")
# Function to extract relevant data
extract_data_p53_RUV <- function(df, name) {
df %>%
filter(Entrez_ID %in% entrez_ids_p53) %>%
mutate(Gene = mapIds(org.Hs.eg.db,
as.character(Entrez_ID),
column = "SYMBOL",
keytype = "ENTREZID",
multiVals = "first"),
Condition = name,
Signif = ifelse(adj.P.Val < 0.05, "*", ""))
}
# Collect all data
# DEG list
deg_list_RUV <- list("DOX_24T" = DOX_24T_R,
"DOX_24R" = DOX_24R_R,
"DOX_144R" = DOX_144R_R
)
# Combine all DEGs and annotate
all_data_p53_RUV <- bind_rows(mapply(extract_data_p53_RUV, deg_list_RUV, names(deg_list_RUV), SIMPLIFY = FALSE)) 'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns
'select()' returned 1:1 mapping between keys and columns#unnecessary to join again as there are not categories on this list of entrezids
# %>%
# left_join(entrez_ids, by = c("Entrez_ID" = "ENTREZID"))
#make a list of these genes for later analysis
# saveRDS(all_data_p53_RUV, "data/new/RUV/p53_targetgenes_RUV.RDS")
# p53_genes <- all_data_p53_RUV %>%
# dplyr::select("SYMBOL", "Entrez_ID")
# saveRDS(p53_genes, "data/new/RUV/p53_genes_symbolentrez.RDS")
# Create matrices
logFC_mat53_RUV <- acast(all_data_p53_RUV, Gene ~ Condition, value.var = "logFC")
signif_mat53_RUV <- acast(all_data_p53_RUV, Gene ~ Condition, value.var = "Signif")
# Desired column order
desired_order <- c("DOX_24T",
"DOX_24R",
"DOX_144R")
logFC_mat_p53_RUV <- logFC_mat53_RUV[, desired_order]
signif_mat_p53_RUV <- signif_mat53_RUV[, desired_order]
# Column annotation
meta_p53_RUV <- str_split_fixed(colnames(logFC_mat_p53_RUV), "_", 3)
meta_p53_RUV <- str_split_fixed(colnames(logFC_mat_p53_RUV), "_", 2)
col_annot_p53_RUV <- HeatmapAnnotation(
Drug = meta_p53_RUV[, 1],
Time = meta_p53_RUV[, 2],
col = list(
Drug = c("DOX" = "#499FBD"),
Time = c("24T" = "#238B45",
"24R" = "#74C476",
"144R" = "#C7E9C0")
),
annotation_height = unit(c(1, 1, 1), "cm")
)
# Draw heatmap
Heatmap(logFC_mat_p53_RUV,
name = "logFC",
top_annotation = col_annot,
cluster_columns = FALSE,
cluster_rows = TRUE,
show_row_names = TRUE,
show_column_names = FALSE,
layer_fun = function(j, i, x, y, width, height, fill) {
grid.text(signif_mat_p53_RUV[cbind(i, j)], x, y, gp = gpar(fontsize = 9))
},
column_title = "P53 Target Genes Expression\nDOX Recovery (RUVs Corrected)",
column_title_gp = gpar(fontsize = 14, fontface = "bold")
)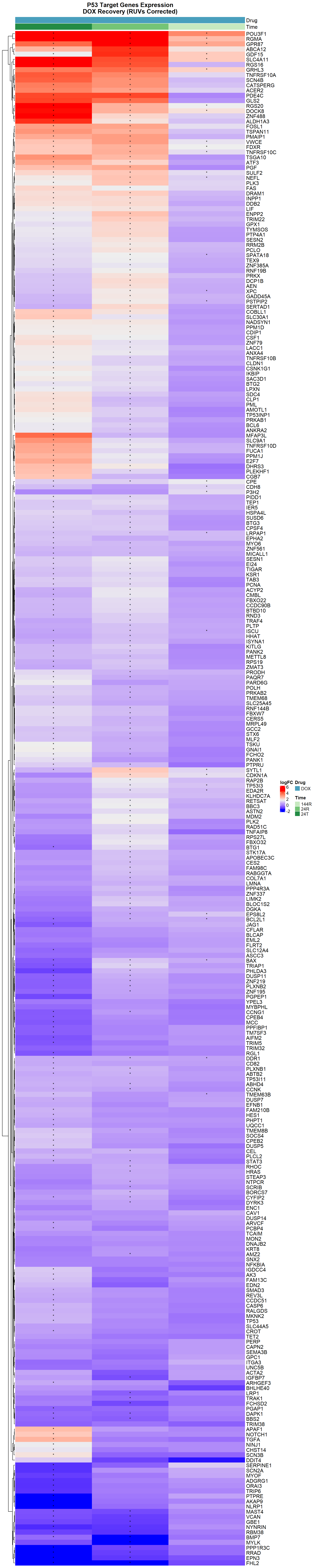
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
##p53 logFC boxplots
#get an overall idea of the logFC of these genes over timepoints
# library(rstatix)
# Read p53 gene list - named p53_genes
# p53_genes <- readRDS("data/new/RUV/p53_genes_symbolentrez.RDS")
#
# # Convert gene symbols to Entrez IDs
# p53_targets <- p53_genes %>%
# mutate(Entrez_ID = mapIds(org.Hs.eg.db,
# keys = SYMBOL,
# column = "ENTREZID",
# keytype = "SYMBOL",
# multiVals = "first"))
#
#p53_target_genes <- na.omit(p53_targets$Entrez_ID)
# saveRDS(p53_target_genes, "data/new/RUV/p53_target_genes_symbolentrez.RDS")
#this already has both SYMBOL and Entrez_ID
p53_target_genes <- readRDS("data/new/RUV/p53_target_genes_symbolentrez.RDS")
all_toptables_RUV <- bind_rows(
DOX_24T_R %>% mutate(Drug = "DOX", Timepoint = "24T"),
DOX_24R_R %>% mutate(Drug = "DOX", Timepoint = "24R"),
DOX_144R_R %>% mutate(Drug = "DOX", Timepoint = "144R"),
)
filtered_toptables_RUV_p53 <- all_toptables_RUV %>%
filter(Entrez_ID %in% p53_target_genes) %>%
mutate(abs_logFC = abs(logFC))
filtered_toptables_RUV_p53 <- filtered_toptables_RUV_p53 %>%
mutate(
Drug = factor(Drug, levels = c("DOX")),
Timepoint = factor(Timepoint, levels = c("24T", "24R", "144R"),
labels = c("Timepoint: 24T", "Timepoint: 24R", "Timepoint: 144R"))
)
wilcox_results_p53 <- filtered_toptables_RUV_p53 %>%
wilcox_test(abs_logFC ~ Timepoint) %>%
adjust_pvalue(method = "bonferroni") %>%
mutate(significance = ifelse(p < 0.05, "*", "")) %>%
mutate(Conditions = paste(group1, "vs", group2, sep = " "))
#make a little thing of your timepoint comparisons
pairs_p53 <- wilcox_results_p53 %>%
dplyr::select(group1, group2) %>%
distinct()
max_per_timepoint_p53 <- filtered_toptables_RUV_p53 %>%
group_by(Timepoint) %>%
summarise(max_logFC = max(abs_logFC, na.rm = TRUE), .groups = "drop")
#Join max values for group1
star_positions_p53 <- pairs_p53 %>%
left_join(max_per_timepoint_p53, by = c("group1" = "Timepoint"))
names(star_positions_p53)[names(star_positions_p53) == "max_logFC"] <- "max1"
#Join max values for group2, with suffixes to avoid name conflict
star_positions_p53 <- star_positions_p53 %>%
left_join(max_per_timepoint_p53, by = c("group2" = "Timepoint"), suffix = c(".group1", ".group2"))
names(star_positions_p53)[names(star_positions_p53) == "max_logFC"] <- "max2"
star_positions_p53 <- star_positions_p53 %>%
mutate(y_pos = pmax(max1, max2, na.rm = TRUE) + 0.2)
#now star positions has the same group1 and group2 as wilcoxon
wilcox_results_plot_p53 <- wilcox_results_p53 %>%
left_join(star_positions_p53, by = c("group1", "group2")) %>%
mutate(x_position = 1.5)
wilcox_results_plot_p53 <- wilcox_results_plot_p53 %>%
mutate(group1 = gsub("Timepoint: ", "", group1),
group2 = gsub("Timepoint: ", "", group2))
wilcox_results_plot_p53 <- wilcox_results_plot_p53 %>%
mutate(x_position = case_when(
group1 == "24T" & group2 == "24R" ~ 1.5,
group1 == "24T" & group2 == "144R" ~ 2,
group1 == "24R" & group2 == "144R" ~ 2.5
))
####logFC####
ggplot(filtered_toptables_RUV_p53, aes(x = Timepoint, y = logFC, fill = Timepoint)) +
geom_boxplot() +
scale_fill_manual(values = c(
"Timepoint: 24T" = "#238B45",
"Timepoint: 24R" = "#74C476",
"Timepoint: 144R" = "#C7E9C0")) +
facet_wrap(~ Drug) +
# geom_text(
# data = wilcox_results_plot_p53,
# aes(x = x_position, y = y_pos, label = significance),
# size = 6, fontface = "bold", color = "black", inherit.aes = FALSE
# ) +
theme_bw() +
xlab("") +
ylab("Log Fold Change") +
ggtitle("Log Fold Change for p53 Target Genes") +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.line = element_line(linewidth = 1.5),
strip.background = element_rect(fill = "gray"),
strip.text = element_text(size = 12, color = "black", face = "bold"),
axis.text.x = element_text(size = 8, color = "black", angle = 0)
)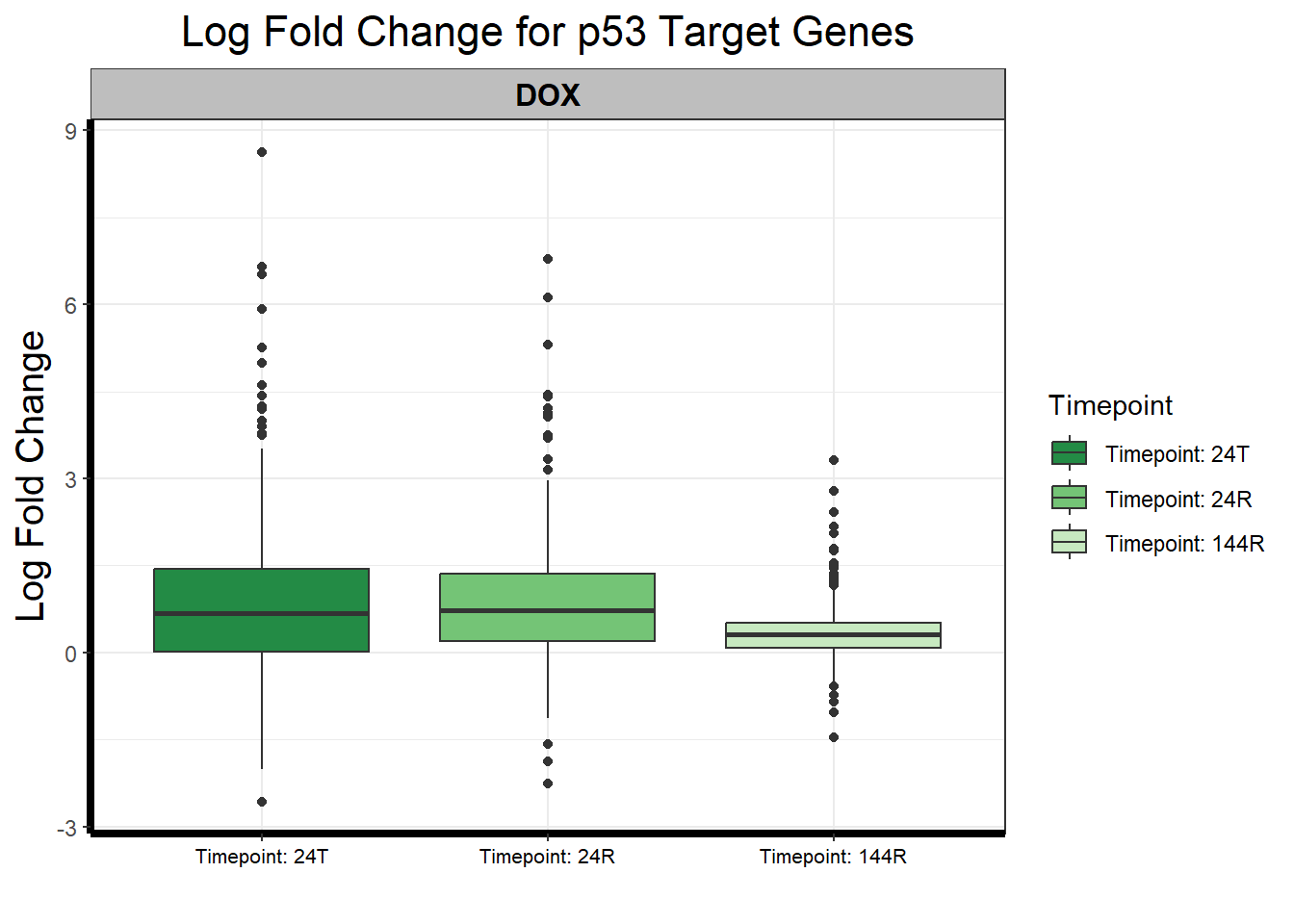
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
####abs logFC####
ggplot(filtered_toptables_RUV_p53, aes(x = Timepoint, y = abs_logFC, fill = Timepoint)) +
geom_boxplot() +
scale_fill_manual(values = c(
"Timepoint: 24T" = "#238B45",
"Timepoint: 24R" = "#74C476",
"Timepoint: 144R" = "#C7E9C0")) +
facet_wrap(~ Drug) +
# geom_text(
# data = wilcox_results_plot_p53,
# aes(x = x_position, y = y_pos, label = significance),
# size = 6, fontface = "bold", color = "black", inherit.aes = FALSE
# ) +
theme_bw() +
xlab("") +
ylab("|Log Fold Change|") +
ggtitle("|Log Fold Change| for p53 Target Genes") +
theme(
plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.line = element_line(linewidth = 1.5),
strip.background = element_rect(fill = "gray"),
strip.text = element_text(size = 12, color = "black", face = "bold"),
axis.text.x = element_text(size = 8, color = "black", angle = 0)
)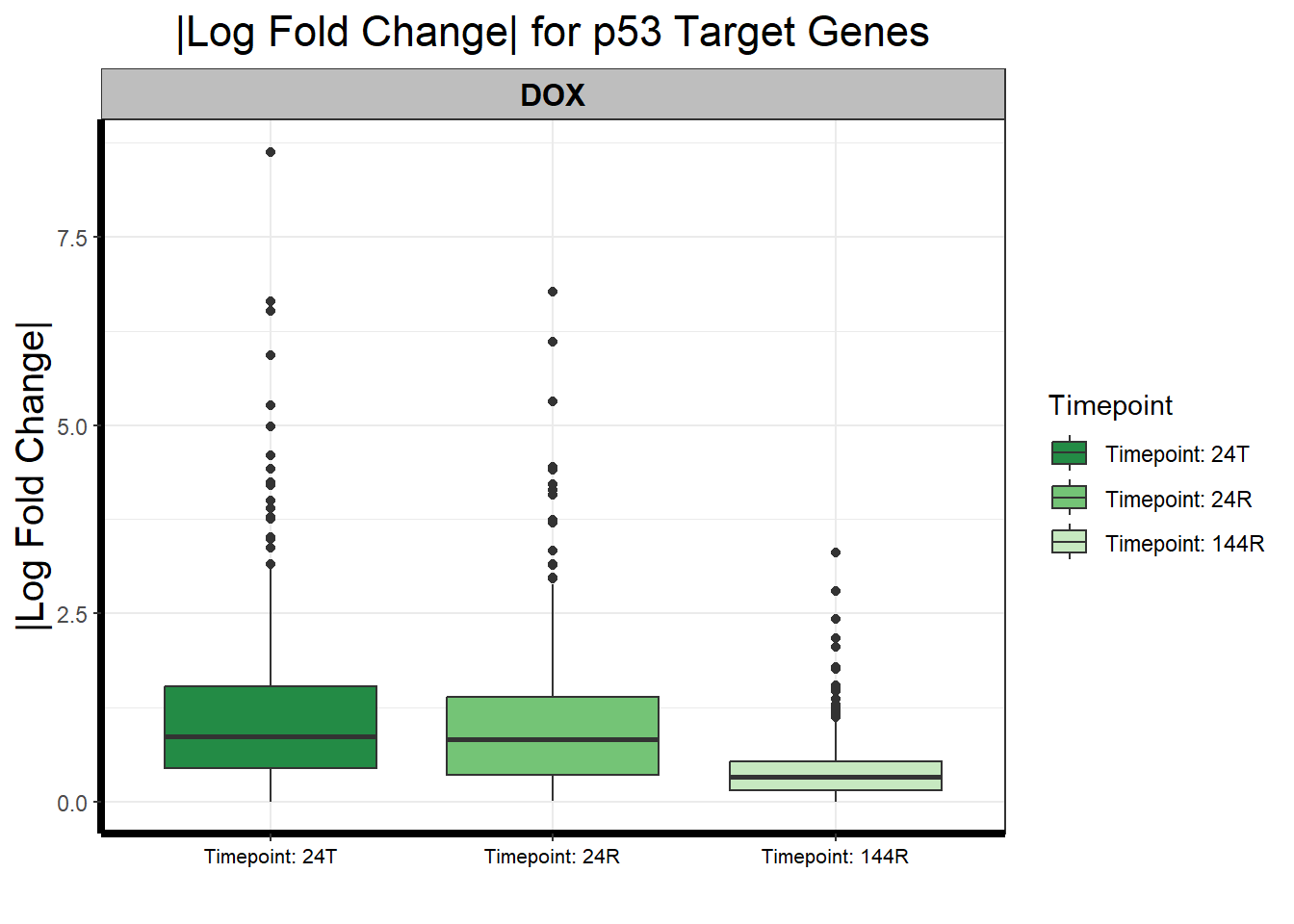
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
###logFC boxplots with all timepoint comparisons wilcoxon
# #try this where each of my timepoints are compared like the wilcoxon test - plot all of these and see which are significant
#
# Comparisons <- tribble(
# ~group1, ~group2,
# "24T", "24R",
# "24T", "144R",
# "24R", "144R"
# )
#
# # Expand your filtered_toptables_RUV_p53 to include comparisons
# filtered_toptables_RUV_p53_comp <- filtered_toptables_RUV_p53 %>%
# # Create a cross join with the comparisons
# crossing(Comparisons) %>%
# # Keep only the rows where the gene's timepoint matches one of the groups in the comparison
# filter(Timepoint %in% c(group1, group2)) %>%
# mutate(
# Conditions = factor(paste(group1, "vs", group2),
# levels = c("24T vs 24R", "24T vs 144R", "24R vs 144R"))
# )
#
#
# # filtered_toptables_RUV_p53_comp <- filtered_toptables_RUV_p53_comp %>%
# # mutate(
# # Drug = factor(Drug, levels = c("DOX")),
# # Timepoint_raw = Timepoint, #save unmod version(24T, 24R, 144R)
# # Timepoint = factor(Timepoint, levels = c("24T", "24R", "144R"),
# # labels = c("24T", "24R", "144R")),
# # Conditions = factor(paste(group1, "vs", group2),
# # levels = c("24T vs 24R", "24T vs 144R", "24R vs 144R"))
# # )
#
# wilcox_results_p53 <- filtered_toptables_RUV_p53_comp %>%
# wilcox_test(abs_logFC ~ Timepoint) %>%
# adjust_pvalue(method = "bonferroni") %>%
# mutate(significance = ifelse(p < 0.05, "*", "")) %>%
# mutate(Comparisons = paste(group1, "vs", group2, sep = " "))
#
# #make a little thing of your timepoint comparisons
# pairs_p53 <- wilcox_results_p53 %>%
# dplyr::select(group1, group2) %>%
# distinct()
#
# max_per_timepoint_p53 <- filtered_toptables_RUV_p53_comp %>%
# group_by(Timepoint) %>%
# summarise(max_logFC = max(abs_logFC, na.rm = TRUE), .groups = "drop")
#
# #Join max values for group1
# star_positions_p53 <- pairs_p53 %>%
# left_join(max_per_timepoint_p53, by = c("group1" = "Timepoint"))
# names(star_positions_p53)[names(star_positions_p53) == "max_logFC"] <- "max1"
#
# #Join max values for group2, with suffixes to avoid name conflict
# star_positions_p53 <- star_positions_p53 %>%
# left_join(max_per_timepoint_p53, by = c("group2" = "Timepoint"), suffix = c(".group1", ".group2"))
# names(star_positions_p53)[names(star_positions_p53) == "max_logFC"] <- "max2"
#
# star_positions_p53 <- star_positions_p53 %>%
# mutate(y_pos = pmax(max1, max2, na.rm = TRUE) + 0.2)
#
# #now star positions has the same group1 and group2 as wilcoxon
#
# wilcox_results_plot_p53 <- wilcox_results_p53 %>%
# left_join(star_positions_p53, by = c("group1", "group2")) %>%
# mutate(x_position = 1.5)
#
# wilcox_results_plot_p53 <- wilcox_results_plot_p53 %>%
# mutate(group1 = gsub("Timepoint: ", "", group1),
# group2 = gsub("Timepoint: ", "", group2))
#
# wilcox_results_plot_p53 <- wilcox_results_plot_p53 %>%
# mutate(
# group1_raw = str_remove(group1, "^Timepoint: "),
# group2_raw = str_remove(group2, "^Timepoint: ")
# )
#
# wilcox_results_plot_p53 <- wilcox_results_plot_p53 %>%
# mutate(x_position = case_when(
# group1 == "24T" & group2 == "24R" ~ 1.5,
# group1 == "24T" & group2 == "144R" ~ 2,
# group1 == "24R" & group2 == "144R" ~ 2.5
# ))
#
# ####logFC####
# ggplot(filtered_toptables_RUV_p53_comp, aes(x = Timepoint, y = logFC, fill = Timepoint)) +
# geom_boxplot() +
# scale_fill_manual(values = c(
# "24T" = "#238B45",
# "24R" = "#74C476",
# "144R" = "#C7E9C0")) +
# facet_wrap(~Conditions) +
# geom_text(
# data = wilcox_results_plot_p53,
# aes(x = x_position, y = y_pos, label = significance),
# size = 6, fontface = "bold", color = "black", inherit.aes = FALSE
# ) +
# theme_bw() +
# xlab("") +
# ylab("Log Fold Change") +
# ggtitle("Log Fold Change for p53 Target Genes") +
# theme(
# plot.title = element_text(size = rel(1.5), hjust = 0.5),
# axis.title = element_text(size = 15, color = "black"),
# axis.line = element_line(linewidth = 1.5),
# strip.background = element_rect(fill = "gray"),
# strip.text = element_text(size = 12, color = "black", face = "bold"),
# axis.text.x = element_text(size = 8, color = "black", angle = 0)
# )
#
# ####abs logFC####
# ggplot(filtered_toptables_RUV_p53_comp, aes(x = Conditions, y = abs_logFC, fill = Timepoint)) +
# geom_boxplot() +
# scale_fill_manual(values = c(
# "24T" = "#238B45",
# "24R" = "#74C476",
# "144R" = "#C7E9C0")) +
# facet_grid(~Conditions) +
# geom_text(
# data = wilcox_results_plot_p53,
# aes(x = x_position, y = y_pos, label = significance),
# size = 6, fontface = "bold", color = "black", inherit.aes = FALSE
# ) +
# theme_bw() +
# xlab("") +
# ylab("|Log Fold Change|") +
# ggtitle("|Log Fold Change| for p53 Target Genes") +
# theme(
# plot.title = element_text(size = rel(1.5), hjust = 0.5),
# axis.title = element_text(size = 15, color = "black"),
# axis.line = element_line(linewidth = 1.5),
# strip.background = element_rect(fill = "gray"),
# strip.text = element_text(size = 12, color = "black", face = "bold"),
# axis.text.x = element_text(size = 8, color = "black", angle = 0)
# )#Read in my DEGs Data
#read in boxplot1 for my log2cpm data
boxplot1 <- read.csv("data/new/filcpm_final_matrix.csv") %>%
as.data.frame()
#this dataframe is unchanged
# Load Toptables
deg_files_ruv <- list.files("data/new/DEGs", pattern = "Toptable_RUV.*\\.csv", full.names = TRUE)
deg_list_ruv <- lapply(deg_files_ruv, read.csv)
names(deg_list_ruv) <- gsub("data/DEGs/Toptable_|\\.csv", "", deg_files_ruv)
# Function to check significance based on **Entrez_ID in the correct sample**
is_significant <- function(gene, drug, conc, timepoint) {
condition <- paste(drug, conc, timepoint, sep = "_")
if (!condition %in% names(deg_list_ruv)) return(FALSE)
toptable <- deg_list[[condition]]
gene_entrez <- boxplot1$ENTREZID[boxplot1$SYMBOL == gene]
if (length(gene_entrez) == 0) return(FALSE)
return(any(gene_entrez %in% toptable$Entrez_ID[toptable$adj.P.Val < 0.05]))
}##Process Data for Plotting RUVs logFC Boxplots
process_gene_data_DDR_RUVs_boxplots <- function(gene) {
gene_data_boxplots_RUVs <- boxplot1 %>% filter(SYMBOL == gene)
# Reshape data
long_data_RUV_DDR <- gene_data_boxplots_RUVs %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2CPM") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6R$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, Indv, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
# Identify significant conditions **per Drug, Conc, and Timepoint**
significance_labels <- long_data_RUV_DDR %>%
distinct(Drug, Timepoint, Indv) %>%
rowwise() %>%
mutate(
max_log2CPM = max(long_data_RUV_DDR$log2CPM[long_data_RUV_DDR$Condition == Condition], na.rm = TRUE),
Significance = ifelse(is_significant(gene, Drug, Timepoint, Indv), "*", "")
) %>%
filter(Significance != "") %>% ungroup()
list(long_data_RUV_DDR = long_data_RUV_DDR, significance_labels = significance_labels)
}
#####now I can go ahead and generate boxplots as requested
#want one for DDR, one for DIC, and one for p53
#genelists:
DDR_genes_ruv <- all_data_DDR_RUV$Gene
DIC_genes_ruv <- all_data_DIC_RUV$Gene
p53_genes_ruv <- all_data_p53_RUV$Gene
# #Generate Boxplots from the above function using our gene lists above
# for (gene in boxplot1) {
# gene_data_boxplots_RUVs <- process_top5_D24R_1(gene)
# p <- ggplot(gene_data_boxplots_RUVs, aes(x = Condition, y = log2cpm, fill = Drug)) +
# geom_boxplot(outlier.shape = NA) +
# geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
# scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
# ggtitle(paste("Log2cpm", gene, "top5DEGs D24R RUVs")) +
# labs(x = "Treatment", y = "log2cpm") +
# theme_bw() +
# theme(
# plot.title = element_text(size = rel(2), hjust = 0.5),
# axis.title = element_text(size = 15, color = "black"),
# axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
# )
# print(p)
# }#Cormotif Function
## Fit limma model using code as it is found in the original cormotif code. It has
## only been modified to add names to the matrix of t values, as well as the
## limma fits
limmafit.default <- function(exprs,groupid,compid) {
limmafits <- list()
compnum <- nrow(compid)
genenum <- nrow(exprs)
limmat <- matrix(0,genenum,compnum)
limmas2 <- rep(0,compnum)
limmadf <- rep(0,compnum)
limmav0 <- rep(0,compnum)
limmag1num <- rep(0,compnum)
limmag2num <- rep(0,compnum)
rownames(limmat) <- rownames(exprs)
colnames(limmat) <- rownames(compid)
names(limmas2) <- rownames(compid)
names(limmadf) <- rownames(compid)
names(limmav0) <- rownames(compid)
names(limmag1num) <- rownames(compid)
names(limmag2num) <- rownames(compid)
for(i in 1:compnum) {
selid1 <- which(groupid == compid[i,1])
selid2 <- which(groupid == compid[i,2])
eset <- new("ExpressionSet", exprs=cbind(exprs[,selid1],exprs[,selid2]))
g1num <- length(selid1)
g2num <- length(selid2)
designmat <- cbind(base=rep(1,(g1num+g2num)), delta=c(rep(0,g1num),rep(1,g2num)))
fit <- lmFit(eset,designmat)
fit <- eBayes(fit)
limmat[,i] <- fit$t[,2]
limmas2[i] <- fit$s2.prior
limmadf[i] <- fit$df.prior
limmav0[i] <- fit$var.prior[2]
limmag1num[i] <- g1num
limmag2num[i] <- g2num
limmafits[[i]] <- fit
# log odds
# w<-sqrt(1+fit$var.prior[2]/(1/g1num+1/g2num))
# log(0.99)+dt(fit$t[1,2],g1num+g2num-2+fit$df.prior,log=TRUE)-log(0.01)-dt(fit$t[1,2]/w, g1num+g2num-2+fit$df.prior, log=TRUE)+log(w)
}
names(limmafits) <- rownames(compid)
limmacompnum<-nrow(compid)
result<-list(t = limmat,
v0 = limmav0,
df0 = limmadf,
s20 = limmas2,
g1num = limmag1num,
g2num = limmag2num,
compnum = limmacompnum,
fits = limmafits)
}
limmafit.counts <-
function (exprs, groupid, compid, norm.factor.method = "TMM", voom.normalize.method = "none")
{
limmafits <- list()
compnum <- nrow(compid)
genenum <- nrow(exprs)
limmat <- matrix(NA,genenum,compnum)
limmas2 <- rep(0,compnum)
limmadf <- rep(0,compnum)
limmav0 <- rep(0,compnum)
limmag1num <- rep(0,compnum)
limmag2num <- rep(0,compnum)
rownames(limmat) <- rownames(exprs)
colnames(limmat) <- rownames(compid)
names(limmas2) <- rownames(compid)
names(limmadf) <- rownames(compid)
names(limmav0) <- rownames(compid)
names(limmag1num) <- rownames(compid)
names(limmag2num) <- rownames(compid)
for (i in 1:compnum) {
message(paste("Running limma for comparision",i,"/",compnum))
selid1 <- which(groupid == compid[i, 1])
selid2 <- which(groupid == compid[i, 2])
# make a new count data frame
counts <- cbind(exprs[, selid1], exprs[, selid2])
# remove NAs
not.nas <- which(apply(counts, 1, function(x) !any(is.na(x))) == TRUE)
# runn voom/limma
d <- DGEList(counts[not.nas,])
d <- calcNormFactors(d, method = norm.factor.method)
g1num <- length(selid1)
g2num <- length(selid2)
designmat <- cbind(base = rep(1, (g1num + g2num)), delta = c(rep(0,
g1num), rep(1, g2num)))
y <- voom(d, designmat, normalize.method = voom.normalize.method)
fit <- lmFit(y, designmat)
fit <- eBayes(fit)
limmafits[[i]] <- fit
limmat[not.nas, i] <- fit$t[, 2]
limmas2[i] <- fit$s2.prior
limmadf[i] <- fit$df.prior
limmav0[i] <- fit$var.prior[2]
limmag1num[i] <- g1num
limmag2num[i] <- g2num
}
limmacompnum <- nrow(compid)
names(limmafits) <- rownames(compid)
result <- list(t = limmat,
v0 = limmav0,
df0 = limmadf,
s20 = limmas2,
g1num = limmag1num,
g2num = limmag2num,
compnum = limmacompnum,
fits = limmafits)
}
limmafit.list <-
function (fitlist, cmp.idx=2)
{
compnum <- length(fitlist)
genes <- c()
for (i in 1:compnum) genes <- unique(c(genes, rownames(fitlist[[i]])))
genenum <- length(genes)
limmat <- matrix(NA,genenum,compnum)
limmas2 <- rep(0,compnum)
limmadf <- rep(0,compnum)
limmav0 <- rep(0,compnum)
limmag1num <- rep(0,compnum)
limmag2num <- rep(0,compnum)
rownames(limmat) <- genes
colnames(limmat) <- names(fitlist)
names(limmas2) <- names(fitlist)
names(limmadf) <- names(fitlist)
names(limmav0) <- names(fitlist)
names(limmag1num) <- names(fitlist)
names(limmag2num) <- names(fitlist)
for (i in 1:compnum) {
this.t <- fitlist[[i]]$t[,cmp.idx]
limmat[names(this.t),i] <- this.t
limmas2[i] <- fitlist[[i]]$s2.prior
limmadf[i] <- fitlist[[i]]$df.prior
limmav0[i] <- fitlist[[i]]$var.prior[cmp.idx]
limmag1num[i] <- sum(fitlist[[i]]$design[,cmp.idx]==0)
limmag2num[i] <- sum(fitlist[[i]]$design[,cmp.idx]==1)
}
limmacompnum <- compnum
result <- list(t = limmat,
v0 = limmav0,
df0 = limmadf,
s20 = limmas2,
g1num = limmag1num,
g2num = limmag2num,
compnum = limmacompnum,
fits = limmafits)
}
## Rank genes based on statistics
generank<-function(x) {
xcol<-ncol(x)
xrow<-nrow(x)
result<-matrix(0,xrow,xcol)
z<-(1:1:xrow)
for(i in 1:xcol) {
y<-sort(x[,i],decreasing=TRUE,na.last=TRUE)
result[,i]<-match(x[,i],y)
result[,i]<-order(result[,i])
}
result
}
## Log-likelihood for moderated t under H0
modt.f0.loglike<-function(x,df) {
a<-dt(x, df, log=TRUE)
result<-as.vector(a)
flag<-which(is.na(result)==TRUE)
result[flag]<-0
result
}
## Log-likelihood for moderated t under H1
## param=c(df,g1num,g2num,v0)
modt.f1.loglike<-function(x,param) {
df<-param[1]
g1num<-param[2]
g2num<-param[3]
v0<-param[4]
w<-sqrt(1+v0/(1/g1num+1/g2num))
dt(x/w, df, log=TRUE)-log(w)
a<-dt(x/w, df, log=TRUE)-log(w)
result<-as.vector(a)
flag<-which(is.na(result)==TRUE)
result[flag]<-0
result
}
## Correlation Motif Fit
cmfit.X<-function(x, type, K=1, tol=1e-3, max.iter=100) {
## initialize
xrow <- nrow(x)
xcol <- ncol(x)
loglike0 <- list()
loglike1 <- list()
p <- rep(1, K)/K
q <- matrix(runif(K * xcol), K, xcol)
q[1, ] <- rep(0.01, xcol)
for (i in 1:xcol) {
f0 <- type[[i]][[1]]
f0param <- type[[i]][[2]]
f1 <- type[[i]][[3]]
f1param <- type[[i]][[4]]
loglike0[[i]] <- f0(x[, i], f0param)
loglike1[[i]] <- f1(x[, i], f1param)
}
condlike <- list()
for (i in 1:xcol) {
condlike[[i]] <- matrix(0, xrow, K)
}
loglike.old <- -1e+10
for (i.iter in 1:max.iter) {
if ((i.iter%%50) == 0) {
print(paste("We have run the first ", i.iter, " iterations for K=",
K, sep = ""))
}
err <- tol + 1
clustlike <- matrix(0, xrow, K)
#templike <- matrix(0, xrow, 2)
templike1 <- rep(0, xrow)
templike2 <- rep(0, xrow)
for (j in 1:K) {
for (i in 1:xcol) {
templike1 <- log(q[j, i]) + loglike1[[i]]
templike2 <- log(1 - q[j, i]) + loglike0[[i]]
tempmax <- Rfast::Pmax(templike1, templike2)
templike1 <- exp(templike1 - tempmax)
templike2 <- exp(templike2 - tempmax)
tempsum <- templike1 + templike2
clustlike[, j] <- clustlike[, j] + tempmax +
log(tempsum)
condlike[[i]][, j] <- templike1/tempsum
}
clustlike[, j] <- clustlike[, j] + log(p[j])
}
#tempmax <- apply(clustlike, 1, max)
tempmax <- Rfast::rowMaxs(clustlike, value=TRUE)
for (j in 1:K) {
clustlike[, j] <- exp(clustlike[, j] - tempmax)
}
#tempsum <- apply(clustlike, 1, sum)
tempsum <- Rfast::rowsums(clustlike)
for (j in 1:K) {
clustlike[, j] <- clustlike[, j]/tempsum
}
#p.new <- (apply(clustlike, 2, sum) + 1)/(xrow + K)
p.new <- (Rfast::colsums(clustlike) + 1)/(xrow + K)
q.new <- matrix(0, K, xcol)
for (j in 1:K) {
clustpsum <- sum(clustlike[, j])
for (i in 1:xcol) {
q.new[j, i] <- (sum(clustlike[, j] * condlike[[i]][,
j]) + 1)/(clustpsum + 2)
}
}
err.p <- max(abs(p.new - p)/p)
err.q <- max(abs(q.new - q)/q)
err <- max(err.p, err.q)
loglike.new <- (sum(tempmax + log(tempsum)) + sum(log(p.new)) +
sum(log(q.new) + log(1 - q.new)))/xrow
p <- p.new
q <- q.new
loglike.old <- loglike.new
if (err < tol) {
break
}
}
clustlike <- matrix(0, xrow, K)
for (j in 1:K) {
for (i in 1:xcol) {
templike1 <- log(q[j, i]) + loglike1[[i]]
templike2 <- log(1 - q[j, i]) + loglike0[[i]]
tempmax <- Rfast::Pmax(templike1, templike2)
templike1 <- exp(templike1 - tempmax)
templike2 <- exp(templike2 - tempmax)
tempsum <- templike1 + templike2
clustlike[, j] <- clustlike[, j] + tempmax + log(tempsum)
condlike[[i]][, j] <- templike1/tempsum
}
clustlike[, j] <- clustlike[, j] + log(p[j])
}
#tempmax <- apply(clustlike, 1, max)
tempmax <- Rfast::rowMaxs(clustlike, value=TRUE)
for (j in 1:K) {
clustlike[, j] <- exp(clustlike[, j] - tempmax)
}
#tempsum <- apply(clustlike, 1, sum)
tempsum <- Rfast::rowsums(clustlike)
for (j in 1:K) {
clustlike[, j] <- clustlike[, j]/tempsum
}
p.post <- matrix(0, xrow, xcol)
for (j in 1:K) {
for (i in 1:xcol) {
p.post[, i] <- p.post[, i] + clustlike[, j] * condlike[[i]][,
j]
}
}
loglike.old <- loglike.old - (sum(log(p)) + sum(log(q) +
log(1 - q)))/xrow
loglike.old <- loglike.old * xrow
result <- list(p.post = p.post, motif.prior = p, motif.q = q,
loglike = loglike.old, clustlike=clustlike, condlike=condlike)
}
## Fit using (0,0,...,0) and (1,1,...,1)
cmfitall<-function(x, type, tol=1e-3, max.iter=100) {
## initialize
xrow<-nrow(x)
xcol<-ncol(x)
loglike0<-list()
loglike1<-list()
p<-0.01
## compute loglikelihood
L0<-matrix(0,xrow,1)
L1<-matrix(0,xrow,1)
for(i in 1:xcol) {
f0<-type[[i]][[1]]
f0param<-type[[i]][[2]]
f1<-type[[i]][[3]]
f1param<-type[[i]][[4]]
loglike0[[i]]<-f0(x[,i],f0param)
loglike1[[i]]<-f1(x[,i],f1param)
L0<-L0+loglike0[[i]]
L1<-L1+loglike1[[i]]
}
## EM algorithm to get MLE of p and q
loglike.old <- -1e10
for(i.iter in 1:max.iter) {
if((i.iter%%50) == 0) {
print(paste("We have run the first ", i.iter, " iterations",sep=""))
}
err<-tol+1
## compute posterior cluster membership
clustlike<-matrix(0,xrow,2)
clustlike[,1]<-log(1-p)+L0
clustlike[,2]<-log(p)+L1
tempmax<-apply(clustlike,1,max)
for(j in 1:2) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
## update motif occurrence rate
for(j in 1:2) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.new<-(sum(clustlike[,2])+1)/(xrow+2)
## evaluate convergence
err<-abs(p.new-p)/p
## evaluate whether the log.likelihood increases
loglike.new<-(sum(tempmax+log(tempsum))+log(p.new)+log(1-p.new))/xrow
loglike.old<-loglike.new
p<-p.new
if(err<tol) {
break;
}
}
## compute posterior p
clustlike<-matrix(0,xrow,2)
clustlike[,1]<-log(1-p)+L0
clustlike[,2]<-log(p)+L1
tempmax<-apply(clustlike,1,max)
for(j in 1:2) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
for(j in 1:2) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.post<-matrix(0,xrow,xcol)
for(i in 1:xcol) {
p.post[,i]<-clustlike[,2]
}
## return
#calculate back loglikelihood
loglike.old<-loglike.old-(log(p)+log(1-p))/xrow
loglike.old<-loglike.old*xrow
result<-list(p.post=p.post, motif.prior=p, loglike=loglike.old)
}
## Fit each dataset separately
cmfitsep<-function(x, type, tol=1e-3, max.iter=100) {
## initialize
xrow<-nrow(x)
xcol<-ncol(x)
loglike0<-list()
loglike1<-list()
p<-0.01*rep(1,xcol)
loglike.final<-rep(0,xcol)
## compute loglikelihood
for(i in 1:xcol) {
f0<-type[[i]][[1]]
f0param<-type[[i]][[2]]
f1<-type[[i]][[3]]
f1param<-type[[i]][[4]]
loglike0[[i]]<-f0(x[,i],f0param)
loglike1[[i]]<-f1(x[,i],f1param)
}
p.post<-matrix(0,xrow,xcol)
## EM algorithm to get MLE of p
for(coli in 1:xcol) {
loglike.old <- -1e10
for(i.iter in 1:max.iter) {
if((i.iter%%50) == 0) {
print(paste("We have run the first ", i.iter, " iterations",sep=""))
}
err<-tol+1
## compute posterior cluster membership
clustlike<-matrix(0,xrow,2)
clustlike[,1]<-log(1-p[coli])+loglike0[[coli]]
clustlike[,2]<-log(p[coli])+loglike1[[coli]]
tempmax<-apply(clustlike,1,max)
for(j in 1:2) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
## evaluate whether the log.likelihood increases
loglike.new<-sum(tempmax+log(tempsum))/xrow
## update motif occurrence rate
for(j in 1:2) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.new<-(sum(clustlike[,2]))/(xrow)
## evaluate convergence
err<-abs(p.new-p[coli])/p[coli]
loglike.old<-loglike.new
p[coli]<-p.new
if(err<tol) {
break;
}
}
## compute posterior p
clustlike<-matrix(0,xrow,2)
clustlike[,1]<-log(1-p[coli])+loglike0[[coli]]
clustlike[,2]<-log(p[coli])+loglike1[[coli]]
tempmax<-apply(clustlike,1,max)
for(j in 1:2) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
for(j in 1:2) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.post[,coli]<-clustlike[,2]
loglike.final[coli]<-loglike.old
}
## return
loglike.final<-loglike.final*xrow
result<-list(p.post=p.post, motif.prior=p, loglike=loglike.final)
}
## Fit the full model
cmfitfull<-function(x, type, tol=1e-3, max.iter=100) {
## initialize
xrow<-nrow(x)
xcol<-ncol(x)
loglike0<-list()
loglike1<-list()
K<-2^xcol
p<-rep(1,K)/K
pattern<-rep(0,xcol)
patid<-matrix(0,K,xcol)
## compute loglikelihood
for(i in 1:xcol) {
f0<-type[[i]][[1]]
f0param<-type[[i]][[2]]
f1<-type[[i]][[3]]
f1param<-type[[i]][[4]]
loglike0[[i]]<-f0(x[,i],f0param)
loglike1[[i]]<-f1(x[,i],f1param)
}
L<-matrix(0,xrow,K)
for(i in 1:K)
{
patid[i,]<-pattern
for(j in 1:xcol) {
if(pattern[j] < 0.5) {
L[,i]<-L[,i]+loglike0[[j]]
} else {
L[,i]<-L[,i]+loglike1[[j]]
}
}
if(i < K) {
pattern[xcol]<-pattern[xcol]+1
j<-xcol
while(pattern[j] > 1) {
pattern[j]<-0
j<-j-1
pattern[j]<-pattern[j]+1
}
}
}
## EM algorithm to get MLE of p and q
loglike.old <- -1e10
for(i.iter in 1:max.iter) {
if((i.iter%%50) == 0) {
print(paste("We have run the first ", i.iter, " iterations",sep=""))
}
err<-tol+1
## compute posterior cluster membership
clustlike<-matrix(0,xrow,K)
for(j in 1:K) {
clustlike[,j]<-log(p[j])+L[,j]
}
tempmax<-apply(clustlike,1,max)
for(j in 1:K) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
## update motif occurrence rate
for(j in 1:K) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.new<-(apply(clustlike,2,sum)+1)/(xrow+K)
## evaluate convergence
err<-max(abs(p.new-p)/p)
## evaluate whether the log.likelihood increases
loglike.new<-(sum(tempmax+log(tempsum))+sum(log(p.new)))/xrow
loglike.old<-loglike.new
p<-p.new
if(err<tol) {
break;
}
}
## compute posterior p
clustlike<-matrix(0,xrow,K)
for(j in 1:K) {
clustlike[,j]<-log(p[j])+L[,j]
}
tempmax<-apply(clustlike,1,max)
for(j in 1:K) {
clustlike[,j]<-exp(clustlike[,j]-tempmax)
}
tempsum<-apply(clustlike,1,sum)
for(j in 1:K) {
clustlike[,j]<-clustlike[,j]/tempsum
}
p.post<-matrix(0,xrow,xcol)
for(j in 1:K) {
for(i in 1:xcol) {
if(patid[j,i] > 0.5) {
p.post[,i]<-p.post[,i]+clustlike[,j]
}
}
}
## return
#calculate back loglikelihood
loglike.old<-loglike.old-sum(log(p))/xrow
loglike.old<-loglike.old*xrow
result<-list(p.post=p.post, motif.prior=p, loglike=loglike.old)
}
generatetype<-function(limfitted)
{
jtype<-list()
df<-limfitted$g1num+limfitted$g2num-2+limfitted$df0
for(j in 1:limfitted$compnum)
{
jtype[[j]]<-list(f0=modt.f0.loglike, f0.param=df[j], f1=modt.f1.loglike, f1.param=c(df[j],limfitted$g1num[j],limfitted$g2num[j],limfitted$v0[j]))
}
jtype
}
cormotiffit <- function(exprs, groupid=NULL, compid=NULL, K=1, tol=1e-3,
max.iter=100, BIC=TRUE, norm.factor.method="TMM",
voom.normalize.method = "none", runtype=c("logCPM","counts","limmafits"), each=3)
{
# first I want to do some typechecking. Input can be either a normalized
# matrix, a count matrix, or a list of limma fits. Dispatch the correct
# limmafit accordingly.
# todo: add some typechecking here
limfitted <- list()
if (runtype=="counts") {DOX_24T_shared_DEGs
limfitted <- limmafit.counts(exprs,groupid,compid, norm.factor.method, voom.normalize.method)
} else if (runtype=="logCPM") {
limfitted <- limmafit.default(exprs,groupid,compid)
} else if (runtype=="limmafits") {
limfitted <- limmafit.list(exprs)
} else {
stop("runtype must be one of 'logCPM', 'counts', or 'limmafits'")
}
jtype<-generatetype(limfitted)
fitresult<-list()
ks <- rep(K, each = each)
fitresult <- bplapply(1:length(ks), function(i, x, type, ks, tol, max.iter) {
cmfit.X(x, type, K = ks[i], tol = tol, max.iter = max.iter)
}, x=limfitted$t, type=jtype, ks=ks, tol=tol, max.iter=max.iter)
best.fitresults <- list()
for (i in 1:length(K)) {
w.k <- which(ks==K[i])
this.bic <- c()
for (j in w.k) this.bic[j] <- -2 * fitresult[[j]]$loglike + (K[i] - 1 + K[i] * limfitted$compnum) * log(dim(limfitted$t)[1])
w.min <- which(this.bic == min(this.bic, na.rm = TRUE))[1]
best.fitresults[[i]] <- fitresult[[w.min]]
}
fitresult <- best.fitresults
bic <- rep(0, length(K))
aic <- rep(0, length(K))
loglike <- rep(0, length(K))
for (i in 1:length(K)) loglike[i] <- fitresult[[i]]$loglike
for (i in 1:length(K)) bic[i] <- -2 * fitresult[[i]]$loglike + (K[i] - 1 + K[i] * limfitted$compnum) * log(dim(limfitted$t)[1])
for (i in 1:length(K)) aic[i] <- -2 * fitresult[[i]]$loglike + 2 * (K[i] - 1 + K[i] * limfitted$compnum)
if(BIC==TRUE) {
bestflag=which(bic==min(bic))
}
else {
bestflag=which(aic==min(aic))
}
result<-list(bestmotif=fitresult[[bestflag]],bic=cbind(K,bic),
aic=cbind(K,aic),loglike=cbind(K,loglike), allmotifs=fitresult)
}
cormotiffitall<-function(exprs,groupid,compid, tol=1e-3, max.iter=100)
{
limfitted<-limmafit(exprs,groupid,compid)
jtype<-generatetype(limfitted)
fitresult<-cmfitall(limfitted$t,type=jtype,tol=1e-3,max.iter=max.iter)
}
cormotiffitsep<-function(exprs,groupid,compid, tol=1e-3, max.iter=100)
{
limfitted<-limmafit(exprs,groupid,compid)
jtype<-generatetype(limfitted)
fitresult<-cmfitsep(limfitted$t,type=jtype,tol=1e-3,max.iter=max.iter)
}
cormotiffitfull<-function(exprs,groupid,compid, tol=1e-3, max.iter=100)
{
limfitted<-limmafit(exprs,groupid,compid)
jtype<-generatetype(limfitted)
fitresult<-cmfitfull(limfitted$t,type=jtype,tol=1e-3,max.iter=max.iter)
}
plotIC<-function(fitted_cormotif)
{
oldpar<-par(mfrow=c(1,2))
plot(fitted_cormotif$bic[,1], fitted_cormotif$bic[,2], type="b",xlab="Motif Number", ylab="BIC", main="BIC")
plot(fitted_cormotif$aic[,1], fitted_cormotif$aic[,2], type="b",xlab="Motif Number", ylab="AIC", main="AIC")
}
plotMotif<-function(fitted_cormotif,title="")
{
layout(matrix(1:2,ncol=2))
u<-1:dim(fitted_cormotif$bestmotif$motif.q)[2]
v<-1:dim(fitted_cormotif$bestmotif$motif.q)[1]
image(u,v,t(fitted_cormotif$bestmotif$motif.q),
col=gray(seq(from=1,to=0,by=-0.1)),xlab="Study",yaxt = "n",
ylab="Corr. Motifs",main=paste(title,"pattern",sep=" "))
axis(2,at=1:length(v))
for(i in 1:(length(u)+1))
{
abline(v=(i-0.5))
}
for(i in 1:(length(v)+1))
{
abline(h=(i-0.5))
}
Ng=10000
if(is.null(fitted_cormotif$bestmotif$p.post)!=TRUE)
Ng=nrow(fitted_cormotif$bestmotif$p.post)
genecount=floor(fitted_cormotif$bestmotif$motif.p*Ng)
NK=nrow(fitted_cormotif$bestmotif$motif.q)
plot(0,0.7,pch=".",xlim=c(0,1.2),ylim=c(0.75,NK+0.25),
frame.plot=FALSE,axes=FALSE,xlab="No. of genes",ylab="", main=paste(title,"frequency",sep=" "))
segments(0,0.7,fitted_cormotif$bestmotif$motif.p[1],0.7)
rect(0,1:NK-0.3,fitted_cormotif$bestmotif$motif.p,1:NK+0.3,
col="dark grey")
mtext(1:NK,at=1:NK,side=2,cex=0.8)
text(fitted_cormotif$bestmotif$motif.p+0.15,1:NK,
labels=floor(fitted_cormotif$bestmotif$motif.p*Ng))
}#Don't load me in if you're using the above function, as it has been modified above
#library(Cormotif)##Input Data for Cormotif Original Matrix
#input the cormotif matrix you're going to use
##this should be tmm normalized log2cpm
#the matrix that I used previously for limma was TMM counts - cpm this
#dge was the name of the DGE list object
cormotif_test <- cpm(dge, log = TRUE)
colnames(cormotif_test) <- (Metadata_2$Final_sample_name)
cormotif_counts <- dge
cormotif_test_df <- cormotif_test %>%
as.data.frame() %>%
rownames_to_column(., var = "Entrez_ID")
#write.csv(cormotif_test, "data/new/Cormotif_test_matrix.csv")
#reorder my test matrix to match the new groupid I've made
#I want my columns to be in this order:
#DOX24T 1-6, DOX24R 1-6, DOX144R 1-6, DMSO24T 1-6, DMSO24R 1-6, DMSO144R 1-6
Cormotif <- read.csv("data/new/Cormotif_matrix_final.csv")
dim(Cormotif)[1] 14319 37#14319 genes across 37 cols (1 is Entrez_ID)
Cormotif_df <- data.frame(Cormotif)
rownames(Cormotif_df) <- Cormotif_df$Entrez_ID
exprs.cormotif <- as.matrix(Cormotif_df[,2:37])
dim(exprs.cormotif)[1] 14319 36#put together my group id and comparison id to make the correct comparisons between experimental conditions
#groupid tells which experimental conditions are grouped together
#compid tells which experimental conditions should be compared against one another
##ie DOX24T vs DMSO24T matched control
groupid_csv <- read.csv("data/new/GroupID.csv")
#now I have to make this into a vector (named vector)
groupid <- c(
DOX_24T_1 = 1,
DOX_24T_2 = 1,
DOX_24T_3 = 1,
DOX_24T_4 = 1,
DOX_24T_5 = 1,
DOX_24T_6 = 1,
DOX_24R_1 = 2,
DOX_24R_2 = 2,
DOX_24R_3 = 2,
DOX_24R_4 = 2,
DOX_24R_5 = 2,
DOX_24R_6 = 2,
DOX_144R_1 = 3,
DOX_144R_2 = 3,
DOX_144R_3 = 3,
DOX_144R_4 = 3,
DOX_144R_5 = 3,
DOX_144R_6 = 3,
DMSO_24T_1 = 4,
DMSO_24T_2 = 4,
DMSO_24T_3 = 4,
DMSO_24T_4 = 4,
DMSO_24T_5 = 4,
DMSO_24T_6 = 4,
DMSO_24R_1 = 5,
DMSO_24R_2 = 5,
DMSO_24R_3 = 5,
DMSO_24R_4 = 5,
DMSO_24R_5 = 5,
DMSO_24R_6 = 5,
DMSO_144R_1 = 6,
DMSO_144R_2 = 6,
DMSO_144R_3 = 6,
DMSO_144R_4 = 6,
DMSO_144R_5 = 6,
DMSO_144R_6 = 6
)
#saveRDS(groupid, "data/new/groupidCormotif.RDS")
compid <- data.frame(Cond1 = c(1, 2, 3), Cond2 = c(4, 5, 6))
#saveRDS(compid, "data/new/compidCormotif.RDS")###Run Cormotif Original Data
#fit Cormotif model
# set.seed(19191)
#only set the seed ONCE
# motif.fitted_new <- cormotiffit(
# exprs = exprs.cormotif,
# groupid = groupid,
# compid = compid,
# K = 1:8,
# max.iter = 1000,
# BIC = TRUE,
# runtype = "logCPM"
# )
# saveRDS(motif.fitted, "data/new/motif.fitted_new_250604.RDS")
motif.fitted <- readRDS("data/new/motif.fitted_final_3motif.RDS")###Plotting Cormotif Original Data
#plot BIC and AIC to see which number of motifs was best for both models
plotIC(motif.fitted)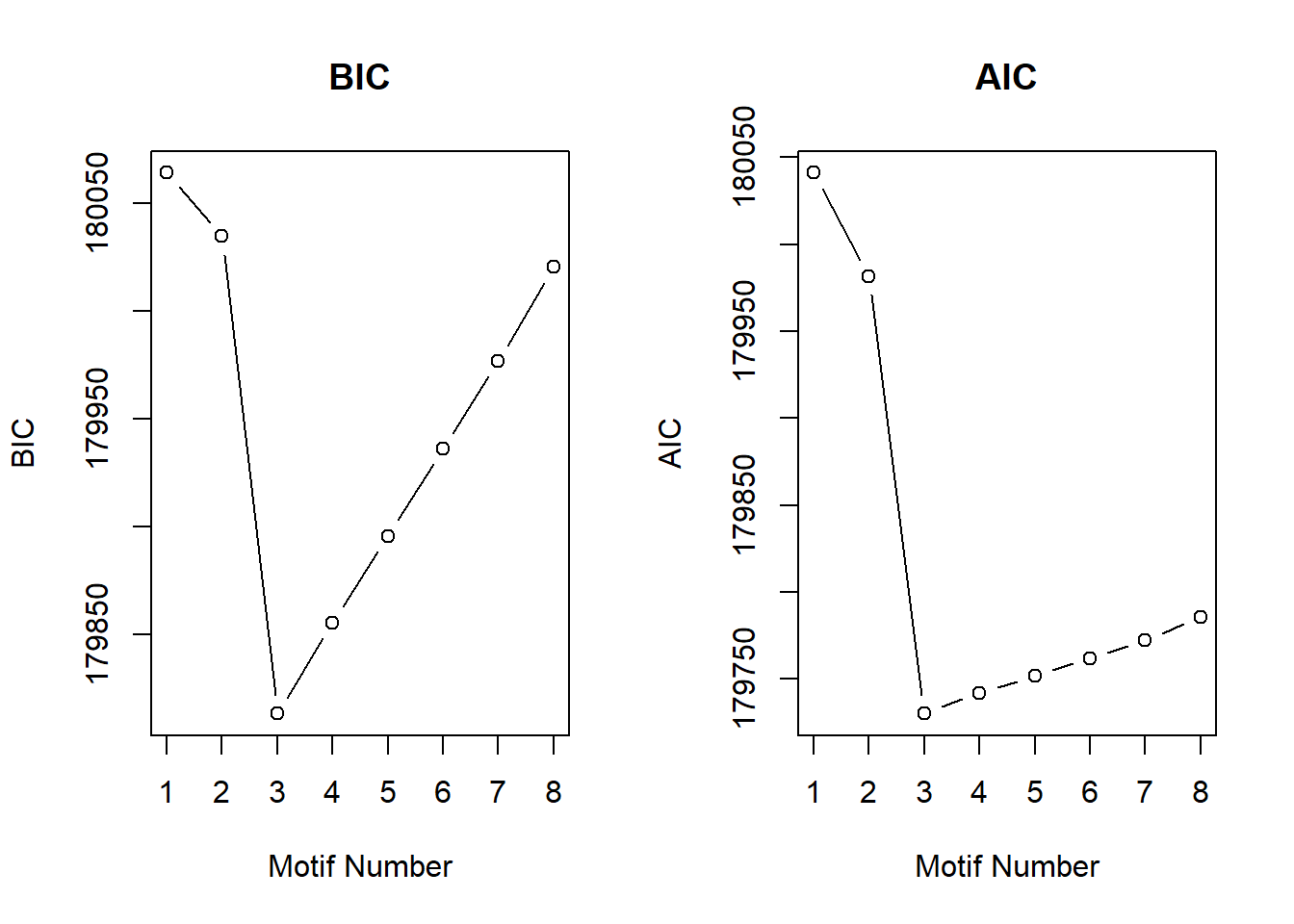
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
motif.fitted$bic K bic
[1,] 1 180063.9
[2,] 2 180034.4
[3,] 3 179813.4
[4,] 4 179855.6
[5,] 5 179895.7
[6,] 6 179936.2
[7,] 7 179976.6
[8,] 8 180020.3#now plot the motifs themselves
plotMotif(motif.fitted, title = "Fitted Motifs for DXR")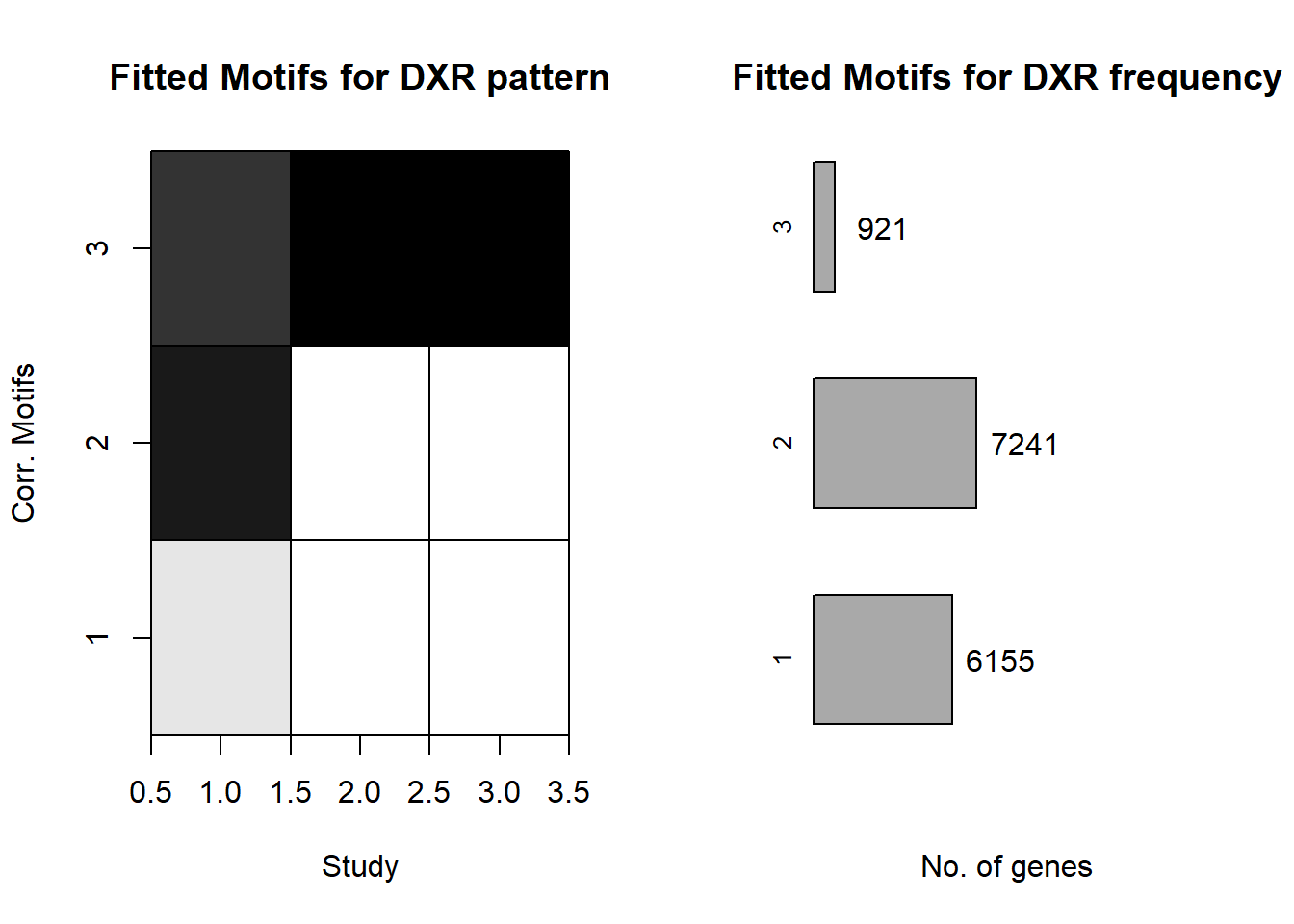
| Version | Author | Date |
|---|---|---|
| 29a5c35 | emmapfort | 2025-05-16 |
#plot the probability legend
myColors <- rev(c("#FFFFFF", "#E6E6E6" ,"#CCCCCC", "#B3B3B3", "#999999", "#808080", "#666666","#4C4C4C", "#333333", "#191919","#000000"))
plot.new()
legend('bottomleft',fill=myColors, legend =rev(c("0", "0.1", "0.2", "0.3", "0.4", "0.5", "0.6", "0.7", "0.8","0.9", "1")), box.col="white",title = "Probability\nlegend", horiz=FALSE,title.cex=.8)
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
###Extract the Gene Probabilities
#extract the posterior probability that these DEGs belong to motifs
gene_prob_all <- motif.fitted$bestmotif$p.post
rownames(gene_prob_all) <- rownames(Cormotif_df)
#assign each gene to a motif with max post prob
assigned_motifs <- apply(gene_prob_all, 1, which.max)
max_probs <- apply(gene_prob_all, 1, max)
#combine these into a dataframe - motif assigned genes (p.post)
motif_assignment_df <- gene_prob_all %>%
as.data.frame() %>%
rownames_to_column("Gene") %>%
mutate(
Assigned_Motif = assigned_motifs[Gene],
Max_Probability = max_probs[Gene]
)
#make some histograms of the unfiltered data from Cormotif p.post
gene_prob_all %>%
as.data.frame() %>%
ggplot(., aes(x = V1))+
geom_histogram(bins = 50)+
xlim(0,1)+
ggtitle("Cormotif Genes p.post Distribution M1")Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).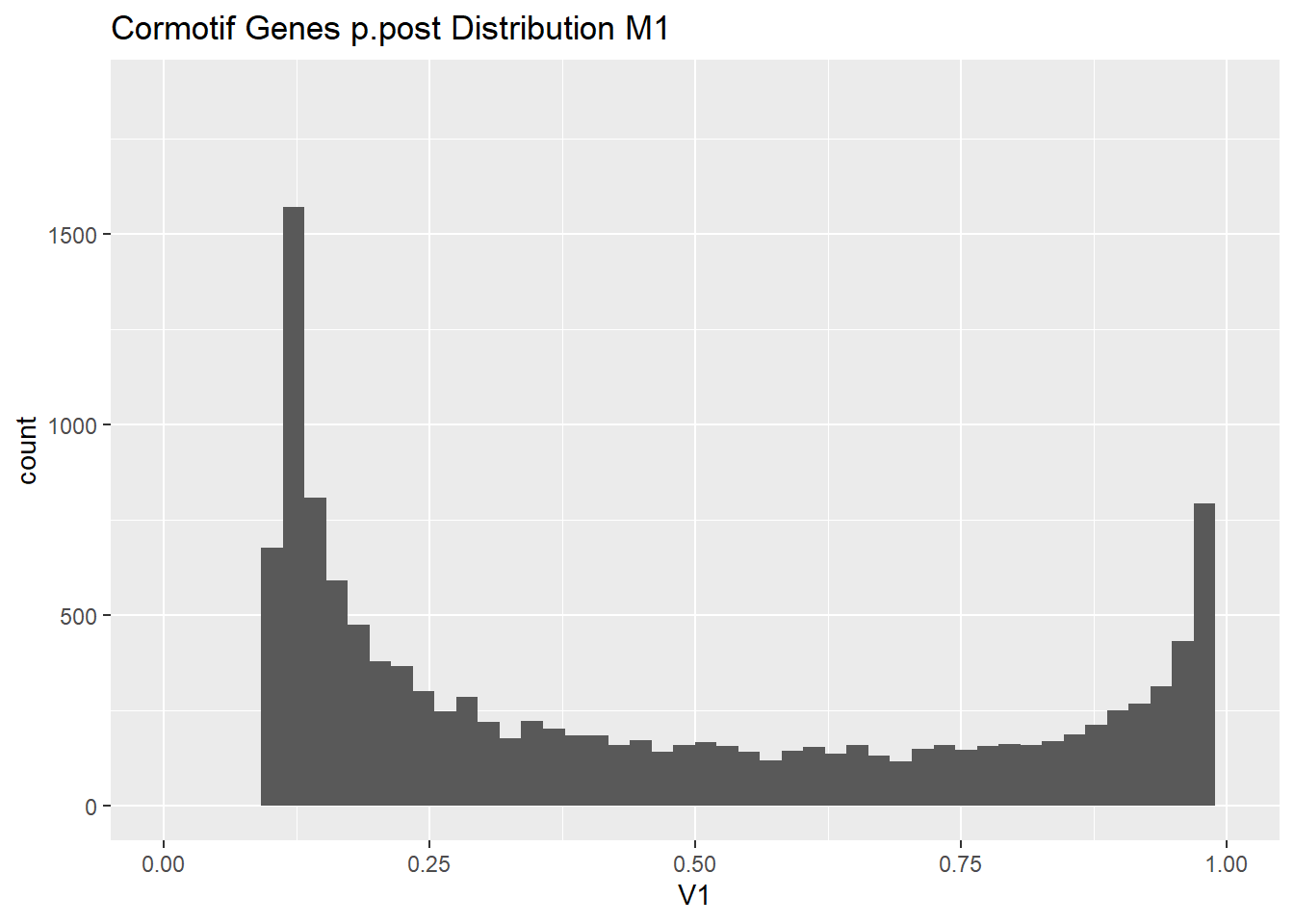
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
gene_prob_all %>%
as.data.frame() %>%
ggplot(., aes(x = V2))+
geom_histogram(bins = 50)+
xlim(0,1)+
ggtitle("Cormotif Genes p.post Distribution M2")Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).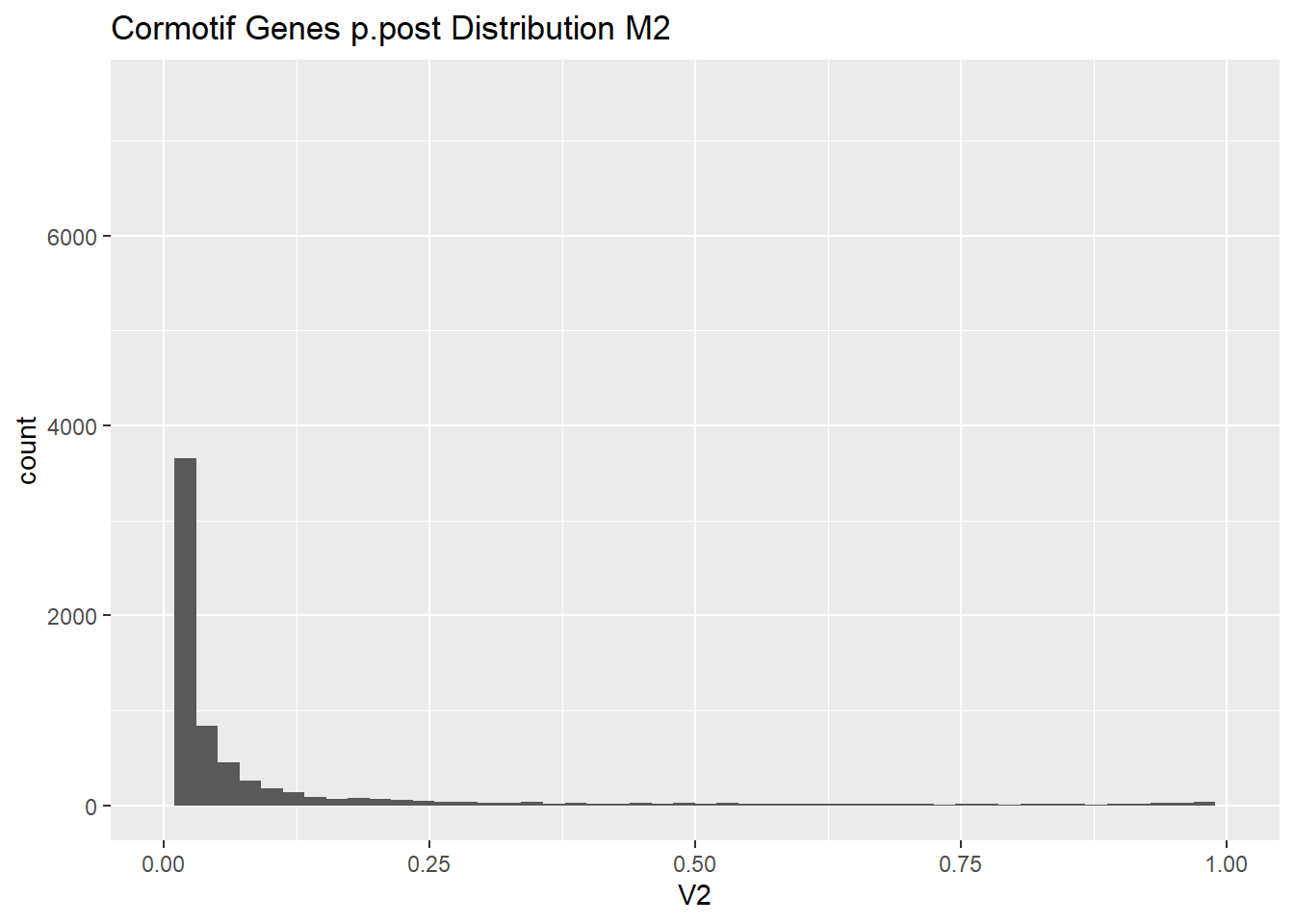
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
gene_prob_all %>%
as.data.frame() %>%
ggplot(., aes(x = V3))+
geom_histogram(bins = 50)+
xlim(0,1)+
ggtitle("Cormotif Genes p.post Distribution M3")Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).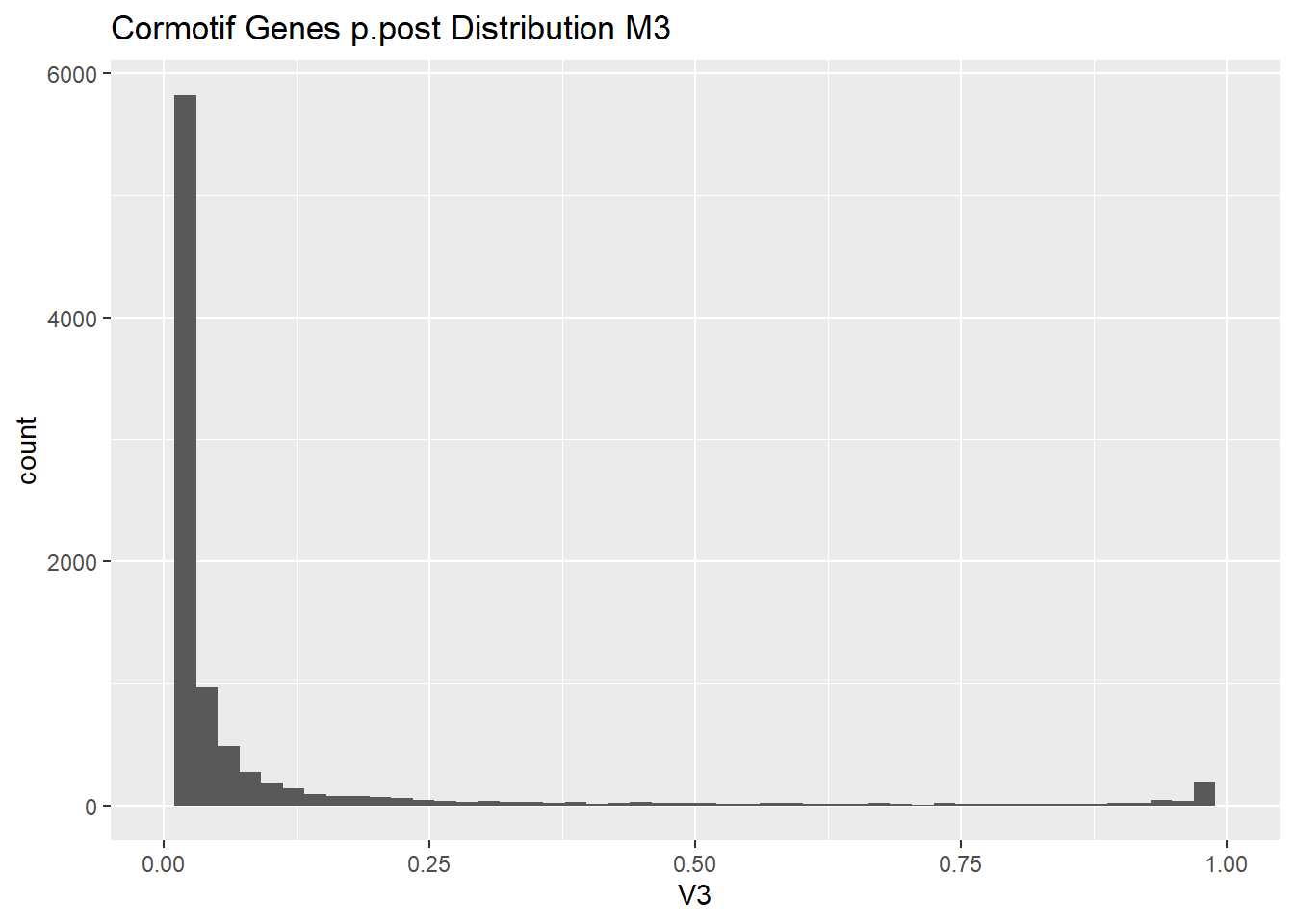
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#now change the probability cutoffs to get the ideal gene set with no overlaps
# Define gene probability groups
prob_all_1 <- rownames(gene_prob_all[(gene_prob_all[,1] >0.05 & gene_prob_all[,1] <0.3 & gene_prob_all[,2] <0.5 & gene_prob_all[,3] <0.5),])
length(prob_all_1)[1] 5675#5675
prob_all_2 <- rownames(gene_prob_all[(gene_prob_all[,1] >0.5 & gene_prob_all[,2] <0.5 & gene_prob_all[,3] <0.5),])
length(prob_all_2)[1] 6353#6353
prob_all_3 <- rownames(gene_prob_all[(gene_prob_all[,1] >0.3 & gene_prob_all[,1] <0.9 & gene_prob_all[,2] >0.5 & gene_prob_all[,3] >0.5),])
length(prob_all_3)[1] 231#231 genes with >0.3 <0.9 1
#compare between motif 1 and 2
mf1_genes <- prob_all_1
mf2_genes <- prob_all_2
mf3_genes <- prob_all_3
vennCor <- list(mf1_genes, mf2_genes, mf3_genes)
ggVennDiagram(
vennCor,
category.names = c("Motif 1", "Motif 2", "Motif 3")
) + ggtitle("Cormotif Specific and Shared DEGs p.post")+
theme(
plot.title = element_text(size = 16, face = "bold"), # Increase title size
text = element_text(size = 16) # Increase text size globally
)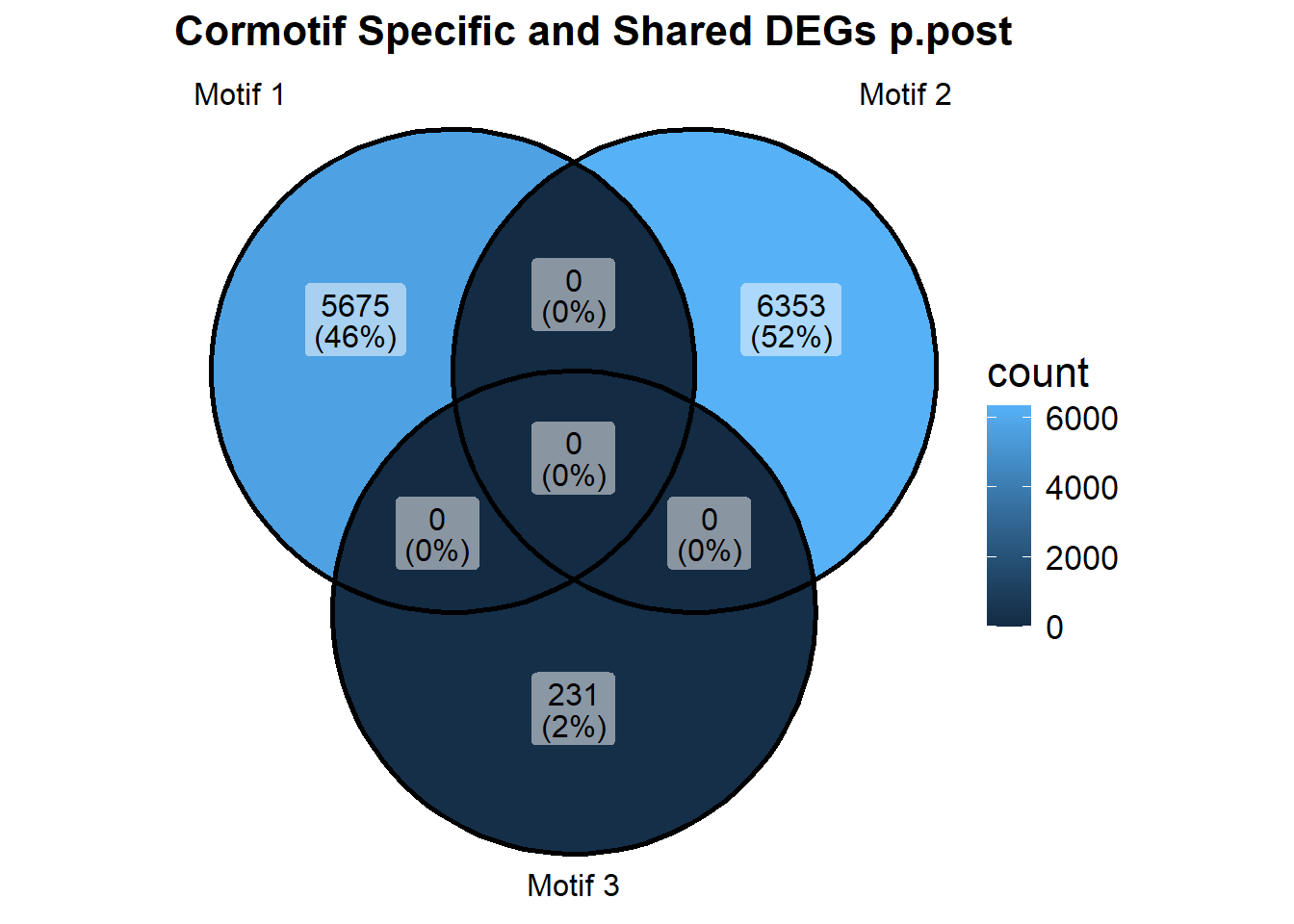
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#extract the cluster likelihood - which DEGs are most likely to be in this cluster
motif_prob <- motif.fitted$bestmotif$clustlike
rownames(motif_prob) <- rownames(gene_prob_all)
#write.csv(motif_prob,"data/new/cormotif_probability_genelist_all.csv")
#make some histograms to look at the distribution of clustlike genes without filtering
motif_prob %>%
as.data.frame() %>%
ggplot(., aes(x = V1))+
geom_histogram(bins = 50)+
xlim(0,1)+
ggtitle("Cormotif Genes clustlike Distribution M1")Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).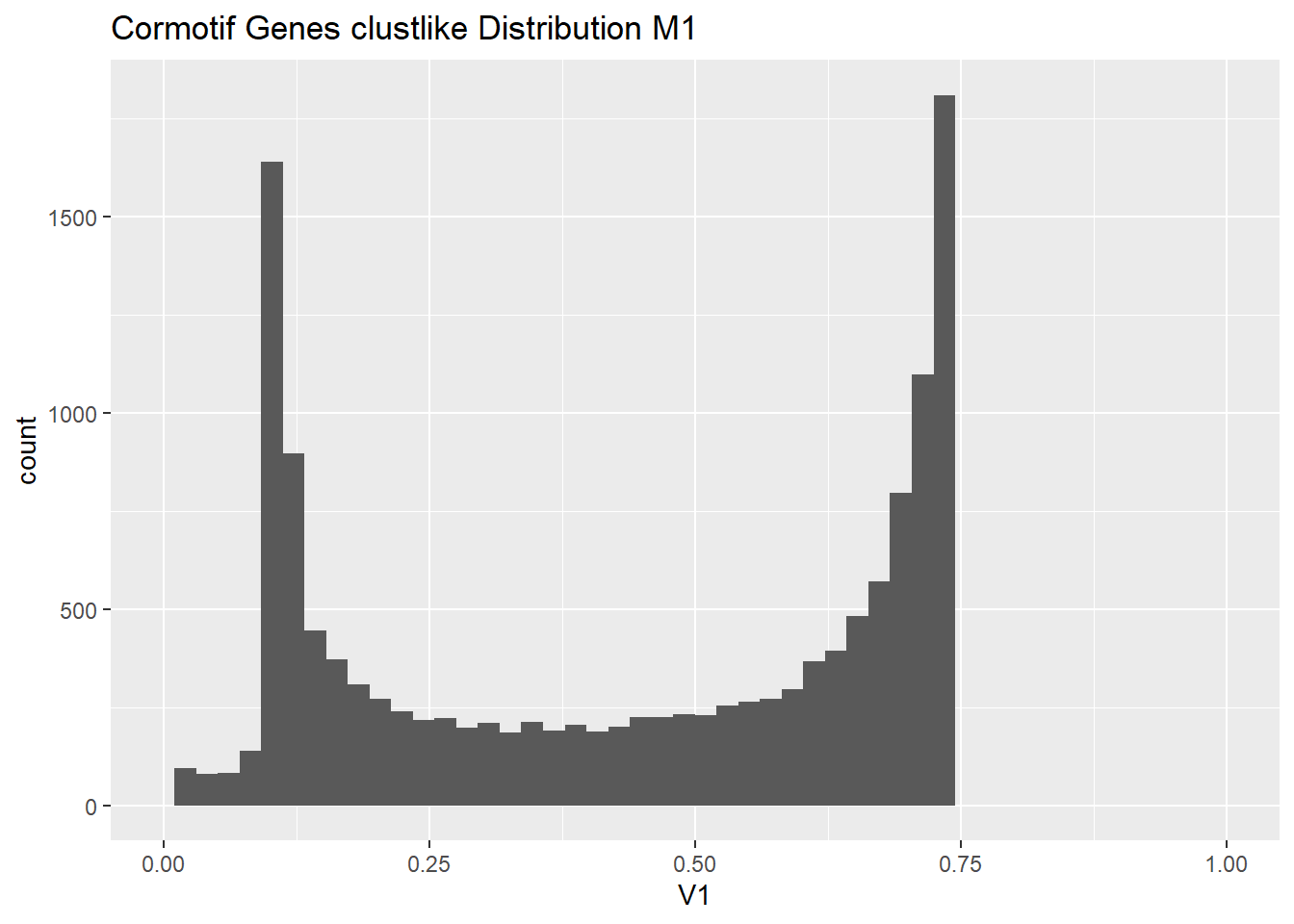
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
motif_prob %>%
as.data.frame() %>%
ggplot(., aes(x = V2))+
geom_histogram(bins = 50)+
xlim(0,1)+
ggtitle("Cormotif Genes clustlike Distribution M2")Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).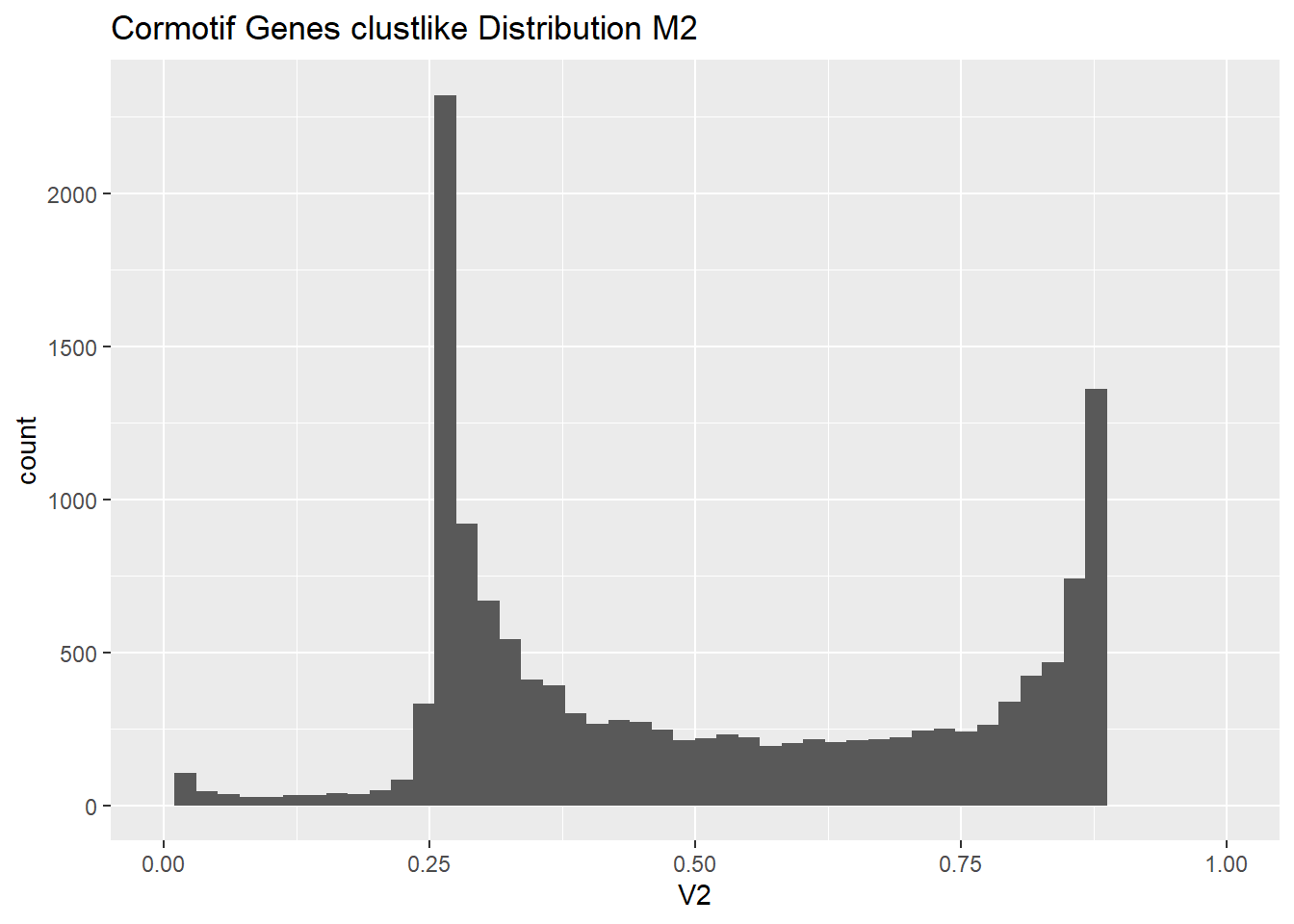
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
motif_prob %>%
as.data.frame() %>%
ggplot(., aes(x = V3))+
geom_histogram(bins = 50)+
xlim(0,1)+
ggtitle("Cormotif Genes clustlike Distribution M3")Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).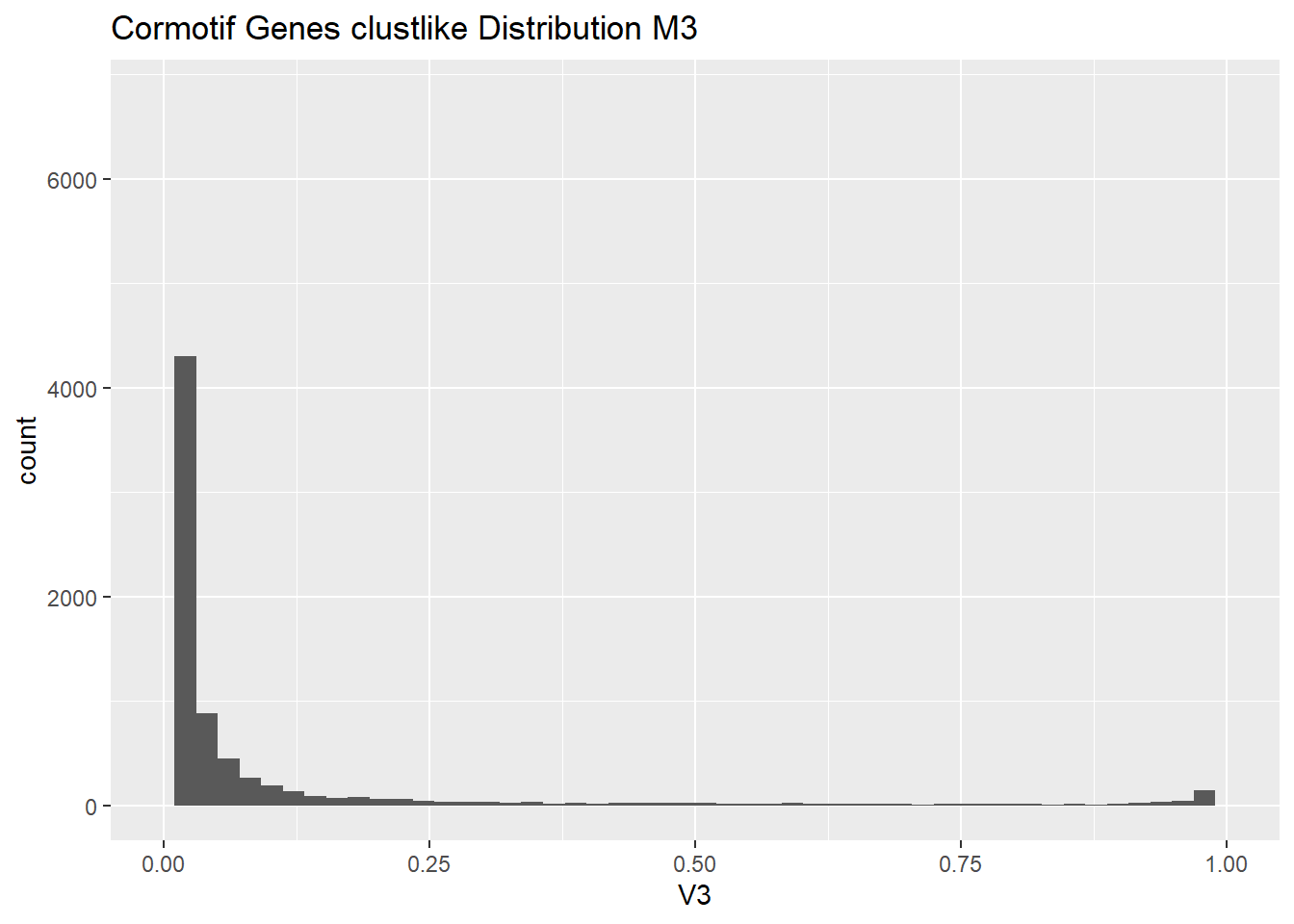
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
####try this Renee's way####
clust1_p <- motif_prob %>%
as.data.frame() %>%
filter(V1>0.5 & V2<0.5 & V3<0.5) %>%
rownames
clust2_p <- motif_prob %>%
as.data.frame() %>%
filter(V2>0.5 & V1<0.5 & V3<0.5) %>%
rownames
clust3_p <- motif_prob %>%
as.data.frame() %>%
filter(V3>0.5 & V1 <0.5 & V2 <0.5) %>%
rownames
length(clust1_p)[1] 6827#6827 (too many)
length(clust2_p)[1] 6474#6474
length(clust3_p)[1] 607#607
mf1_genes_p <- clust1_p
mf2_genes_p <- clust2_p
mf3_genes_p <- clust3_p
vennCor <- list(mf1_genes_p, mf2_genes_p, mf3_genes_p)
ggVennDiagram(
vennCor,
category.names = c("Motif 1", "Motif 2", "Motif 3")
) + ggtitle("Cormotif Specific and Shared DEGs clustlike")+
theme(
plot.title = element_text(size = 16, face = "bold"), # Increase title size
text = element_text(size = 16) # Increase text size globally
)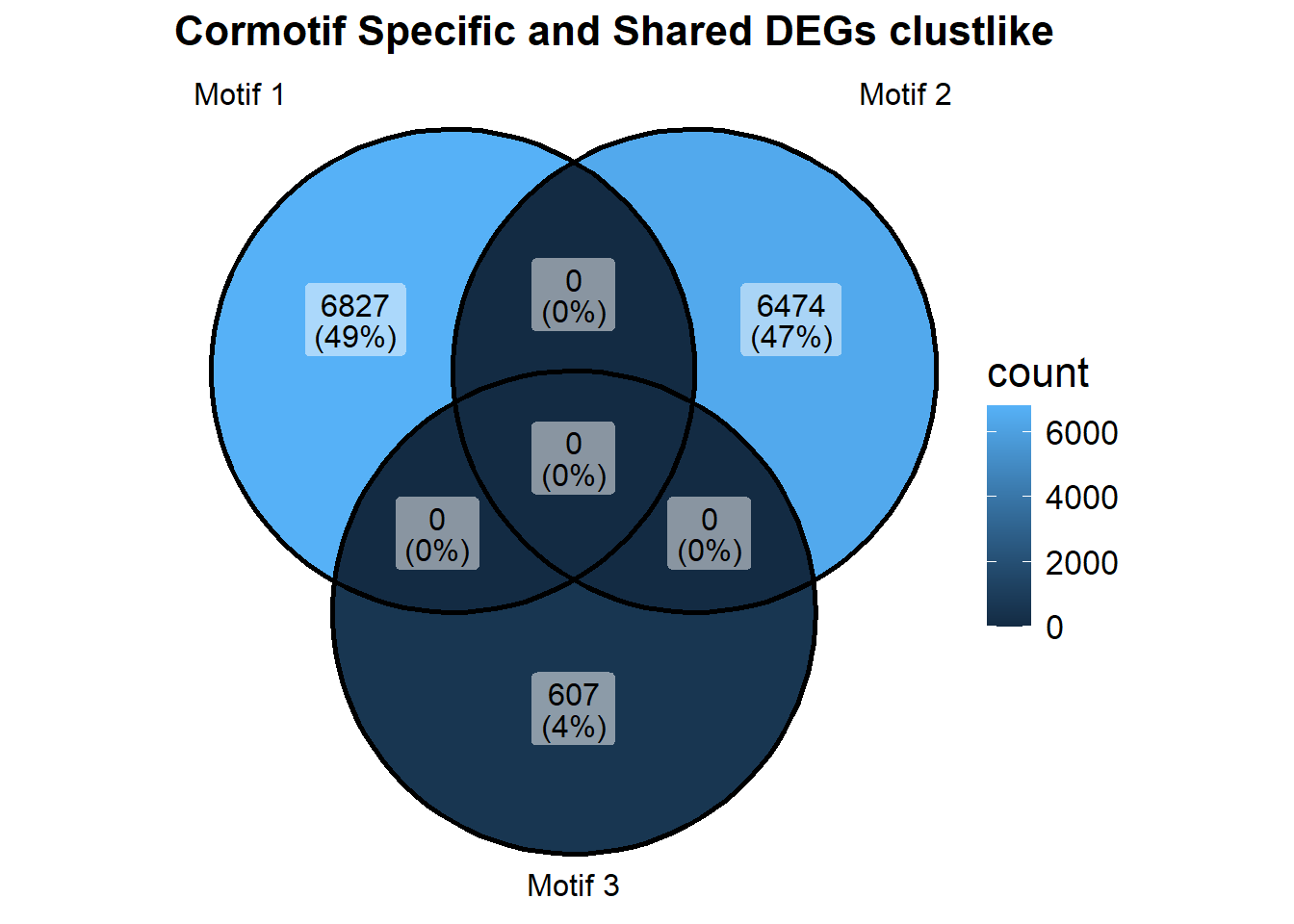
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
###Use Clustlike and P.post together for Cormotif
#begin with the clustlike cutoffs and then overlay the p.post to check if the genes are present in both
#these filters are the combination of the above analysis to get a gene set that fits in both p.post and clustlike
####motif 1####
#filter 1: clustlike
clust1_genes <- motif_prob %>%
as.data.frame() %>%
filter(V1>0.5 & V2<0.5 & V3<0.5) %>%
rownames
### Filter 2: Gene-level posterior pattern
prob_filtered_genes_1 <- rownames(gene_prob_all[(gene_prob_all[,1] >0.05 & gene_prob_all[,1] <0.3 & gene_prob_all[,2] <0.5 & gene_prob_all[,3] <0.5),])
### Final intersection of both filters
final_genes_1 <- intersect(clust1_genes, prob_filtered_genes_1)
cat("Number of genes passing both filters:", length(final_genes_1), "\n")Number of genes passing both filters: 5638 #5638 genes pass both filters here
####motif 2####
#filter 1: clustlike
clust2_genes <- motif_prob %>%
as.data.frame() %>%
filter(V2>0.5 & V1<0.5 & V3<0.5) %>%
rownames
### Filter 2: Gene-level posterior pattern
prob_filtered_genes_2 <- rownames(gene_prob_all[(gene_prob_all[,1] >0.5 & gene_prob_all[,2] <0.5 & gene_prob_all[,3] <0.5),])
### Final intersection of both filters
final_genes_2 <- intersect(clust2_genes, prob_filtered_genes_2)
cat("Number of genes passing both filters:", length(final_genes_2), "\n")Number of genes passing both filters: 6218 #6218 genes pass both filters here
####motif3####
#filter 1: clustlike
clust3_genes <- motif_prob %>%
as.data.frame() %>%
filter(V3 > 0.5 & V1 < 0.5 & V2 < 0.5) %>%
rownames()
### Filter 2: Gene-level posterior pattern (Motif 2 & 3 high, Motif 1 intermediate)
prob_filtered_genes <- rownames(gene_prob_all[
gene_prob_all[,1] > 0.3 & gene_prob_all[,1] < 0.9 &
gene_prob_all[,2] > 0.5 &
gene_prob_all[,3] > 0.5, ])
### Final intersection of both filters
final_genes_3 <- intersect(clust3_genes, prob_filtered_genes)
cat("Number of genes passing both filters:", length(final_genes_3), "\n")Number of genes passing both filters: 231 #231 genes pass both filters here
#what is the proportion of my genes that are included?
5638+6218+231[1] 12087#12087
(12087/14319)*100[1] 84.41232#84.4% of my genes are represented here out of the 14319 original
#want to find out which genes are not assigned to motifs
final_genes_list <- union(final_genes_1, union(final_genes_2, final_genes_3))
length(final_genes_list)[1] 12087#12087 genes as found above
initial_genes_list <- rownames(filcpm_matrix)
length(initial_genes_list)[1] 14319#14319 genes as usual
unassigned_genes_list <- setdiff(initial_genes_list, final_genes_list)
length(unassigned_genes_list)[1] 2232#2232 genes not assigned - correct number
#saveRDS(unassigned_genes_list, "data/new/Cormotif/unassigned_genes_Cormotif.RDS")
#now, ensure that none of these genes are shared amongst categories
vennCor_final <- list(final_genes_1, final_genes_2, final_genes_3)
ggVennDiagram(
vennCor_final,
category.names = c("Motif 1", "Motif 2", "Motif 3")
) + ggtitle("Cormotif Specific and Shared DEGs clustlike + p.post")+
theme(
plot.title = element_text(size = 16, face = "bold"), # Increase title size
text = element_text(size = 16) # Increase text size globally
)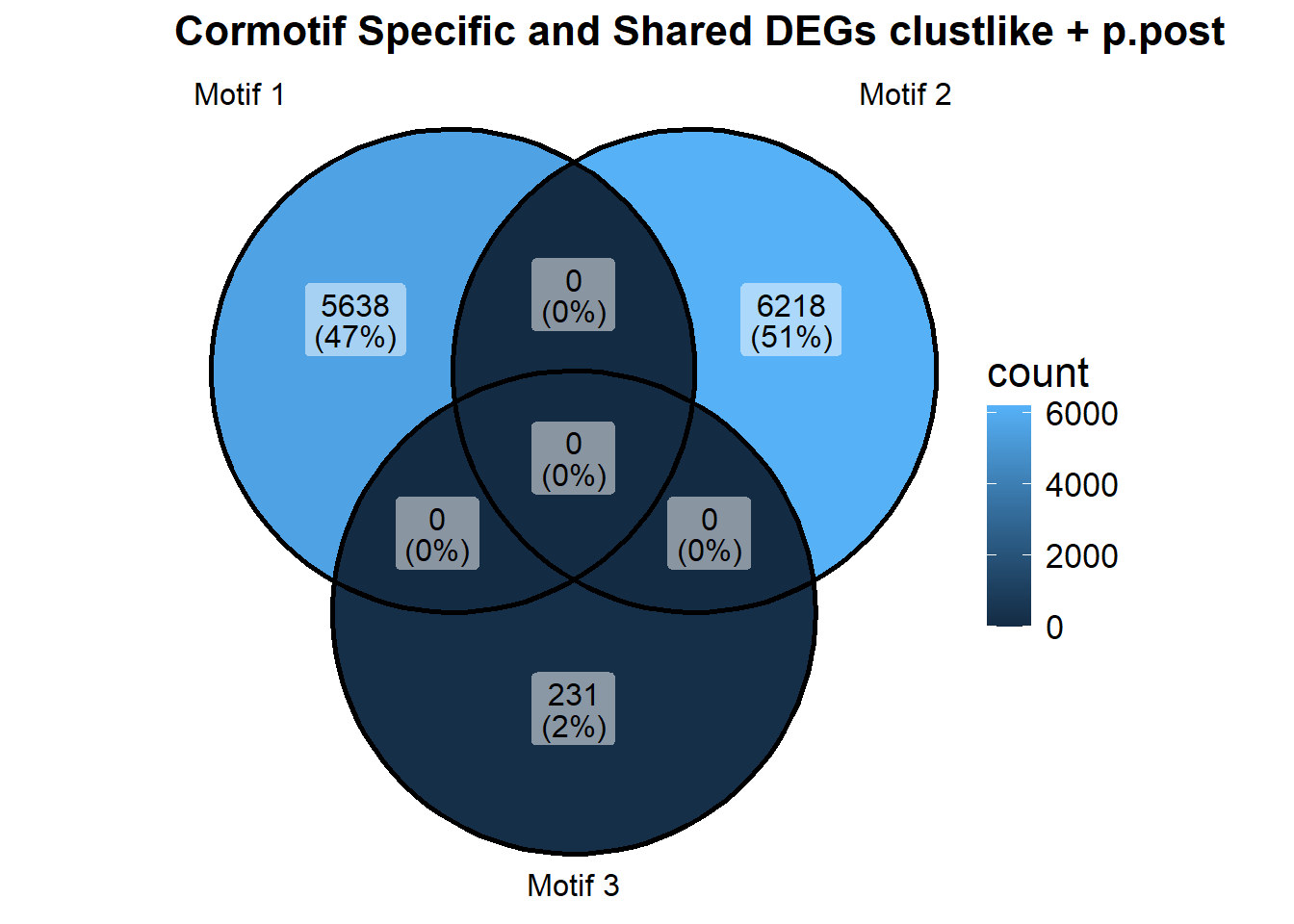
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#no overlapping genes found in these gene sets
clusterdata_dxr_fin <- data.frame(
Category = c("Motif 1", "Motif 2", "Motif 3"),
Value = c(length(final_genes_1), length(final_genes_2), length(final_genes_3))
)
piecolors_dxr_fin <- c("Motif 1" = "#007896",
"Motif 2" = "#58508D",
"Motif 3" = "#BC5090")
#make a piechart of these distributions
clusterdata_dxr_fin %>% ggplot(aes(x = "", y = Value, fill = Category))+
geom_bar(width = 1, stat = "identity")+
coord_polar("y", start = 0)+
geom_text(aes(label = Value),
position = position_stack(vjust = 0.5),
size = 4, color = "black")+
labs(title = "Distribution of Gene Clusters Identified By Cormotif", x = NULL, y = NULL)+
theme_void()+
scale_fill_manual(values = piecolors_dxr_fin)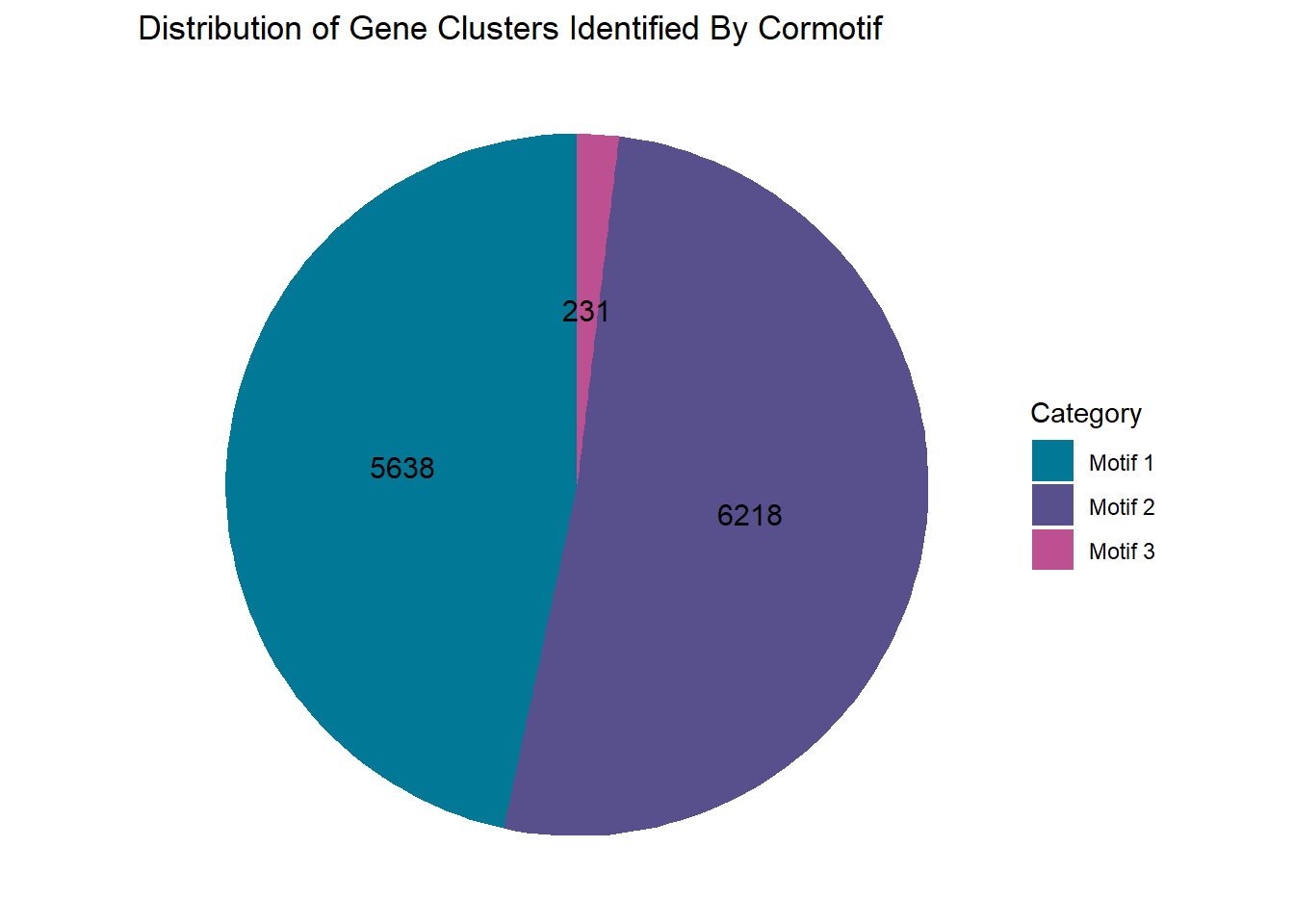
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#now, continue by plotting the logFC of all of these gene sets per motif, as well as look at some random genes for log2cpm to see if the patterns are reflected in the example genes###Plot logFC of Clustlike vs P.post Cormotif
####clustlike logFC of initial set####
##motif 1
length(mf1_genes_p)[1] 6827##motif 2
length(mf2_genes_p)[1] 6474##motif 3
length(mf3_genes_p)[1] 607#Combine the toptables I have from pairwise analysis into a single dataframe
d24_toptable_dxr <- Toptable_V.D24T %>%
rownames_to_column(var = "entrezgene_ID") %>%
mutate(Time = "24")
d24r_toptable_dxr <- Toptable_V.D24R %>%
rownames_to_column(var = "entrezgene_ID") %>%
mutate(Time = "24R")
d144r_toptable_dxr <- Toptable_V.D144R %>%
rownames_to_column(var = "entrezgene_ID") %>%
mutate(Time = "144R")
combined_toptables_dxr <- bind_rows(
d24_toptable_dxr,
d24r_toptable_dxr,
d144r_toptable_dxr)
#Filter the data based on each motif
filt_toptable_dxr_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% mf1_genes_p) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC for all genes in Motif 1 (n=6827)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 2
filt_toptable_2_dxr_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% final_genes_2) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC for all genes in Motif 2 (n=6218)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 3
filt_toptable_3_dxr_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% final_genes_3) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC for all genes in Motif 3 (n=231)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#plots
filt_toptable_dxr_fin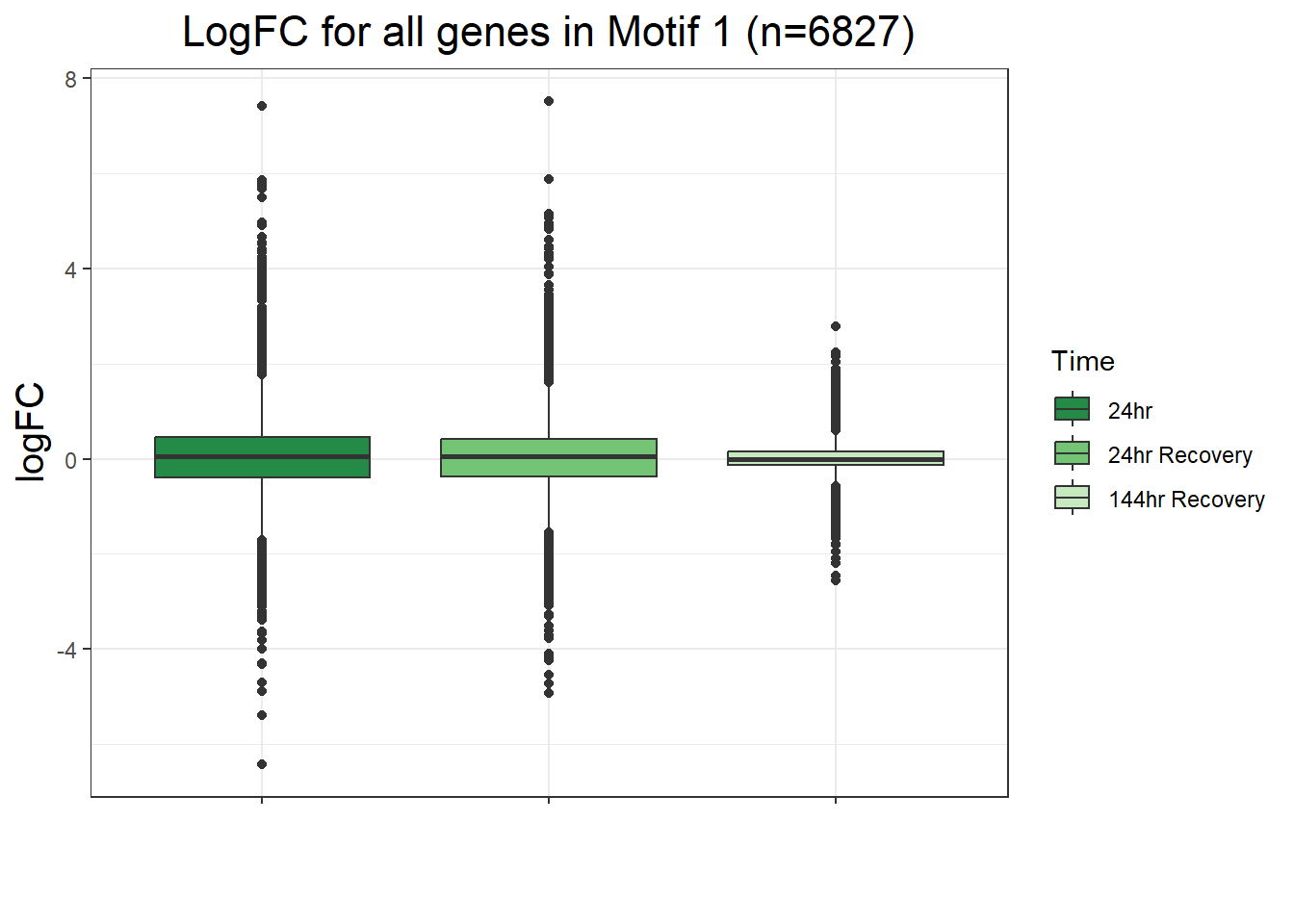
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
filt_toptable_2_dxr_fin 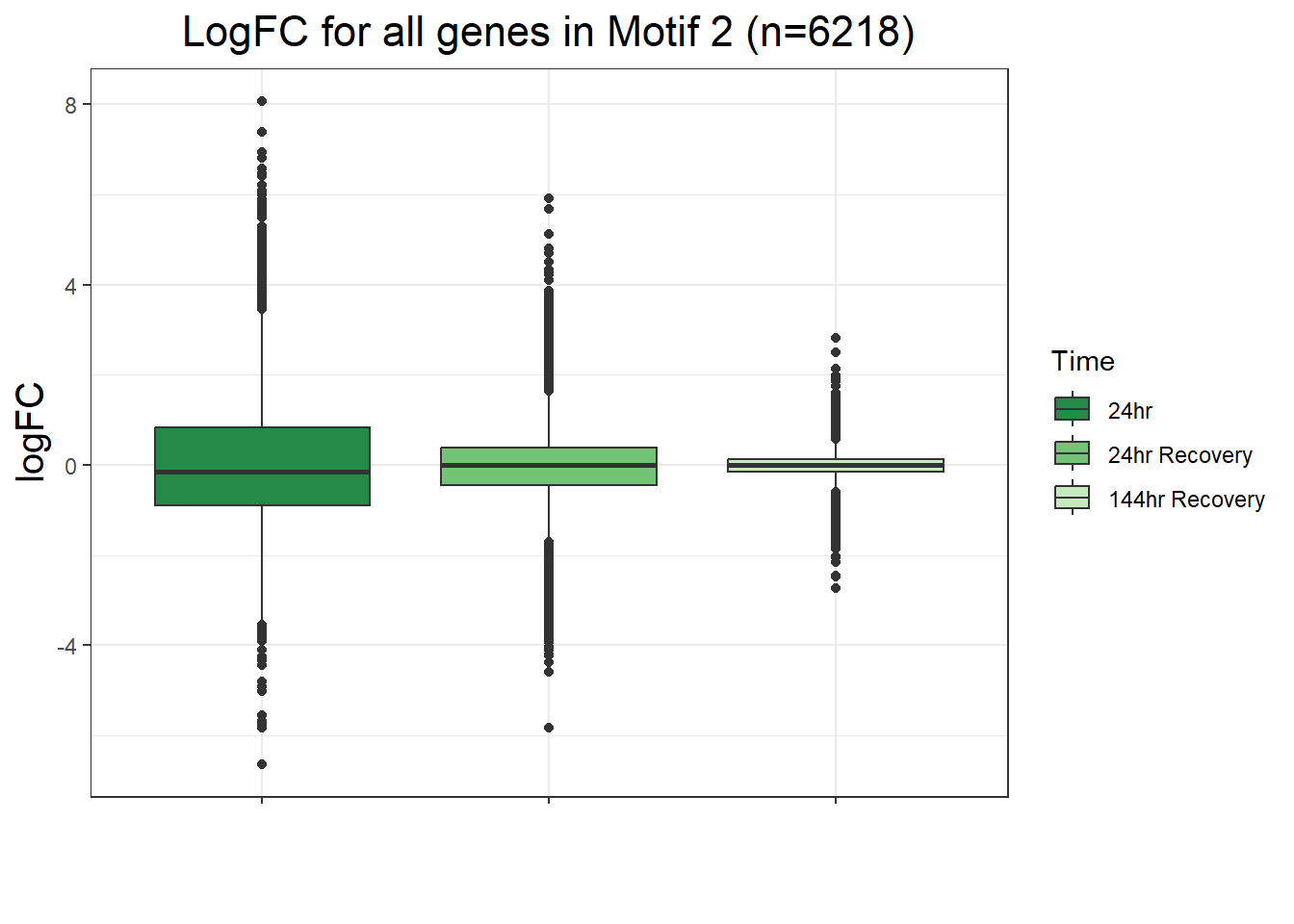
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
filt_toptable_3_dxr_fin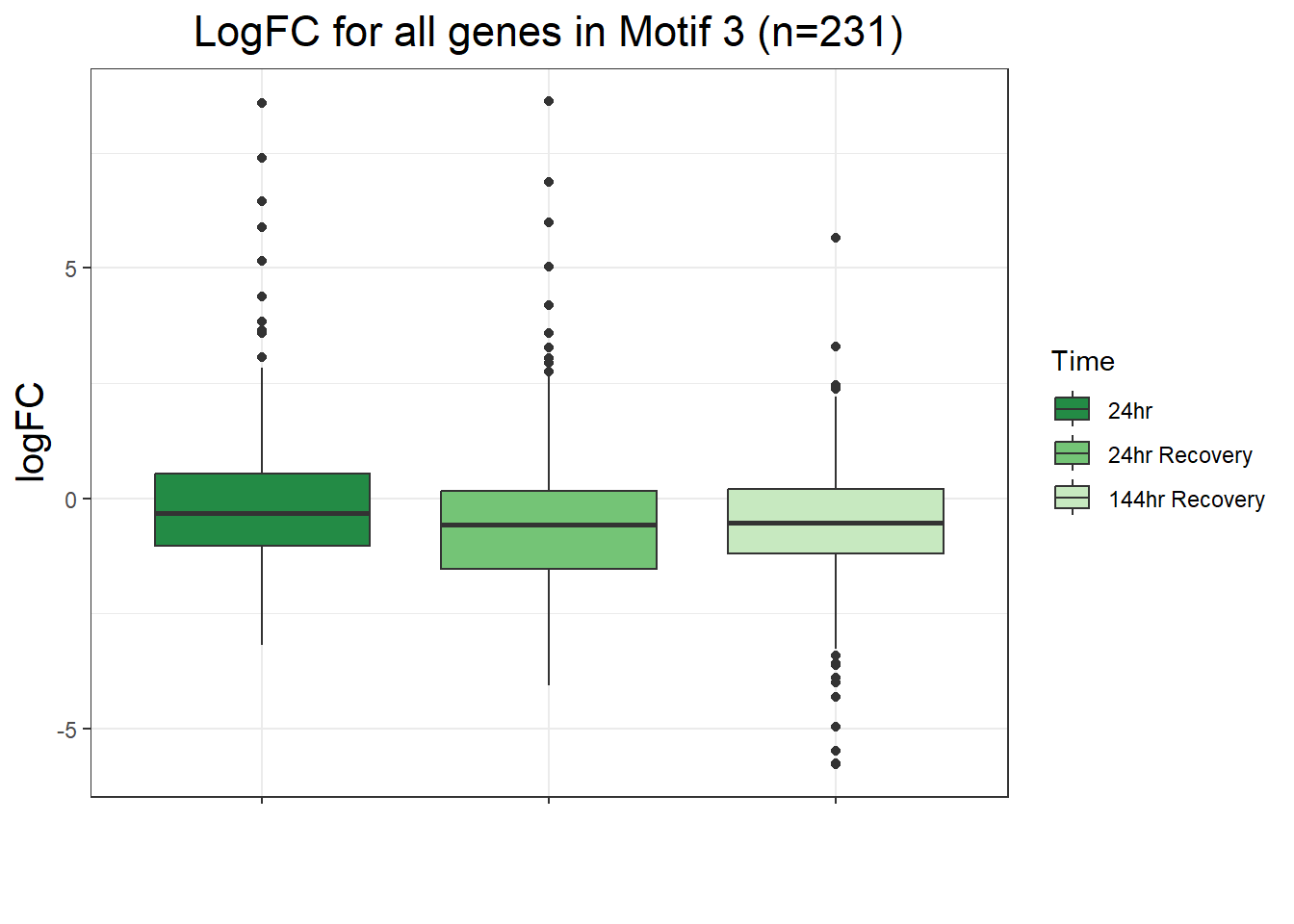
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#now plot the abs logFC for each of these too
filt_toptable_abs_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% final_genes_1) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = absFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("|logFC|")+
theme_bw()+
ggtitle("Abs LogFC for all genes in Motif 1 (n=5638)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 2
filt_toptable_2_abs_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% final_genes_2) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = absFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("|logFC|")+
theme_bw()+
ggtitle("Abs LogFC for all genes in Motif 2 (n=6218)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 2
filt_toptable_3_abs_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% final_genes_3) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = absFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("|logFC|")+
theme_bw()+
ggtitle("Abs LogFC for all genes in Motif 3 (n=231)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#plots
filt_toptable_abs_fin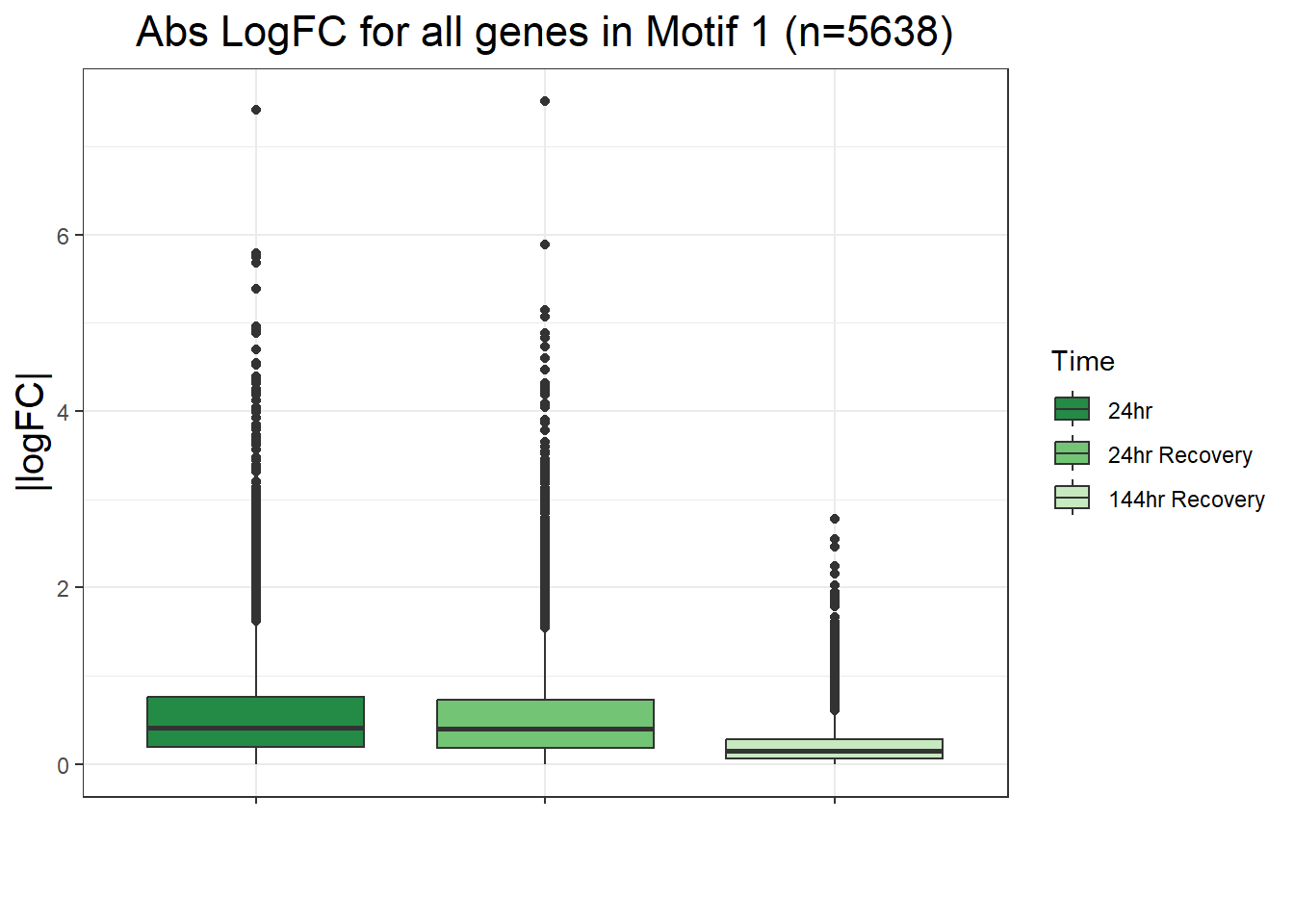
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
filt_toptable_2_abs_fin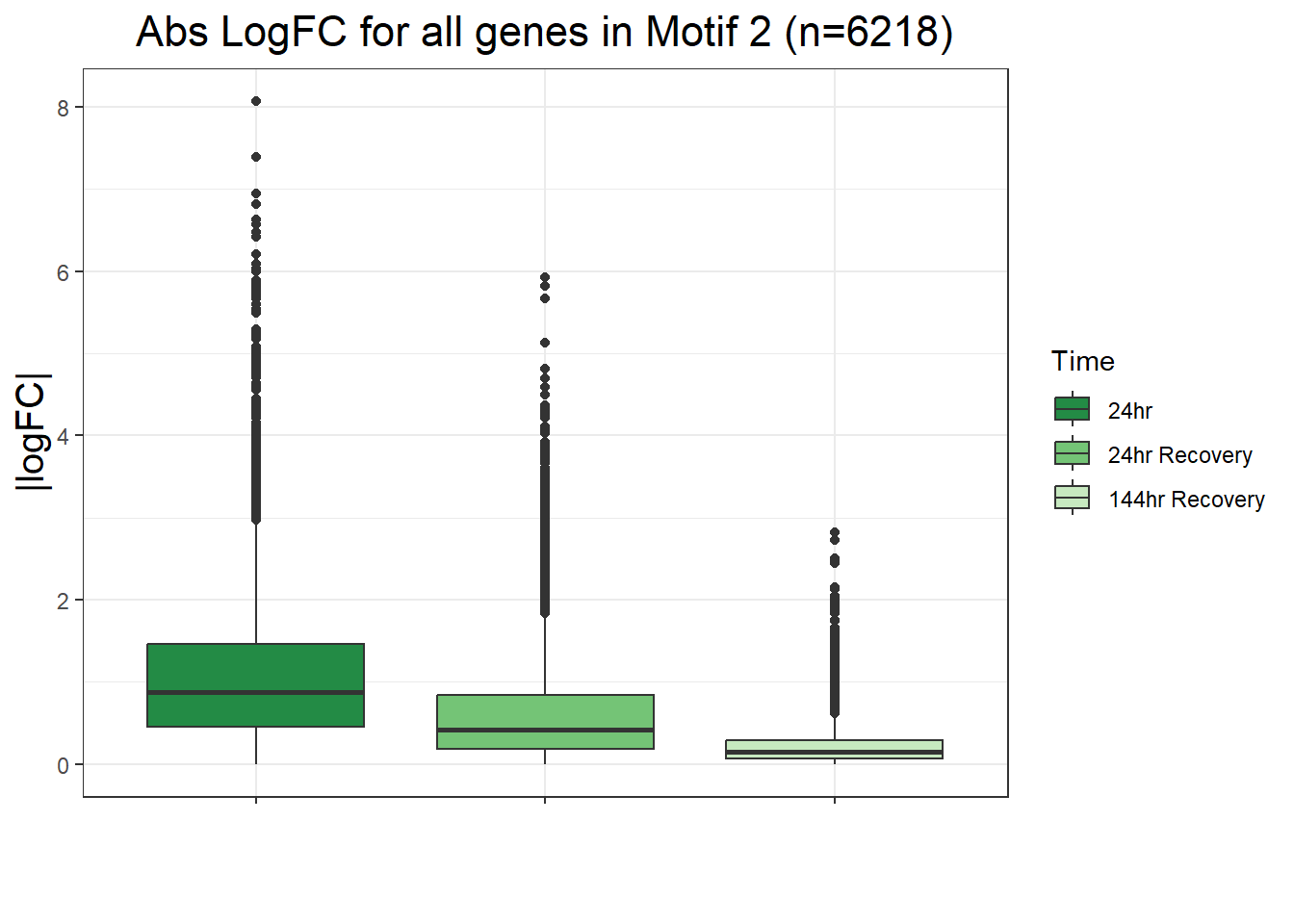
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
filt_toptable_3_abs_fin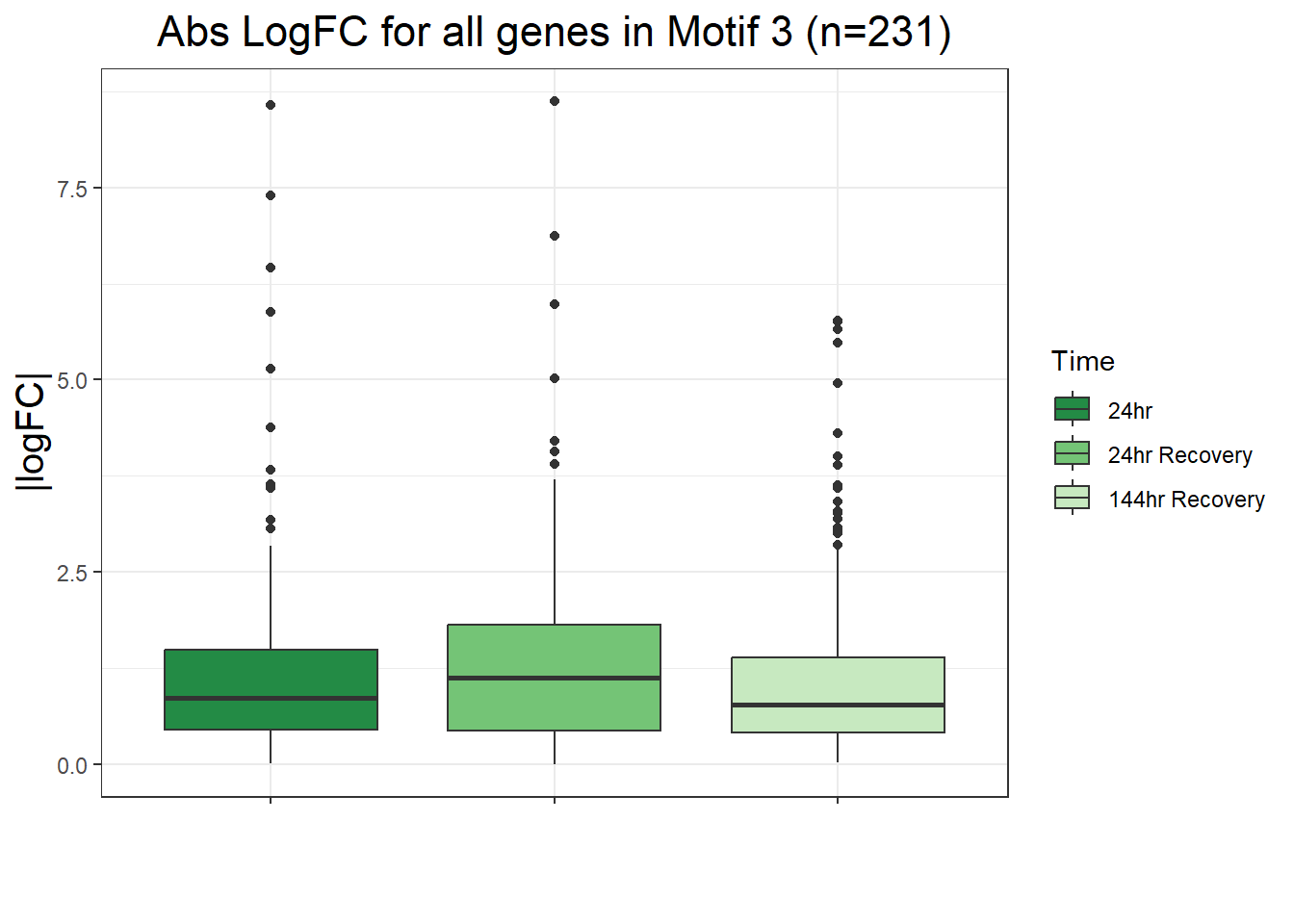
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
###Plot logFC of All Motifs
##motif 1
motif1_genes_fin <- final_genes_1
length(motif1_genes_fin)[1] 5638##motif 2
motif2_genes_fin <- final_genes_2
length(motif2_genes_fin)[1] 6218##motif 3
motif3_genes_fin <- final_genes_3
length(motif3_genes_fin)[1] 231#Combine the toptables I have from pairwise analysis into a single dataframe
d24_toptable_dxr <- Toptable_V.D24T %>%
rownames_to_column(var = "entrezgene_ID") %>%
mutate(Time = "24")
d24r_toptable_dxr <- Toptable_V.D24R %>%
rownames_to_column(var = "entrezgene_ID") %>%
mutate(Time = "24R")
d144r_toptable_dxr <- Toptable_V.D144R %>%
rownames_to_column(var = "entrezgene_ID") %>%
mutate(Time = "144R")
combined_toptables_dxr <- bind_rows(
d24_toptable_dxr,
d24r_toptable_dxr,
d144r_toptable_dxr)
#Filter the data based on each motif
filt_toptable_dxr_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% final_genes_1) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC for all genes in Motif 1 (n=5638)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 2
filt_toptable_2_dxr_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% final_genes_2) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC for all genes in Motif 2 (n=6218)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 3
filt_toptable_3_dxr_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% final_genes_3) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC for all genes in Motif 3 (n=231)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#plots
filt_toptable_dxr_fin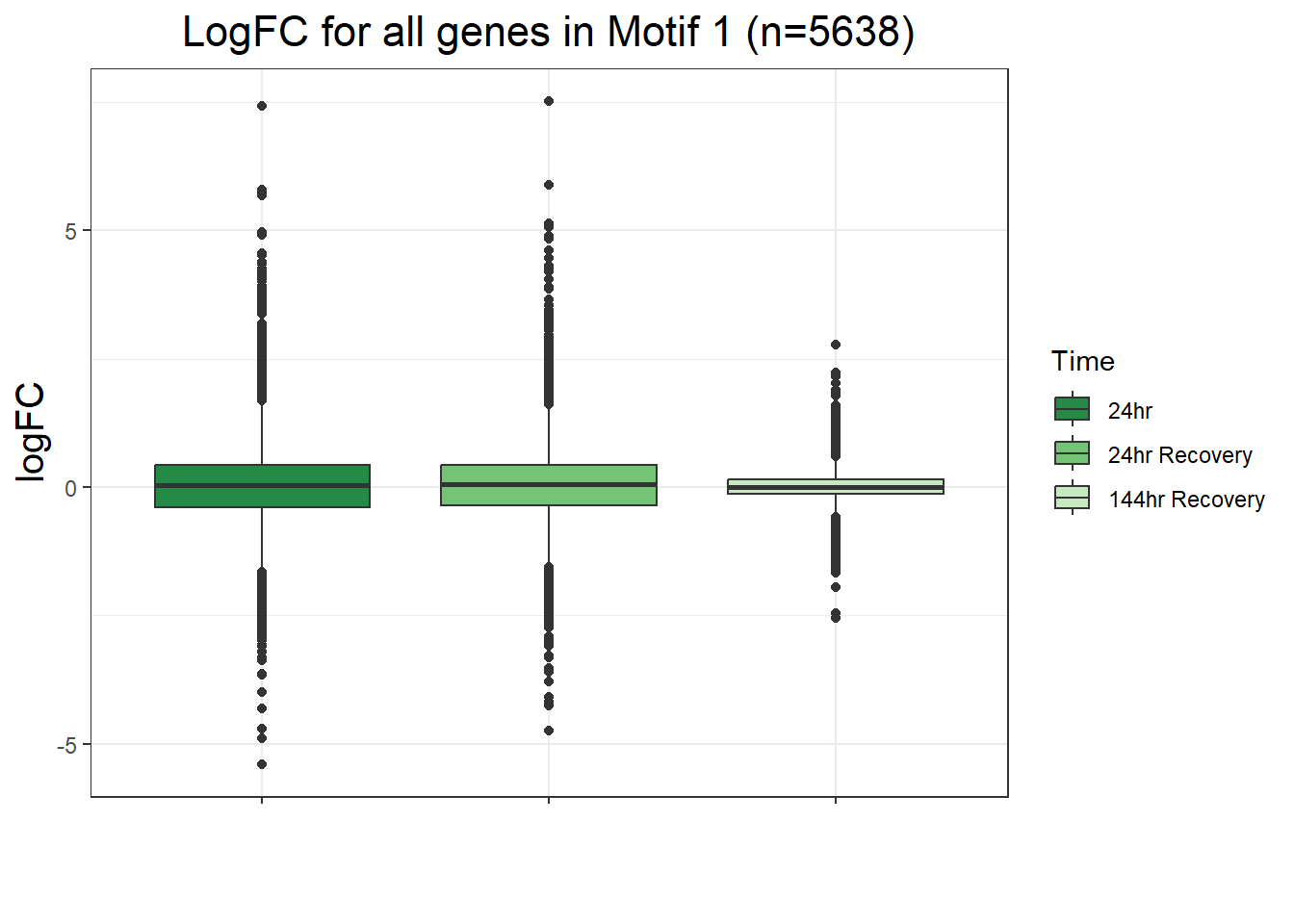
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
filt_toptable_2_dxr_fin 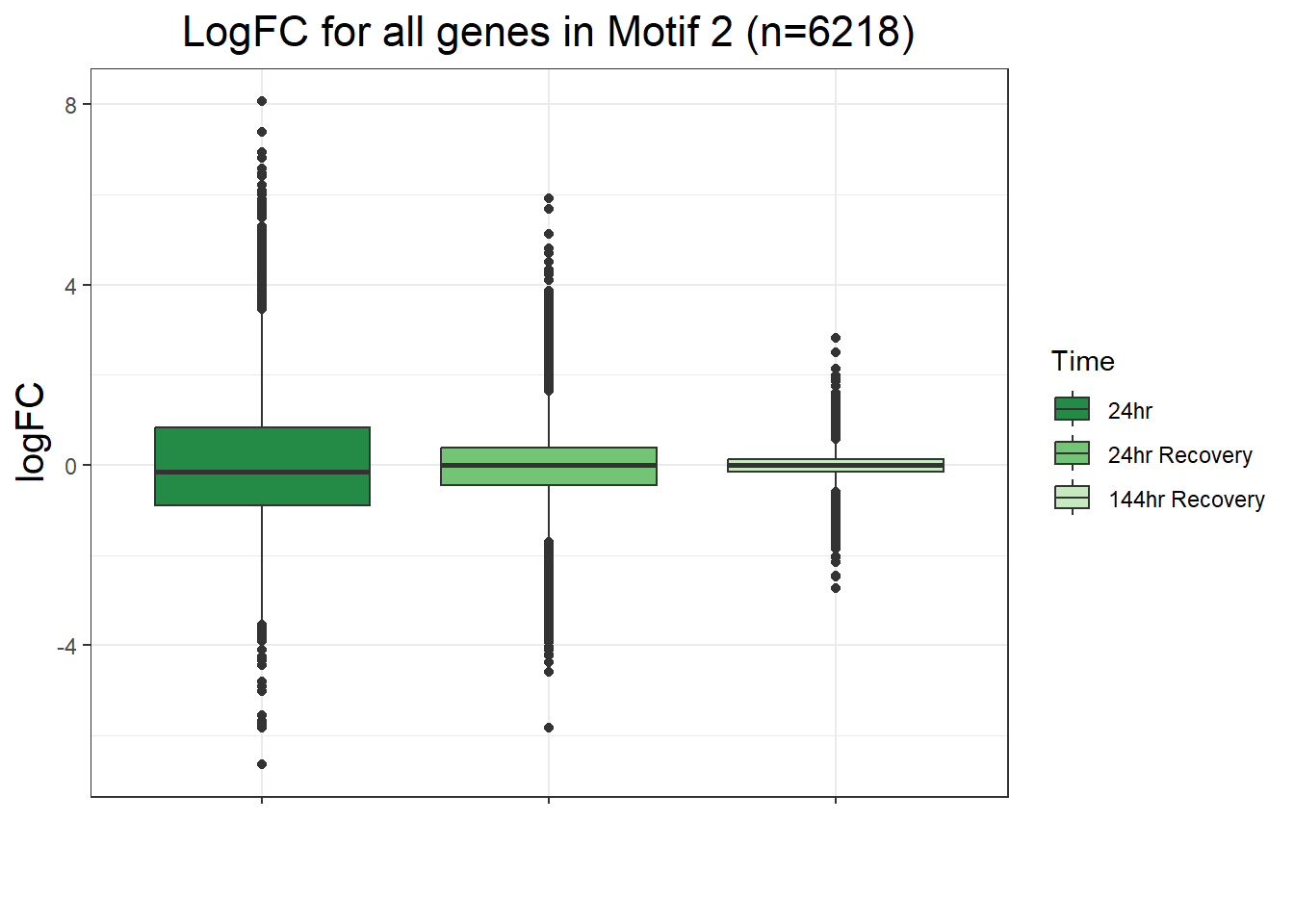
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
filt_toptable_3_dxr_fin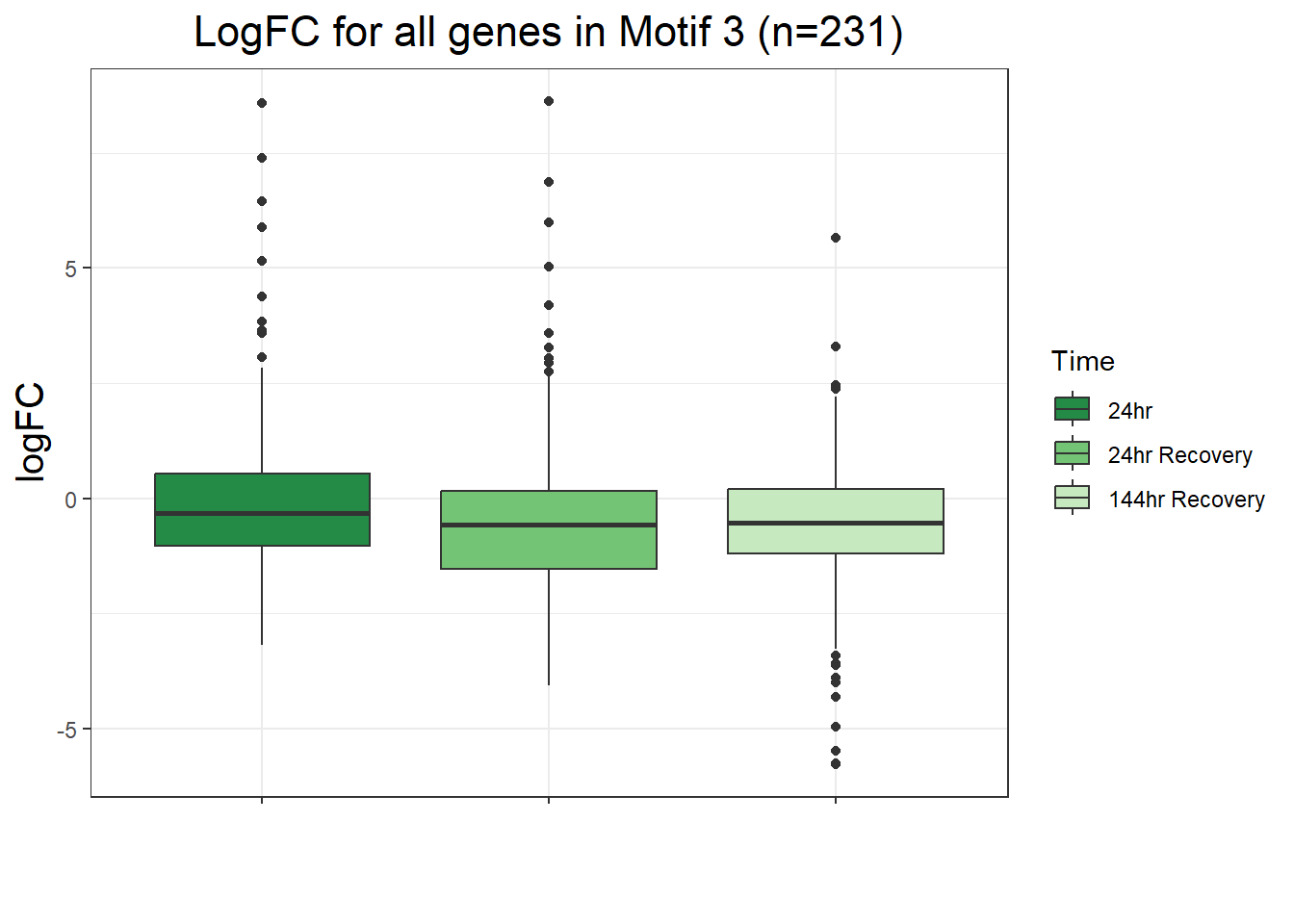
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#now plot the abs logFC for each of these too
filt_toptable_abs_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% final_genes_1) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = absFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("|logFC|")+
theme_bw()+
ggtitle("Abs LogFC for all genes in Motif 1 (n=5638)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 2
filt_toptable_2_abs_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% final_genes_2) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = absFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("|logFC|")+
theme_bw()+
ggtitle("Abs LogFC for all genes in Motif 2 (n=6218)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 2
filt_toptable_3_abs_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% final_genes_3) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = absFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("|logFC|")+
theme_bw()+
ggtitle("Abs LogFC for all genes in Motif 3 (n=231)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#plots
filt_toptable_abs_fin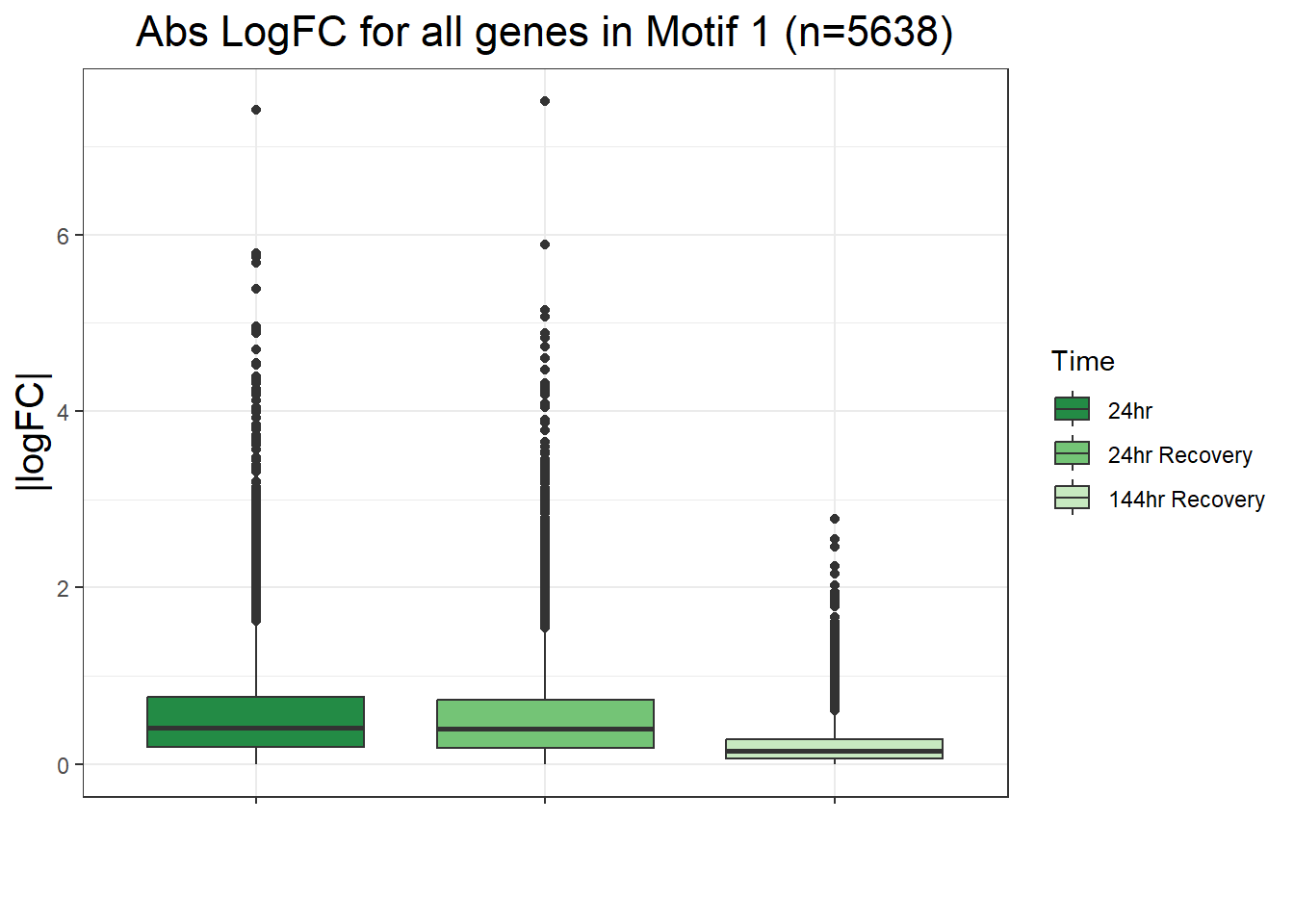
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
filt_toptable_2_abs_fin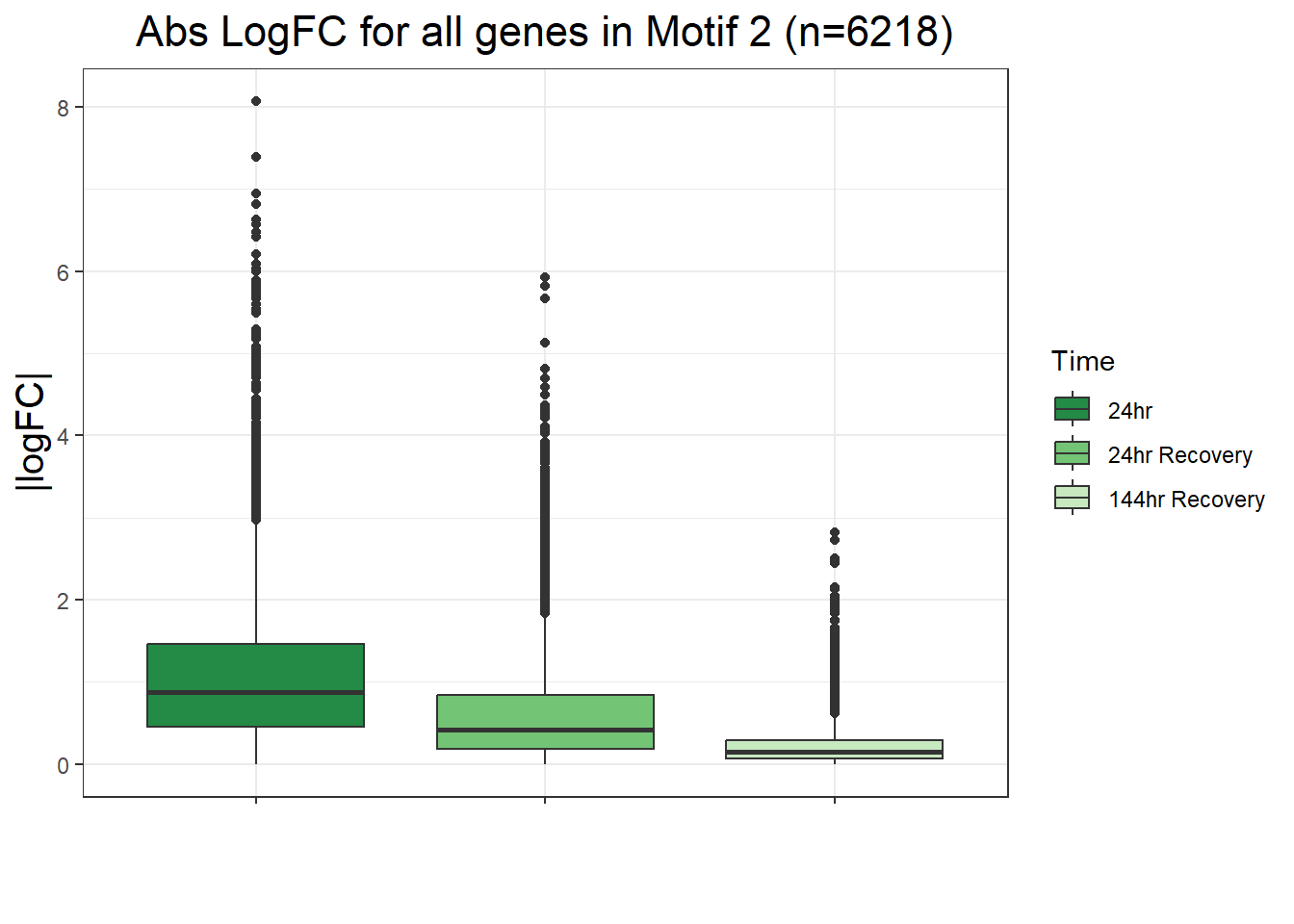
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
filt_toptable_3_abs_fin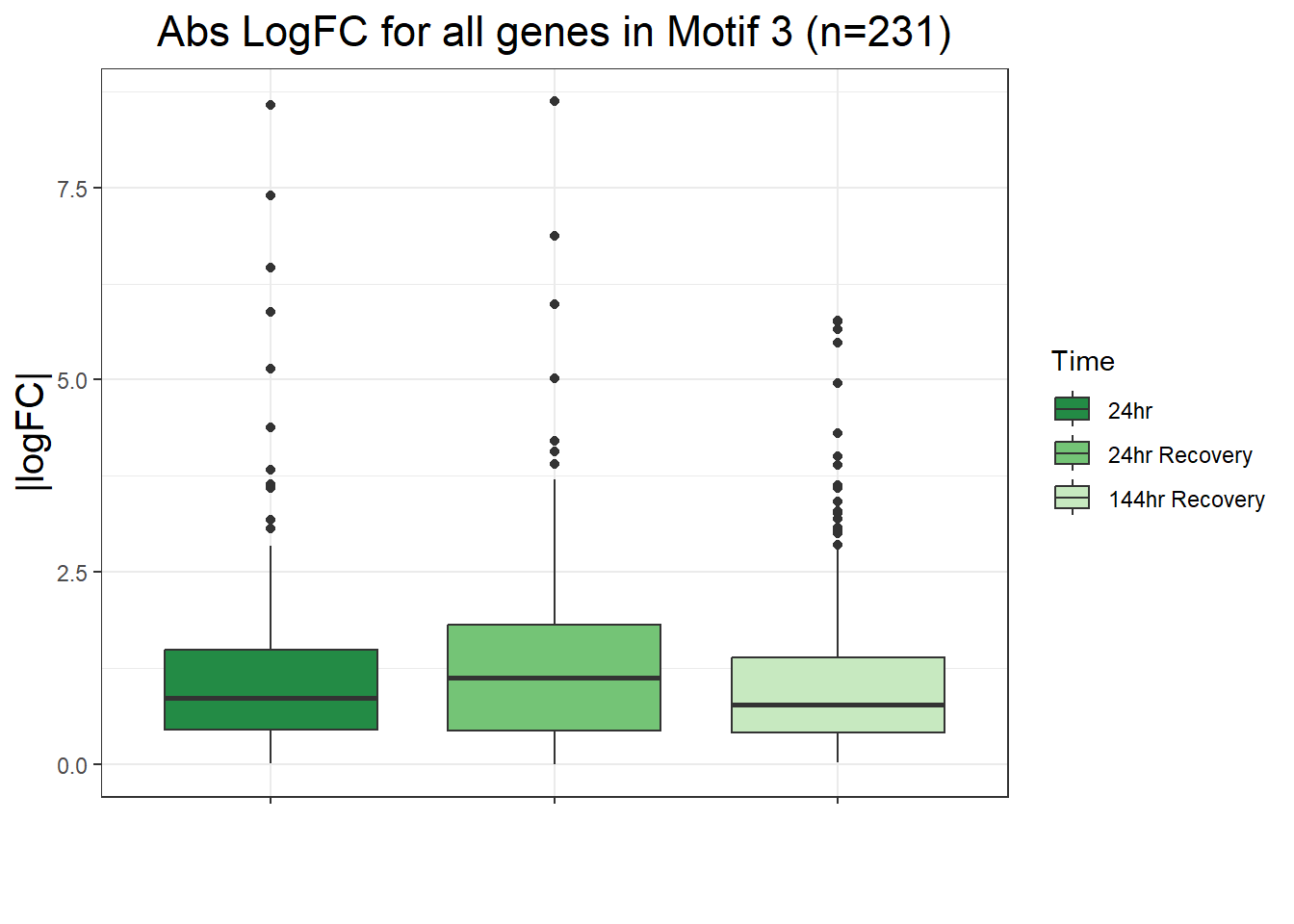
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
###Put together log2cpm Genes for Each Motif
#use the final genes from cross-verifying with clustlike + p.post
#Load in my count matrices
# final_genes_1
# final_genes_2
# final_genes_3
#these are all the genes that are present in each category, make them into dataframes by filtering out the rest of the genes in my main boxplot df
boxplots_cormotif <- boxplot1
motif1_genes_fin <- boxplots_cormotif[boxplots_cormotif$Entrez_ID %in% final_genes_1,]
dim(motif1_genes_fin)[1] 5638 44#5638 genes in 44 cols
motif2_genes_fin <- boxplots_cormotif[boxplots_cormotif$Entrez_ID %in% final_genes_2,]
dim(motif2_genes_fin)[1] 6218 44#6218 genes in 44 cols
motif3_genes_fin <- boxplots_cormotif[boxplots_cormotif$Entrez_ID %in% final_genes_3,]
dim(motif3_genes_fin)[1] 231 44#231 genes in 44 cols
#pull out some random genes for each using sample
m1_genes_fin <- motif1_genes_fin[sample(nrow(motif1_genes_fin), 3), , drop = FALSE]
#Define gene list(s)
#these are the genes pulled out by m1_genes
initial_test_genes1_fin <- c(m1_genes_fin$SYMBOL)
#Add more gene symbols as needed or add more categories
#Now put in the function I want to use to generate boxplots of genes
process_gene_data_1_fin <- function(gene) {
gene_data <- motif1_genes_fin %>% filter(SYMBOL == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2CPM") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6REP$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#Generate Boxplots from the above function using our gene list above
for (gene in initial_test_genes1_fin) {
gene_data <- process_gene_data_1_fin(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2CPM, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2CPM Expression of", gene, "Motif 1")) +
labs(x = "Treatment", y = "log2CPM") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}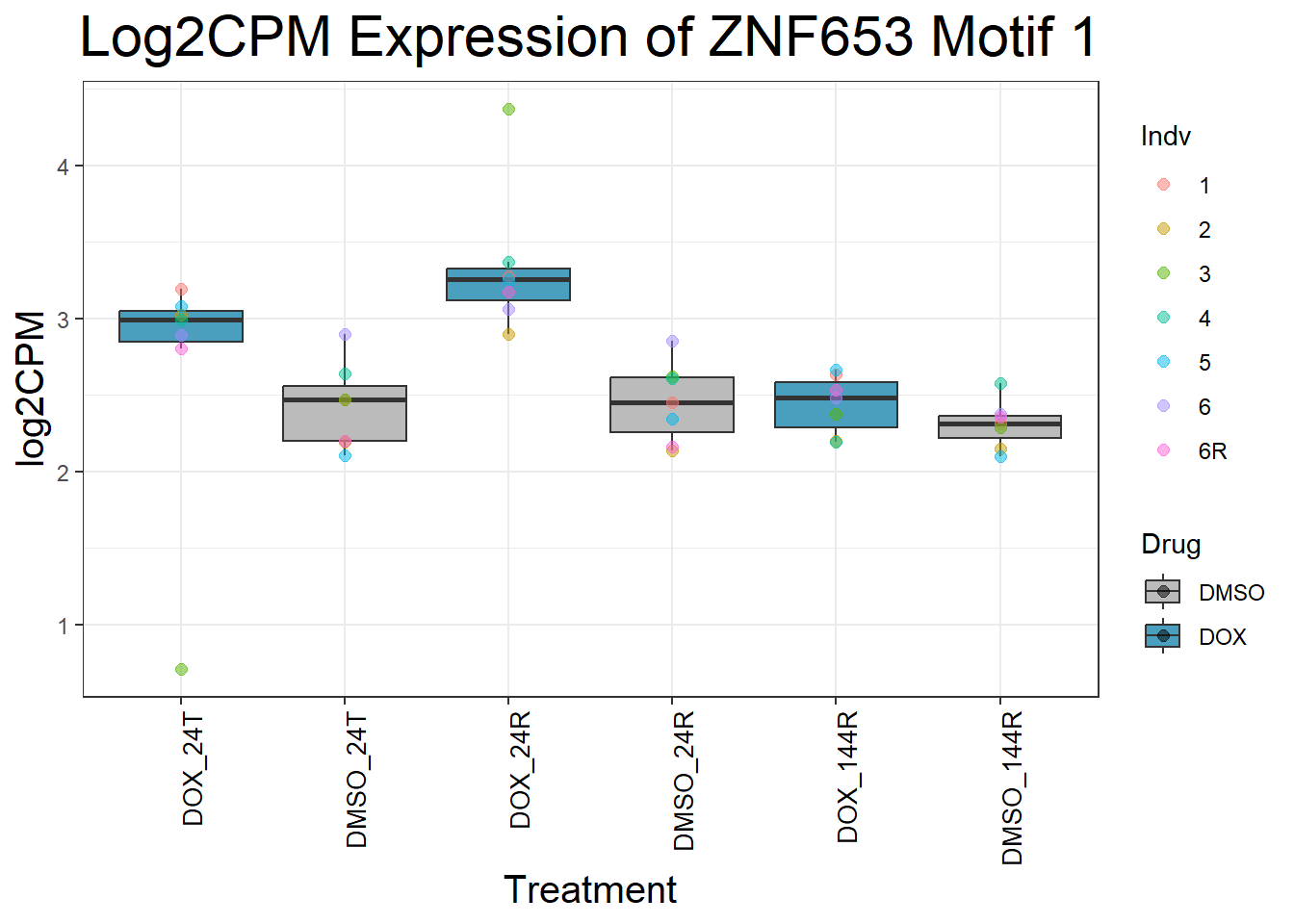
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
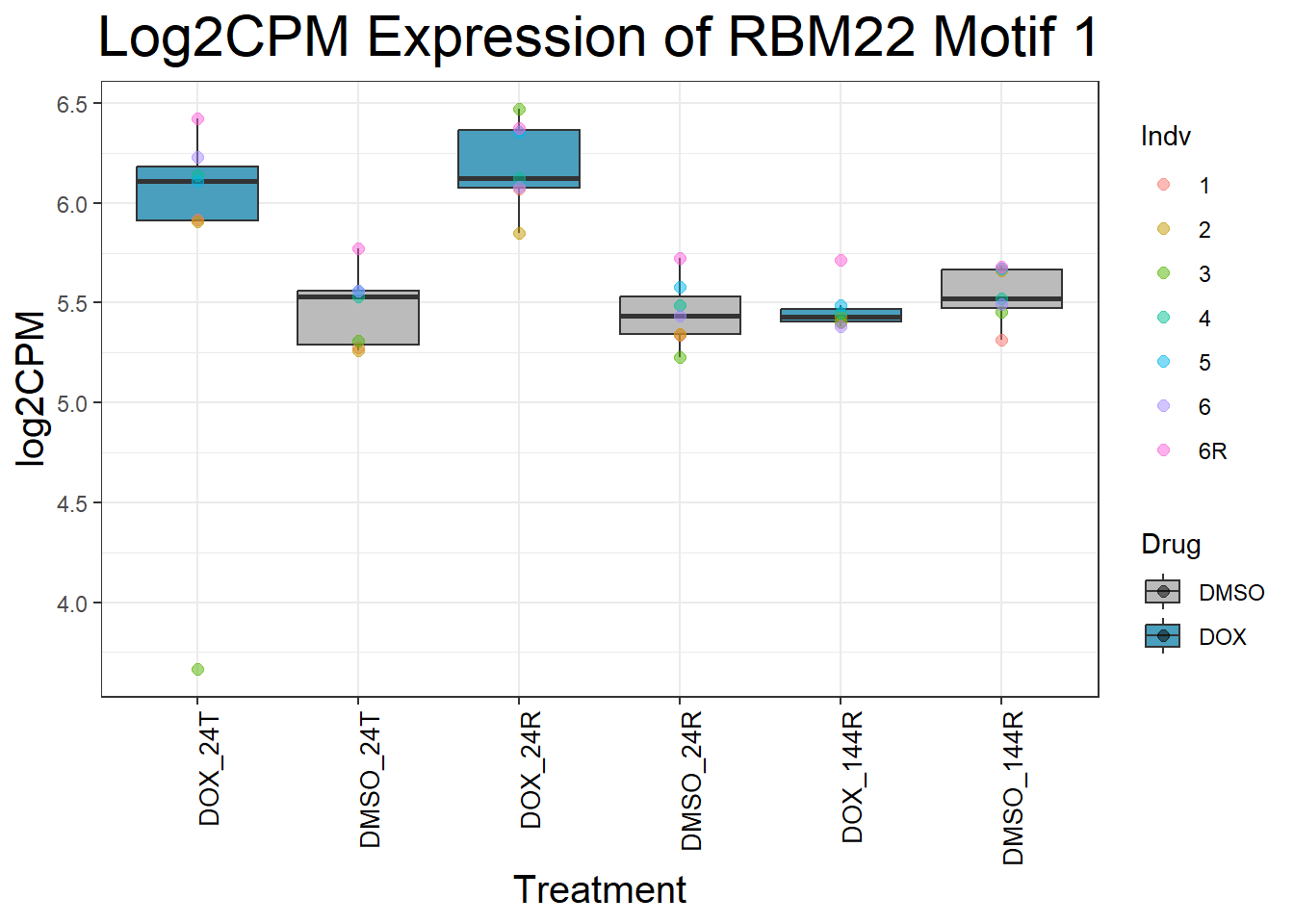
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
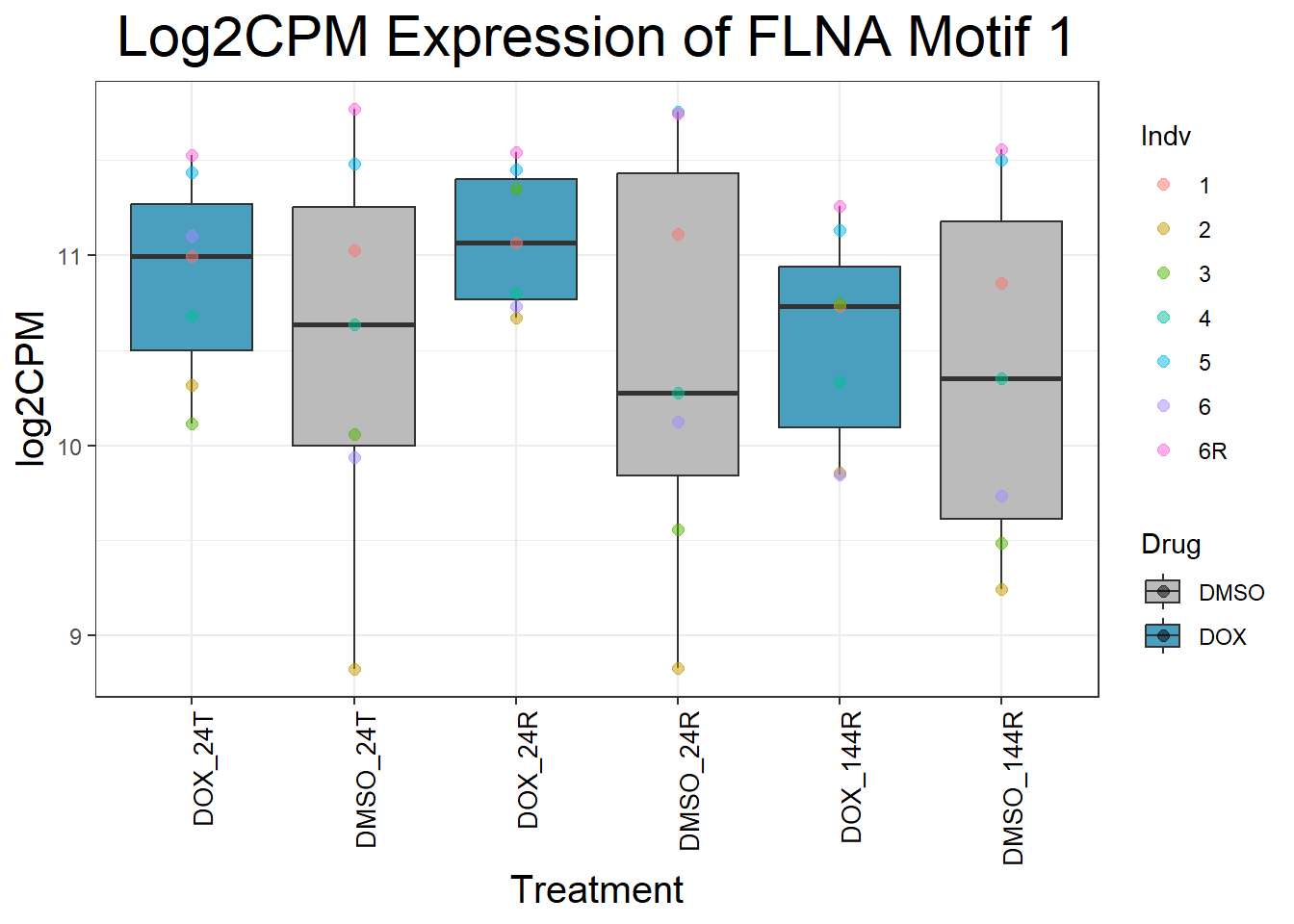
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####motif 2#####
m2_genes_fin <- motif2_genes_fin[sample(nrow(motif2_genes_fin), 3), , drop = FALSE]
#Define gene list(s)
#these are the genes pulled out by m1_genes
initial_test_genes2_fin <- c(m2_genes_fin$SYMBOL)
#Add more gene symbols as needed or add more categories
#Now put in the function I want to use to generate boxplots of genes
process_gene_datam2_fin <- function(gene) {
gene_data <- motif2_genes_fin %>% filter(SYMBOL == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2CPM") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6REP$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#this function is saved under process_gene_data so I will save as an R object
#saveRDS(process_gene_data, "data/new/process_gene_data_funct.RDS")
#Generate Boxplots from the above function using our gene list above
for (gene in initial_test_genes2_fin) {
gene_data <- process_gene_datam2_fin(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2CPM, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2CPM Expression of", gene, "Motif 2")) +
labs(x = "Treatment", y = "log2CPM") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}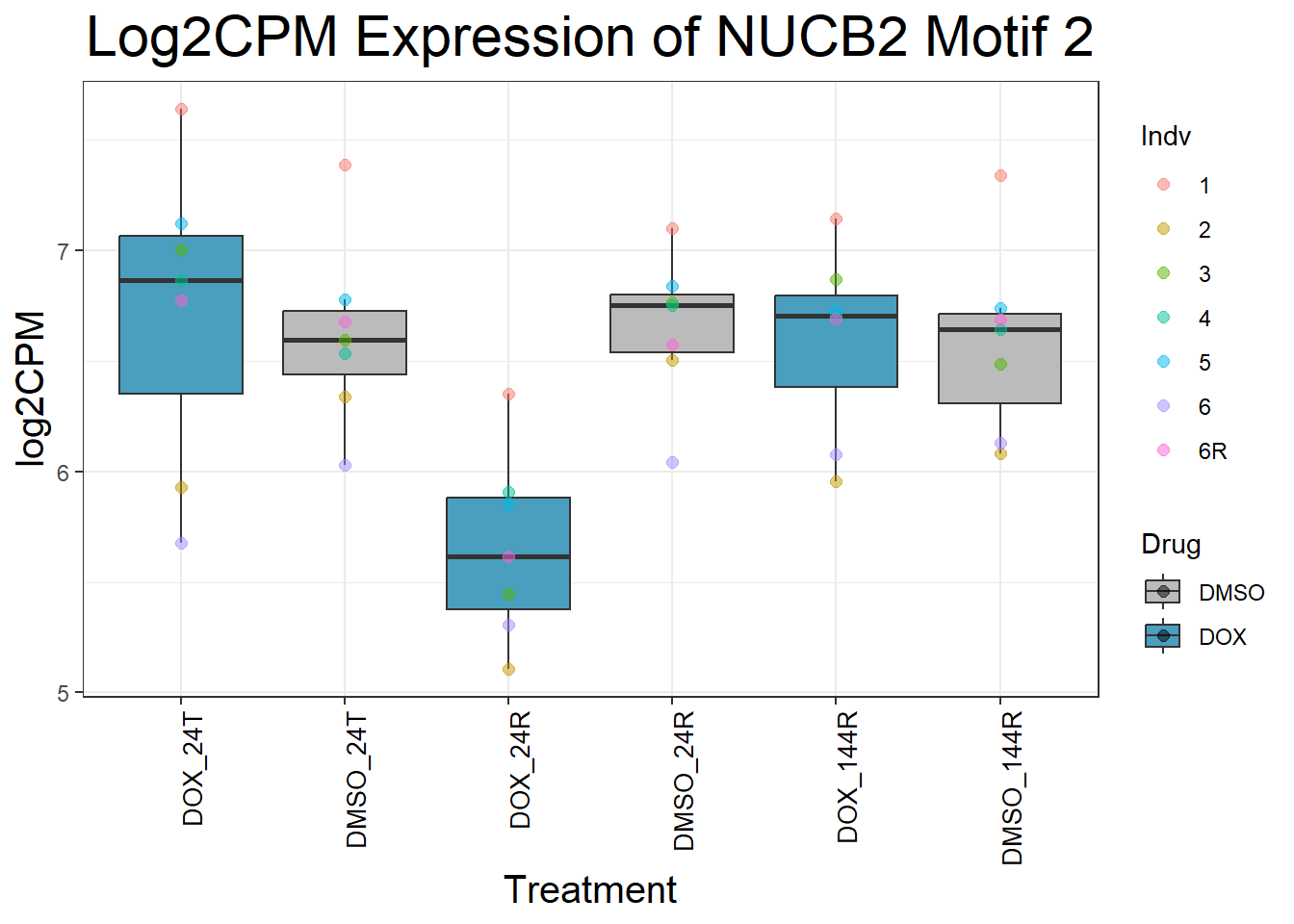
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
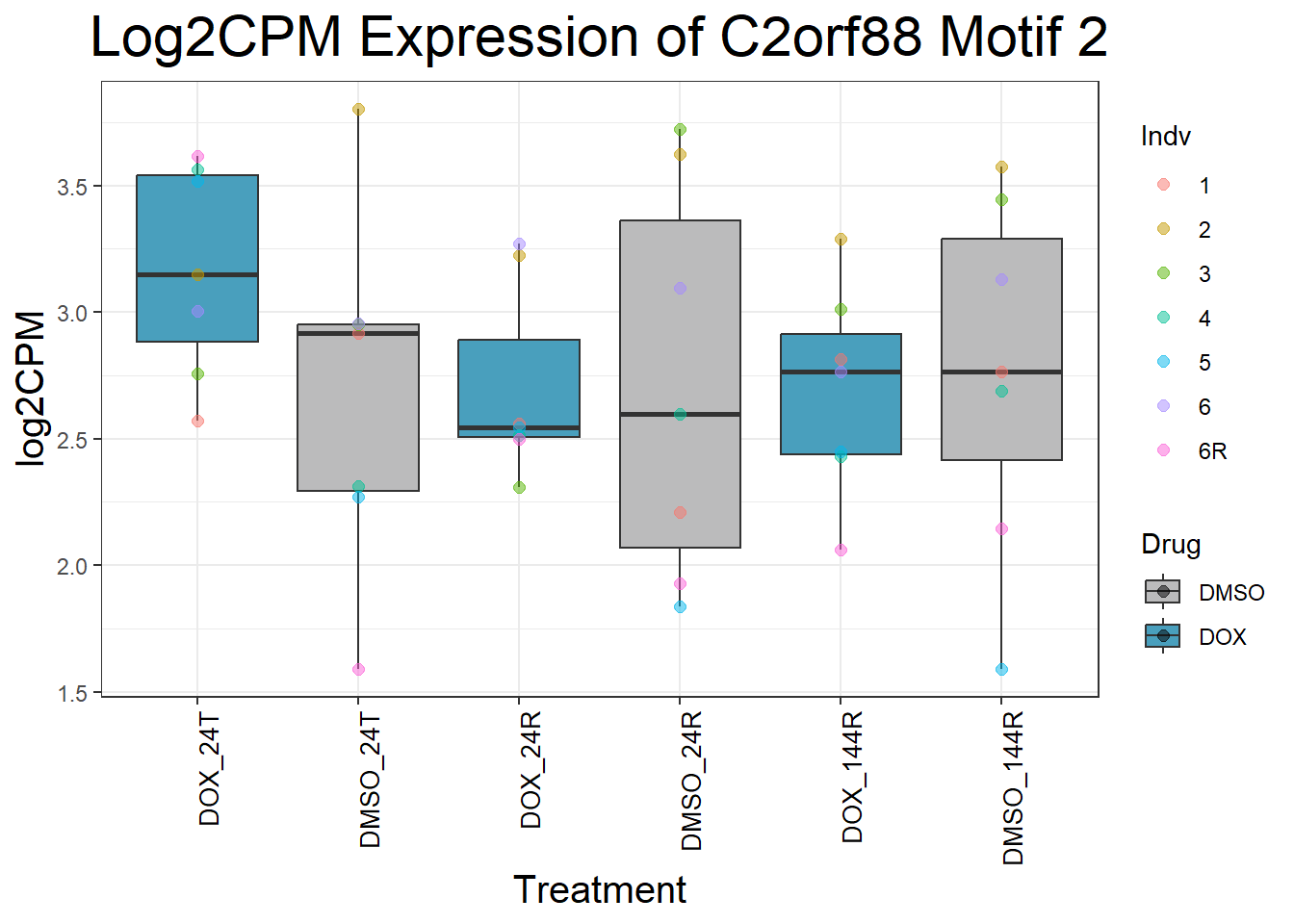
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
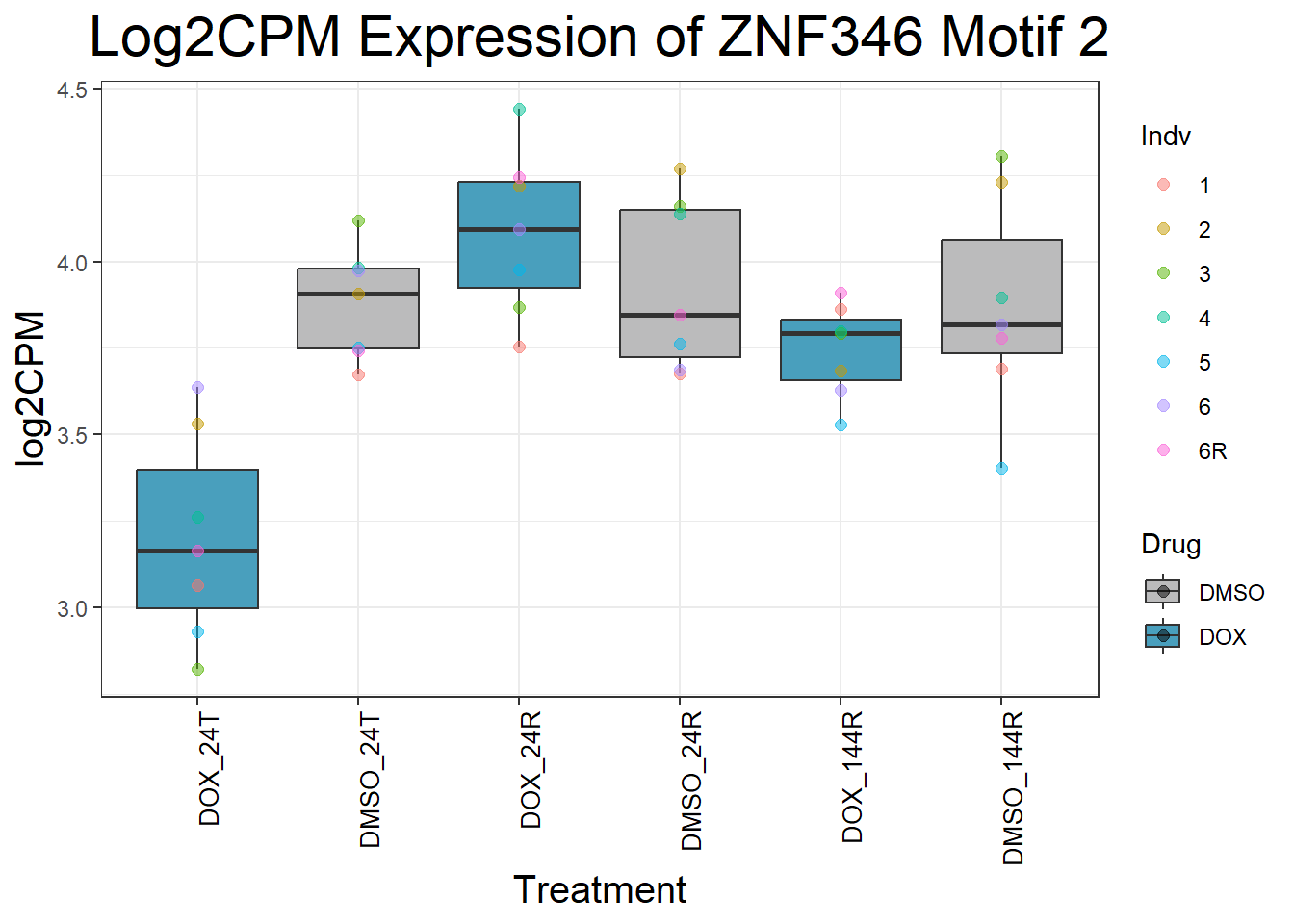
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####motif 3#####
m3_genes_fin <- motif3_genes_fin[sample(nrow(motif3_genes_fin), 3), , drop = FALSE]
#Define gene list(s)
#these are the genes pulled out by m3_genes
initial_test_genes3_fin <- c(m3_genes_fin$SYMBOL)
#Add more gene symbols as needed or add more categories
#Now put in the function I want to use to generate boxplots of genes
process_gene_datam3_fin <- function(gene) {
gene_data <- motif3_genes_fin %>% filter(SYMBOL == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2CPM") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6REP$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#Generate Boxplots from the above function using our gene list above
for (gene in initial_test_genes3_fin) {
gene_data <- process_gene_datam3_fin(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2CPM, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2CPM Expression of", gene, "Motif 3")) +
labs(x = "Treatment", y = "log2CPM") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}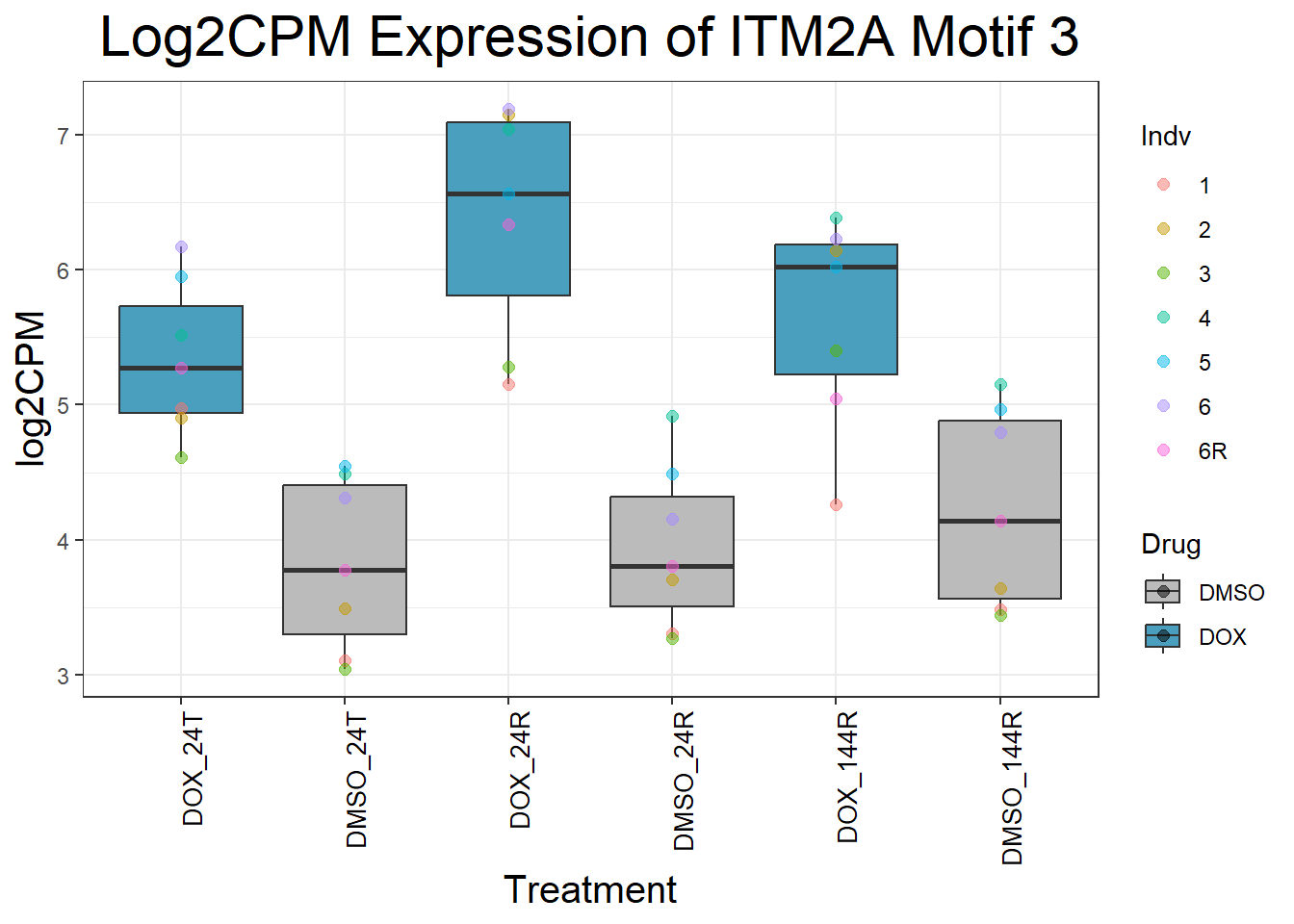
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
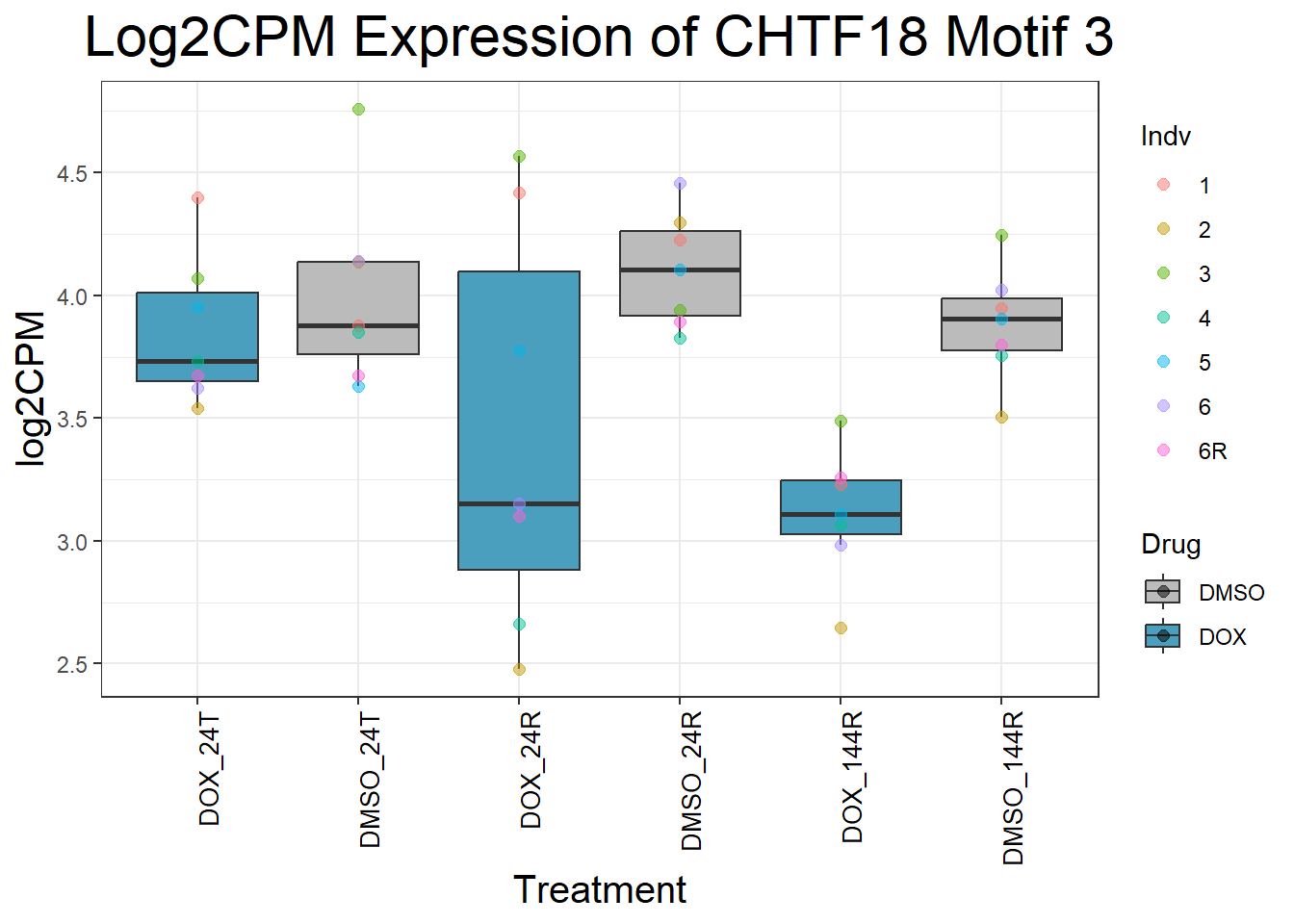
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
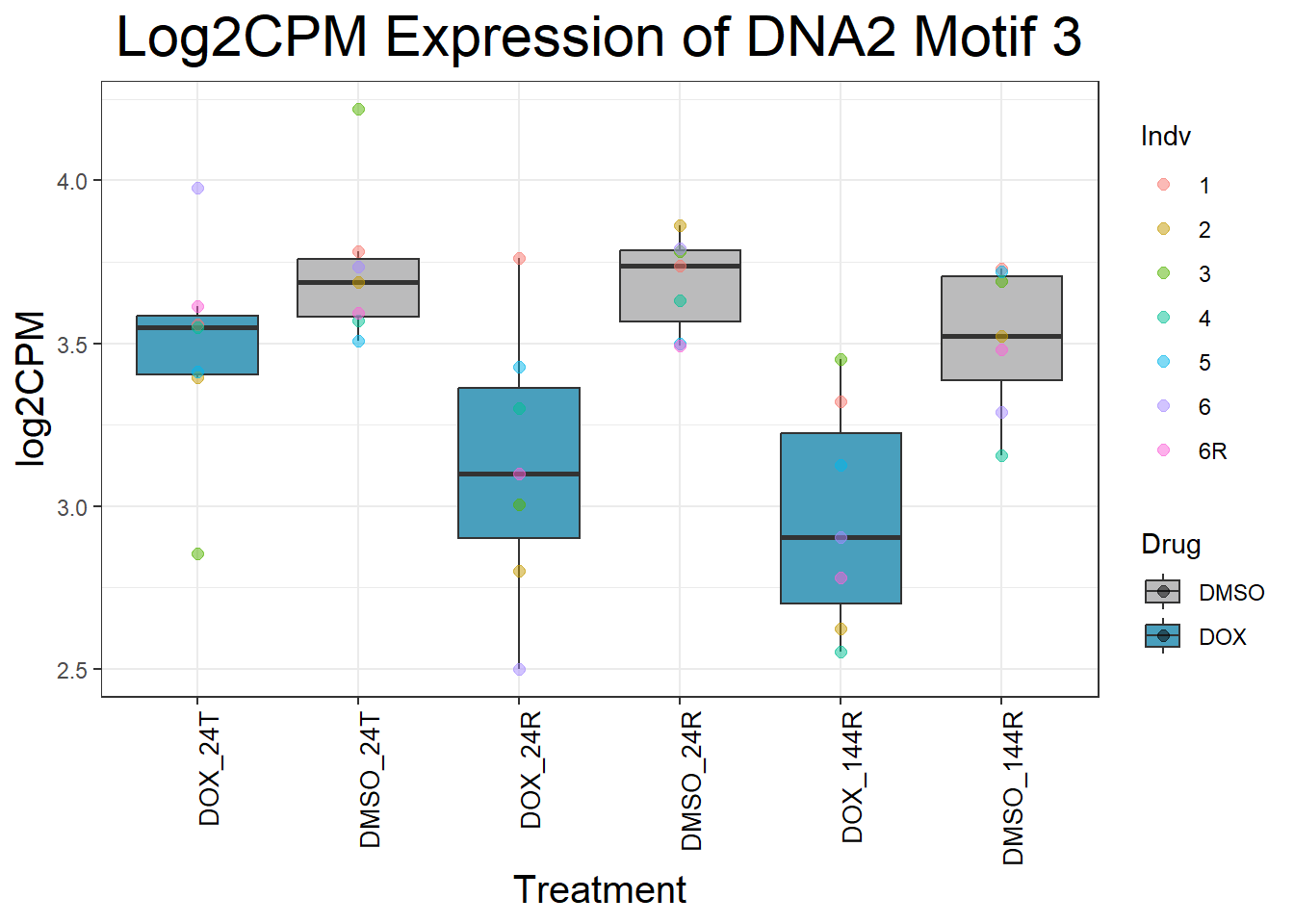
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
###GO/KEGG Final Genes Cormotif
#now take these final gene sets for each motif - clustlike + p.post consensus genes
# final_genes_1
# final_genes_2
# final_genes_3
library(gprofiler2)
#####Motif 1 Gene Set#####
# motif1_genes_matrix <- as.matrix(final_genes_1)
# colnames(motif1_genes_matrix) <- c("entrezgene_ID")
# saveRDS(motif1_genes_matrix, "data/new/motif1_genes_matrix.RDS")
motif1_genes_matrix <- readRDS("data/new/motif1_genes_matrix.RDS")
length(motif1_genes_matrix)[1] 5638#5638 genes in this set for motif 1
motif1_mat_GOKEGG <- gost(query = motif1_genes_matrix,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
motif1_GOKEGG_genes <- gostplot(motif1_mat_GOKEGG, capped = FALSE, interactive = TRUE)
motif1_GOKEGG_genestable_motif1_GOKEGG_genes <- motif1_mat_GOKEGG$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_motif1_GOKEGG_genes %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0019538 | protein metabolic process | 1530 | 4721 | 5.534e-67 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 1663 | 5390 | 3.258e-57 |
| GO:BP | GO:0046907 | intracellular transport | 559 | 1381 | 2.736e-50 |
| GO:BP | GO:0051179 | localization | 1668 | 5555 | 1.485e-48 |
| GO:BP | GO:0006810 | transport | 1376 | 4407 | 1.007e-47 |
| GO:BP | GO:0051649 | establishment of localization in cell | 732 | 2010 | 1.114e-46 |
| GO:BP | GO:0051641 | cellular localization | 1179 | 3661 | 1.114e-46 |
| GO:BP | GO:0043412 | macromolecule modification | 1004 | 3030 | 2.043e-44 |
| GO:BP | GO:0009056 | catabolic process | 896 | 2639 | 1.174e-43 |
| GO:BP | GO:0006996 | organelle organization | 1146 | 3594 | 8.587e-43 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 547 | 1417 | 3.742e-42 |
| GO:BP | GO:0036211 | protein modification process | 940 | 2846 | 1.631e-40 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1802 | 6264 | 2.887e-40 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1717 | 5920 | 5.625e-40 |
| GO:BP | GO:0071705 | nitrogen compound transport | 682 | 1923 | 7.303e-39 |
| GO:BP | GO:0033036 | macromolecule localization | 1034 | 3228 | 8.231e-39 |
| GO:BP | GO:0008104 | protein localization | 907 | 2770 | 2.723e-37 |
| GO:BP | GO:0051234 | establishment of localization | 1460 | 4928 | 2.723e-37 |
| GO:BP | GO:0065007 | biological regulation | 3264 | 12743 | 5.102e-37 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 909 | 2782 | 5.102e-37 |
| GO:BP | GO:0015031 | protein transport | 539 | 1444 | 7.520e-37 |
| GO:BP | GO:0030163 | protein catabolic process | 411 | 1030 | 7.217e-35 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 1208 | 3990 | 9.411e-34 |
| GO:BP | GO:0050789 | regulation of biological process | 3152 | 12336 | 1.383e-32 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 632 | 1855 | 6.070e-30 |
| GO:BP | GO:0050794 | regulation of cellular process | 3051 | 11946 | 7.799e-30 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 1112 | 3687 | 1.116e-29 |
| GO:BP | GO:0023051 | regulation of signaling | 1057 | 3478 | 2.621e-29 |
| GO:BP | GO:0019222 | regulation of metabolic process | 1925 | 7035 | 7.415e-29 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 320 | 786 | 1.349e-28 |
| GO:BP | GO:0010646 | regulation of cell communication | 1055 | 3486 | 1.735e-28 |
| GO:BP | GO:0009966 | regulation of signal transduction | 938 | 3034 | 2.826e-28 |
| GO:BP | GO:0035556 | intracellular signal transduction | 919 | 2965 | 4.949e-28 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1180 | 3993 | 7.051e-28 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1075 | 3597 | 7.801e-27 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 332 | 853 | 1.464e-25 |
| GO:BP | GO:0033554 | cellular response to stress | 608 | 1830 | 1.673e-25 |
| GO:BP | GO:0006886 | intracellular protein transport | 281 | 686 | 1.736e-25 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 989 | 3288 | 2.466e-25 |
| GO:BP | GO:0006351 | DNA-templated transcription | 1055 | 3549 | 2.639e-25 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 542 | 1592 | 2.707e-25 |
| GO:BP | GO:0043687 | post-translational protein modification | 382 | 1027 | 3.545e-25 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 366 | 975 | 6.494e-25 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 1021 | 3428 | 1.141e-24 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 1015 | 3409 | 1.926e-24 |
| GO:BP | GO:0009894 | regulation of catabolic process | 383 | 1040 | 2.572e-24 |
| GO:BP | GO:0045184 | establishment of protein localization | 629 | 1937 | 1.233e-23 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 753 | 2407 | 1.472e-23 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 753 | 2410 | 2.126e-23 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 759 | 2433 | 2.126e-23 |
| GO:BP | GO:0016567 | protein ubiquitination | 300 | 768 | 2.274e-23 |
| GO:BP | GO:0007275 | multicellular organism development | 1335 | 4727 | 4.486e-23 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 630 | 1953 | 6.777e-23 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 1757 | 6492 | 1.313e-22 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 255 | 628 | 1.754e-22 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 257 | 638 | 4.424e-22 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 258 | 643 | 6.896e-22 |
| GO:BP | GO:0032502 | developmental process | 1762 | 6553 | 3.521e-21 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 225 | 543 | 4.552e-21 |
| GO:BP | GO:0048731 | system development | 1157 | 4053 | 5.120e-21 |
| GO:BP | GO:0048856 | anatomical structure development | 1626 | 5997 | 1.318e-20 |
| GO:BP | GO:0065008 | regulation of biological quality | 876 | 2947 | 2.560e-20 |
| GO:BP | GO:0006508 | proteolysis | 487 | 1486 | 9.924e-19 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 538 | 1680 | 1.784e-18 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1572 | 5834 | 2.263e-18 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 629 | 2036 | 7.568e-18 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 439 | 1326 | 1.258e-17 |
| GO:BP | GO:0007005 | mitochondrion organization | 182 | 435 | 1.998e-17 |
| GO:BP | GO:0007399 | nervous system development | 773 | 2604 | 2.204e-17 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 594 | 1912 | 2.472e-17 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 799 | 2711 | 3.596e-17 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1514 | 5629 | 4.185e-17 |
| GO:BP | GO:0006915 | apoptotic process | 595 | 1923 | 5.906e-17 |
| GO:BP | GO:0008219 | cell death | 612 | 1991 | 8.342e-17 |
| GO:BP | GO:0006412 | translation | 268 | 727 | 8.342e-17 |
| GO:BP | GO:0012501 | programmed cell death | 610 | 1987 | 1.280e-16 |
| GO:BP | GO:0023057 | negative regulation of signaling | 464 | 1435 | 1.333e-16 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 464 | 1436 | 1.522e-16 |
| GO:BP | GO:0006914 | autophagy | 228 | 597 | 3.069e-16 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 228 | 597 | 3.069e-16 |
| GO:BP | GO:0006950 | response to stress | 1097 | 3948 | 1.435e-15 |
| GO:BP | GO:0032879 | regulation of localization | 615 | 2029 | 1.810e-15 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 754 | 2578 | 3.323e-15 |
| GO:BP | GO:0002181 | cytoplasmic translation | 87 | 165 | 3.824e-15 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 783 | 2696 | 4.504e-15 |
| GO:BP | GO:0044238 | primary metabolic process | 3034 | 12342 | 8.204e-15 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 366 | 1105 | 1.400e-14 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 754 | 2593 | 1.443e-14 |
| GO:BP | GO:0031399 | regulation of protein modification process | 349 | 1049 | 3.584e-14 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 244 | 677 | 6.161e-14 |
| GO:BP | GO:0060341 | regulation of cellular localization | 338 | 1013 | 6.722e-14 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 345 | 1039 | 7.145e-14 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 1531 | 5809 | 9.620e-14 |
| GO:BP | GO:0050793 | regulation of developmental process | 714 | 2457 | 1.252e-13 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 555 | 1835 | 1.252e-13 |
| GO:BP | GO:0016236 | macroautophagy | 149 | 362 | 1.600e-13 |
| GO:BP | GO:0016197 | endosomal transport | 126 | 290 | 1.958e-13 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 680 | 2331 | 2.795e-13 |
| GO:BP | GO:0033365 | protein localization to organelle | 388 | 1209 | 3.515e-13 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 164 | 414 | 3.652e-13 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1487 | 5644 | 3.673e-13 |
| GO:BP | GO:0016310 | phosphorylation | 417 | 1320 | 4.750e-13 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 132 | 312 | 4.981e-13 |
| GO:BP | GO:0032880 | regulation of protein localization | 305 | 907 | 5.496e-13 |
| GO:BP | GO:0023056 | positive regulation of signaling | 541 | 1796 | 6.337e-13 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 98 | 209 | 7.318e-13 |
| GO:BP | GO:0065009 | regulation of molecular function | 462 | 1496 | 9.281e-13 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 459 | 1489 | 1.584e-12 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 538 | 1795 | 2.000e-12 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 193 | 519 | 2.041e-12 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 234 | 662 | 2.374e-12 |
| GO:BP | GO:0051716 | cellular response to stimulus | 1886 | 7376 | 2.391e-12 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 481 | 1578 | 2.697e-12 |
| GO:BP | GO:0022008 | neurogenesis | 531 | 1771 | 2.707e-12 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 483 | 1588 | 3.454e-12 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 330 | 1011 | 3.454e-12 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 200 | 546 | 3.617e-12 |
| GO:BP | GO:0042254 | ribosome biogenesis | 133 | 323 | 4.207e-12 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 116 | 270 | 5.421e-12 |
| GO:BP | GO:0048468 | cell development | 808 | 2876 | 5.534e-12 |
| GO:BP | GO:0030030 | cell projection organization | 493 | 1631 | 5.545e-12 |
| GO:BP | GO:0010468 | regulation of gene expression | 1450 | 5536 | 7.571e-12 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 465 | 1529 | 1.058e-11 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 146 | 369 | 1.089e-11 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 219 | 619 | 1.314e-11 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 384 | 1223 | 1.377e-11 |
| GO:BP | GO:0006468 | protein phosphorylation | 385 | 1227 | 1.408e-11 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 430 | 1400 | 1.844e-11 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 344 | 1077 | 2.300e-11 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 507 | 1699 | 2.392e-11 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 2111 | 8393 | 2.472e-11 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 506 | 1697 | 2.887e-11 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 445 | 1462 | 3.049e-11 |
| GO:BP | GO:1901698 | response to nitrogen compound | 344 | 1080 | 3.335e-11 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 456 | 1506 | 3.664e-11 |
| GO:BP | GO:0000209 | protein polyubiquitination | 117 | 280 | 3.726e-11 |
| GO:BP | GO:0006974 | DNA damage response | 297 | 908 | 4.518e-11 |
| GO:BP | GO:0051049 | regulation of transport | 481 | 1607 | 5.892e-11 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 351 | 1113 | 7.941e-11 |
| GO:BP | GO:0044281 | small molecule metabolic process | 523 | 1776 | 1.039e-10 |
| GO:BP | GO:0030154 | cell differentiation | 1178 | 4437 | 1.092e-10 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 312 | 970 | 1.120e-10 |
| GO:BP | GO:0048869 | cellular developmental process | 1178 | 4438 | 1.152e-10 |
| GO:BP | GO:0050896 | response to stimulus | 2243 | 8999 | 1.155e-10 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 315 | 983 | 1.419e-10 |
| GO:BP | GO:0031647 | regulation of protein stability | 132 | 335 | 1.959e-10 |
| GO:BP | GO:0080134 | regulation of response to stress | 421 | 1387 | 1.992e-10 |
| GO:BP | GO:0070925 | organelle assembly | 331 | 1046 | 2.156e-10 |
| GO:BP | GO:0033043 | regulation of organelle organization | 357 | 1148 | 3.608e-10 |
| GO:BP | GO:0034504 | protein localization to nucleus | 126 | 318 | 3.811e-10 |
| GO:BP | GO:0061024 | membrane organization | 269 | 821 | 4.339e-10 |
| GO:BP | GO:0048699 | generation of neurons | 457 | 1536 | 5.774e-10 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 753 | 2713 | 6.013e-10 |
| GO:BP | GO:0006417 | regulation of translation | 144 | 380 | 6.102e-10 |
| GO:BP | GO:0051169 | nuclear transport | 130 | 334 | 7.349e-10 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 130 | 334 | 7.349e-10 |
| GO:BP | GO:0010970 | transport along microtubule | 81 | 178 | 7.371e-10 |
| GO:BP | GO:0030182 | neuron differentiation | 434 | 1451 | 8.690e-10 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 227 | 673 | 8.923e-10 |
| GO:BP | GO:0010256 | endomembrane system organization | 210 | 613 | 1.065e-09 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 231 | 689 | 1.128e-09 |
| GO:BP | GO:0007010 | cytoskeleton organization | 453 | 1529 | 1.332e-09 |
| GO:BP | GO:0099111 | microtubule-based transport | 94 | 220 | 1.366e-09 |
| GO:BP | GO:0010506 | regulation of autophagy | 139 | 367 | 1.373e-09 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 100 | 239 | 1.373e-09 |
| GO:BP | GO:0009987 | cellular process | 4645 | 20247 | 1.467e-09 |
| GO:BP | GO:0070848 | response to growth factor | 239 | 721 | 1.721e-09 |
| GO:BP | GO:0030029 | actin filament-based process | 267 | 825 | 2.039e-09 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 101 | 244 | 2.261e-09 |
| GO:BP | GO:0030334 | regulation of cell migration | 303 | 960 | 2.288e-09 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 87 | 200 | 2.317e-09 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 221 | 658 | 2.428e-09 |
| GO:BP | GO:0048666 | neuron development | 360 | 1177 | 2.610e-09 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 391 | 1296 | 2.636e-09 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 403 | 1343 | 2.844e-09 |
| GO:BP | GO:0007049 | cell cycle | 485 | 1663 | 3.017e-09 |
| GO:BP | GO:0031175 | neuron projection development | 319 | 1023 | 3.142e-09 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 216 | 642 | 3.278e-09 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 98 | 236 | 3.354e-09 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 217 | 646 | 3.448e-09 |
| GO:BP | GO:0030162 | regulation of proteolysis | 149 | 406 | 3.628e-09 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 386 | 1280 | 3.628e-09 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 287 | 905 | 4.067e-09 |
| GO:BP | GO:2000145 | regulation of cell motility | 318 | 1022 | 4.277e-09 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 153 | 421 | 4.581e-09 |
| GO:BP | GO:0048513 | animal organ development | 837 | 3085 | 4.755e-09 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 239 | 729 | 5.152e-09 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 205 | 606 | 5.652e-09 |
| GO:BP | GO:0051648 | vesicle localization | 92 | 219 | 5.797e-09 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 85 | 197 | 5.797e-09 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 292 | 928 | 6.818e-09 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 71 | 155 | 7.717e-09 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 277 | 874 | 9.013e-09 |
| GO:BP | GO:0016477 | cell migration | 449 | 1534 | 9.035e-09 |
| GO:BP | GO:0040012 | regulation of locomotion | 328 | 1067 | 9.549e-09 |
| GO:BP | GO:0036503 | ERAD pathway | 55 | 109 | 1.001e-08 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 133 | 357 | 1.129e-08 |
| GO:BP | GO:0034330 | cell junction organization | 265 | 834 | 1.672e-08 |
| GO:BP | GO:0009725 | response to hormone | 284 | 907 | 2.096e-08 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 346 | 1144 | 2.398e-08 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 79 | 184 | 3.291e-08 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 83 | 197 | 3.719e-08 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 70 | 157 | 4.263e-08 |
| GO:BP | GO:0016043 | cellular component organization | 2028 | 8184 | 4.448e-08 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 52 | 104 | 4.551e-08 |
| GO:BP | GO:0016072 | rRNA metabolic process | 104 | 266 | 4.666e-08 |
| GO:BP | GO:0051640 | organelle localization | 205 | 620 | 4.666e-08 |
| GO:BP | GO:0016032 | viral process | 150 | 423 | 4.666e-08 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 270 | 861 | 4.666e-08 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 609 | 2192 | 5.557e-08 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 50 | 99 | 5.944e-08 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 171 | 499 | 6.126e-08 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 127 | 345 | 6.385e-08 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 165 | 478 | 6.495e-08 |
| GO:BP | GO:0006364 | rRNA processing | 91 | 225 | 6.825e-08 |
| GO:BP | GO:0006401 | RNA catabolic process | 118 | 315 | 7.309e-08 |
| GO:BP | GO:0007015 | actin filament organization | 160 | 461 | 7.323e-08 |
| GO:BP | GO:0008088 | axo-dendritic transport | 44 | 83 | 7.930e-08 |
| GO:BP | GO:0141188 | nucleic acid catabolic process | 126 | 343 | 8.372e-08 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 180 | 535 | 1.084e-07 |
| GO:BP | GO:0006979 | response to oxidative stress | 142 | 400 | 1.123e-07 |
| GO:BP | GO:0051604 | protein maturation | 181 | 539 | 1.143e-07 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 196 | 594 | 1.232e-07 |
| GO:BP | GO:0032501 | multicellular organismal process | 1823 | 7322 | 1.565e-07 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 171 | 505 | 1.575e-07 |
| GO:BP | GO:0007169 | cell surface receptor protein tyrosine kinase signaling pathway | 205 | 629 | 1.641e-07 |
| GO:BP | GO:0006259 | DNA metabolic process | 305 | 1005 | 1.641e-07 |
| GO:BP | GO:0051656 | establishment of organelle localization | 167 | 491 | 1.652e-07 |
| GO:BP | GO:0006986 | response to unfolded protein | 63 | 140 | 1.672e-07 |
| GO:BP | GO:0051170 | import into nucleus | 73 | 171 | 1.722e-07 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 101 | 262 | 1.786e-07 |
| GO:BP | GO:0023052 | signaling | 1635 | 6515 | 1.874e-07 |
| GO:BP | GO:0006839 | mitochondrial transport | 75 | 178 | 2.099e-07 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 116 | 314 | 2.364e-07 |
| GO:BP | GO:0006606 | protein import into nucleus | 71 | 166 | 2.492e-07 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 126 | 349 | 2.632e-07 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 170 | 505 | 2.686e-07 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 272 | 884 | 2.749e-07 |
| GO:BP | GO:0007017 | microtubule-based process | 302 | 999 | 2.889e-07 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 87 | 218 | 2.998e-07 |
| GO:BP | GO:0000902 | cell morphogenesis | 301 | 996 | 3.145e-07 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 219 | 686 | 3.191e-07 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 129 | 361 | 3.476e-07 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 449 | 1576 | 3.603e-07 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 84 | 209 | 3.605e-07 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 153 | 446 | 3.618e-07 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 102 | 269 | 3.898e-07 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 222 | 701 | 5.187e-07 |
| GO:BP | GO:0006897 | endocytosis | 224 | 709 | 5.534e-07 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 471 | 1669 | 5.618e-07 |
| GO:BP | GO:0007154 | cell communication | 1635 | 6540 | 5.717e-07 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 108 | 292 | 6.825e-07 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 206 | 645 | 8.252e-07 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 60 | 136 | 8.731e-07 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 147 | 430 | 8.791e-07 |
| GO:BP | GO:0000278 | mitotic cell cycle | 271 | 892 | 1.004e-06 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 255 | 831 | 1.004e-06 |
| GO:BP | GO:0051050 | positive regulation of transport | 259 | 847 | 1.095e-06 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 107 | 291 | 1.107e-06 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 97 | 257 | 1.111e-06 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 241 | 779 | 1.128e-06 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 176 | 537 | 1.171e-06 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 203 | 637 | 1.216e-06 |
| GO:BP | GO:0040011 | locomotion | 364 | 1255 | 1.246e-06 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 52 | 113 | 1.316e-06 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 254 | 830 | 1.347e-06 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 82 | 208 | 1.384e-06 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 89 | 232 | 1.662e-06 |
| GO:BP | GO:0032535 | regulation of cellular component size | 125 | 356 | 1.710e-06 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 70 | 170 | 1.792e-06 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 286 | 956 | 1.924e-06 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 182 | 563 | 1.994e-06 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 111 | 308 | 2.013e-06 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 286 | 957 | 2.117e-06 |
| GO:BP | GO:0006325 | chromatin organization | 267 | 884 | 2.129e-06 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 281 | 938 | 2.164e-06 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 362 | 1256 | 2.706e-06 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 781 | 2956 | 2.709e-06 |
| GO:BP | GO:0022402 | cell cycle process | 368 | 1280 | 2.766e-06 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 162 | 492 | 2.766e-06 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 162 | 493 | 3.197e-06 |
| GO:BP | GO:0048870 | cell motility | 496 | 1793 | 3.321e-06 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 201 | 638 | 3.442e-06 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 98 | 266 | 3.442e-06 |
| GO:BP | GO:0072359 | circulatory system development | 333 | 1145 | 3.445e-06 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 172 | 531 | 3.790e-06 |
| GO:BP | GO:0045727 | positive regulation of translation | 59 | 138 | 3.959e-06 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 115 | 326 | 4.139e-06 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 126 | 365 | 4.223e-06 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 95 | 257 | 4.390e-06 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 35 | 67 | 4.473e-06 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 78 | 201 | 5.890e-06 |
| GO:BP | GO:0008152 | metabolic process | 3338 | 14136 | 6.117e-06 |
| GO:BP | GO:0035295 | tube development | 320 | 1101 | 6.440e-06 |
| GO:BP | GO:0098657 | import into cell | 274 | 922 | 6.481e-06 |
| GO:BP | GO:0000423 | mitophagy | 36 | 71 | 7.744e-06 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 104 | 291 | 7.744e-06 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 110 | 312 | 7.744e-06 |
| GO:BP | GO:0071806 | protein transmembrane transport | 37 | 74 | 8.286e-06 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 65 | 160 | 8.726e-06 |
| GO:BP | GO:0016482 | cytosolic transport | 59 | 141 | 9.299e-06 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 50 | 113 | 9.681e-06 |
| GO:BP | GO:0030031 | cell projection assembly | 192 | 613 | 9.878e-06 |
| GO:BP | GO:0042592 | homeostatic process | 478 | 1736 | 9.933e-06 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 111 | 317 | 9.933e-06 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 34 | 66 | 9.971e-06 |
| GO:BP | GO:0051338 | regulation of transferase activity | 161 | 498 | 1.031e-05 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 79 | 207 | 1.049e-05 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 742 | 2819 | 1.061e-05 |
| GO:BP | GO:0098930 | axonal transport | 35 | 69 | 1.074e-05 |
| GO:BP | GO:0050821 | protein stabilization | 84 | 224 | 1.074e-05 |
| GO:BP | GO:0001944 | vasculature development | 231 | 762 | 1.099e-05 |
| GO:BP | GO:0008283 | cell population proliferation | 546 | 2015 | 1.205e-05 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 188 | 600 | 1.231e-05 |
| GO:BP | GO:0006338 | chromatin remodeling | 221 | 725 | 1.245e-05 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 110 | 315 | 1.265e-05 |
| GO:BP | GO:0050808 | synapse organization | 178 | 563 | 1.291e-05 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 161 | 500 | 1.318e-05 |
| GO:BP | GO:0016050 | vesicle organization | 127 | 376 | 1.337e-05 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 42 | 90 | 1.397e-05 |
| GO:BP | GO:0009165 | nucleotide biosynthetic process | 85 | 229 | 1.471e-05 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 50 | 115 | 1.750e-05 |
| GO:BP | GO:0061572 | actin filament bundle organization | 65 | 163 | 1.800e-05 |
| GO:BP | GO:0043434 | response to peptide hormone | 144 | 440 | 1.868e-05 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 71 | 183 | 1.889e-05 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 46 | 103 | 1.923e-05 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 43 | 94 | 1.985e-05 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 337 | 1182 | 2.069e-05 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 160 | 500 | 2.124e-05 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 341 | 1199 | 2.273e-05 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 50 | 116 | 2.339e-05 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 174 | 553 | 2.347e-05 |
| GO:BP | GO:0045333 | cellular respiration | 88 | 242 | 2.498e-05 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 44 | 98 | 2.742e-05 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 119 | 352 | 2.778e-05 |
| GO:BP | GO:0032456 | endocytic recycling | 40 | 86 | 2.779e-05 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 148 | 458 | 2.889e-05 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 206 | 676 | 2.959e-05 |
| GO:BP | GO:0042255 | ribosome assembly | 32 | 63 | 3.028e-05 |
| GO:BP | GO:0009060 | aerobic respiration | 75 | 199 | 3.304e-05 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 207 | 681 | 3.356e-05 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 54 | 130 | 3.419e-05 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 204 | 670 | 3.510e-05 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 46 | 105 | 3.560e-05 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 68 | 176 | 3.649e-05 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 52 | 124 | 3.760e-05 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 112 | 329 | 3.810e-05 |
| GO:BP | GO:0006164 | purine nucleotide biosynthetic process | 73 | 193 | 3.824e-05 |
| GO:BP | GO:0051726 | regulation of cell cycle | 311 | 1087 | 3.830e-05 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 85 | 234 | 3.830e-05 |
| GO:BP | GO:0051223 | regulation of protein transport | 143 | 442 | 3.916e-05 |
| GO:BP | GO:0007165 | signal transduction | 1486 | 6002 | 3.927e-05 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 126 | 380 | 4.014e-05 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 126 | 380 | 4.014e-05 |
| GO:BP | GO:0060322 | head development | 239 | 806 | 4.014e-05 |
| GO:BP | GO:0070482 | response to oxygen levels | 116 | 344 | 4.122e-05 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 223 | 745 | 4.297e-05 |
| GO:BP | GO:0019080 | viral gene expression | 47 | 109 | 4.557e-05 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 174 | 559 | 4.628e-05 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 123 | 371 | 5.307e-05 |
| GO:BP | GO:0035239 | tube morphogenesis | 257 | 879 | 5.335e-05 |
| GO:BP | GO:0001568 | blood vessel development | 219 | 732 | 5.472e-05 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 139 | 430 | 5.587e-05 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 26 | 48 | 6.131e-05 |
| GO:BP | GO:0051668 | localization within membrane | 268 | 924 | 6.389e-05 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 32 | 65 | 6.792e-05 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 55 | 136 | 6.917e-05 |
| GO:BP | GO:0007249 | canonical NF-kappaB signal transduction | 105 | 308 | 7.097e-05 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 124 | 377 | 7.385e-05 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 124 | 377 | 7.385e-05 |
| GO:BP | GO:0034329 | cell junction assembly | 158 | 503 | 7.507e-05 |
| GO:BP | GO:0006403 | RNA localization | 74 | 200 | 7.905e-05 |
| GO:BP | GO:0006281 | DNA repair | 189 | 621 | 8.209e-05 |
| GO:BP | GO:0009142 | nucleoside triphosphate biosynthetic process | 52 | 127 | 8.214e-05 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 180 | 587 | 8.399e-05 |
| GO:BP | GO:0043542 | endothelial cell migration | 80 | 221 | 8.448e-05 |
| GO:BP | GO:0075522 | IRES-dependent viral translational initiation | 10 | 11 | 9.040e-05 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 391 | 1418 | 9.146e-05 |
| GO:BP | GO:0007033 | vacuole organization | 85 | 239 | 9.454e-05 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 171 | 554 | 9.454e-05 |
| GO:BP | GO:0019058 | viral life cycle | 105 | 310 | 9.562e-05 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 41 | 93 | 9.947e-05 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 122 | 372 | 1.007e-04 |
| GO:BP | GO:0000266 | mitochondrial fission | 23 | 41 | 1.017e-04 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 62 | 161 | 1.078e-04 |
| GO:BP | GO:0009152 | purine ribonucleotide biosynthetic process | 62 | 161 | 1.078e-04 |
| GO:BP | GO:0031648 | protein destabilization | 27 | 52 | 1.084e-04 |
| GO:BP | GO:0006402 | mRNA catabolic process | 91 | 261 | 1.094e-04 |
| GO:BP | GO:0007417 | central nervous system development | 301 | 1061 | 1.112e-04 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 45 | 106 | 1.153e-04 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 61 | 158 | 1.153e-04 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 114 | 344 | 1.175e-04 |
| GO:BP | GO:0000165 | MAPK cascade | 220 | 744 | 1.175e-04 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 216 | 729 | 1.231e-04 |
| GO:BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | 49 | 119 | 1.233e-04 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 95 | 276 | 1.238e-04 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 198 | 660 | 1.276e-04 |
| GO:BP | GO:0006629 | lipid metabolic process | 383 | 1391 | 1.278e-04 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 89 | 255 | 1.290e-04 |
| GO:BP | GO:0043122 | regulation of canonical NF-kappaB signal transduction | 98 | 287 | 1.290e-04 |
| GO:BP | GO:0046903 | secretion | 283 | 992 | 1.308e-04 |
| GO:BP | GO:0009888 | tissue development | 540 | 2032 | 1.372e-04 |
| GO:BP | GO:0051258 | protein polymerization | 97 | 284 | 1.411e-04 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 37 | 82 | 1.438e-04 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 110 | 331 | 1.440e-04 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 34 | 73 | 1.443e-04 |
| GO:BP | GO:0043487 | regulation of RNA stability | 71 | 193 | 1.443e-04 |
| GO:BP | GO:0007178 | cell surface receptor protein serine/threonine kinase signaling pathway | 119 | 364 | 1.468e-04 |
| GO:BP | GO:0051896 | regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 91 | 263 | 1.482e-04 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 191 | 635 | 1.514e-04 |
| GO:BP | GO:0061061 | muscle structure development | 206 | 693 | 1.552e-04 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 90 | 260 | 1.621e-04 |
| GO:BP | GO:0006282 | regulation of DNA repair | 80 | 225 | 1.683e-04 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 55 | 140 | 1.736e-04 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 91 | 264 | 1.741e-04 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 89 | 257 | 1.768e-04 |
| GO:BP | GO:0040007 | growth | 266 | 929 | 1.801e-04 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 115 | 351 | 1.827e-04 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 104 | 311 | 1.830e-04 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 54 | 137 | 1.844e-04 |
| GO:BP | GO:0007420 | brain development | 221 | 754 | 2.010e-04 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 33 | 71 | 2.010e-04 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 35 | 77 | 2.010e-04 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 44 | 105 | 2.028e-04 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 89 | 258 | 2.063e-04 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 860 | 3375 | 2.076e-04 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 43 | 102 | 2.116e-04 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 73 | 202 | 2.116e-04 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 83 | 237 | 2.133e-04 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 116 | 356 | 2.161e-04 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 162 | 528 | 2.181e-04 |
| GO:BP | GO:0001666 | response to hypoxia | 101 | 302 | 2.380e-04 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 39 | 90 | 2.489e-04 |
| GO:BP | GO:0032868 | response to insulin | 92 | 270 | 2.523e-04 |
| GO:BP | GO:0006399 | tRNA metabolic process | 75 | 210 | 2.560e-04 |
| GO:BP | GO:0001701 | in utero embryonic development | 131 | 413 | 2.634e-04 |
| GO:BP | GO:1901875 | positive regulation of post-translational protein modification | 52 | 132 | 2.649e-04 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 82 | 235 | 2.758e-04 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 33 | 72 | 2.804e-04 |
| GO:BP | GO:0055085 | transmembrane transport | 426 | 1578 | 2.804e-04 |
| GO:BP | GO:0030010 | establishment of cell polarity | 60 | 159 | 2.804e-04 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 181 | 603 | 2.804e-04 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 89 | 260 | 2.804e-04 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 64 | 173 | 3.060e-04 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 106 | 322 | 3.067e-04 |
| GO:BP | GO:0009790 | embryo development | 316 | 1135 | 3.073e-04 |
| GO:BP | GO:0001525 | angiogenesis | 166 | 547 | 3.248e-04 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 158 | 517 | 3.422e-04 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 85 | 247 | 3.425e-04 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 62 | 167 | 3.604e-04 |
| GO:BP | GO:0006457 | protein folding | 77 | 219 | 3.604e-04 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 47 | 117 | 3.668e-04 |
| GO:BP | GO:0007034 | vacuolar transport | 64 | 174 | 3.693e-04 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 251 | 879 | 3.693e-04 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 64 | 174 | 3.693e-04 |
| GO:BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | 46 | 114 | 3.883e-04 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 61 | 164 | 3.883e-04 |
| GO:BP | GO:0036010 | protein localization to endosome | 17 | 28 | 3.938e-04 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 195 | 661 | 4.086e-04 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 71 | 199 | 4.282e-04 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 165 | 546 | 4.317e-04 |
| GO:BP | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 99 | 299 | 4.323e-04 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 51 | 131 | 4.442e-04 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 24 | 47 | 4.495e-04 |
| GO:BP | GO:0050792 | regulation of viral process | 59 | 158 | 4.547e-04 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 37 | 86 | 4.714e-04 |
| GO:BP | GO:0048545 | response to steroid hormone | 110 | 340 | 4.756e-04 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 101 | 307 | 4.756e-04 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 77 | 221 | 4.994e-04 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 55 | 145 | 5.074e-04 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 97 | 293 | 5.158e-04 |
| GO:BP | GO:0051168 | nuclear export | 62 | 169 | 5.262e-04 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 33 | 74 | 5.277e-04 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 57 | 152 | 5.311e-04 |
| GO:BP | GO:0061462 | protein localization to lysosome | 28 | 59 | 5.327e-04 |
| GO:BP | GO:1900078 | positive regulation of cellular response to insulin stimulus | 16 | 26 | 5.327e-04 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 85 | 250 | 5.367e-04 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 188 | 637 | 5.367e-04 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 40 | 96 | 5.367e-04 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 48 | 122 | 5.393e-04 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 54 | 142 | 5.393e-04 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 333 | 1212 | 5.453e-04 |
| GO:BP | GO:0046034 | ATP metabolic process | 79 | 229 | 5.700e-04 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 22 | 42 | 5.708e-04 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 103 | 316 | 5.876e-04 |
| GO:BP | GO:0034644 | cellular response to UV | 38 | 90 | 5.876e-04 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 67 | 187 | 5.894e-04 |
| GO:BP | GO:0051236 | establishment of RNA localization | 60 | 163 | 6.099e-04 |
| GO:BP | GO:0034063 | stress granule assembly | 19 | 34 | 6.119e-04 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 121 | 383 | 6.129e-04 |
| GO:BP | GO:0141193 | nuclear receptor-mediated signaling pathway | 64 | 177 | 6.329e-04 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 126 | 402 | 6.329e-04 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 64 | 177 | 6.329e-04 |
| GO:BP | GO:0019725 | cellular homeostasis | 238 | 835 | 6.389e-04 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 70 | 198 | 6.389e-04 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 68 | 191 | 6.389e-04 |
| GO:BP | GO:0021543 | pallium development | 70 | 198 | 6.389e-04 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 119 | 376 | 6.408e-04 |
| GO:BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | 45 | 113 | 6.447e-04 |
| GO:BP | GO:0007040 | lysosome organization | 45 | 113 | 6.447e-04 |
| GO:BP | GO:0080171 | lytic vacuole organization | 45 | 113 | 6.447e-04 |
| GO:BP | GO:0050658 | RNA transport | 59 | 160 | 6.476e-04 |
| GO:BP | GO:0050657 | nucleic acid transport | 59 | 160 | 6.476e-04 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 24 | 48 | 6.511e-04 |
| GO:BP | GO:0015986 | proton motive force-driven ATP synthesis | 34 | 78 | 6.706e-04 |
| GO:BP | GO:0009615 | response to virus | 129 | 414 | 6.765e-04 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 65 | 181 | 6.821e-04 |
| GO:BP | GO:0048009 | insulin-like growth factor receptor signaling pathway | 25 | 51 | 6.821e-04 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 31 | 69 | 7.173e-04 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 88 | 263 | 7.173e-04 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 31 | 69 | 7.173e-04 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 801 | 3157 | 7.173e-04 |
| GO:BP | GO:0007041 | lysosomal transport | 52 | 137 | 7.537e-04 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 42 | 104 | 7.745e-04 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 61 | 168 | 7.987e-04 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 73 | 210 | 7.996e-04 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 85 | 253 | 8.119e-04 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 18 | 32 | 8.347e-04 |
| GO:BP | GO:0043549 | regulation of kinase activity | 133 | 431 | 8.347e-04 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 167 | 561 | 8.415e-04 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 60 | 165 | 8.659e-04 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 40 | 98 | 8.695e-04 |
| GO:BP | GO:0019079 | viral genome replication | 50 | 131 | 8.745e-04 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 447 | 1685 | 8.834e-04 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 66 | 186 | 8.844e-04 |
| GO:BP | GO:0009411 | response to UV | 57 | 155 | 9.168e-04 |
| GO:BP | GO:0043401 | steroid hormone receptor signaling pathway | 52 | 138 | 9.213e-04 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 71 | 204 | 9.482e-04 |
| GO:BP | GO:0007507 | heart development | 178 | 605 | 9.584e-04 |
| GO:BP | GO:0099518 | vesicle cytoskeletal trafficking | 29 | 64 | 1.011e-03 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 26 | 55 | 1.011e-03 |
| GO:BP | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | 26 | 55 | 1.011e-03 |
| GO:BP | GO:0072665 | protein localization to vacuole | 37 | 89 | 1.019e-03 |
| GO:BP | GO:0033002 | muscle cell proliferation | 72 | 208 | 1.019e-03 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 41 | 102 | 1.042e-03 |
| GO:BP | GO:0003174 | mitral valve development | 10 | 13 | 1.047e-03 |
| GO:BP | GO:0009314 | response to radiation | 134 | 437 | 1.049e-03 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 200 | 692 | 1.056e-03 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 50 | 132 | 1.069e-03 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 80 | 237 | 1.079e-03 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 122 | 392 | 1.086e-03 |
| GO:BP | GO:0060537 | muscle tissue development | 132 | 430 | 1.113e-03 |
| GO:BP | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway | 9 | 11 | 1.113e-03 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 34 | 80 | 1.170e-03 |
| GO:BP | GO:0014706 | striated muscle tissue development | 126 | 408 | 1.206e-03 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 70 | 202 | 1.208e-03 |
| GO:BP | GO:0010586 | miRNA metabolic process | 42 | 106 | 1.233e-03 |
| GO:BP | GO:0009451 | RNA modification | 62 | 174 | 1.239e-03 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 441 | 1667 | 1.250e-03 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 53 | 143 | 1.257e-03 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 32 | 74 | 1.260e-03 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 71 | 206 | 1.292e-03 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 86 | 260 | 1.292e-03 |
| GO:BP | GO:0043009 | chordate embryonic development | 194 | 671 | 1.293e-03 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 41 | 103 | 1.305e-03 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 41 | 103 | 1.305e-03 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 69 | 199 | 1.305e-03 |
| GO:BP | GO:0000723 | telomere maintenance | 59 | 164 | 1.328e-03 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 106 | 334 | 1.341e-03 |
| GO:BP | GO:0170036 | import into the mitochondrion | 24 | 50 | 1.367e-03 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 72 | 210 | 1.373e-03 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 82 | 246 | 1.379e-03 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 28 | 62 | 1.381e-03 |
| GO:BP | GO:1901799 | negative regulation of proteasomal protein catabolic process | 26 | 56 | 1.395e-03 |
| GO:BP | GO:0051028 | mRNA transport | 49 | 130 | 1.395e-03 |
| GO:BP | GO:0044770 | cell cycle phase transition | 158 | 532 | 1.403e-03 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 88 | 268 | 1.403e-03 |
| GO:BP | GO:0032940 | secretion by cell | 240 | 854 | 1.412e-03 |
| GO:BP | GO:0005977 | glycogen metabolic process | 35 | 84 | 1.427e-03 |
| GO:BP | GO:1901652 | response to peptide | 267 | 962 | 1.427e-03 |
| GO:BP | GO:0030041 | actin filament polymerization | 58 | 161 | 1.427e-03 |
| GO:BP | GO:0043123 | positive regulation of canonical NF-kappaB signal transduction | 73 | 214 | 1.449e-03 |
| GO:BP | GO:0032259 | methylation | 79 | 236 | 1.550e-03 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 16 | 28 | 1.574e-03 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 16 | 28 | 1.574e-03 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 16 | 28 | 1.574e-03 |
| GO:BP | GO:0000028 | ribosomal small subunit assembly | 12 | 18 | 1.582e-03 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 105 | 332 | 1.632e-03 |
| GO:BP | GO:0140352 | export from cell | 258 | 928 | 1.644e-03 |
| GO:BP | GO:0035357 | peroxisome proliferator activated receptor signaling pathway | 14 | 23 | 1.713e-03 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 60 | 169 | 1.713e-03 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 36 | 88 | 1.755e-03 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 21 | 42 | 1.761e-03 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 296 | 1082 | 1.784e-03 |
| GO:BP | GO:0006903 | vesicle targeting | 29 | 66 | 1.824e-03 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 29 | 66 | 1.824e-03 |
| GO:BP | GO:0048041 | focal adhesion assembly | 35 | 85 | 1.852e-03 |
| GO:BP | GO:0010257 | NADH dehydrogenase complex assembly | 28 | 63 | 1.864e-03 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 17 | 31 | 1.864e-03 |
| GO:BP | GO:0032981 | mitochondrial respiratory chain complex I assembly | 28 | 63 | 1.864e-03 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 95 | 296 | 1.865e-03 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 42 | 108 | 1.878e-03 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 42 | 108 | 1.878e-03 |
| GO:BP | GO:0006513 | protein monoubiquitination | 23 | 48 | 1.878e-03 |
| GO:BP | GO:0043149 | stress fiber assembly | 42 | 108 | 1.878e-03 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 50 | 135 | 1.884e-03 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 52 | 142 | 1.940e-03 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 93 | 289 | 1.948e-03 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 81 | 245 | 1.950e-03 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 41 | 105 | 2.013e-03 |
| GO:BP | GO:0034497 | protein localization to phagophore assembly site | 11 | 16 | 2.017e-03 |
| GO:BP | GO:0017148 | negative regulation of translation | 49 | 132 | 2.040e-03 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 72 | 213 | 2.119e-03 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 40 | 102 | 2.161e-03 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 43 | 112 | 2.163e-03 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 43 | 112 | 2.163e-03 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 144 | 483 | 2.203e-03 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 48 | 129 | 2.209e-03 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 73 | 217 | 2.240e-03 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 101 | 320 | 2.273e-03 |
| GO:BP | GO:1901524 | regulation of mitophagy | 19 | 37 | 2.288e-03 |
| GO:BP | GO:0071514 | genomic imprinting | 13 | 21 | 2.293e-03 |
| GO:BP | GO:0040008 | regulation of growth | 175 | 604 | 2.319e-03 |
| GO:BP | GO:0044042 | glucan metabolic process | 35 | 86 | 2.341e-03 |
| GO:BP | GO:0048589 | developmental growth | 190 | 663 | 2.341e-03 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 60 | 171 | 2.355e-03 |
| GO:BP | GO:0060816 | random inactivation of X chromosome | 10 | 14 | 2.534e-03 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 21 | 43 | 2.555e-03 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 21 | 43 | 2.555e-03 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 53 | 147 | 2.563e-03 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 46 | 123 | 2.581e-03 |
| GO:BP | GO:0034097 | response to cytokine | 261 | 947 | 2.581e-03 |
| GO:BP | GO:0021537 | telencephalon development | 91 | 284 | 2.612e-03 |
| GO:BP | GO:0007155 | cell adhesion | 404 | 1530 | 2.672e-03 |
| GO:BP | GO:0007029 | endoplasmic reticulum organization | 40 | 103 | 2.673e-03 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 40 | 103 | 2.673e-03 |
| GO:BP | GO:0051897 | positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 64 | 186 | 2.687e-03 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 32 | 77 | 2.759e-03 |
| GO:BP | GO:0048729 | tissue morphogenesis | 177 | 614 | 2.760e-03 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 52 | 144 | 2.767e-03 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 52 | 144 | 2.767e-03 |
| GO:BP | GO:0097581 | lamellipodium organization | 36 | 90 | 2.784e-03 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 56 | 158 | 2.791e-03 |
| GO:BP | GO:0051402 | neuron apoptotic process | 94 | 296 | 2.887e-03 |
| GO:BP | GO:0046628 | positive regulation of insulin receptor signaling pathway | 14 | 24 | 2.976e-03 |
| GO:BP | GO:0003183 | mitral valve morphogenesis | 9 | 12 | 3.032e-03 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 96 | 304 | 3.070e-03 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 96 | 304 | 3.070e-03 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 41 | 107 | 3.070e-03 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 38 | 97 | 3.070e-03 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 41 | 107 | 3.070e-03 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 206 | 730 | 3.096e-03 |
| GO:BP | GO:0001649 | osteoblast differentiation | 77 | 234 | 3.132e-03 |
| GO:BP | GO:0033993 | response to lipid | 256 | 930 | 3.132e-03 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 98 | 312 | 3.267e-03 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 28 | 65 | 3.286e-03 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 40 | 104 | 3.291e-03 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 148 | 503 | 3.291e-03 |
| GO:BP | GO:0048227 | plasma membrane to endosome transport | 7 | 8 | 3.354e-03 |
| GO:BP | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway | 7 | 8 | 3.354e-03 |
| GO:BP | GO:0030518 | nuclear receptor-mediated steroid hormone signaling pathway | 45 | 121 | 3.358e-03 |
| GO:BP | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway | 8 | 10 | 3.379e-03 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 19 | 38 | 3.379e-03 |
| GO:BP | GO:0060284 | regulation of cell development | 235 | 847 | 3.391e-03 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 79 | 242 | 3.412e-03 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 61 | 177 | 3.441e-03 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 47 | 128 | 3.441e-03 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 20 | 41 | 3.521e-03 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 51 | 142 | 3.556e-03 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 21 | 44 | 3.615e-03 |
| GO:BP | GO:0051646 | mitochondrion localization | 24 | 53 | 3.622e-03 |
| GO:BP | GO:0016049 | cell growth | 143 | 485 | 3.698e-03 |
| GO:BP | GO:0014812 | muscle cell migration | 35 | 88 | 3.713e-03 |
| GO:BP | GO:0031929 | TOR signaling | 58 | 167 | 3.817e-03 |
| GO:BP | GO:0021987 | cerebral cortex development | 48 | 132 | 3.817e-03 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 52 | 146 | 3.880e-03 |
| GO:BP | GO:0007030 | Golgi organization | 54 | 153 | 3.880e-03 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 229 | 825 | 3.880e-03 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 115 | 378 | 3.910e-03 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 45 | 122 | 4.051e-03 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 66 | 196 | 4.056e-03 |
| GO:BP | GO:0007063 | regulation of sister chromatid cohesion | 11 | 17 | 4.064e-03 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 108 | 352 | 4.170e-03 |
| GO:BP | GO:0030832 | regulation of actin filament length | 53 | 150 | 4.232e-03 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 148 | 506 | 4.244e-03 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 28 | 66 | 4.288e-03 |
| GO:BP | GO:0009306 | protein secretion | 115 | 379 | 4.331e-03 |
| GO:BP | GO:0001558 | regulation of cell growth | 122 | 406 | 4.428e-03 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 65 | 193 | 4.428e-03 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 27 | 63 | 4.471e-03 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 41 | 109 | 4.579e-03 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 71 | 215 | 4.583e-03 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 80 | 248 | 4.583e-03 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 50 | 140 | 4.586e-03 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 52 | 147 | 4.586e-03 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 38 | 99 | 4.662e-03 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 25 | 57 | 4.792e-03 |
| GO:BP | GO:0010761 | fibroblast migration | 24 | 54 | 4.930e-03 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 19 | 39 | 4.930e-03 |
| GO:BP | GO:0051882 | mitochondrial depolarization | 14 | 25 | 4.932e-03 |
| GO:BP | GO:0032543 | mitochondrial translation | 47 | 130 | 4.949e-03 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 34 | 86 | 4.969e-03 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 23 | 51 | 5.002e-03 |
| GO:BP | GO:0008089 | anterograde axonal transport | 23 | 51 | 5.002e-03 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 98 | 316 | 5.018e-03 |
| GO:BP | GO:0006476 | protein deacetylation | 20 | 42 | 5.018e-03 |
| GO:BP | GO:0030900 | forebrain development | 126 | 423 | 5.136e-03 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 99 | 320 | 5.146e-03 |
| GO:BP | GO:0061564 | axon development | 153 | 528 | 5.203e-03 |
| GO:BP | GO:0010720 | positive regulation of cell development | 135 | 458 | 5.234e-03 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 29 | 70 | 5.263e-03 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 29 | 70 | 5.263e-03 |
| GO:BP | GO:0050807 | regulation of synapse organization | 96 | 309 | 5.309e-03 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 10 | 15 | 5.309e-03 |
| GO:BP | GO:0019827 | stem cell population maintenance | 61 | 180 | 5.364e-03 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 48 | 134 | 5.373e-03 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 48 | 134 | 5.373e-03 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 78 | 242 | 5.378e-03 |
| GO:BP | GO:0042776 | proton motive force-driven mitochondrial ATP synthesis | 28 | 67 | 5.508e-03 |
| GO:BP | GO:0006517 | protein deglycosylation | 12 | 20 | 5.508e-03 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 28 | 67 | 5.508e-03 |
| GO:BP | GO:0043603 | amide metabolic process | 134 | 455 | 5.620e-03 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 115 | 382 | 5.752e-03 |
| GO:BP | GO:0033059 | cellular pigmentation | 27 | 64 | 5.794e-03 |
| GO:BP | GO:0031667 | response to nutrient levels | 148 | 510 | 5.881e-03 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 119 | 398 | 6.146e-03 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 16 | 31 | 6.154e-03 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 16 | 31 | 6.154e-03 |
| GO:BP | GO:0046685 | response to arsenic-containing substance | 16 | 31 | 6.154e-03 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 16 | 31 | 6.154e-03 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 252 | 925 | 6.163e-03 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 61 | 181 | 6.182e-03 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 44 | 121 | 6.270e-03 |
| GO:BP | GO:0098732 | macromolecule deacylation | 25 | 58 | 6.292e-03 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 52 | 149 | 6.294e-03 |
| GO:BP | GO:0010934 | macrophage cytokine production | 18 | 37 | 6.848e-03 |
| GO:BP | GO:0060271 | cilium assembly | 120 | 403 | 6.848e-03 |
| GO:BP | GO:0010935 | regulation of macrophage cytokine production | 18 | 37 | 6.848e-03 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 51 | 146 | 6.877e-03 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 22 | 49 | 6.877e-03 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 51 | 146 | 6.877e-03 |
| GO:BP | GO:0061028 | establishment of endothelial barrier | 22 | 49 | 6.877e-03 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 49 | 139 | 6.925e-03 |
| GO:BP | GO:0007416 | synapse assembly | 82 | 259 | 6.984e-03 |
| GO:BP | GO:0048489 | synaptic vesicle transport | 20 | 43 | 7.005e-03 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 116 | 388 | 7.048e-03 |
| GO:BP | GO:0007517 | muscle organ development | 110 | 365 | 7.054e-03 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 56 | 164 | 7.095e-03 |
| GO:BP | GO:0031503 | protein-containing complex localization | 70 | 215 | 7.343e-03 |
| GO:BP | GO:0072657 | protein localization to membrane | 227 | 827 | 7.351e-03 |
| GO:BP | GO:0043473 | pigmentation | 42 | 115 | 7.376e-03 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 11 | 18 | 7.471e-03 |
| GO:BP | GO:0007032 | endosome organization | 37 | 98 | 7.471e-03 |
| GO:BP | GO:2000641 | regulation of early endosome to late endosome transport | 11 | 18 | 7.471e-03 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 50 | 143 | 7.471e-03 |
| GO:BP | GO:0001522 | pseudouridine synthesis | 11 | 18 | 7.471e-03 |
| GO:BP | GO:0097494 | regulation of vesicle size | 11 | 18 | 7.471e-03 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 37 | 98 | 7.471e-03 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 130 | 443 | 7.595e-03 |
| GO:BP | GO:0046777 | protein autophosphorylation | 60 | 179 | 7.738e-03 |
| GO:BP | GO:0019081 | viral translation | 14 | 26 | 7.763e-03 |
| GO:BP | GO:0070198 | protein localization to chromosome, telomeric region | 14 | 26 | 7.763e-03 |
| GO:BP | GO:0007549 | sex-chromosome dosage compensation | 14 | 26 | 7.763e-03 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 72 | 223 | 7.898e-03 |
| GO:BP | GO:0031032 | actomyosin structure organization | 69 | 212 | 7.934e-03 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 51 | 147 | 7.974e-03 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 51 | 147 | 7.974e-03 |
| GO:BP | GO:0042455 | ribonucleoside biosynthetic process | 8 | 11 | 8.519e-03 |
| GO:BP | GO:0046129 | purine ribonucleoside biosynthetic process | 8 | 11 | 8.519e-03 |
| GO:BP | GO:0042451 | purine nucleoside biosynthetic process | 8 | 11 | 8.519e-03 |
| GO:BP | GO:0090140 | regulation of mitochondrial fission | 15 | 29 | 8.565e-03 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 15 | 29 | 8.565e-03 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 52 | 151 | 8.567e-03 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 32 | 82 | 8.740e-03 |
| GO:BP | GO:0032200 | telomere organization | 63 | 191 | 9.040e-03 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 28 | 69 | 9.053e-03 |
| GO:BP | GO:0051301 | cell division | 184 | 658 | 9.080e-03 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 92 | 299 | 9.080e-03 |
| GO:BP | GO:0045088 | regulation of innate immune response | 131 | 449 | 9.098e-03 |
| GO:BP | GO:0035601 | protein deacylation | 22 | 50 | 9.205e-03 |
| GO:BP | GO:0098727 | maintenance of cell number | 61 | 184 | 9.366e-03 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 31 | 79 | 9.366e-03 |
| GO:BP | GO:0106070 | regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 12 | 21 | 9.373e-03 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 21 | 47 | 9.432e-03 |
| GO:BP | GO:0043045 | epigenetic programming of gene expression | 17 | 35 | 9.452e-03 |
| GO:BP | GO:0006516 | glycoprotein catabolic process | 18 | 38 | 9.624e-03 |
| GO:BP | GO:0006413 | translational initiation | 45 | 127 | 9.624e-03 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 18 | 38 | 9.624e-03 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 19 | 41 | 9.653e-03 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 30 | 76 | 1.002e-02 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 33 | 86 | 1.004e-02 |
| GO:BP | GO:0030200 | heparan sulfate proteoglycan catabolic process | 7 | 9 | 1.004e-02 |
| GO:BP | GO:0015886 | heme transport | 7 | 9 | 1.004e-02 |
| GO:BP | GO:1990481 | mRNA pseudouridine synthesis | 7 | 9 | 1.004e-02 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 92 | 300 | 1.004e-02 |
| GO:BP | GO:0030167 | proteoglycan catabolic process | 10 | 16 | 1.009e-02 |
| GO:BP | GO:0044849 | estrous cycle | 10 | 16 | 1.009e-02 |
| GO:BP | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining | 10 | 16 | 1.009e-02 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 94 | 308 | 1.044e-02 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 35 | 93 | 1.055e-02 |
| GO:BP | GO:0003281 | ventricular septum development | 29 | 73 | 1.067e-02 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 13 | 24 | 1.086e-02 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 82 | 263 | 1.088e-02 |
| GO:BP | GO:0007409 | axonogenesis | 134 | 463 | 1.096e-02 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 125 | 428 | 1.108e-02 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 24 | 57 | 1.118e-02 |
| GO:BP | GO:0048511 | rhythmic process | 93 | 305 | 1.134e-02 |
| GO:BP | GO:0043393 | regulation of protein binding | 41 | 114 | 1.134e-02 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 34 | 90 | 1.140e-02 |
| GO:BP | GO:0045444 | fat cell differentiation | 76 | 241 | 1.148e-02 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 23 | 54 | 1.171e-02 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 73 | 230 | 1.174e-02 |
| GO:BP | GO:0042594 | response to starvation | 70 | 219 | 1.197e-02 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 14 | 27 | 1.198e-02 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 22 | 51 | 1.212e-02 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 22 | 51 | 1.212e-02 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 61 | 186 | 1.212e-02 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 22 | 51 | 1.212e-02 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 22 | 51 | 1.212e-02 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 27 | 67 | 1.212e-02 |
| GO:BP | GO:0044091 | membrane biogenesis | 27 | 67 | 1.212e-02 |
| GO:BP | GO:1905037 | autophagosome organization | 44 | 125 | 1.230e-02 |
| GO:BP | GO:1903533 | regulation of protein targeting | 30 | 77 | 1.239e-02 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 168 | 599 | 1.249e-02 |
| GO:BP | GO:0042908 | xenobiotic transport | 21 | 48 | 1.257e-02 |
| GO:BP | GO:0051960 | regulation of nervous system development | 132 | 457 | 1.264e-02 |
| GO:BP | GO:1902600 | proton transmembrane transport | 62 | 190 | 1.266e-02 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 35 | 94 | 1.275e-02 |
| GO:BP | GO:0048524 | positive regulation of viral process | 26 | 64 | 1.281e-02 |
| GO:BP | GO:0045064 | T-helper 2 cell differentiation | 11 | 19 | 1.289e-02 |
| GO:BP | GO:0031623 | receptor internalization | 47 | 136 | 1.306e-02 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 158 | 560 | 1.309e-02 |
| GO:BP | GO:0045048 | protein insertion into ER membrane | 16 | 33 | 1.309e-02 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 158 | 560 | 1.309e-02 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 77 | 246 | 1.311e-02 |
| GO:BP | GO:1990776 | response to angiotensin | 19 | 42 | 1.313e-02 |
| GO:BP | GO:0006901 | vesicle coating | 18 | 39 | 1.319e-02 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 29 | 74 | 1.319e-02 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 18 | 39 | 1.319e-02 |
| GO:BP | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 18 | 39 | 1.319e-02 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 18 | 39 | 1.319e-02 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 17 | 36 | 1.319e-02 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 18 | 39 | 1.319e-02 |
| GO:BP | GO:0035358 | regulation of peroxisome proliferator activated receptor signaling pathway | 9 | 14 | 1.343e-02 |
| GO:BP | GO:0007018 | microtubule-based movement | 137 | 478 | 1.400e-02 |
| GO:BP | GO:0051098 | regulation of binding | 68 | 213 | 1.400e-02 |
| GO:BP | GO:0030032 | lamellipodium assembly | 28 | 71 | 1.412e-02 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 90 | 296 | 1.430e-02 |
| GO:BP | GO:0060840 | artery development | 38 | 105 | 1.438e-02 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 51 | 151 | 1.438e-02 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 42 | 119 | 1.445e-02 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 40 | 112 | 1.445e-02 |
| GO:BP | GO:0009416 | response to light stimulus | 97 | 323 | 1.460e-02 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 49 | 144 | 1.477e-02 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 82 | 266 | 1.482e-02 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 23 | 55 | 1.501e-02 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 23 | 55 | 1.501e-02 |
| GO:BP | GO:0061157 | mRNA destabilization | 37 | 102 | 1.565e-02 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 50 | 148 | 1.573e-02 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 22 | 52 | 1.580e-02 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 22 | 52 | 1.580e-02 |
| GO:BP | GO:0010212 | response to ionizing radiation | 48 | 141 | 1.615e-02 |
| GO:BP | GO:0034605 | cellular response to heat | 26 | 65 | 1.615e-02 |
| GO:BP | GO:0042060 | wound healing | 125 | 433 | 1.659e-02 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 13 | 25 | 1.665e-02 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 44 | 127 | 1.686e-02 |
| GO:BP | GO:0043414 | macromolecule methylation | 44 | 127 | 1.686e-02 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 42 | 120 | 1.707e-02 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 197 | 720 | 1.707e-02 |
| GO:BP | GO:0050779 | RNA destabilization | 38 | 106 | 1.715e-02 |
| GO:BP | GO:0007020 | microtubule nucleation | 20 | 46 | 1.715e-02 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 49 | 145 | 1.715e-02 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 152 | 540 | 1.715e-02 |
| GO:BP | GO:0019318 | hexose metabolic process | 71 | 226 | 1.729e-02 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 230 | 854 | 1.741e-02 |
| GO:BP | GO:0070085 | glycosylation | 76 | 245 | 1.756e-02 |
| GO:BP | GO:0047497 | mitochondrion transport along microtubule | 14 | 28 | 1.756e-02 |
| GO:BP | GO:0046112 | nucleobase biosynthetic process | 10 | 17 | 1.756e-02 |
| GO:BP | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining | 14 | 28 | 1.756e-02 |
| GO:BP | GO:0044790 | suppression of viral release by host | 10 | 17 | 1.756e-02 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 19 | 43 | 1.756e-02 |
| GO:BP | GO:0036508 | protein alpha-1,2-demannosylation | 10 | 17 | 1.756e-02 |
| GO:BP | GO:0036507 | protein demannosylation | 10 | 17 | 1.756e-02 |
| GO:BP | GO:0035672 | oligopeptide transmembrane transport | 10 | 17 | 1.756e-02 |
| GO:BP | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated | 14 | 28 | 1.756e-02 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 28 | 72 | 1.756e-02 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 14 | 28 | 1.756e-02 |
| GO:BP | GO:0001503 | ossification | 124 | 430 | 1.756e-02 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 72 | 230 | 1.756e-02 |
| GO:BP | GO:0045628 | regulation of T-helper 2 cell differentiation | 8 | 12 | 1.763e-02 |
| GO:BP | GO:0031293 | membrane protein intracellular domain proteolysis | 8 | 12 | 1.763e-02 |
| GO:BP | GO:0010172 | embryonic body morphogenesis | 8 | 12 | 1.763e-02 |
| GO:BP | GO:0050847 | progesterone receptor signaling pathway | 8 | 12 | 1.763e-02 |
| GO:BP | GO:0060606 | tube closure | 33 | 89 | 1.764e-02 |
| GO:BP | GO:0043543 | protein acylation | 33 | 89 | 1.764e-02 |
| GO:BP | GO:1903900 | regulation of viral life cycle | 45 | 131 | 1.764e-02 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 50 | 149 | 1.776e-02 |
| GO:BP | GO:0022411 | cellular component disassembly | 128 | 446 | 1.786e-02 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 73 | 234 | 1.790e-02 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 73 | 234 | 1.790e-02 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 16 | 34 | 1.806e-02 |
| GO:BP | GO:0051654 | establishment of mitochondrion localization | 16 | 34 | 1.806e-02 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 16 | 34 | 1.806e-02 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 37 | 103 | 1.819e-02 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 30 | 79 | 1.825e-02 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 243 | 909 | 1.914e-02 |
| GO:BP | GO:0003013 | circulatory system process | 167 | 602 | 1.965e-02 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 77 | 250 | 1.988e-02 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 38 | 107 | 2.000e-02 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 47 | 139 | 2.002e-02 |
| GO:BP | GO:0008033 | tRNA processing | 47 | 139 | 2.002e-02 |
| GO:BP | GO:1990089 | response to nerve growth factor | 22 | 53 | 2.011e-02 |
| GO:BP | GO:0030097 | hemopoiesis | 259 | 976 | 2.090e-02 |
| GO:BP | GO:0042221 | response to chemical | 953 | 3909 | 2.112e-02 |
| GO:BP | GO:0044788 | modulation by host of viral process | 21 | 50 | 2.127e-02 |
| GO:BP | GO:0002682 | regulation of immune system process | 405 | 1581 | 2.127e-02 |
| GO:BP | GO:0006082 | organic acid metabolic process | 244 | 915 | 2.128e-02 |
| GO:BP | GO:0042692 | muscle cell differentiation | 117 | 405 | 2.128e-02 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 28 | 73 | 2.138e-02 |
| GO:BP | GO:0000045 | autophagosome assembly | 41 | 118 | 2.138e-02 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 41 | 118 | 2.138e-02 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 100 | 339 | 2.160e-02 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 100 | 339 | 2.160e-02 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 138 | 488 | 2.169e-02 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 64 | 202 | 2.173e-02 |
| GO:BP | GO:0048878 | chemical homeostasis | 275 | 1043 | 2.227e-02 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 30 | 80 | 2.243e-02 |
| GO:BP | GO:0030100 | regulation of endocytosis | 89 | 297 | 2.243e-02 |
| GO:BP | GO:0007051 | spindle organization | 65 | 206 | 2.243e-02 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 52 | 158 | 2.255e-02 |
| GO:BP | GO:0032401 | establishment of melanosome localization | 12 | 23 | 2.262e-02 |
| GO:BP | GO:0060052 | neurofilament cytoskeleton organization | 7 | 10 | 2.262e-02 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 7 | 10 | 2.262e-02 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 42 | 122 | 2.262e-02 |
| GO:BP | GO:1904667 | negative regulation of ubiquitin protein ligase activity | 7 | 10 | 2.262e-02 |
| GO:BP | GO:0043124 | negative regulation of canonical NF-kappaB signal transduction | 24 | 60 | 2.262e-02 |
| GO:BP | GO:0180012 | co-transcriptional RNA 3’-end processing, cleavage and polyadenylation pathway | 7 | 10 | 2.262e-02 |
| GO:BP | GO:0006104 | succinyl-CoA metabolic process | 7 | 10 | 2.262e-02 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 7 | 10 | 2.262e-02 |
| GO:BP | GO:0000963 | mitochondrial RNA processing | 12 | 23 | 2.262e-02 |
| GO:BP | GO:0048194 | Golgi vesicle budding | 7 | 10 | 2.262e-02 |
| GO:BP | GO:0035443 | tripeptide transmembrane transport | 7 | 10 | 2.262e-02 |
| GO:BP | GO:1902513 | regulation of organelle transport along microtubule | 7 | 10 | 2.262e-02 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 214 | 794 | 2.262e-02 |
| GO:BP | GO:0010907 | positive regulation of glucose metabolic process | 19 | 44 | 2.286e-02 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 92 | 309 | 2.315e-02 |
| GO:BP | GO:0030641 | regulation of cellular pH | 34 | 94 | 2.319e-02 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 38 | 108 | 2.321e-02 |
| GO:BP | GO:0003279 | cardiac septum development | 38 | 108 | 2.321e-02 |
| GO:BP | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 9 | 15 | 2.373e-02 |
| GO:BP | GO:0061684 | chaperone-mediated autophagy | 9 | 15 | 2.373e-02 |
| GO:BP | GO:0106057 | negative regulation of calcineurin-mediated signaling | 9 | 15 | 2.373e-02 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 9 | 15 | 2.373e-02 |
| GO:BP | GO:0106072 | negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway | 9 | 15 | 2.373e-02 |
| GO:BP | GO:0061614 | miRNA transcription | 29 | 77 | 2.383e-02 |
| GO:BP | GO:0009267 | cellular response to starvation | 57 | 177 | 2.409e-02 |
| GO:BP | GO:0060765 | regulation of androgen receptor signaling pathway | 14 | 29 | 2.468e-02 |
| GO:BP | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 14 | 29 | 2.468e-02 |
| GO:BP | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 14 | 29 | 2.468e-02 |
| GO:BP | GO:0006970 | response to osmotic stress | 31 | 84 | 2.468e-02 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 16 | 35 | 2.475e-02 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 33 | 91 | 2.511e-02 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 33 | 91 | 2.511e-02 |
| GO:BP | GO:0006486 | protein glycosylation | 70 | 226 | 2.526e-02 |
| GO:BP | GO:0044782 | cilium organization | 123 | 431 | 2.526e-02 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 70 | 226 | 2.526e-02 |
| GO:BP | GO:0009743 | response to carbohydrate | 75 | 245 | 2.546e-02 |
| GO:BP | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway | 56 | 174 | 2.625e-02 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 42 | 123 | 2.625e-02 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 50 | 152 | 2.643e-02 |
| GO:BP | GO:1901342 | regulation of vasculature development | 89 | 299 | 2.662e-02 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 236 | 887 | 2.675e-02 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 210 | 781 | 2.680e-02 |
| GO:BP | GO:0002064 | epithelial cell development | 68 | 219 | 2.680e-02 |
| GO:BP | GO:0001843 | neural tube closure | 32 | 88 | 2.730e-02 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 34 | 95 | 2.755e-02 |
| GO:BP | GO:0016358 | dendrite development | 74 | 242 | 2.771e-02 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 129 | 456 | 2.786e-02 |
| GO:BP | GO:0009605 | response to external stimulus | 601 | 2415 | 2.794e-02 |
| GO:BP | GO:0006695 | cholesterol biosynthetic process | 24 | 61 | 2.799e-02 |
| GO:BP | GO:1902653 | secondary alcohol biosynthetic process | 24 | 61 | 2.799e-02 |
| GO:BP | GO:0071478 | cellular response to radiation | 58 | 182 | 2.806e-02 |
| GO:BP | GO:0061724 | lipophagy | 6 | 8 | 2.806e-02 |
| GO:BP | GO:0051607 | defense response to virus | 94 | 319 | 2.806e-02 |
| GO:BP | GO:2000643 | positive regulation of early endosome to late endosome transport | 6 | 8 | 2.806e-02 |
| GO:BP | GO:0099173 | postsynapse organization | 75 | 246 | 2.806e-02 |
| GO:BP | GO:0034775 | glutathione transmembrane transport | 6 | 8 | 2.806e-02 |
| GO:BP | GO:0070973 | protein localization to endoplasmic reticulum exit site | 6 | 8 | 2.806e-02 |
| GO:BP | GO:0006302 | double-strand break repair | 94 | 319 | 2.806e-02 |
| GO:BP | GO:0034331 | cell junction maintenance | 20 | 48 | 2.808e-02 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 20 | 48 | 2.808e-02 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 20 | 48 | 2.808e-02 |
| GO:BP | GO:0019896 | axonal transport of mitochondrion | 10 | 18 | 2.814e-02 |
| GO:BP | GO:0003158 | endothelium development | 46 | 138 | 2.815e-02 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 46 | 138 | 2.815e-02 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 55 | 171 | 2.821e-02 |
| GO:BP | GO:0009611 | response to wounding | 157 | 568 | 2.828e-02 |
| GO:BP | GO:0043489 | RNA stabilization | 29 | 78 | 2.856e-02 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 39 | 113 | 2.884e-02 |
| GO:BP | GO:0043200 | response to amino acid | 44 | 131 | 2.913e-02 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 37 | 106 | 2.931e-02 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 19 | 45 | 2.942e-02 |
| GO:BP | GO:0030209 | dermatan sulfate proteoglycan catabolic process | 4 | 4 | 2.942e-02 |
| GO:BP | GO:1990180 | mitochondrial tRNA 3’-end processing | 4 | 4 | 2.942e-02 |
| GO:BP | GO:0009791 | post-embryonic development | 33 | 92 | 2.942e-02 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 26 | 68 | 2.942e-02 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 33 | 92 | 2.942e-02 |
| GO:BP | GO:0035329 | hippo signaling | 23 | 58 | 2.942e-02 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 26 | 68 | 2.942e-02 |
| GO:BP | GO:0090148 | membrane fission | 19 | 45 | 2.942e-02 |
| GO:BP | GO:0090261 | positive regulation of inclusion body assembly | 4 | 4 | 2.942e-02 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 73 | 239 | 2.942e-02 |
| GO:BP | GO:0000964 | mitochondrial RNA 5’-end processing | 4 | 4 | 2.942e-02 |
| GO:BP | GO:0035196 | miRNA processing | 19 | 45 | 2.942e-02 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 28 | 75 | 3.078e-02 |
| GO:BP | GO:0003170 | heart valve development | 28 | 75 | 3.078e-02 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 48 | 146 | 3.080e-02 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 51 | 157 | 3.080e-02 |
| GO:BP | GO:0044321 | response to leptin | 11 | 21 | 3.104e-02 |
| GO:BP | GO:0008361 | regulation of cell size | 58 | 183 | 3.106e-02 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 22 | 55 | 3.142e-02 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 22 | 55 | 3.142e-02 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 34 | 96 | 3.151e-02 |
| GO:BP | GO:2000354 | regulation of ovarian follicle development | 5 | 6 | 3.151e-02 |
| GO:BP | GO:1901387 | positive regulation of voltage-gated calcium channel activity | 5 | 6 | 3.151e-02 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 17 | 39 | 3.151e-02 |
| GO:BP | GO:0048200 | Golgi transport vesicle coating | 5 | 6 | 3.151e-02 |
| GO:BP | GO:0036295 | cellular response to increased oxygen levels | 8 | 13 | 3.151e-02 |
| GO:BP | GO:0031347 | regulation of defense response | 216 | 809 | 3.151e-02 |
| GO:BP | GO:0048205 | COPI coating of Golgi vesicle | 5 | 6 | 3.151e-02 |
| GO:BP | GO:0061795 | Golgi lumen acidification | 8 | 13 | 3.151e-02 |
| GO:BP | GO:0090646 | mitochondrial tRNA processing | 8 | 13 | 3.151e-02 |
| GO:BP | GO:0051276 | chromosome organization | 158 | 574 | 3.151e-02 |
| GO:BP | GO:1902525 | regulation of protein monoubiquitination | 5 | 6 | 3.151e-02 |
| GO:BP | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission | 5 | 6 | 3.151e-02 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 5 | 6 | 3.151e-02 |
| GO:BP | GO:0032486 | Rap protein signal transduction | 8 | 13 | 3.151e-02 |
| GO:BP | GO:1904386 | response to L-phenylalanine derivative | 5 | 6 | 3.151e-02 |
| GO:BP | GO:0098935 | dendritic transport | 8 | 13 | 3.151e-02 |
| GO:BP | GO:0035964 | COPI-coated vesicle budding | 5 | 6 | 3.151e-02 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 5 | 6 | 3.151e-02 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 143 | 514 | 3.158e-02 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 12 | 24 | 3.238e-02 |
| GO:BP | GO:0051905 | establishment of pigment granule localization | 12 | 24 | 3.238e-02 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 12 | 24 | 3.238e-02 |
| GO:BP | GO:0097066 | response to thyroid hormone | 12 | 24 | 3.238e-02 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 16 | 36 | 3.238e-02 |
| GO:BP | GO:0016485 | protein processing | 69 | 225 | 3.267e-02 |
| GO:BP | GO:0002090 | regulation of receptor internalization | 27 | 72 | 3.267e-02 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 53 | 165 | 3.267e-02 |
| GO:BP | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 15 | 33 | 3.310e-02 |
| GO:BP | GO:0048199 | vesicle targeting, to, from or within Golgi | 15 | 33 | 3.310e-02 |
| GO:BP | GO:0006929 | substrate-dependent cell migration | 13 | 27 | 3.316e-02 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 13 | 27 | 3.316e-02 |
| GO:BP | GO:0048771 | tissue remodeling | 57 | 180 | 3.316e-02 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 75 | 248 | 3.316e-02 |
| GO:BP | GO:1901654 | response to ketone | 70 | 229 | 3.322e-02 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 70 | 229 | 3.322e-02 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 14 | 30 | 3.329e-02 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 29 | 79 | 3.344e-02 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 251 | 954 | 3.371e-02 |
| GO:BP | GO:0006006 | glucose metabolic process | 58 | 184 | 3.413e-02 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 45 | 136 | 3.415e-02 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 45 | 136 | 3.415e-02 |
| GO:BP | GO:0016126 | sterol biosynthetic process | 26 | 69 | 3.527e-02 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 43 | 129 | 3.559e-02 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 49 | 151 | 3.592e-02 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 137 | 492 | 3.592e-02 |
| GO:BP | GO:0060612 | adipose tissue development | 23 | 59 | 3.595e-02 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 207 | 775 | 3.615e-02 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 207 | 775 | 3.615e-02 |
| GO:BP | GO:0051865 | protein autoubiquitination | 28 | 76 | 3.634e-02 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 28 | 76 | 3.634e-02 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 190 | 706 | 3.661e-02 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 19 | 46 | 3.685e-02 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 19 | 46 | 3.685e-02 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 19 | 46 | 3.685e-02 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 19 | 46 | 3.685e-02 |
| GO:BP | GO:0048525 | negative regulation of viral process | 30 | 83 | 3.685e-02 |
| GO:BP | GO:0033627 | cell adhesion mediated by integrin | 30 | 83 | 3.685e-02 |
| GO:BP | GO:0051453 | regulation of intracellular pH | 32 | 90 | 3.690e-02 |
| GO:BP | GO:0045216 | cell-cell junction organization | 66 | 215 | 3.743e-02 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 77 | 257 | 3.746e-02 |
| GO:BP | GO:0061450 | trophoblast cell migration | 9 | 16 | 3.794e-02 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 9 | 16 | 3.794e-02 |
| GO:BP | GO:0038083 | peptidyl-tyrosine autophosphorylation | 9 | 16 | 3.794e-02 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 9 | 16 | 3.794e-02 |
| GO:BP | GO:0045651 | positive regulation of macrophage differentiation | 9 | 16 | 3.794e-02 |
| GO:BP | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 9 | 16 | 3.794e-02 |
| GO:BP | GO:0030033 | microvillus assembly | 9 | 16 | 3.794e-02 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 142 | 513 | 3.800e-02 |
| GO:BP | GO:0023061 | signal release | 139 | 501 | 3.800e-02 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 107 | 374 | 3.804e-02 |
| GO:BP | GO:0006473 | protein acetylation | 22 | 56 | 3.816e-02 |
| GO:BP | GO:0022900 | electron transport chain | 42 | 126 | 3.835e-02 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 18 | 43 | 3.866e-02 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 18 | 43 | 3.866e-02 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 35 | 101 | 3.922e-02 |
| GO:BP | GO:0090398 | cellular senescence | 35 | 101 | 3.922e-02 |
| GO:BP | GO:1902414 | protein localization to cell junction | 40 | 119 | 3.965e-02 |
| GO:BP | GO:0039531 | regulation of cytoplasmic pattern recognition receptor signaling pathway | 40 | 119 | 3.965e-02 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 40 | 119 | 3.965e-02 |
| GO:BP | GO:0014020 | primary neural tube formation | 33 | 94 | 3.970e-02 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 76 | 254 | 4.030e-02 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 17 | 40 | 4.050e-02 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 24 | 63 | 4.050e-02 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 17 | 40 | 4.050e-02 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 17 | 40 | 4.050e-02 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 137 | 494 | 4.050e-02 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 24 | 63 | 4.050e-02 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 24 | 63 | 4.050e-02 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 21 | 53 | 4.071e-02 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 78 | 262 | 4.131e-02 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 86 | 293 | 4.150e-02 |
| GO:BP | GO:0090085 | regulation of protein deubiquitination | 7 | 11 | 4.168e-02 |
| GO:BP | GO:0048490 | anterograde synaptic vesicle transport | 10 | 19 | 4.168e-02 |
| GO:BP | GO:0006307 | DNA alkylation repair | 7 | 11 | 4.168e-02 |
| GO:BP | GO:0070213 | protein auto-ADP-ribosylation | 7 | 11 | 4.168e-02 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 26 | 70 | 4.168e-02 |
| GO:BP | GO:0006607 | NLS-bearing protein import into nucleus | 10 | 19 | 4.168e-02 |
| GO:BP | GO:0002158 | osteoclast proliferation | 7 | 11 | 4.168e-02 |
| GO:BP | GO:0006144 | purine nucleobase metabolic process | 10 | 19 | 4.168e-02 |
| GO:BP | GO:0045945 | positive regulation of transcription by RNA polymerase III | 10 | 19 | 4.168e-02 |
| GO:BP | GO:1905383 | protein localization to presynapse | 7 | 11 | 4.168e-02 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 47 | 145 | 4.168e-02 |
| GO:BP | GO:0034284 | response to monosaccharide | 67 | 220 | 4.168e-02 |
| GO:BP | GO:0001101 | response to acid chemical | 47 | 145 | 4.168e-02 |
| GO:BP | GO:0051580 | regulation of neurotransmitter uptake | 10 | 19 | 4.168e-02 |
| GO:BP | GO:0099517 | synaptic vesicle transport along microtubule | 10 | 19 | 4.168e-02 |
| GO:BP | GO:0016479 | negative regulation of transcription by RNA polymerase I | 7 | 11 | 4.168e-02 |
| GO:BP | GO:0099514 | synaptic vesicle cytoskeletal transport | 10 | 19 | 4.168e-02 |
| GO:BP | GO:0006857 | oligopeptide transport | 10 | 19 | 4.168e-02 |
| GO:BP | GO:0099536 | synaptic signaling | 214 | 807 | 4.197e-02 |
| GO:BP | GO:0046785 | microtubule polymerization | 34 | 98 | 4.202e-02 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 120 | 427 | 4.202e-02 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 271 | 1042 | 4.237e-02 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 142 | 515 | 4.242e-02 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 23 | 60 | 4.321e-02 |
| GO:BP | GO:0009112 | nucleobase metabolic process | 15 | 34 | 4.335e-02 |
| GO:BP | GO:0140058 | neuron projection arborization | 15 | 34 | 4.335e-02 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 60 | 194 | 4.378e-02 |
| GO:BP | GO:0060429 | epithelium development | 318 | 1238 | 4.384e-02 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 11 | 22 | 4.394e-02 |
| GO:BP | GO:0009746 | response to hexose | 65 | 213 | 4.394e-02 |
| GO:BP | GO:0060008 | Sertoli cell differentiation | 11 | 22 | 4.394e-02 |
| GO:BP | GO:0006907 | pinocytosis | 11 | 22 | 4.394e-02 |
| GO:BP | GO:0032402 | melanosome transport | 11 | 22 | 4.394e-02 |
| GO:BP | GO:0019082 | viral protein processing | 14 | 31 | 4.442e-02 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 14 | 31 | 4.442e-02 |
| GO:BP | GO:0000470 | maturation of LSU-rRNA | 12 | 25 | 4.464e-02 |
| GO:BP | GO:0061339 | establishment or maintenance of monopolar cell polarity | 12 | 25 | 4.464e-02 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 46 | 142 | 4.464e-02 |
| GO:BP | GO:0036296 | response to increased oxygen levels | 13 | 28 | 4.464e-02 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 53 | 168 | 4.464e-02 |
| GO:BP | GO:0045624 | positive regulation of T-helper cell differentiation | 12 | 25 | 4.464e-02 |
| GO:BP | GO:0060155 | platelet dense granule organization | 12 | 25 | 4.464e-02 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 13 | 28 | 4.464e-02 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 13 | 28 | 4.464e-02 |
| GO:BP | GO:0032400 | melanosome localization | 12 | 25 | 4.464e-02 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 13 | 28 | 4.464e-02 |
| GO:BP | GO:0033598 | mammary gland epithelial cell proliferation | 13 | 28 | 4.464e-02 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 12 | 25 | 4.464e-02 |
| GO:BP | GO:0035994 | response to muscle stretch | 12 | 25 | 4.464e-02 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 13 | 28 | 4.464e-02 |
| GO:BP | GO:0071396 | cellular response to lipid | 166 | 613 | 4.516e-02 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 19 | 47 | 4.523e-02 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 33 | 95 | 4.523e-02 |
| GO:BP | GO:0043038 | amino acid activation | 19 | 47 | 4.523e-02 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 27 | 74 | 4.549e-02 |
| GO:BP | GO:0045824 | negative regulation of innate immune response | 31 | 88 | 4.574e-02 |
| GO:BP | GO:0001510 | RNA methylation | 29 | 81 | 4.575e-02 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 29 | 81 | 4.575e-02 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 22 | 57 | 4.575e-02 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 68 | 225 | 4.575e-02 |
| GO:BP | GO:0051851 | host-mediated perturbation of symbiont process | 36 | 106 | 4.720e-02 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 55 | 176 | 4.720e-02 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 55 | 176 | 4.720e-02 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 123 | 441 | 4.783e-02 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 18 | 44 | 4.814e-02 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 24 | 64 | 4.814e-02 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 34 | 99 | 4.837e-02 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 56 | 180 | 4.847e-02 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 52 | 165 | 4.851e-02 |
| GO:BP | GO:0051899 | membrane depolarization | 32 | 92 | 4.929e-02 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 32 | 92 | 4.929e-02 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 26 | 71 | 4.932e-02 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 26 | 71 | 4.932e-02 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 26 | 71 | 4.932e-02 |
| GO:BP | GO:0048863 | stem cell differentiation | 73 | 245 | 4.935e-02 |
| GO:BP | GO:0003205 | cardiac chamber development | 53 | 169 | 4.985e-02 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 53 | 169 | 4.985e-02 |
| KEGG | KEGG:05132 | Salmonella infection | 111 | 249 | 1.529e-06 |
| KEGG | KEGG:04144 | Endocytosis | 107 | 248 | 1.215e-05 |
| KEGG | KEGG:04510 | Focal adhesion | 89 | 202 | 3.360e-05 |
| KEGG | KEGG:05012 | Parkinson disease | 109 | 265 | 8.427e-05 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 65 | 140 | 8.950e-05 |
| KEGG | KEGG:04142 | Lysosome | 61 | 131 | 1.195e-04 |
| KEGG | KEGG:03010 | Ribosome | 69 | 153 | 1.195e-04 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 172 | 474 | 5.852e-04 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 92 | 231 | 1.201e-03 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 70 | 168 | 1.782e-03 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 40 | 84 | 2.170e-03 |
| KEGG | KEGG:05010 | Alzheimer disease | 138 | 382 | 3.556e-03 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 39 | 84 | 4.141e-03 |
| KEGG | KEGG:05016 | Huntington disease | 113 | 305 | 4.141e-03 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 80 | 205 | 5.120e-03 |
| KEGG | KEGG:04148 | Efferocytosis | 63 | 156 | 7.401e-03 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 50 | 118 | 7.421e-03 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 32 | 68 | 9.001e-03 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 172 | 506 | 1.108e-02 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 128 | 363 | 1.108e-02 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 43 | 100 | 1.108e-02 |
| KEGG | KEGG:05020 | Prion disease | 98 | 271 | 1.669e-02 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 76 | 202 | 1.669e-02 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 22 | 44 | 1.896e-02 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 54 | 137 | 2.360e-02 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 33 | 76 | 2.861e-02 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 77 | 210 | 3.027e-02 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 48 | 121 | 3.181e-02 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 82 | 227 | 3.240e-02 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 59 | 155 | 3.240e-02 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 65 | 174 | 3.345e-02 |
| KEGG | KEGG:04668 | TNF signaling pathway | 45 | 113 | 3.418e-02 |
| KEGG | KEGG:04714 | Thermogenesis | 83 | 232 | 3.776e-02 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 23 | 50 | 4.167e-02 |
| KEGG | KEGG:05212 | Pancreatic cancer | 32 | 76 | 4.401e-02 |
# write.csv(table_motif1_GOKEGG_genes, "data/new/table_motif1_GOKEGG_genes.csv")
#GO:BP
table_motif1_genes_GOBP <- table_motif1_GOKEGG_genes %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
# saveRDS(table_motif1_genes_GOBP, "data/table_motif1_genes_GOBP.RDS")
table_motif1_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("Cormotif Motif 1 Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))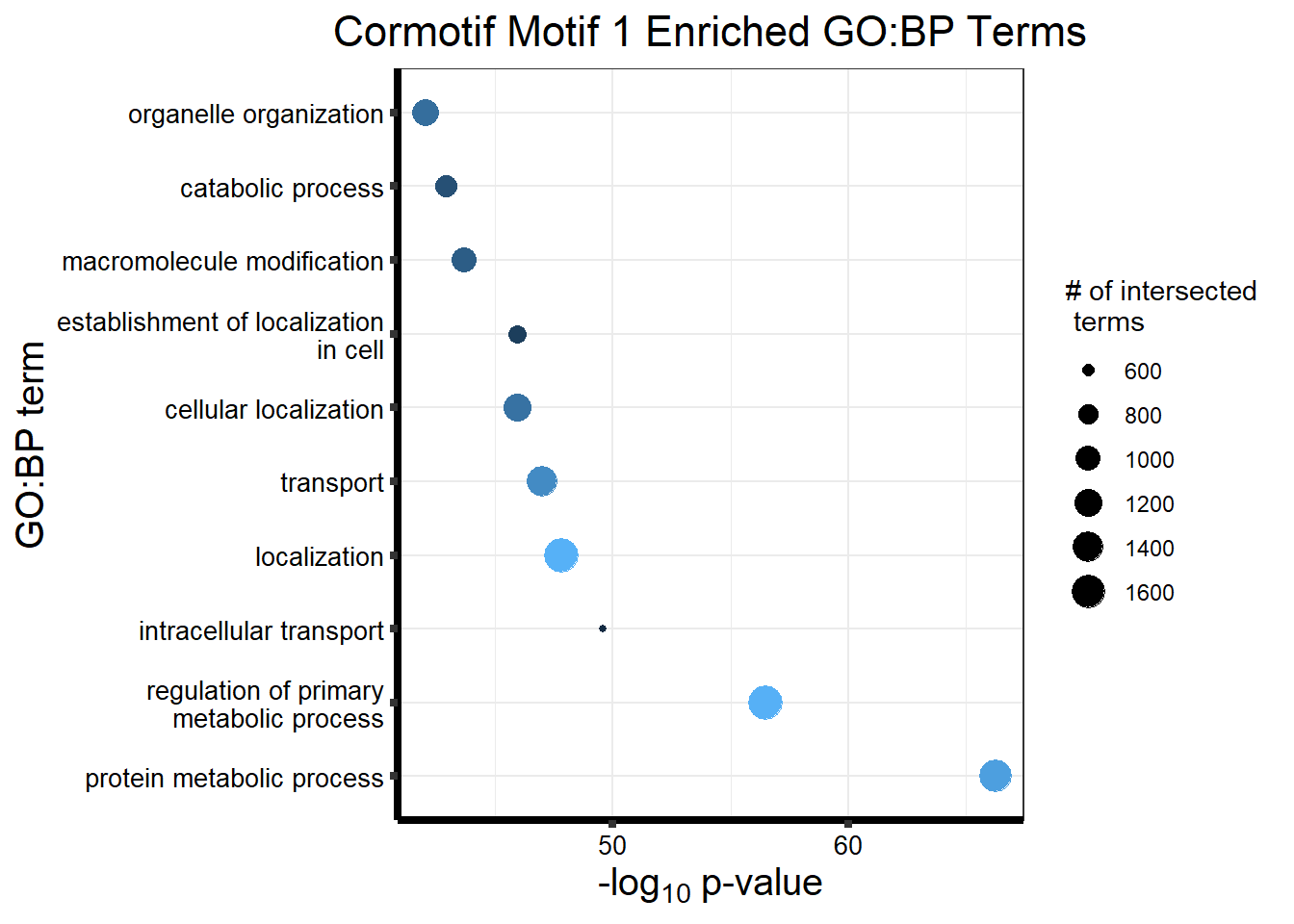
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_motif1_genes_KEGG <- table_motif1_GOKEGG_genes %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_motif1_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("Cormotif Motif 1 DEGs Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))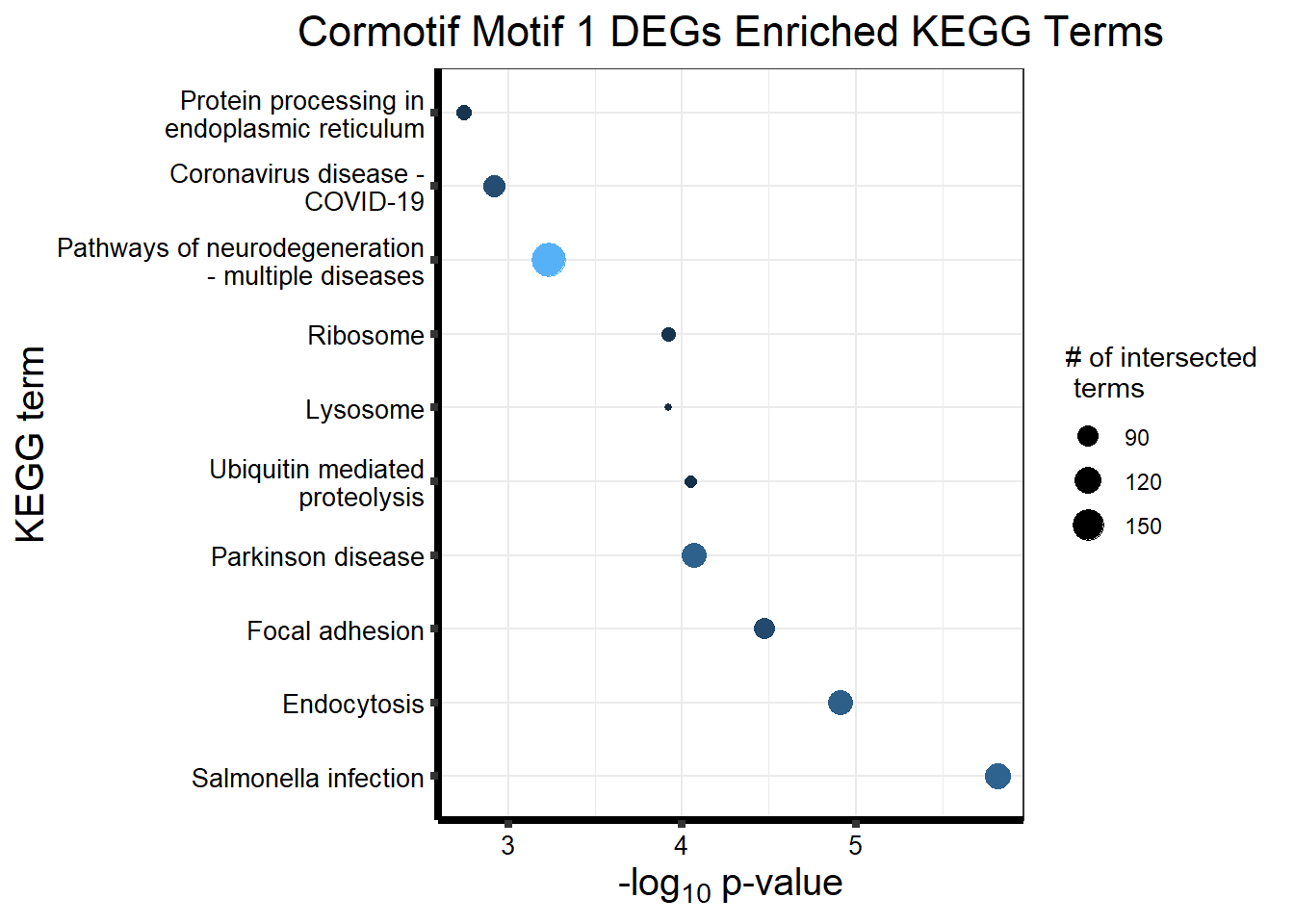
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####Motif 2 Genes#####
# motif2_genes_matrix <- as.matrix(final_genes_2)
# colnames(motif2_genes_matrix) <- c("entrezgene_ID")
# saveRDS(motif2_genes_matrix, "data/new/motif2_genes_matrix.RDS")
motif2_genes_matrix <- readRDS("data/new/motif2_genes_matrix.RDS")
length(motif2_genes_matrix)[1] 6218#6218 genes in this set for motif 2
motif2_mat_GOKEGG <- gost(query = motif2_genes_matrix,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
motif2_GOKEGG_genes <- gostplot(motif2_mat_GOKEGG, capped = FALSE, interactive = TRUE)
motif2_GOKEGG_genestable_motif2_GOKEGG_genes <- motif2_mat_GOKEGG$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_motif2_GOKEGG_genes %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0080090 | regulation of primary metabolic process | 1774 | 5390 | 3.227e-49 |
| GO:BP | GO:0019538 | protein metabolic process | 1570 | 4721 | 7.436e-45 |
| GO:BP | GO:0048518 | positive regulation of biological process | 1969 | 6264 | 1.103e-40 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 1874 | 5920 | 3.472e-40 |
| GO:BP | GO:0050789 | regulation of biological process | 3502 | 12336 | 4.738e-40 |
| GO:BP | GO:0065007 | biological regulation | 3594 | 12743 | 5.343e-39 |
| GO:BP | GO:0050794 | regulation of cellular process | 3398 | 11946 | 2.649e-38 |
| GO:BP | GO:0006996 | organelle organization | 1221 | 3594 | 1.195e-37 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 883 | 2433 | 3.931e-37 |
| GO:BP | GO:0043412 | macromolecule modification | 1047 | 3030 | 1.239e-34 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 1314 | 3990 | 1.034e-33 |
| GO:BP | GO:0007275 | multicellular organism development | 1515 | 4727 | 2.679e-33 |
| GO:BP | GO:0048856 | anatomical structure development | 1846 | 5997 | 3.191e-31 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 1217 | 3687 | 3.740e-31 |
| GO:BP | GO:0006351 | DNA-templated transcription | 1177 | 3549 | 6.798e-31 |
| GO:BP | GO:0032502 | developmental process | 1975 | 6553 | 3.668e-28 |
| GO:BP | GO:0036211 | protein modification process | 964 | 2846 | 6.886e-28 |
| GO:BP | GO:0048731 | system development | 1302 | 4053 | 6.886e-28 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 1119 | 3409 | 4.575e-27 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 1124 | 3428 | 5.033e-27 |
| GO:BP | GO:0009056 | catabolic process | 895 | 2639 | 8.883e-26 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1161 | 3597 | 4.474e-25 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 825 | 2407 | 4.595e-25 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 825 | 2410 | 6.885e-25 |
| GO:BP | GO:0019222 | regulation of metabolic process | 2071 | 7035 | 1.584e-23 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1259 | 3993 | 4.220e-23 |
| GO:BP | GO:0010646 | regulation of cell communication | 1117 | 3486 | 1.193e-22 |
| GO:BP | GO:0023051 | regulation of signaling | 1110 | 3478 | 8.795e-22 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1057 | 3288 | 1.007e-21 |
| GO:BP | GO:0009966 | regulation of signal transduction | 985 | 3034 | 1.697e-21 |
| GO:BP | GO:0033554 | cellular response to stress | 641 | 1830 | 1.914e-21 |
| GO:BP | GO:0035556 | intracellular signal transduction | 962 | 2965 | 8.139e-21 |
| GO:BP | GO:0030030 | cell projection organization | 579 | 1631 | 1.174e-20 |
| GO:BP | GO:0051179 | localization | 1663 | 5555 | 1.281e-20 |
| GO:BP | GO:0051716 | cellular response to stimulus | 2137 | 7376 | 2.177e-20 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 887 | 2711 | 3.869e-20 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 691 | 2036 | 2.503e-19 |
| GO:BP | GO:0006810 | transport | 1347 | 4407 | 3.580e-19 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 882 | 2713 | 4.127e-19 |
| GO:BP | GO:0007399 | nervous system development | 851 | 2604 | 4.771e-19 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 559 | 1588 | 5.185e-19 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 843 | 2578 | 6.143e-19 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 1895 | 6492 | 1.101e-18 |
| GO:BP | GO:0051641 | cellular localization | 1141 | 3661 | 1.275e-18 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 626 | 1835 | 6.459e-18 |
| GO:BP | GO:0033043 | regulation of organelle organization | 423 | 1148 | 6.646e-18 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 381 | 1011 | 7.423e-18 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 1660 | 5629 | 1.786e-17 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 399 | 1077 | 2.758e-17 |
| GO:BP | GO:0007507 | heart development | 250 | 605 | 6.291e-17 |
| GO:BP | GO:0022008 | neurogenesis | 600 | 1771 | 2.225e-16 |
| GO:BP | GO:0044281 | small molecule metabolic process | 601 | 1776 | 2.646e-16 |
| GO:BP | GO:0048519 | negative regulation of biological process | 1705 | 5834 | 2.646e-16 |
| GO:BP | GO:0044238 | primary metabolic process | 3350 | 12342 | 2.817e-16 |
| GO:BP | GO:0048869 | cellular developmental process | 1335 | 4438 | 3.013e-16 |
| GO:BP | GO:0030154 | cell differentiation | 1334 | 4437 | 3.781e-16 |
| GO:BP | GO:0072359 | circulatory system development | 412 | 1145 | 1.953e-15 |
| GO:BP | GO:0048699 | generation of neurons | 526 | 1536 | 3.587e-15 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 573 | 1699 | 3.778e-15 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 572 | 1697 | 4.537e-15 |
| GO:BP | GO:0009894 | regulation of catabolic process | 378 | 1040 | 7.529e-15 |
| GO:BP | GO:0033036 | macromolecule localization | 997 | 3228 | 1.274e-14 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 614 | 1855 | 1.902e-14 |
| GO:BP | GO:0040007 | growth | 342 | 929 | 3.007e-14 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 872 | 2782 | 3.257e-14 |
| GO:BP | GO:0051649 | establishment of localization in cell | 656 | 2010 | 4.283e-14 |
| GO:BP | GO:0051234 | establishment of localization | 1449 | 4928 | 4.417e-14 |
| GO:BP | GO:0046907 | intracellular transport | 475 | 1381 | 5.601e-14 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 918 | 2956 | 5.997e-14 |
| GO:BP | GO:0050896 | response to stimulus | 2498 | 8999 | 6.284e-14 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 485 | 1417 | 6.809e-14 |
| GO:BP | GO:0008104 | protein localization | 866 | 2770 | 7.996e-14 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 845 | 2696 | 9.883e-14 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 356 | 983 | 1.004e-13 |
| GO:BP | GO:0050793 | regulation of developmental process | 778 | 2457 | 1.340e-13 |
| GO:BP | GO:0032879 | regulation of localization | 658 | 2029 | 1.524e-13 |
| GO:BP | GO:0030182 | neuron differentiation | 493 | 1451 | 1.549e-13 |
| GO:BP | GO:0032501 | multicellular organismal process | 2067 | 7322 | 1.601e-13 |
| GO:BP | GO:0007010 | cytoskeleton organization | 513 | 1529 | 4.898e-13 |
| GO:BP | GO:0012501 | programmed cell death | 642 | 1987 | 8.067e-13 |
| GO:BP | GO:0008219 | cell death | 643 | 1991 | 8.384e-13 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 810 | 2593 | 9.995e-13 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 1668 | 5809 | 1.185e-12 |
| GO:BP | GO:0048468 | cell development | 887 | 2876 | 1.242e-12 |
| GO:BP | GO:0061061 | muscle structure development | 263 | 693 | 1.422e-12 |
| GO:BP | GO:0065008 | regulation of biological quality | 905 | 2947 | 1.961e-12 |
| GO:BP | GO:0031175 | neuron projection development | 362 | 1023 | 2.285e-12 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 88 | 168 | 2.661e-12 |
| GO:BP | GO:0006915 | apoptotic process | 620 | 1923 | 3.674e-12 |
| GO:BP | GO:0016049 | cell growth | 196 | 485 | 3.812e-12 |
| GO:BP | GO:0007154 | cell communication | 1851 | 6540 | 6.232e-12 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 525 | 1592 | 6.232e-12 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 384 | 1105 | 6.795e-12 |
| GO:BP | GO:0008033 | tRNA processing | 76 | 139 | 7.709e-12 |
| GO:BP | GO:0030029 | actin filament-based process | 300 | 825 | 9.511e-12 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 865 | 2819 | 9.879e-12 |
| GO:BP | GO:0022411 | cellular component disassembly | 182 | 446 | 1.035e-11 |
| GO:BP | GO:0048666 | neuron development | 404 | 1177 | 1.107e-11 |
| GO:BP | GO:0023052 | signaling | 1842 | 6515 | 1.110e-11 |
| GO:BP | GO:0006399 | tRNA metabolic process | 102 | 210 | 1.236e-11 |
| GO:BP | GO:0048513 | animal organ development | 935 | 3085 | 2.029e-11 |
| GO:BP | GO:0070925 | organelle assembly | 363 | 1046 | 3.930e-11 |
| GO:BP | GO:0061024 | membrane organization | 296 | 821 | 4.274e-11 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 259 | 701 | 6.300e-11 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1609 | 5644 | 6.478e-11 |
| GO:BP | GO:0040008 | regulation of growth | 229 | 604 | 7.040e-11 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 620 | 1953 | 8.137e-11 |
| GO:BP | GO:0009790 | embryo development | 387 | 1135 | 9.965e-11 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 2313 | 8393 | 9.965e-11 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 500 | 1529 | 1.054e-10 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 116 | 257 | 1.348e-10 |
| GO:BP | GO:0015031 | protein transport | 475 | 1444 | 1.464e-10 |
| GO:BP | GO:0042692 | muscle cell differentiation | 165 | 405 | 1.507e-10 |
| GO:BP | GO:0006259 | DNA metabolic process | 348 | 1005 | 1.519e-10 |
| GO:BP | GO:0060537 | muscle tissue development | 173 | 430 | 1.519e-10 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 283 | 786 | 1.519e-10 |
| GO:BP | GO:0032543 | mitochondrial translation | 70 | 130 | 1.605e-10 |
| GO:BP | GO:0042254 | ribosome biogenesis | 138 | 323 | 1.629e-10 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 606 | 1912 | 1.923e-10 |
| GO:BP | GO:0060047 | heart contraction | 111 | 244 | 2.034e-10 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 209 | 546 | 2.229e-10 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 200 | 519 | 3.229e-10 |
| GO:BP | GO:0016072 | rRNA metabolic process | 118 | 266 | 3.253e-10 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 238 | 643 | 3.805e-10 |
| GO:BP | GO:0009888 | tissue development | 637 | 2032 | 4.351e-10 |
| GO:BP | GO:0030163 | protein catabolic process | 353 | 1030 | 4.368e-10 |
| GO:BP | GO:0006281 | DNA repair | 231 | 621 | 4.379e-10 |
| GO:BP | GO:0006950 | response to stress | 1155 | 3948 | 4.691e-10 |
| GO:BP | GO:0006974 | DNA damage response | 317 | 908 | 4.691e-10 |
| GO:BP | GO:0007165 | signal transduction | 1693 | 6002 | 4.834e-10 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 242 | 658 | 5.056e-10 |
| GO:BP | GO:0003015 | heart process | 113 | 253 | 5.079e-10 |
| GO:BP | GO:0023057 | negative regulation of signaling | 469 | 1435 | 5.236e-10 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 469 | 1436 | 5.847e-10 |
| GO:BP | GO:0044057 | regulation of system process | 209 | 553 | 8.034e-10 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 488 | 1506 | 8.038e-10 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 537 | 1680 | 8.038e-10 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 223 | 599 | 8.657e-10 |
| GO:BP | GO:0051049 | regulation of transport | 516 | 1607 | 9.573e-10 |
| GO:BP | GO:0010468 | regulation of gene expression | 1568 | 5536 | 1.205e-09 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 234 | 638 | 1.446e-09 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 261 | 729 | 1.852e-09 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 210 | 561 | 1.890e-09 |
| GO:BP | GO:0014706 | striated muscle tissue development | 162 | 408 | 2.139e-09 |
| GO:BP | GO:0006936 | muscle contraction | 143 | 349 | 2.210e-09 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 434 | 1326 | 2.527e-09 |
| GO:BP | GO:0000902 | cell morphogenesis | 339 | 996 | 2.634e-09 |
| GO:BP | GO:0065009 | regulation of molecular function | 482 | 1496 | 2.639e-09 |
| GO:BP | GO:0001558 | regulation of cell growth | 161 | 406 | 2.674e-09 |
| GO:BP | GO:0006400 | tRNA modification | 54 | 95 | 2.707e-09 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 234 | 642 | 2.725e-09 |
| GO:BP | GO:0071705 | nitrogen compound transport | 601 | 1923 | 2.827e-09 |
| GO:BP | GO:0003012 | muscle system process | 171 | 438 | 2.850e-09 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 299 | 861 | 3.187e-09 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 80 | 165 | 3.912e-09 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 183 | 478 | 3.983e-09 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 471 | 1462 | 4.339e-09 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 226 | 619 | 4.713e-09 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 234 | 646 | 5.146e-09 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 453 | 1400 | 5.178e-09 |
| GO:BP | GO:0042592 | homeostatic process | 547 | 1736 | 5.616e-09 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 200 | 535 | 5.722e-09 |
| GO:BP | GO:0072524 | pyridine-containing compound metabolic process | 81 | 169 | 5.859e-09 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 244 | 681 | 6.688e-09 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 228 | 628 | 6.825e-09 |
| GO:BP | GO:0009987 | cellular process | 5127 | 20247 | 6.964e-09 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 245 | 686 | 8.613e-09 |
| GO:BP | GO:0048589 | developmental growth | 238 | 663 | 9.014e-09 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 241 | 673 | 9.014e-09 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 103 | 234 | 9.357e-09 |
| GO:BP | GO:0019362 | pyridine nucleotide metabolic process | 78 | 162 | 9.357e-09 |
| GO:BP | GO:0046496 | nicotinamide nucleotide metabolic process | 78 | 162 | 9.357e-09 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 410 | 1256 | 1.156e-08 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 241 | 676 | 1.425e-08 |
| GO:BP | GO:0006412 | translation | 256 | 727 | 1.574e-08 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 236 | 660 | 1.579e-08 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 217 | 597 | 1.595e-08 |
| GO:BP | GO:0006914 | autophagy | 217 | 597 | 1.595e-08 |
| GO:BP | GO:0008016 | regulation of heart contraction | 92 | 204 | 1.807e-08 |
| GO:BP | GO:0006364 | rRNA processing | 99 | 225 | 2.076e-08 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 474 | 1489 | 2.193e-08 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 706 | 2331 | 2.360e-08 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 165 | 430 | 2.450e-08 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 150 | 383 | 2.874e-08 |
| GO:BP | GO:0034330 | cell junction organization | 286 | 834 | 3.199e-08 |
| GO:BP | GO:0006941 | striated muscle contraction | 86 | 189 | 3.793e-08 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 325 | 970 | 4.020e-08 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 413 | 1280 | 4.794e-08 |
| GO:BP | GO:0099173 | postsynapse organization | 105 | 246 | 4.794e-08 |
| GO:BP | GO:0090257 | regulation of muscle system process | 100 | 231 | 4.794e-08 |
| GO:BP | GO:0043687 | post-translational protein modification | 341 | 1027 | 4.794e-08 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 214 | 594 | 4.794e-08 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 123 | 301 | 4.847e-08 |
| GO:BP | GO:0051726 | regulation of cell cycle | 358 | 1087 | 5.052e-08 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 254 | 729 | 5.325e-08 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 69 | 142 | 5.667e-08 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 417 | 1296 | 5.719e-08 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 172 | 458 | 6.609e-08 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 204 | 563 | 6.633e-08 |
| GO:BP | GO:0050808 | synapse organization | 204 | 563 | 6.633e-08 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 190 | 517 | 6.801e-08 |
| GO:BP | GO:0006629 | lipid metabolic process | 443 | 1391 | 7.351e-08 |
| GO:BP | GO:0016310 | phosphorylation | 423 | 1320 | 7.451e-08 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 52 | 97 | 8.041e-08 |
| GO:BP | GO:0006302 | double-strand break repair | 128 | 319 | 8.240e-08 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 151 | 392 | 8.785e-08 |
| GO:BP | GO:0055001 | muscle cell development | 88 | 198 | 9.288e-08 |
| GO:BP | GO:0043414 | macromolecule methylation | 63 | 127 | 9.917e-08 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 197 | 543 | 1.089e-07 |
| GO:BP | GO:0006508 | proteolysis | 468 | 1486 | 1.254e-07 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 394 | 1223 | 1.354e-07 |
| GO:BP | GO:0080134 | regulation of response to stress | 440 | 1387 | 1.407e-07 |
| GO:BP | GO:0051604 | protein maturation | 195 | 539 | 1.668e-07 |
| GO:BP | GO:0045184 | establishment of protein localization | 591 | 1937 | 2.055e-07 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 492 | 1578 | 2.129e-07 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 180 | 493 | 3.037e-07 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 385 | 1199 | 3.115e-07 |
| GO:BP | GO:0043009 | chordate embryonic development | 233 | 671 | 3.333e-07 |
| GO:BP | GO:0060341 | regulation of cellular localization | 332 | 1013 | 3.398e-07 |
| GO:BP | GO:0006325 | chromatin organization | 295 | 884 | 3.398e-07 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 286 | 853 | 3.460e-07 |
| GO:BP | GO:0048729 | tissue morphogenesis | 216 | 614 | 3.497e-07 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 321 | 975 | 3.548e-07 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 339 | 1039 | 3.937e-07 |
| GO:BP | GO:0055002 | striated muscle cell development | 77 | 171 | 4.309e-07 |
| GO:BP | GO:0009451 | RNA modification | 78 | 174 | 4.402e-07 |
| GO:BP | GO:0030031 | cell projection assembly | 215 | 613 | 4.913e-07 |
| GO:BP | GO:0016043 | cellular component organization | 2216 | 8184 | 4.969e-07 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 209 | 593 | 5.023e-07 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 549 | 1795 | 5.073e-07 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 238 | 692 | 5.599e-07 |
| GO:BP | GO:0006886 | intracellular protein transport | 236 | 686 | 6.260e-07 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 120 | 304 | 7.139e-07 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 120 | 304 | 7.139e-07 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 294 | 887 | 7.139e-07 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 134 | 349 | 7.927e-07 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 210 | 600 | 8.444e-07 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 154 | 414 | 8.444e-07 |
| GO:BP | GO:0007049 | cell cycle | 511 | 1663 | 8.507e-07 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 180 | 500 | 9.001e-07 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 73 | 162 | 9.353e-07 |
| GO:BP | GO:0023056 | positive regulation of signaling | 547 | 1796 | 9.843e-07 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 121 | 309 | 1.030e-06 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 61 | 128 | 1.055e-06 |
| GO:BP | GO:0001701 | in utero embryonic development | 153 | 413 | 1.244e-06 |
| GO:BP | GO:0003007 | heart morphogenesis | 107 | 266 | 1.268e-06 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 163 | 446 | 1.278e-06 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 421 | 1343 | 1.337e-06 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 104 | 257 | 1.354e-06 |
| GO:BP | GO:0060485 | mesenchyme development | 124 | 320 | 1.372e-06 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 365 | 1144 | 1.488e-06 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 276 | 831 | 1.507e-06 |
| GO:BP | GO:0006082 | organic acid metabolic process | 300 | 915 | 1.550e-06 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 41 | 75 | 1.660e-06 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 80 | 185 | 1.789e-06 |
| GO:BP | GO:0061564 | axon development | 187 | 528 | 1.789e-06 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 180 | 505 | 1.904e-06 |
| GO:BP | GO:0072526 | pyridine-containing compound catabolic process | 51 | 102 | 1.965e-06 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 652 | 2192 | 2.008e-06 |
| GO:BP | GO:0035239 | tube morphogenesis | 289 | 879 | 2.024e-06 |
| GO:BP | GO:0006282 | regulation of DNA repair | 93 | 225 | 2.085e-06 |
| GO:BP | GO:1903522 | regulation of blood circulation | 103 | 256 | 2.120e-06 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 63 | 136 | 2.224e-06 |
| GO:BP | GO:0007409 | axonogenesis | 167 | 463 | 2.283e-06 |
| GO:BP | GO:0001510 | RNA methylation | 43 | 81 | 2.345e-06 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 297 | 909 | 2.500e-06 |
| GO:BP | GO:0030239 | myofibril assembly | 41 | 76 | 2.603e-06 |
| GO:BP | GO:0032880 | regulation of protein localization | 296 | 907 | 2.954e-06 |
| GO:BP | GO:0006468 | protein phosphorylation | 386 | 1227 | 3.144e-06 |
| GO:BP | GO:0016567 | protein ubiquitination | 256 | 768 | 3.144e-06 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 175 | 492 | 3.269e-06 |
| GO:BP | GO:0019751 | polyol metabolic process | 53 | 109 | 3.369e-06 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 120 | 312 | 3.381e-06 |
| GO:BP | GO:0008152 | metabolic process | 3683 | 14136 | 3.437e-06 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 225 | 662 | 3.488e-06 |
| GO:BP | GO:0048878 | chemical homeostasis | 334 | 1043 | 3.542e-06 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 69 | 155 | 3.547e-06 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 507 | 1667 | 3.639e-06 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 482 | 1576 | 3.679e-06 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 63 | 138 | 4.086e-06 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 132 | 352 | 4.284e-06 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 100 | 250 | 4.307e-06 |
| GO:BP | GO:0031399 | regulation of protein modification process | 335 | 1049 | 4.479e-06 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 20 | 27 | 4.767e-06 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 71 | 162 | 4.814e-06 |
| GO:BP | GO:0019364 | pyridine nucleotide catabolic process | 49 | 99 | 4.866e-06 |
| GO:BP | GO:0070848 | response to growth factor | 241 | 721 | 5.546e-06 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 133 | 357 | 5.789e-06 |
| GO:BP | GO:0016358 | dendrite development | 97 | 242 | 5.789e-06 |
| GO:BP | GO:0003013 | circulatory system process | 206 | 602 | 6.588e-06 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 100 | 252 | 6.588e-06 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 98 | 246 | 6.986e-06 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 96 | 240 | 7.388e-06 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 145 | 398 | 7.684e-06 |
| GO:BP | GO:0035295 | tube development | 348 | 1101 | 7.716e-06 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 436 | 1418 | 7.740e-06 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 120 | 317 | 8.474e-06 |
| GO:BP | GO:0032790 | ribosome disassembly | 26 | 41 | 8.512e-06 |
| GO:BP | GO:0051051 | negative regulation of transport | 149 | 412 | 8.637e-06 |
| GO:BP | GO:0006457 | protein folding | 89 | 219 | 8.677e-06 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 59 | 129 | 8.839e-06 |
| GO:BP | GO:0007169 | cell surface receptor protein tyrosine kinase signaling pathway | 213 | 629 | 9.551e-06 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 101 | 257 | 9.551e-06 |
| GO:BP | GO:0048588 | developmental cell growth | 91 | 226 | 1.029e-05 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 21 | 30 | 1.029e-05 |
| GO:BP | GO:0019318 | hexose metabolic process | 91 | 226 | 1.029e-05 |
| GO:BP | GO:0006402 | mRNA catabolic process | 102 | 261 | 1.131e-05 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 183 | 528 | 1.145e-05 |
| GO:BP | GO:0030258 | lipid modification | 81 | 196 | 1.270e-05 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 306 | 957 | 1.298e-05 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 77 | 184 | 1.336e-05 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 125 | 336 | 1.367e-05 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 74 | 175 | 1.367e-05 |
| GO:BP | GO:0006414 | translational elongation | 41 | 80 | 1.416e-05 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 135 | 369 | 1.424e-05 |
| GO:BP | GO:0031032 | actomyosin structure organization | 86 | 212 | 1.433e-05 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 98 | 250 | 1.570e-05 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 60 | 134 | 1.643e-05 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 305 | 956 | 1.644e-05 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 157 | 443 | 1.644e-05 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 502 | 1669 | 1.649e-05 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 229 | 689 | 1.651e-05 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 101 | 260 | 1.711e-05 |
| GO:BP | GO:0046032 | ADP catabolic process | 47 | 97 | 1.719e-05 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 123 | 331 | 1.744e-05 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 85 | 210 | 1.809e-05 |
| GO:BP | GO:0009191 | ribonucleoside diphosphate catabolic process | 49 | 103 | 1.960e-05 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 175 | 505 | 1.960e-05 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 56 | 123 | 1.960e-05 |
| GO:BP | GO:0030488 | tRNA methylation | 25 | 40 | 1.974e-05 |
| GO:BP | GO:0050807 | regulation of synapse organization | 116 | 309 | 1.974e-05 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 187 | 546 | 1.986e-05 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 58 | 129 | 2.047e-05 |
| GO:BP | GO:0007005 | mitochondrion organization | 154 | 435 | 2.174e-05 |
| GO:BP | GO:0006096 | glycolytic process | 46 | 95 | 2.247e-05 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 119 | 320 | 2.453e-05 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 119 | 320 | 2.453e-05 |
| GO:BP | GO:0051640 | organelle localization | 208 | 620 | 2.514e-05 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 56 | 124 | 2.619e-05 |
| GO:BP | GO:0045927 | positive regulation of growth | 99 | 256 | 2.676e-05 |
| GO:BP | GO:0046031 | ADP metabolic process | 49 | 104 | 2.714e-05 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 98 | 253 | 2.766e-05 |
| GO:BP | GO:1903008 | organelle disassembly | 37 | 71 | 2.771e-05 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 137 | 380 | 2.771e-05 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 224 | 677 | 2.985e-05 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 101 | 263 | 2.985e-05 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 100 | 260 | 3.100e-05 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 128 | 351 | 3.112e-05 |
| GO:BP | GO:0060173 | limb development | 79 | 194 | 3.150e-05 |
| GO:BP | GO:0048736 | appendage development | 79 | 194 | 3.150e-05 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 206 | 616 | 3.514e-05 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 95 | 245 | 3.724e-05 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 117 | 316 | 3.726e-05 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 134 | 372 | 3.728e-05 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 62 | 143 | 3.733e-05 |
| GO:BP | GO:0072344 | rescue of stalled ribosome | 22 | 34 | 3.733e-05 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 16 | 21 | 3.945e-05 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 51 | 111 | 4.029e-05 |
| GO:BP | GO:0048638 | regulation of developmental growth | 115 | 310 | 4.029e-05 |
| GO:BP | GO:0032535 | regulation of cellular component size | 129 | 356 | 4.029e-05 |
| GO:BP | GO:0032259 | methylation | 92 | 236 | 4.065e-05 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 135 | 376 | 4.081e-05 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 118 | 320 | 4.137e-05 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 71 | 171 | 4.534e-05 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 56 | 126 | 4.564e-05 |
| GO:BP | GO:0009137 | purine nucleoside diphosphate catabolic process | 47 | 100 | 4.583e-05 |
| GO:BP | GO:0009181 | purine ribonucleoside diphosphate catabolic process | 47 | 100 | 4.583e-05 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 58 | 132 | 4.670e-05 |
| GO:BP | GO:0050890 | cognition | 120 | 328 | 5.256e-05 |
| GO:BP | GO:0006006 | glucose metabolic process | 75 | 184 | 5.312e-05 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 287 | 905 | 5.421e-05 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 104 | 276 | 5.421e-05 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 172 | 503 | 5.540e-05 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 23 | 37 | 5.693e-05 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 131 | 365 | 5.753e-05 |
| GO:BP | GO:0051261 | protein depolymerization | 54 | 121 | 5.802e-05 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 296 | 938 | 5.856e-05 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 68 | 163 | 5.901e-05 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 56 | 127 | 5.964e-05 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 46 | 98 | 5.967e-05 |
| GO:BP | GO:0016236 | macroautophagy | 130 | 362 | 5.973e-05 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 64 | 151 | 6.072e-05 |
| GO:BP | GO:0099536 | synaptic signaling | 259 | 807 | 6.096e-05 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 293 | 928 | 6.172e-05 |
| GO:BP | GO:0008015 | blood circulation | 176 | 518 | 6.426e-05 |
| GO:BP | GO:0001944 | vasculature development | 246 | 762 | 6.683e-05 |
| GO:BP | GO:0006486 | protein glycosylation | 88 | 226 | 6.717e-05 |
| GO:BP | GO:0009134 | nucleoside diphosphate catabolic process | 49 | 107 | 6.717e-05 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 88 | 226 | 6.717e-05 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 186 | 553 | 6.894e-05 |
| GO:BP | GO:0019395 | fatty acid oxidation | 50 | 110 | 6.897e-05 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 22 | 35 | 6.971e-05 |
| GO:BP | GO:0009135 | purine nucleoside diphosphate metabolic process | 52 | 116 | 7.241e-05 |
| GO:BP | GO:0009179 | purine ribonucleoside diphosphate metabolic process | 52 | 116 | 7.241e-05 |
| GO:BP | GO:0035108 | limb morphogenesis | 65 | 155 | 7.574e-05 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 65 | 155 | 7.574e-05 |
| GO:BP | GO:0035107 | appendage morphogenesis | 65 | 155 | 7.574e-05 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 84 | 214 | 7.588e-05 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 94 | 246 | 7.813e-05 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 114 | 311 | 7.836e-05 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 70 | 171 | 8.930e-05 |
| GO:BP | GO:0016311 | dephosphorylation | 85 | 218 | 9.003e-05 |
| GO:BP | GO:1901698 | response to nitrogen compound | 334 | 1080 | 9.218e-05 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 68 | 165 | 9.218e-05 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 51 | 114 | 9.403e-05 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 94 | 247 | 9.403e-05 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 98 | 260 | 9.550e-05 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 61 | 144 | 9.980e-05 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 279 | 884 | 1.051e-04 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 27 | 48 | 1.052e-04 |
| GO:BP | GO:0016050 | vesicle organization | 133 | 376 | 1.052e-04 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 323 | 1042 | 1.054e-04 |
| GO:BP | GO:0086018 | SA node cell to atrial cardiac muscle cell signaling | 9 | 9 | 1.061e-04 |
| GO:BP | GO:0086015 | SA node cell action potential | 9 | 9 | 1.061e-04 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 203 | 616 | 1.077e-04 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 90 | 235 | 1.085e-04 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 110 | 300 | 1.098e-04 |
| GO:BP | GO:0030334 | regulation of cell migration | 300 | 960 | 1.114e-04 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 200 | 606 | 1.124e-04 |
| GO:BP | GO:0019725 | cellular homeostasis | 265 | 835 | 1.124e-04 |
| GO:BP | GO:0007417 | central nervous system development | 328 | 1061 | 1.125e-04 |
| GO:BP | GO:0070085 | glycosylation | 93 | 245 | 1.151e-04 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 112 | 307 | 1.166e-04 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 133 | 377 | 1.187e-04 |
| GO:BP | GO:2000145 | regulation of cell motility | 317 | 1022 | 1.187e-04 |
| GO:BP | GO:0044782 | cilium organization | 149 | 431 | 1.187e-04 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 146 | 421 | 1.213e-04 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 52 | 118 | 1.236e-04 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 107 | 291 | 1.237e-04 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 61 | 145 | 1.237e-04 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 277 | 879 | 1.237e-04 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 218 | 670 | 1.237e-04 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 73 | 182 | 1.239e-04 |
| GO:BP | GO:0032409 | regulation of transporter activity | 73 | 182 | 1.239e-04 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 57 | 133 | 1.253e-04 |
| GO:BP | GO:0007610 | behavior | 219 | 674 | 1.293e-04 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 131 | 371 | 1.293e-04 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 34 | 67 | 1.306e-04 |
| GO:BP | GO:0040012 | regulation of locomotion | 329 | 1067 | 1.336e-04 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 67 | 164 | 1.421e-04 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 275 | 874 | 1.490e-04 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 48 | 107 | 1.512e-04 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 89 | 234 | 1.591e-04 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 261 | 825 | 1.631e-04 |
| GO:BP | GO:0141188 | nucleic acid catabolic process | 122 | 343 | 1.776e-04 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 45 | 99 | 1.900e-04 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 168 | 500 | 1.950e-04 |
| GO:BP | GO:0006310 | DNA recombination | 123 | 347 | 1.959e-04 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 248 | 781 | 2.031e-04 |
| GO:BP | GO:0006470 | protein dephosphorylation | 60 | 144 | 2.031e-04 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 119 | 334 | 2.059e-04 |
| GO:BP | GO:0051169 | nuclear transport | 119 | 334 | 2.059e-04 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 58 | 138 | 2.066e-04 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 95 | 255 | 2.107e-04 |
| GO:BP | GO:0003205 | cardiac chamber development | 68 | 169 | 2.172e-04 |
| GO:BP | GO:0006338 | chromatin remodeling | 232 | 725 | 2.175e-04 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 24 | 42 | 2.259e-04 |
| GO:BP | GO:0097352 | autophagosome maturation | 32 | 63 | 2.293e-04 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 32 | 63 | 2.293e-04 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 42 | 91 | 2.294e-04 |
| GO:BP | GO:0098657 | import into cell | 287 | 922 | 2.294e-04 |
| GO:BP | GO:0010256 | endomembrane system organization | 200 | 613 | 2.333e-04 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 122 | 345 | 2.344e-04 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 64 | 157 | 2.354e-04 |
| GO:BP | GO:0061337 | cardiac conduction | 43 | 94 | 2.366e-04 |
| GO:BP | GO:0007611 | learning or memory | 103 | 282 | 2.378e-04 |
| GO:BP | GO:0009611 | response to wounding | 187 | 568 | 2.429e-04 |
| GO:BP | GO:0048870 | cell motility | 524 | 1793 | 2.498e-04 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 165 | 492 | 2.516e-04 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 60 | 145 | 2.516e-04 |
| GO:BP | GO:0060271 | cilium assembly | 139 | 403 | 2.529e-04 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 69 | 173 | 2.568e-04 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 230 | 720 | 2.614e-04 |
| GO:BP | GO:0006979 | response to oxidative stress | 138 | 400 | 2.661e-04 |
| GO:BP | GO:0034329 | cell junction assembly | 168 | 503 | 2.697e-04 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 39 | 83 | 2.718e-04 |
| GO:BP | GO:0030199 | collagen fibril organization | 34 | 69 | 2.740e-04 |
| GO:BP | GO:0043603 | amide metabolic process | 154 | 455 | 2.768e-04 |
| GO:BP | GO:0006417 | regulation of translation | 132 | 380 | 2.769e-04 |
| GO:BP | GO:0046329 | negative regulation of JNK cascade | 21 | 35 | 2.774e-04 |
| GO:BP | GO:0099068 | postsynapse assembly | 41 | 89 | 2.948e-04 |
| GO:BP | GO:0001568 | blood vessel development | 233 | 732 | 2.948e-04 |
| GO:BP | GO:0031503 | protein-containing complex localization | 82 | 215 | 2.948e-04 |
| GO:BP | GO:0046173 | polyol biosynthetic process | 31 | 61 | 2.951e-04 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 245 | 775 | 3.020e-04 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 245 | 775 | 3.020e-04 |
| GO:BP | GO:0016477 | cell migration | 453 | 1534 | 3.315e-04 |
| GO:BP | GO:0007017 | microtubule-based process | 307 | 999 | 3.346e-04 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 47 | 107 | 3.383e-04 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 232 | 730 | 3.403e-04 |
| GO:BP | GO:0048284 | organelle fusion | 66 | 165 | 3.403e-04 |
| GO:BP | GO:0034440 | lipid oxidation | 50 | 116 | 3.428e-04 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 65 | 162 | 3.487e-04 |
| GO:BP | GO:0006403 | RNA localization | 77 | 200 | 3.545e-04 |
| GO:BP | GO:0007517 | muscle organ development | 127 | 365 | 3.555e-04 |
| GO:BP | GO:0045333 | cellular respiration | 90 | 242 | 3.557e-04 |
| GO:BP | GO:0043647 | inositol phosphate metabolic process | 24 | 43 | 3.583e-04 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 357 | 1182 | 3.583e-04 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 40 | 87 | 3.843e-04 |
| GO:BP | GO:0045214 | sarcomere organization | 27 | 51 | 3.939e-04 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 57 | 138 | 4.139e-04 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 43 | 96 | 4.181e-04 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 43 | 96 | 4.181e-04 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 55 | 132 | 4.248e-04 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 36 | 76 | 4.415e-04 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 50 | 117 | 4.420e-04 |
| GO:BP | GO:0030811 | regulation of nucleotide catabolic process | 28 | 54 | 4.523e-04 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 28 | 54 | 4.523e-04 |
| GO:BP | GO:0033121 | regulation of purine nucleotide catabolic process | 28 | 54 | 4.523e-04 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 88 | 237 | 4.587e-04 |
| GO:BP | GO:0010506 | regulation of autophagy | 127 | 367 | 4.619e-04 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 259 | 830 | 4.622e-04 |
| GO:BP | GO:0006739 | NADP metabolic process | 23 | 41 | 4.675e-04 |
| GO:BP | GO:0032868 | response to insulin | 98 | 270 | 4.697e-04 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 34 | 71 | 5.490e-04 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 175 | 534 | 5.598e-04 |
| GO:BP | GO:0022402 | cell cycle process | 382 | 1280 | 5.614e-04 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 79 | 209 | 5.684e-04 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 18 | 29 | 5.709e-04 |
| GO:BP | GO:0043062 | extracellular structure organization | 115 | 328 | 5.718e-04 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 35 | 74 | 5.788e-04 |
| GO:BP | GO:0006066 | alcohol metabolic process | 125 | 362 | 5.857e-04 |
| GO:BP | GO:0006401 | RNA catabolic process | 111 | 315 | 6.091e-04 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 329 | 1087 | 6.109e-04 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 72 | 187 | 6.186e-04 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 75 | 197 | 6.677e-04 |
| GO:BP | GO:0000278 | mitotic cell cycle | 275 | 892 | 6.687e-04 |
| GO:BP | GO:0007416 | synapse assembly | 94 | 259 | 6.687e-04 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 87 | 236 | 6.700e-04 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 79 | 210 | 6.750e-04 |
| GO:BP | GO:0060322 | head development | 251 | 806 | 6.947e-04 |
| GO:BP | GO:0002027 | regulation of heart rate | 44 | 101 | 7.376e-04 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 72 | 188 | 7.488e-04 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 60 | 150 | 7.496e-04 |
| GO:BP | GO:0008283 | cell population proliferation | 578 | 2015 | 7.566e-04 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 125 | 364 | 7.591e-04 |
| GO:BP | GO:0051168 | nuclear export | 66 | 169 | 7.634e-04 |
| GO:BP | GO:0044282 | small molecule catabolic process | 127 | 371 | 7.760e-04 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 58 | 144 | 7.861e-04 |
| GO:BP | GO:0030198 | extracellular matrix organization | 114 | 327 | 7.879e-04 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 75 | 198 | 7.923e-04 |
| GO:BP | GO:0051236 | establishment of RNA localization | 64 | 163 | 8.130e-04 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 56 | 138 | 8.205e-04 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 56 | 138 | 8.205e-04 |
| GO:BP | GO:0050657 | nucleic acid transport | 63 | 160 | 8.327e-04 |
| GO:BP | GO:0050658 | RNA transport | 63 | 160 | 8.327e-04 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 144 | 430 | 8.327e-04 |
| GO:BP | GO:0001503 | ossification | 144 | 430 | 8.327e-04 |
| GO:BP | GO:0030162 | regulation of proteolysis | 137 | 406 | 8.420e-04 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 103 | 291 | 9.018e-04 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 198 | 620 | 9.181e-04 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 194 | 606 | 9.325e-04 |
| GO:BP | GO:0001947 | heart looping | 32 | 67 | 9.384e-04 |
| GO:BP | GO:0061157 | mRNA destabilization | 44 | 102 | 9.470e-04 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 65 | 167 | 9.515e-04 |
| GO:BP | GO:0008361 | regulation of cell size | 70 | 183 | 9.578e-04 |
| GO:BP | GO:0048511 | rhythmic process | 107 | 305 | 9.731e-04 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 64 | 164 | 9.808e-04 |
| GO:BP | GO:0009303 | rRNA transcription | 22 | 40 | 9.808e-04 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 129 | 380 | 1.004e-03 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 109 | 312 | 1.005e-03 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 18 | 30 | 1.010e-03 |
| GO:BP | GO:0000725 | recombinational repair | 73 | 193 | 1.014e-03 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 114 | 329 | 1.014e-03 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 34 | 73 | 1.024e-03 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 34 | 73 | 1.024e-03 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 20 | 35 | 1.024e-03 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 20 | 35 | 1.024e-03 |
| GO:BP | GO:0030048 | actin filament-based movement | 55 | 136 | 1.024e-03 |
| GO:BP | GO:0046034 | ATP metabolic process | 84 | 229 | 1.036e-03 |
| GO:BP | GO:0007033 | vacuole organization | 87 | 239 | 1.058e-03 |
| GO:BP | GO:0006740 | NADPH regeneration | 15 | 23 | 1.086e-03 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 51 | 124 | 1.115e-03 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 167 | 513 | 1.133e-03 |
| GO:BP | GO:0007178 | cell surface receptor protein serine/threonine kinase signaling pathway | 124 | 364 | 1.139e-03 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 69 | 181 | 1.182e-03 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 172 | 531 | 1.185e-03 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 170 | 524 | 1.187e-03 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 94 | 263 | 1.193e-03 |
| GO:BP | GO:0050779 | RNA destabilization | 45 | 106 | 1.193e-03 |
| GO:BP | GO:0006664 | glycolipid metabolic process | 45 | 106 | 1.193e-03 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 57 | 143 | 1.197e-03 |
| GO:BP | GO:0006897 | endocytosis | 222 | 709 | 1.231e-03 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 167 | 514 | 1.248e-03 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 32 | 68 | 1.275e-03 |
| GO:BP | GO:0030100 | regulation of endocytosis | 104 | 297 | 1.275e-03 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 54 | 134 | 1.295e-03 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 92 | 257 | 1.309e-03 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 66 | 172 | 1.313e-03 |
| GO:BP | GO:0051258 | protein polymerization | 100 | 284 | 1.358e-03 |
| GO:BP | GO:1905037 | autophagosome organization | 51 | 125 | 1.387e-03 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 172 | 533 | 1.443e-03 |
| GO:BP | GO:0051926 | negative regulation of calcium ion transport | 29 | 60 | 1.499e-03 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 37 | 83 | 1.501e-03 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 89 | 248 | 1.516e-03 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 22 | 41 | 1.516e-03 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 95 | 268 | 1.516e-03 |
| GO:BP | GO:1903509 | liposaccharide metabolic process | 45 | 107 | 1.521e-03 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 44 | 104 | 1.532e-03 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 108 | 312 | 1.553e-03 |
| GO:BP | GO:0001654 | eye development | 138 | 415 | 1.571e-03 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 30 | 63 | 1.584e-03 |
| GO:BP | GO:0060561 | apoptotic process involved in morphogenesis | 16 | 26 | 1.623e-03 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 179 | 559 | 1.626e-03 |
| GO:BP | GO:0060429 | epithelium development | 366 | 1238 | 1.654e-03 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 31 | 66 | 1.669e-03 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 59 | 151 | 1.679e-03 |
| GO:BP | GO:0006506 | GPI anchor biosynthetic process | 18 | 31 | 1.708e-03 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 83 | 229 | 1.726e-03 |
| GO:BP | GO:0043487 | regulation of RNA stability | 72 | 193 | 1.743e-03 |
| GO:BP | GO:0009225 | nucleotide-sugar metabolic process | 23 | 44 | 1.743e-03 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 23 | 44 | 1.743e-03 |
| GO:BP | GO:0051050 | positive regulation of transport | 259 | 847 | 1.774e-03 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 94 | 266 | 1.821e-03 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 117 | 344 | 1.830e-03 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 102 | 293 | 1.833e-03 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 200 | 635 | 1.833e-03 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 47 | 114 | 1.854e-03 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 145 | 441 | 1.869e-03 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 66 | 174 | 1.870e-03 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 55 | 139 | 1.889e-03 |
| GO:BP | GO:0140352 | export from cell | 281 | 928 | 1.897e-03 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 45 | 108 | 1.903e-03 |
| GO:BP | GO:0007623 | circadian rhythm | 77 | 210 | 1.928e-03 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 54 | 136 | 1.940e-03 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 38 | 87 | 1.958e-03 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 148 | 452 | 1.965e-03 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 280 | 925 | 1.979e-03 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 105 | 304 | 2.034e-03 |
| GO:BP | GO:0051028 | mRNA transport | 52 | 130 | 2.048e-03 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 29 | 61 | 2.049e-03 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 75 | 204 | 2.107e-03 |
| GO:BP | GO:0006839 | mitochondrial transport | 67 | 178 | 2.128e-03 |
| GO:BP | GO:0006734 | NADH metabolic process | 19 | 34 | 2.128e-03 |
| GO:BP | GO:0006812 | monoatomic cation transport | 319 | 1069 | 2.145e-03 |
| GO:BP | GO:0060972 | left/right pattern formation | 57 | 146 | 2.145e-03 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 30 | 64 | 2.149e-03 |
| GO:BP | GO:0071985 | multivesicular body sorting pathway | 25 | 50 | 2.155e-03 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 230 | 745 | 2.198e-03 |
| GO:BP | GO:0030001 | metal ion transport | 268 | 883 | 2.237e-03 |
| GO:BP | GO:0000045 | autophagosome assembly | 48 | 118 | 2.245e-03 |
| GO:BP | GO:0086070 | SA node cell to atrial cardiac muscle cell communication | 9 | 11 | 2.259e-03 |
| GO:BP | GO:0048251 | elastic fiber assembly | 9 | 11 | 2.259e-03 |
| GO:BP | GO:0001941 | postsynaptic membrane organization | 22 | 42 | 2.259e-03 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 80 | 221 | 2.266e-03 |
| GO:BP | GO:0051960 | regulation of nervous system development | 149 | 457 | 2.271e-03 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 65 | 172 | 2.283e-03 |
| GO:BP | GO:0045444 | fat cell differentiation | 86 | 241 | 2.283e-03 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 202 | 645 | 2.298e-03 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 26 | 53 | 2.329e-03 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 33 | 73 | 2.362e-03 |
| GO:BP | GO:0150063 | visual system development | 138 | 419 | 2.366e-03 |
| GO:BP | GO:0043434 | response to peptide hormone | 144 | 440 | 2.380e-03 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 125 | 374 | 2.390e-03 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 34 | 76 | 2.405e-03 |
| GO:BP | GO:0050773 | regulation of dendrite development | 42 | 100 | 2.450e-03 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 27 | 56 | 2.491e-03 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 39 | 91 | 2.491e-03 |
| GO:BP | GO:0141137 | positive regulation of gene expression, epigenetic | 27 | 56 | 2.491e-03 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 14 | 22 | 2.494e-03 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 121 | 361 | 2.625e-03 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 73 | 199 | 2.664e-03 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 49 | 122 | 2.689e-03 |
| GO:BP | GO:0180046 | GPI anchored protein biosynthesis | 18 | 32 | 2.733e-03 |
| GO:BP | GO:0006505 | GPI anchor metabolic process | 18 | 32 | 2.733e-03 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 18 | 32 | 2.733e-03 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 113 | 334 | 2.791e-03 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 54 | 138 | 2.840e-03 |
| GO:BP | GO:0048675 | axon extension | 46 | 113 | 2.873e-03 |
| GO:BP | GO:0016054 | organic acid catabolic process | 87 | 246 | 2.873e-03 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 87 | 246 | 2.873e-03 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 46 | 113 | 2.873e-03 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 185 | 587 | 2.892e-03 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 71 | 193 | 2.892e-03 |
| GO:BP | GO:0032940 | secretion by cell | 259 | 854 | 2.892e-03 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 21 | 40 | 2.925e-03 |
| GO:BP | GO:0000209 | protein polyubiquitination | 97 | 280 | 2.985e-03 |
| GO:BP | GO:0042180 | ketone metabolic process | 77 | 213 | 2.997e-03 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 109 | 321 | 3.021e-03 |
| GO:BP | GO:0035265 | organ growth | 62 | 164 | 3.030e-03 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 218 | 706 | 3.030e-03 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 66 | 177 | 3.043e-03 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 57 | 148 | 3.056e-03 |
| GO:BP | GO:0060996 | dendritic spine development | 42 | 101 | 3.056e-03 |
| GO:BP | GO:0009799 | specification of symmetry | 57 | 148 | 3.056e-03 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 51 | 129 | 3.076e-03 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 40 | 95 | 3.129e-03 |
| GO:BP | GO:0046620 | regulation of organ growth | 38 | 89 | 3.162e-03 |
| GO:BP | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway | 65 | 174 | 3.162e-03 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 36 | 83 | 3.162e-03 |
| GO:BP | GO:0055006 | cardiac cell development | 37 | 86 | 3.162e-03 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 50 | 126 | 3.162e-03 |
| GO:BP | GO:0031667 | response to nutrient levels | 163 | 510 | 3.183e-03 |
| GO:BP | GO:0090174 | organelle membrane fusion | 49 | 123 | 3.246e-03 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 75 | 207 | 3.250e-03 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 55 | 142 | 3.257e-03 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 22 | 43 | 3.259e-03 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 242 | 794 | 3.259e-03 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 189 | 603 | 3.301e-03 |
| GO:BP | GO:0048880 | sensory system development | 139 | 426 | 3.335e-03 |
| GO:BP | GO:0006887 | exocytosis | 120 | 360 | 3.390e-03 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 74 | 204 | 3.392e-03 |
| GO:BP | GO:0000165 | MAPK cascade | 228 | 744 | 3.455e-03 |
| GO:BP | GO:0042391 | regulation of membrane potential | 148 | 458 | 3.474e-03 |
| GO:BP | GO:0046039 | GTP metabolic process | 15 | 25 | 3.476e-03 |
| GO:BP | GO:0048668 | collateral sprouting | 17 | 30 | 3.478e-03 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 82 | 231 | 3.602e-03 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 114 | 340 | 3.686e-03 |
| GO:BP | GO:0006816 | calcium ion transport | 141 | 434 | 3.690e-03 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 107 | 316 | 3.717e-03 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 75 | 208 | 3.769e-03 |
| GO:BP | GO:0055085 | transmembrane transport | 453 | 1578 | 3.890e-03 |
| GO:BP | GO:0048863 | stem cell differentiation | 86 | 245 | 3.915e-03 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 24 | 49 | 3.932e-03 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 63 | 169 | 4.050e-03 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 37 | 87 | 4.050e-03 |
| GO:BP | GO:0006094 | gluconeogenesis | 36 | 84 | 4.057e-03 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 36 | 84 | 4.057e-03 |
| GO:BP | GO:0006906 | vesicle fusion | 48 | 121 | 4.073e-03 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 70 | 192 | 4.077e-03 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 73 | 202 | 4.135e-03 |
| GO:BP | GO:0007030 | Golgi organization | 58 | 153 | 4.174e-03 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 47 | 118 | 4.183e-03 |
| GO:BP | GO:0045445 | myoblast differentiation | 46 | 115 | 4.304e-03 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 46 | 115 | 4.304e-03 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 57 | 150 | 4.335e-03 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 57 | 150 | 4.335e-03 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 45 | 112 | 4.415e-03 |
| GO:BP | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 11 | 16 | 4.501e-03 |
| GO:BP | GO:0006811 | monoatomic ion transport | 373 | 1282 | 4.619e-03 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 43 | 106 | 4.644e-03 |
| GO:BP | GO:0016073 | snRNA metabolic process | 27 | 58 | 4.649e-03 |
| GO:BP | GO:0048732 | gland development | 146 | 454 | 4.667e-03 |
| GO:BP | GO:0043010 | camera-type eye development | 120 | 363 | 4.707e-03 |
| GO:BP | GO:0042060 | wound healing | 140 | 433 | 4.726e-03 |
| GO:BP | GO:0038202 | TORC1 signaling | 42 | 103 | 4.729e-03 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 97 | 284 | 4.864e-03 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 40 | 97 | 4.926e-03 |
| GO:BP | GO:0006301 | postreplication repair | 19 | 36 | 4.971e-03 |
| GO:BP | GO:0106027 | neuron projection organization | 39 | 94 | 5.004e-03 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 480 | 1685 | 5.101e-03 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 263 | 877 | 5.111e-03 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 47 | 119 | 5.117e-03 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 31 | 70 | 5.122e-03 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 31 | 70 | 5.122e-03 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 23 | 47 | 5.129e-03 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 33 | 76 | 5.197e-03 |
| GO:BP | GO:0090153 | regulation of sphingolipid biosynthetic process | 10 | 14 | 5.232e-03 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 922 | 3375 | 5.232e-03 |
| GO:BP | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation | 10 | 14 | 5.232e-03 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 46 | 116 | 5.232e-03 |
| GO:BP | GO:0015780 | nucleotide-sugar transmembrane transport | 10 | 14 | 5.232e-03 |
| GO:BP | GO:0051259 | protein complex oligomerization | 93 | 271 | 5.232e-03 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 236 | 779 | 5.299e-03 |
| GO:BP | GO:0014074 | response to purine-containing compound | 56 | 148 | 5.332e-03 |
| GO:BP | GO:0046683 | response to organophosphorus | 51 | 132 | 5.383e-03 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 24 | 50 | 5.455e-03 |
| GO:BP | GO:0007420 | brain development | 229 | 754 | 5.486e-03 |
| GO:BP | GO:0048568 | embryonic organ development | 148 | 463 | 5.548e-03 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 59 | 158 | 5.591e-03 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 25 | 53 | 5.771e-03 |
| GO:BP | GO:0072033 | renal vesicle formation | 7 | 8 | 5.791e-03 |
| GO:BP | GO:0018202 | peptidyl-histidine modification | 7 | 8 | 5.791e-03 |
| GO:BP | GO:0022038 | corpus callosum development | 15 | 26 | 5.834e-03 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 109 | 327 | 5.908e-03 |
| GO:BP | GO:0045926 | negative regulation of growth | 80 | 228 | 5.964e-03 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 26 | 56 | 6.014e-03 |
| GO:BP | GO:0051651 | maintenance of location in cell | 74 | 208 | 6.061e-03 |
| GO:BP | GO:0007032 | endosome organization | 40 | 98 | 6.083e-03 |
| GO:BP | GO:1904861 | excitatory synapse assembly | 21 | 42 | 6.177e-03 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 27 | 59 | 6.211e-03 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 92 | 269 | 6.211e-03 |
| GO:BP | GO:0031125 | rRNA 3’-end processing | 8 | 10 | 6.220e-03 |
| GO:BP | GO:0060541 | respiratory system development | 79 | 225 | 6.245e-03 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 38 | 92 | 6.301e-03 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 56 | 149 | 6.315e-03 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 326 | 1113 | 6.325e-03 |
| GO:BP | GO:0007015 | actin filament organization | 147 | 461 | 6.370e-03 |
| GO:BP | GO:0043543 | protein acylation | 37 | 89 | 6.382e-03 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 29 | 65 | 6.455e-03 |
| GO:BP | GO:0022406 | membrane docking | 36 | 86 | 6.455e-03 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 63 | 172 | 6.455e-03 |
| GO:BP | GO:0040011 | locomotion | 364 | 1255 | 6.519e-03 |
| GO:BP | GO:0060419 | heart growth | 35 | 83 | 6.519e-03 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 55 | 146 | 6.541e-03 |
| GO:BP | GO:1903578 | regulation of ATP metabolic process | 32 | 74 | 6.600e-03 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 22 | 45 | 6.623e-03 |
| GO:BP | GO:0018279 | protein N-linked glycosylation via asparagine | 22 | 45 | 6.623e-03 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 117 | 356 | 6.643e-03 |
| GO:BP | GO:0009725 | response to hormone | 270 | 907 | 6.643e-03 |
| GO:BP | GO:0042594 | response to starvation | 77 | 219 | 6.832e-03 |
| GO:BP | GO:0051648 | vesicle localization | 77 | 219 | 6.832e-03 |
| GO:BP | GO:0009060 | aerobic respiration | 71 | 199 | 6.925e-03 |
| GO:BP | GO:0110154 | RNA decapping | 12 | 19 | 7.010e-03 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 53 | 140 | 7.075e-03 |
| GO:BP | GO:0043500 | muscle adaptation | 42 | 105 | 7.075e-03 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 42 | 105 | 7.075e-03 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 16 | 29 | 7.107e-03 |
| GO:BP | GO:0051156 | glucose 6-phosphate metabolic process | 16 | 29 | 7.107e-03 |
| GO:BP | GO:0007219 | Notch signaling pathway | 67 | 186 | 7.214e-03 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 67 | 186 | 7.214e-03 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 109 | 329 | 7.246e-03 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 52 | 137 | 7.336e-03 |
| GO:BP | GO:0007423 | sensory organ development | 192 | 624 | 7.381e-03 |
| GO:BP | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 14 | 24 | 7.444e-03 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 39 | 96 | 7.616e-03 |
| GO:BP | GO:0001889 | liver development | 55 | 147 | 7.727e-03 |
| GO:BP | GO:0016042 | lipid catabolic process | 116 | 354 | 7.759e-03 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 38 | 93 | 7.777e-03 |
| GO:BP | GO:0030097 | hemopoiesis | 288 | 976 | 7.816e-03 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 62 | 170 | 7.816e-03 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 58 | 157 | 8.000e-03 |
| GO:BP | GO:0035050 | embryonic heart tube development | 36 | 87 | 8.074e-03 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 84 | 244 | 8.104e-03 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 84 | 244 | 8.104e-03 |
| GO:BP | GO:0010038 | response to metal ion | 115 | 351 | 8.133e-03 |
| GO:BP | GO:0031929 | TOR signaling | 61 | 167 | 8.133e-03 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 76 | 217 | 8.133e-03 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 61 | 167 | 8.133e-03 |
| GO:BP | GO:0061448 | connective tissue development | 98 | 292 | 8.133e-03 |
| GO:BP | GO:0060420 | regulation of heart growth | 27 | 60 | 8.133e-03 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 27 | 60 | 8.133e-03 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 34 | 81 | 8.224e-03 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 34 | 81 | 8.224e-03 |
| GO:BP | GO:0099022 | vesicle tethering | 17 | 32 | 8.300e-03 |
| GO:BP | GO:0010212 | response to ionizing radiation | 53 | 141 | 8.303e-03 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 29 | 66 | 8.314e-03 |
| GO:BP | GO:0007155 | cell adhesion | 436 | 1530 | 8.319e-03 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 31 | 72 | 8.357e-03 |
| GO:BP | GO:0042221 | response to chemical | 1056 | 3909 | 8.375e-03 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 307 | 1048 | 8.541e-03 |
| GO:BP | GO:0043649 | dicarboxylic acid catabolic process | 11 | 17 | 8.607e-03 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 56 | 151 | 8.607e-03 |
| GO:BP | GO:1901096 | regulation of autophagosome maturation | 11 | 17 | 8.607e-03 |
| GO:BP | GO:0090205 | positive regulation of cholesterol metabolic process | 11 | 17 | 8.607e-03 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 52 | 138 | 8.607e-03 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 41 | 103 | 8.773e-03 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 41 | 103 | 8.773e-03 |
| GO:BP | GO:0007411 | axon guidance | 82 | 238 | 8.839e-03 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 89 | 262 | 8.970e-03 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 62 | 171 | 8.971e-03 |
| GO:BP | GO:1990138 | neuron projection extension | 62 | 171 | 8.971e-03 |
| GO:BP | GO:0039531 | regulation of cytoplasmic pattern recognition receptor signaling pathway | 46 | 119 | 9.049e-03 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 15 | 27 | 9.138e-03 |
| GO:BP | GO:0009226 | nucleotide-sugar biosynthetic process | 15 | 27 | 9.138e-03 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 104 | 314 | 9.198e-03 |
| GO:BP | GO:0051235 | maintenance of location | 76 | 218 | 9.198e-03 |
| GO:BP | GO:0016180 | snRNA processing | 18 | 35 | 9.315e-03 |
| GO:BP | GO:0046320 | regulation of fatty acid oxidation | 18 | 35 | 9.315e-03 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 18 | 35 | 9.315e-03 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 18 | 35 | 9.315e-03 |
| GO:BP | GO:0034312 | diol biosynthetic process | 13 | 22 | 9.393e-03 |
| GO:BP | GO:0035493 | SNARE complex assembly | 13 | 22 | 9.393e-03 |
| GO:BP | GO:0006390 | mitochondrial transcription | 13 | 22 | 9.393e-03 |
| GO:BP | GO:0016197 | endosomal transport | 97 | 290 | 9.393e-03 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 13 | 22 | 9.393e-03 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 254 | 854 | 9.414e-03 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 23 | 49 | 9.447e-03 |
| GO:BP | GO:0006497 | protein lipidation | 23 | 49 | 9.447e-03 |
| GO:BP | GO:0007613 | memory | 49 | 129 | 9.591e-03 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 109 | 332 | 9.733e-03 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 24 | 52 | 9.841e-03 |
| GO:BP | GO:0007254 | JNK cascade | 63 | 175 | 9.849e-03 |
| GO:BP | GO:0097485 | neuron projection guidance | 82 | 239 | 9.912e-03 |
| GO:BP | GO:0022900 | electron transport chain | 48 | 126 | 9.972e-03 |
| GO:BP | GO:0001890 | placenta development | 56 | 152 | 9.977e-03 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 35 | 85 | 9.988e-03 |
| GO:BP | GO:0007249 | canonical NF-kappaB signal transduction | 102 | 308 | 1.009e-02 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 104 | 315 | 1.014e-02 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 71 | 202 | 1.020e-02 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 19 | 38 | 1.021e-02 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 42 | 107 | 1.022e-02 |
| GO:BP | GO:0071248 | cellular response to metal ion | 68 | 192 | 1.028e-02 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 156 | 499 | 1.029e-02 |
| GO:BP | GO:0043462 | regulation of ATP-dependent activity | 27 | 61 | 1.044e-02 |
| GO:BP | GO:0051284 | positive regulation of sequestering of calcium ion | 10 | 15 | 1.045e-02 |
| GO:BP | GO:0072175 | epithelial tube formation | 51 | 136 | 1.045e-02 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 28 | 64 | 1.051e-02 |
| GO:BP | GO:0090130 | tissue migration | 41 | 104 | 1.052e-02 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 136 | 428 | 1.056e-02 |
| GO:BP | GO:0046856 | phosphatidylinositol dephosphorylation | 16 | 30 | 1.063e-02 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 70 | 199 | 1.065e-02 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 40 | 101 | 1.084e-02 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 128 | 400 | 1.091e-02 |
| GO:BP | GO:0032509 | endosome transport via multivesicular body sorting pathway | 20 | 41 | 1.100e-02 |
| GO:BP | GO:0001666 | response to hypoxia | 100 | 302 | 1.112e-02 |
| GO:BP | GO:0070482 | response to oxygen levels | 112 | 344 | 1.129e-02 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 63 | 176 | 1.132e-02 |
| GO:BP | GO:0001525 | angiogenesis | 169 | 547 | 1.141e-02 |
| GO:BP | GO:0003231 | cardiac ventricle development | 48 | 127 | 1.186e-02 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 194 | 638 | 1.189e-02 |
| GO:BP | GO:0006098 | pentose-phosphate shunt | 12 | 20 | 1.205e-02 |
| GO:BP | GO:0033194 | response to hydroperoxide | 12 | 20 | 1.205e-02 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 17 | 33 | 1.212e-02 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 17 | 33 | 1.212e-02 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 17 | 33 | 1.212e-02 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 166 | 537 | 1.215e-02 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 55 | 150 | 1.215e-02 |
| GO:BP | GO:0099643 | signal release from synapse | 55 | 150 | 1.215e-02 |
| GO:BP | GO:0034097 | response to cytokine | 278 | 947 | 1.223e-02 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 22 | 47 | 1.225e-02 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 98 | 296 | 1.227e-02 |
| GO:BP | GO:0051402 | neuron apoptotic process | 98 | 296 | 1.227e-02 |
| GO:BP | GO:0007612 | learning | 58 | 160 | 1.231e-02 |
| GO:BP | GO:0019827 | stem cell population maintenance | 64 | 180 | 1.240e-02 |
| GO:BP | GO:1902817 | negative regulation of protein localization to microtubule | 5 | 5 | 1.248e-02 |
| GO:BP | GO:1902816 | regulation of protein localization to microtubule | 5 | 5 | 1.248e-02 |
| GO:BP | GO:0086046 | membrane depolarization during SA node cell action potential | 5 | 5 | 1.248e-02 |
| GO:BP | GO:0019042 | viral latency | 5 | 5 | 1.248e-02 |
| GO:BP | GO:0097114 | NMDA glutamate receptor clustering | 5 | 5 | 1.248e-02 |
| GO:BP | GO:0097061 | dendritic spine organization | 34 | 83 | 1.250e-02 |
| GO:BP | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation | 9 | 13 | 1.250e-02 |
| GO:BP | GO:2000303 | regulation of ceramide biosynthetic process | 9 | 13 | 1.250e-02 |
| GO:BP | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport | 9 | 13 | 1.250e-02 |
| GO:BP | GO:0007019 | microtubule depolymerization | 23 | 50 | 1.258e-02 |
| GO:BP | GO:0009267 | cellular response to starvation | 63 | 177 | 1.294e-02 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 24 | 53 | 1.294e-02 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 32 | 77 | 1.295e-02 |
| GO:BP | GO:0007389 | pattern specification process | 149 | 477 | 1.313e-02 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 135 | 427 | 1.313e-02 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 25 | 56 | 1.317e-02 |
| GO:BP | GO:0043542 | endothelial cell migration | 76 | 221 | 1.317e-02 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 49 | 131 | 1.326e-02 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 28 | 65 | 1.339e-02 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 27 | 62 | 1.339e-02 |
| GO:BP | GO:0098727 | maintenance of cell number | 65 | 184 | 1.346e-02 |
| GO:BP | GO:0000723 | telomere maintenance | 59 | 164 | 1.350e-02 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 110 | 339 | 1.359e-02 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 110 | 339 | 1.359e-02 |
| GO:BP | GO:0072001 | renal system development | 108 | 332 | 1.360e-02 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 89 | 266 | 1.382e-02 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 67 | 191 | 1.396e-02 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 82 | 242 | 1.396e-02 |
| GO:BP | GO:0097553 | calcium ion transmembrane import into cytosol | 64 | 181 | 1.412e-02 |
| GO:BP | GO:0060323 | head morphogenesis | 19 | 39 | 1.424e-02 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 170 | 554 | 1.466e-02 |
| GO:BP | GO:0035148 | tube formation | 54 | 148 | 1.468e-02 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 81 | 239 | 1.473e-02 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 57 | 158 | 1.486e-02 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 60 | 168 | 1.491e-02 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 20 | 42 | 1.511e-02 |
| GO:BP | GO:1901652 | response to peptide | 281 | 962 | 1.511e-02 |
| GO:BP | GO:0001501 | skeletal system development | 166 | 540 | 1.511e-02 |
| GO:BP | GO:0045540 | regulation of cholesterol biosynthetic process | 13 | 23 | 1.511e-02 |
| GO:BP | GO:0106118 | regulation of sterol biosynthetic process | 13 | 23 | 1.511e-02 |
| GO:BP | GO:0046519 | sphingoid metabolic process | 13 | 23 | 1.511e-02 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 11 | 18 | 1.512e-02 |
| GO:BP | GO:0006743 | ubiquinone metabolic process | 11 | 18 | 1.512e-02 |
| GO:BP | GO:0046903 | secretion | 289 | 992 | 1.529e-02 |
| GO:BP | GO:0097107 | postsynaptic density assembly | 16 | 31 | 1.566e-02 |
| GO:BP | GO:0034311 | diol metabolic process | 16 | 31 | 1.566e-02 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 33 | 81 | 1.573e-02 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 21 | 45 | 1.573e-02 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 21 | 45 | 1.573e-02 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 21 | 45 | 1.573e-02 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 52 | 142 | 1.597e-02 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 52 | 142 | 1.597e-02 |
| GO:BP | GO:0000480 | endonucleolytic cleavage in 5’-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 6 | 7 | 1.614e-02 |
| GO:BP | GO:0060710 | chorio-allantoic fusion | 6 | 7 | 1.614e-02 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 48 | 129 | 1.614e-02 |
| GO:BP | GO:0097680 | double-strand break repair via classical nonhomologous end joining | 7 | 9 | 1.614e-02 |
| GO:BP | GO:0048033 | heme O metabolic process | 7 | 9 | 1.614e-02 |
| GO:BP | GO:0017183 | protein histidyl modification to diphthamide | 6 | 7 | 1.614e-02 |
| GO:BP | GO:0014883 | transition between fast and slow fiber | 6 | 7 | 1.614e-02 |
| GO:BP | GO:0048034 | heme O biosynthetic process | 7 | 9 | 1.614e-02 |
| GO:BP | GO:0000467 | exonucleolytic trimming to generate mature 3’-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 7 | 9 | 1.614e-02 |
| GO:BP | GO:0043604 | amide biosynthetic process | 61 | 172 | 1.614e-02 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 31 | 75 | 1.625e-02 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 30 | 72 | 1.653e-02 |
| GO:BP | GO:0007267 | cell-cell signaling | 382 | 1342 | 1.666e-02 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 47 | 126 | 1.680e-02 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 47 | 126 | 1.680e-02 |
| GO:BP | GO:0046777 | protein autophosphorylation | 63 | 179 | 1.680e-02 |
| GO:BP | GO:0060324 | face development | 24 | 54 | 1.680e-02 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 78 | 230 | 1.690e-02 |
| GO:BP | GO:0050982 | detection of mechanical stimulus | 25 | 57 | 1.694e-02 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 26 | 60 | 1.695e-02 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 26 | 60 | 1.695e-02 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 26 | 60 | 1.695e-02 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 17 | 34 | 1.704e-02 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 50 | 136 | 1.728e-02 |
| GO:BP | GO:0046660 | female sex differentiation | 46 | 123 | 1.750e-02 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 53 | 146 | 1.762e-02 |
| GO:BP | GO:0019985 | translesion synthesis | 14 | 26 | 1.780e-02 |
| GO:BP | GO:0018345 | protein palmitoylation | 14 | 26 | 1.780e-02 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 14 | 26 | 1.780e-02 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 192 | 637 | 1.792e-02 |
| GO:BP | GO:0062014 | negative regulation of small molecule metabolic process | 41 | 107 | 1.795e-02 |
| GO:BP | GO:0031647 | regulation of protein stability | 108 | 335 | 1.798e-02 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis | 18 | 37 | 1.841e-02 |
| GO:BP | GO:0044770 | cell cycle phase transition | 163 | 532 | 1.852e-02 |
| GO:BP | GO:0050821 | protein stabilization | 76 | 224 | 1.869e-02 |
| GO:BP | GO:0048041 | focal adhesion assembly | 34 | 85 | 1.874e-02 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 48 | 130 | 1.891e-02 |
| GO:BP | GO:1902307 | positive regulation of sodium ion transmembrane transport | 10 | 16 | 1.904e-02 |
| GO:BP | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | 10 | 16 | 1.904e-02 |
| GO:BP | GO:0098974 | postsynaptic actin cytoskeleton organization | 10 | 16 | 1.904e-02 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 33 | 82 | 1.923e-02 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 51 | 140 | 1.929e-02 |
| GO:BP | GO:0007369 | gastrulation | 68 | 197 | 1.935e-02 |
| GO:BP | GO:1903034 | regulation of response to wounding | 60 | 170 | 1.938e-02 |
| GO:BP | GO:0006670 | sphingosine metabolic process | 12 | 21 | 1.938e-02 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 19 | 40 | 1.942e-02 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 54 | 150 | 1.943e-02 |
| GO:BP | GO:0021915 | neural tube development | 57 | 160 | 1.944e-02 |
| GO:BP | GO:0010565 | regulation of ketone metabolic process | 47 | 127 | 1.964e-02 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 32 | 79 | 1.964e-02 |
| GO:BP | GO:0051260 | protein homooligomerization | 70 | 204 | 1.976e-02 |
| GO:BP | GO:0033555 | multicellular organismal response to stress | 38 | 98 | 1.994e-02 |
| GO:BP | GO:0046785 | microtubule polymerization | 38 | 98 | 1.994e-02 |
| GO:BP | GO:0043122 | regulation of canonical NF-kappaB signal transduction | 94 | 287 | 2.001e-02 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 20 | 43 | 2.018e-02 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 15 | 29 | 2.018e-02 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 15 | 29 | 2.018e-02 |
| GO:BP | GO:0042552 | myelination | 56 | 157 | 2.036e-02 |
| GO:BP | GO:0010631 | epithelial cell migration | 37 | 95 | 2.064e-02 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 29 | 70 | 2.084e-02 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 29 | 70 | 2.084e-02 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 29 | 70 | 2.084e-02 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 28 | 67 | 2.119e-02 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 22 | 49 | 2.129e-02 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 52 | 144 | 2.129e-02 |
| GO:BP | GO:0003279 | cardiac septum development | 41 | 108 | 2.130e-02 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 27 | 64 | 2.139e-02 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 27 | 64 | 2.139e-02 |
| GO:BP | GO:0009299 | mRNA transcription | 23 | 52 | 2.151e-02 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 73 | 215 | 2.155e-02 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 24 | 55 | 2.160e-02 |
| GO:BP | GO:0048278 | vesicle docking | 25 | 58 | 2.160e-02 |
| GO:BP | GO:0045778 | positive regulation of ossification | 24 | 55 | 2.160e-02 |
| GO:BP | GO:0007626 | locomotory behavior | 75 | 222 | 2.188e-02 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 88 | 267 | 2.191e-02 |
| GO:BP | GO:0035418 | protein localization to synapse | 35 | 89 | 2.191e-02 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 35 | 89 | 2.191e-02 |
| GO:BP | GO:2000279 | negative regulation of DNA biosynthetic process | 16 | 32 | 2.214e-02 |
| GO:BP | GO:0014850 | response to muscle activity | 16 | 32 | 2.214e-02 |
| GO:BP | GO:0016482 | cytosolic transport | 51 | 141 | 2.215e-02 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 34 | 86 | 2.264e-02 |
| GO:BP | GO:0010970 | transport along microtubule | 62 | 178 | 2.274e-02 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 103 | 320 | 2.288e-02 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 274 | 944 | 2.302e-02 |
| GO:BP | GO:1902993 | positive regulation of amyloid precursor protein catabolic process | 13 | 24 | 2.311e-02 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 43 | 115 | 2.316e-02 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 33 | 83 | 2.331e-02 |
| GO:BP | GO:0036151 | phosphatidylcholine acyl-chain remodeling | 9 | 14 | 2.379e-02 |
| GO:BP | GO:0002087 | regulation of respiratory gaseous exchange by nervous system process | 9 | 14 | 2.379e-02 |
| GO:BP | GO:0046322 | negative regulation of fatty acid oxidation | 9 | 14 | 2.379e-02 |
| GO:BP | GO:0043648 | dicarboxylic acid metabolic process | 38 | 99 | 2.379e-02 |
| GO:BP | GO:0050905 | neuromuscular process | 61 | 175 | 2.386e-02 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 32 | 80 | 2.396e-02 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 42 | 112 | 2.414e-02 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 52 | 145 | 2.454e-02 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 31 | 77 | 2.465e-02 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 37 | 96 | 2.470e-02 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 77 | 230 | 2.479e-02 |
| GO:BP | GO:0090201 | negative regulation of release of cytochrome c from mitochondria | 11 | 19 | 2.492e-02 |
| GO:BP | GO:1903830 | magnesium ion transmembrane transport | 11 | 19 | 2.492e-02 |
| GO:BP | GO:0060039 | pericardium development | 11 | 19 | 2.492e-02 |
| GO:BP | GO:0014819 | regulation of skeletal muscle contraction | 11 | 19 | 2.492e-02 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 18 | 38 | 2.506e-02 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 41 | 109 | 2.506e-02 |
| GO:BP | GO:0098698 | postsynaptic specialization assembly | 18 | 38 | 2.506e-02 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 18 | 38 | 2.506e-02 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 30 | 74 | 2.509e-02 |
| GO:BP | GO:0061025 | membrane fusion | 65 | 189 | 2.509e-02 |
| GO:BP | GO:0051216 | cartilage development | 72 | 213 | 2.527e-02 |
| GO:BP | GO:0099111 | microtubule-based transport | 74 | 220 | 2.566e-02 |
| GO:BP | GO:0062208 | positive regulation of pattern recognition receptor signaling pathway | 29 | 71 | 2.570e-02 |
| GO:BP | GO:1902414 | protein localization to cell junction | 44 | 119 | 2.600e-02 |
| GO:BP | GO:1902742 | apoptotic process involved in development | 19 | 41 | 2.607e-02 |
| GO:BP | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway | 14 | 27 | 2.620e-02 |
| GO:BP | GO:0018904 | ether metabolic process | 14 | 27 | 2.620e-02 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 14 | 27 | 2.620e-02 |
| GO:BP | GO:0034504 | protein localization to nucleus | 102 | 318 | 2.620e-02 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 28 | 68 | 2.620e-02 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 64 | 186 | 2.632e-02 |
| GO:BP | GO:0008366 | axon ensheathment | 56 | 159 | 2.651e-02 |
| GO:BP | GO:0007272 | ensheathment of neurons | 56 | 159 | 2.651e-02 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 27 | 65 | 2.660e-02 |
| GO:BP | GO:0006260 | DNA replication | 92 | 283 | 2.667e-02 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 53 | 149 | 2.668e-02 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 26 | 62 | 2.698e-02 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 21 | 47 | 2.717e-02 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 25 | 59 | 2.728e-02 |
| GO:BP | GO:0001822 | kidney development | 103 | 322 | 2.730e-02 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 68 | 200 | 2.740e-02 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 24 | 56 | 2.741e-02 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 24 | 56 | 2.741e-02 |
| GO:BP | GO:0170034 | L-amino acid biosynthetic process | 23 | 53 | 2.745e-02 |
| GO:BP | GO:1900542 | regulation of purine nucleotide metabolic process | 33 | 84 | 2.806e-02 |
| GO:BP | GO:0030516 | regulation of axon extension | 33 | 84 | 2.806e-02 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 42 | 113 | 2.818e-02 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 15 | 30 | 2.886e-02 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 45 | 123 | 2.899e-02 |
| GO:BP | GO:0002218 | activation of innate immune response | 98 | 305 | 2.910e-02 |
| GO:BP | GO:2001046 | positive regulation of integrin-mediated signaling pathway | 8 | 12 | 2.931e-02 |
| GO:BP | GO:0106120 | positive regulation of sterol biosynthetic process | 8 | 12 | 2.931e-02 |
| GO:BP | GO:0043508 | negative regulation of JUN kinase activity | 8 | 12 | 2.931e-02 |
| GO:BP | GO:0045542 | positive regulation of cholesterol biosynthetic process | 8 | 12 | 2.931e-02 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 88 | 270 | 2.950e-02 |
| GO:BP | GO:0140861 | DNA repair-dependent chromatin remodeling | 12 | 22 | 2.990e-02 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 31 | 78 | 2.990e-02 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 103 | 323 | 2.990e-02 |
| GO:BP | GO:0045932 | negative regulation of muscle contraction | 12 | 22 | 2.990e-02 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 12 | 22 | 2.990e-02 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 12 | 22 | 2.990e-02 |
| GO:BP | GO:0035264 | multicellular organism growth | 56 | 160 | 3.022e-02 |
| GO:BP | GO:0019217 | regulation of fatty acid metabolic process | 36 | 94 | 3.029e-02 |
| GO:BP | GO:0060021 | roof of mouth development | 36 | 94 | 3.029e-02 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 68 | 201 | 3.078e-02 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 16 | 33 | 3.088e-02 |
| GO:BP | GO:0055088 | lipid homeostasis | 58 | 167 | 3.122e-02 |
| GO:BP | GO:0006405 | RNA export from nucleus | 35 | 91 | 3.154e-02 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 29 | 72 | 3.155e-02 |
| GO:BP | GO:0043549 | regulation of kinase activity | 133 | 431 | 3.173e-02 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 39 | 104 | 3.198e-02 |
| GO:BP | GO:0044065 | regulation of respiratory system process | 10 | 17 | 3.198e-02 |
| GO:BP | GO:1901018 | positive regulation of potassium ion transmembrane transporter activity | 10 | 17 | 3.198e-02 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 10 | 17 | 3.198e-02 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 39 | 104 | 3.198e-02 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 52 | 147 | 3.206e-02 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 28 | 69 | 3.220e-02 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 28 | 69 | 3.220e-02 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 28 | 69 | 3.220e-02 |
| GO:BP | GO:0006779 | porphyrin-containing compound biosynthetic process | 17 | 36 | 3.235e-02 |
| GO:BP | GO:0033014 | tetrapyrrole biosynthetic process | 17 | 36 | 3.235e-02 |
| GO:BP | GO:1902003 | regulation of amyloid-beta formation | 17 | 36 | 3.235e-02 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 248 | 854 | 3.280e-02 |
| GO:BP | GO:0006836 | neurotransmitter transport | 71 | 212 | 3.312e-02 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 18 | 39 | 3.348e-02 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 26 | 63 | 3.348e-02 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 51 | 144 | 3.348e-02 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 45 | 124 | 3.348e-02 |
| GO:BP | GO:0032205 | negative regulation of telomere maintenance | 18 | 39 | 3.348e-02 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 18 | 39 | 3.348e-02 |
| GO:BP | GO:0046677 | response to antibiotic | 18 | 39 | 3.348e-02 |
| GO:BP | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway | 26 | 63 | 3.348e-02 |
| GO:BP | GO:0061053 | somite development | 33 | 85 | 3.356e-02 |
| GO:BP | GO:0006140 | regulation of nucleotide metabolic process | 33 | 85 | 3.356e-02 |
| GO:BP | GO:2000209 | regulation of anoikis | 13 | 25 | 3.402e-02 |
| GO:BP | GO:0099172 | presynapse organization | 25 | 60 | 3.404e-02 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 56 | 161 | 3.418e-02 |
| GO:BP | GO:0043270 | positive regulation of monoatomic ion transport | 68 | 202 | 3.422e-02 |
| GO:BP | GO:0031099 | regeneration | 68 | 202 | 3.422e-02 |
| GO:BP | GO:0090132 | epithelium migration | 37 | 98 | 3.439e-02 |
| GO:BP | GO:0001755 | neural crest cell migration | 24 | 57 | 3.442e-02 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 24 | 57 | 3.442e-02 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 20 | 45 | 3.468e-02 |
| GO:BP | GO:0090148 | membrane fission | 20 | 45 | 3.468e-02 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 23 | 54 | 3.470e-02 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 23 | 54 | 3.470e-02 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 21 | 48 | 3.478e-02 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 21 | 48 | 3.478e-02 |
| GO:BP | GO:0097106 | postsynaptic density organization | 21 | 48 | 3.478e-02 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 21 | 48 | 3.478e-02 |
| GO:BP | GO:0019673 | GDP-mannose metabolic process | 7 | 10 | 3.508e-02 |
| GO:BP | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity | 7 | 10 | 3.508e-02 |
| GO:BP | GO:0097119 | postsynaptic density protein 95 clustering | 7 | 10 | 3.508e-02 |
| GO:BP | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand | 7 | 10 | 3.508e-02 |
| GO:BP | GO:0010720 | positive regulation of cell development | 140 | 458 | 3.547e-02 |
| GO:BP | GO:0097120 | receptor localization to synapse | 31 | 79 | 3.568e-02 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 69 | 206 | 3.626e-02 |
| GO:BP | GO:0051656 | establishment of organelle localization | 149 | 491 | 3.638e-02 |
| GO:BP | GO:0003002 | regionalization | 133 | 433 | 3.639e-02 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 30 | 76 | 3.693e-02 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 35 | 92 | 3.711e-02 |
| GO:BP | GO:0039529 | RIG-I signaling pathway | 14 | 28 | 3.725e-02 |
| GO:BP | GO:0009314 | response to radiation | 134 | 437 | 3.753e-02 |
| GO:BP | GO:0001508 | action potential | 59 | 172 | 3.766e-02 |
| GO:BP | GO:0050954 | sensory perception of mechanical stimulus | 66 | 196 | 3.771e-02 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 29 | 73 | 3.810e-02 |
| GO:BP | GO:0072093 | metanephric renal vesicle formation | 4 | 4 | 3.850e-02 |
| GO:BP | GO:1900451 | positive regulation of glutamate receptor signaling pathway | 4 | 4 | 3.850e-02 |
| GO:BP | GO:1903278 | positive regulation of sodium ion export across plasma membrane | 4 | 4 | 3.850e-02 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 11 | 20 | 3.850e-02 |
| GO:BP | GO:0042560 | pteridine-containing compound catabolic process | 4 | 4 | 3.850e-02 |
| GO:BP | GO:0009397 | folic acid-containing compound catabolic process | 4 | 4 | 3.850e-02 |
| GO:BP | GO:0048597 | post-embryonic camera-type eye morphogenesis | 4 | 4 | 3.850e-02 |
| GO:BP | GO:1903276 | regulation of sodium ion export across plasma membrane | 4 | 4 | 3.850e-02 |
| GO:BP | GO:0015693 | magnesium ion transport | 11 | 20 | 3.850e-02 |
| GO:BP | GO:1902262 | apoptotic process involved in blood vessel morphogenesis | 4 | 4 | 3.850e-02 |
| GO:BP | GO:0061003 | positive regulation of dendritic spine morphogenesis | 11 | 20 | 3.850e-02 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 28 | 70 | 3.899e-02 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 28 | 70 | 3.899e-02 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 53 | 152 | 3.926e-02 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 41 | 112 | 3.966e-02 |
| GO:BP | GO:0033209 | tumor necrosis factor-mediated signaling pathway | 41 | 112 | 3.966e-02 |
| GO:BP | GO:0016226 | iron-sulfur cluster assembly | 15 | 31 | 3.966e-02 |
| GO:BP | GO:0031163 | metallo-sulfur cluster assembly | 15 | 31 | 3.966e-02 |
| GO:BP | GO:0048864 | stem cell development | 33 | 86 | 3.983e-02 |
| GO:BP | GO:0043248 | proteasome assembly | 9 | 15 | 4.063e-02 |
| GO:BP | GO:0046520 | sphingoid biosynthetic process | 9 | 15 | 4.063e-02 |
| GO:BP | GO:0014874 | response to stimulus involved in regulation of muscle adaptation | 9 | 15 | 4.063e-02 |
| GO:BP | GO:0046512 | sphingosine biosynthetic process | 9 | 15 | 4.063e-02 |
| GO:BP | GO:0006538 | glutamate catabolic process | 6 | 8 | 4.071e-02 |
| GO:BP | GO:0045650 | negative regulation of macrophage differentiation | 6 | 8 | 4.071e-02 |
| GO:BP | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway | 6 | 8 | 4.071e-02 |
| GO:BP | GO:0003344 | pericardium morphogenesis | 6 | 8 | 4.071e-02 |
| GO:BP | GO:1904026 | regulation of collagen fibril organization | 6 | 8 | 4.071e-02 |
| GO:BP | GO:0006785 | heme B biosynthetic process | 6 | 8 | 4.071e-02 |
| GO:BP | GO:0043504 | mitochondrial DNA repair | 6 | 8 | 4.071e-02 |
| GO:BP | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling | 6 | 8 | 4.071e-02 |
| GO:BP | GO:0016139 | glycoside catabolic process | 6 | 8 | 4.071e-02 |
| GO:BP | GO:0015959 | diadenosine polyphosphate metabolic process | 6 | 8 | 4.071e-02 |
| GO:BP | GO:0031077 | post-embryonic camera-type eye development | 6 | 8 | 4.071e-02 |
| GO:BP | GO:0046492 | heme B metabolic process | 6 | 8 | 4.071e-02 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 79 | 242 | 4.073e-02 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 52 | 149 | 4.084e-02 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 342 | 1212 | 4.104e-02 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 40 | 109 | 4.104e-02 |
| GO:BP | GO:0042063 | gliogenesis | 110 | 352 | 4.108e-02 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 16 | 34 | 4.108e-02 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 16 | 34 | 4.108e-02 |
| GO:BP | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway | 16 | 34 | 4.108e-02 |
| GO:BP | GO:0062125 | regulation of mitochondrial gene expression | 16 | 34 | 4.108e-02 |
| GO:BP | GO:0006536 | glutamate metabolic process | 16 | 34 | 4.108e-02 |
| GO:BP | GO:0030879 | mammary gland development | 49 | 139 | 4.124e-02 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 66 | 197 | 4.135e-02 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 36 | 96 | 4.135e-02 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 36 | 96 | 4.135e-02 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 54 | 156 | 4.184e-02 |
| GO:BP | GO:0033365 | protein localization to organelle | 341 | 1209 | 4.228e-02 |
| GO:BP | GO:0014032 | neural crest cell development | 31 | 80 | 4.229e-02 |
| GO:BP | GO:0045324 | late endosome to vacuole transport | 17 | 37 | 4.244e-02 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 51 | 146 | 4.270e-02 |
| GO:BP | GO:0010586 | miRNA metabolic process | 39 | 106 | 4.272e-02 |
| GO:BP | GO:0035176 | social behavior | 23 | 55 | 4.292e-02 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 35 | 93 | 4.309e-02 |
| GO:BP | GO:0055092 | sterol homeostasis | 35 | 93 | 4.309e-02 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 35 | 93 | 4.309e-02 |
| GO:BP | GO:0002071 | glandular epithelial cell maturation | 5 | 6 | 4.312e-02 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 21 | 49 | 4.312e-02 |
| GO:BP | GO:0003283 | atrial septum development | 12 | 23 | 4.312e-02 |
| GO:BP | GO:1990116 | ribosome-associated ubiquitin-dependent protein catabolic process | 5 | 6 | 4.312e-02 |
| GO:BP | GO:0003415 | chondrocyte hypertrophy | 5 | 6 | 4.312e-02 |
| GO:BP | GO:0000389 | mRNA 3’-splice site recognition | 5 | 6 | 4.312e-02 |
| GO:BP | GO:0051096 | positive regulation of helicase activity | 5 | 6 | 4.312e-02 |
| GO:BP | GO:0010171 | body morphogenesis | 22 | 52 | 4.312e-02 |
| GO:BP | GO:0031999 | negative regulation of fatty acid beta-oxidation | 5 | 6 | 4.312e-02 |
| GO:BP | GO:0021999 | neural plate anterior/posterior regionalization | 5 | 6 | 4.312e-02 |
| GO:BP | GO:0019320 | hexose catabolic process | 20 | 46 | 4.312e-02 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 19 | 43 | 4.312e-02 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 19 | 43 | 4.312e-02 |
| GO:BP | GO:0015961 | diadenosine polyphosphate catabolic process | 5 | 6 | 4.312e-02 |
| GO:BP | GO:0010837 | regulation of keratinocyte proliferation | 20 | 46 | 4.312e-02 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 53 | 153 | 4.312e-02 |
| GO:BP | GO:0010332 | response to gamma radiation | 22 | 52 | 4.312e-02 |
| GO:BP | GO:0009298 | GDP-mannose biosynthetic process | 5 | 6 | 4.312e-02 |
| GO:BP | GO:0009066 | aspartate family amino acid metabolic process | 19 | 43 | 4.312e-02 |
| GO:BP | GO:1901016 | regulation of potassium ion transmembrane transporter activity | 18 | 40 | 4.312e-02 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 20 | 46 | 4.312e-02 |
| GO:BP | GO:1901020 | negative regulation of calcium ion transmembrane transporter activity | 12 | 23 | 4.312e-02 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 117 | 378 | 4.312e-02 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 48 | 136 | 4.312e-02 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 12 | 23 | 4.312e-02 |
| GO:BP | GO:0046929 | negative regulation of neurotransmitter secretion | 5 | 6 | 4.312e-02 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 30 | 77 | 4.312e-02 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 69 | 208 | 4.343e-02 |
| GO:BP | GO:0007029 | endoplasmic reticulum organization | 38 | 103 | 4.385e-02 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 34 | 90 | 4.412e-02 |
| GO:BP | GO:1901264 | carbohydrate derivative transport | 34 | 90 | 4.412e-02 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 29 | 74 | 4.449e-02 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 197 | 671 | 4.457e-02 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 47 | 133 | 4.457e-02 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 44 | 123 | 4.482e-02 |
| GO:BP | GO:0032200 | telomere organization | 64 | 191 | 4.492e-02 |
| GO:BP | GO:0030832 | regulation of actin filament length | 52 | 150 | 4.536e-02 |
| GO:BP | GO:0030324 | lung development | 66 | 198 | 4.536e-02 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 37 | 100 | 4.584e-02 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 59 | 174 | 4.607e-02 |
| GO:BP | GO:0008360 | regulation of cell shape | 49 | 140 | 4.628e-02 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 61 | 181 | 4.681e-02 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 46 | 130 | 4.689e-02 |
| GO:BP | GO:0070129 | regulation of mitochondrial translation | 13 | 26 | 4.696e-02 |
| GO:BP | GO:0060669 | embryonic placenta morphogenesis | 13 | 26 | 4.696e-02 |
| GO:BP | GO:0006515 | protein quality control for misfolded or incompletely synthesized proteins | 13 | 26 | 4.696e-02 |
| GO:BP | GO:0006275 | regulation of DNA replication | 43 | 120 | 4.696e-02 |
| GO:BP | GO:0006007 | glucose catabolic process | 13 | 26 | 4.696e-02 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 27 | 68 | 4.733e-02 |
| GO:BP | GO:0060284 | regulation of cell development | 244 | 847 | 4.733e-02 |
| GO:BP | GO:0071396 | cellular response to lipid | 181 | 613 | 4.756e-02 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 51 | 147 | 4.759e-02 |
| GO:BP | GO:0006970 | response to osmotic stress | 32 | 84 | 4.769e-02 |
| GO:BP | GO:0030323 | respiratory tube development | 67 | 202 | 4.799e-02 |
| GO:BP | GO:0006520 | amino acid metabolic process | 94 | 297 | 4.805e-02 |
| GO:BP | GO:0072578 | neurotransmitter-gated ion channel clustering | 10 | 18 | 4.858e-02 |
| GO:BP | GO:0010544 | negative regulation of platelet activation | 10 | 18 | 4.858e-02 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 10 | 18 | 4.858e-02 |
| GO:BP | GO:0061621 | canonical glycolysis | 10 | 18 | 4.858e-02 |
| GO:BP | GO:0061718 | glucose catabolic process to pyruvate | 10 | 18 | 4.858e-02 |
| GO:BP | GO:0016032 | viral process | 129 | 423 | 4.860e-02 |
| GO:BP | GO:0030534 | adult behavior | 53 | 154 | 4.863e-02 |
| GO:BP | GO:0030509 | BMP signaling pathway | 53 | 154 | 4.863e-02 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 39 | 107 | 4.863e-02 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 161 | 540 | 4.918e-02 |
| GO:BP | GO:0051301 | cell division | 193 | 658 | 4.918e-02 |
| GO:BP | GO:0140029 | exocytic process | 31 | 81 | 4.937e-02 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 31 | 81 | 4.937e-02 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 86 | 269 | 4.953e-02 |
| GO:BP | GO:0014020 | primary neural tube formation | 35 | 94 | 4.963e-02 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 25 | 62 | 4.980e-02 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 14 | 29 | 4.988e-02 |
| KEGG | KEGG:01100 | Metabolic pathways | 516 | 1537 | 3.303e-03 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 47 | 97 | 3.303e-03 |
| KEGG | KEGG:04140 | Autophagy - animal | 72 | 165 | 3.303e-03 |
| KEGG | KEGG:03018 | RNA degradation | 40 | 79 | 3.303e-03 |
| KEGG | KEGG:04260 | Cardiac muscle contraction | 43 | 87 | 3.303e-03 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 65 | 153 | 8.789e-03 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 28 | 53 | 8.789e-03 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 183 | 506 | 8.789e-03 |
| KEGG | KEGG:01200 | Carbon metabolism | 52 | 115 | 8.789e-03 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 45 | 97 | 8.789e-03 |
| KEGG | KEGG:04360 | Axon guidance | 74 | 181 | 1.167e-02 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 34 | 70 | 1.167e-02 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 46 | 102 | 1.167e-02 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 69 | 168 | 1.330e-02 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 68 | 166 | 1.480e-02 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 34 | 73 | 2.436e-02 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 20 | 37 | 2.436e-02 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 112 | 301 | 2.436e-02 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 51 | 121 | 2.546e-02 |
| KEGG | KEGG:04931 | Insulin resistance | 46 | 108 | 2.998e-02 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 24 | 48 | 2.998e-02 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 27 | 56 | 3.004e-02 |
| KEGG | KEGG:04137 | Mitophagy - animal | 44 | 103 | 3.126e-02 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 65 | 164 | 3.126e-02 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 15 | 26 | 3.126e-02 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 24 | 49 | 3.430e-02 |
| KEGG | KEGG:05131 | Shigellosis | 92 | 246 | 3.480e-02 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 61 | 154 | 3.812e-02 |
| KEGG | KEGG:05213 | Endometrial cancer | 27 | 58 | 4.281e-02 |
| KEGG | KEGG:01524 | Platinum drug resistance | 32 | 72 | 4.407e-02 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 129 | 363 | 4.407e-02 |
| KEGG | KEGG:03040 | Spliceosome | 67 | 174 | 4.814e-02 |
| KEGG | KEGG:04714 | Thermogenesis | 86 | 232 | 4.941e-02 |
| KEGG | KEGG:05412 | Arrhythmogenic right ventricular cardiomyopathy | 36 | 84 | 4.941e-02 |
# write.csv(table_motif2_GOKEGG_genes, "data/new/table_motif2_GOKEGG_genes.csv")
#GO:BP
table_motif2_genes_GOBP <- table_motif2_GOKEGG_genes %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
# saveRDS(table_motif2_genes_GOBP, "data/table_motif2_genes_GOBP.RDS")
table_motif2_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("Cormotif Motif 2 Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))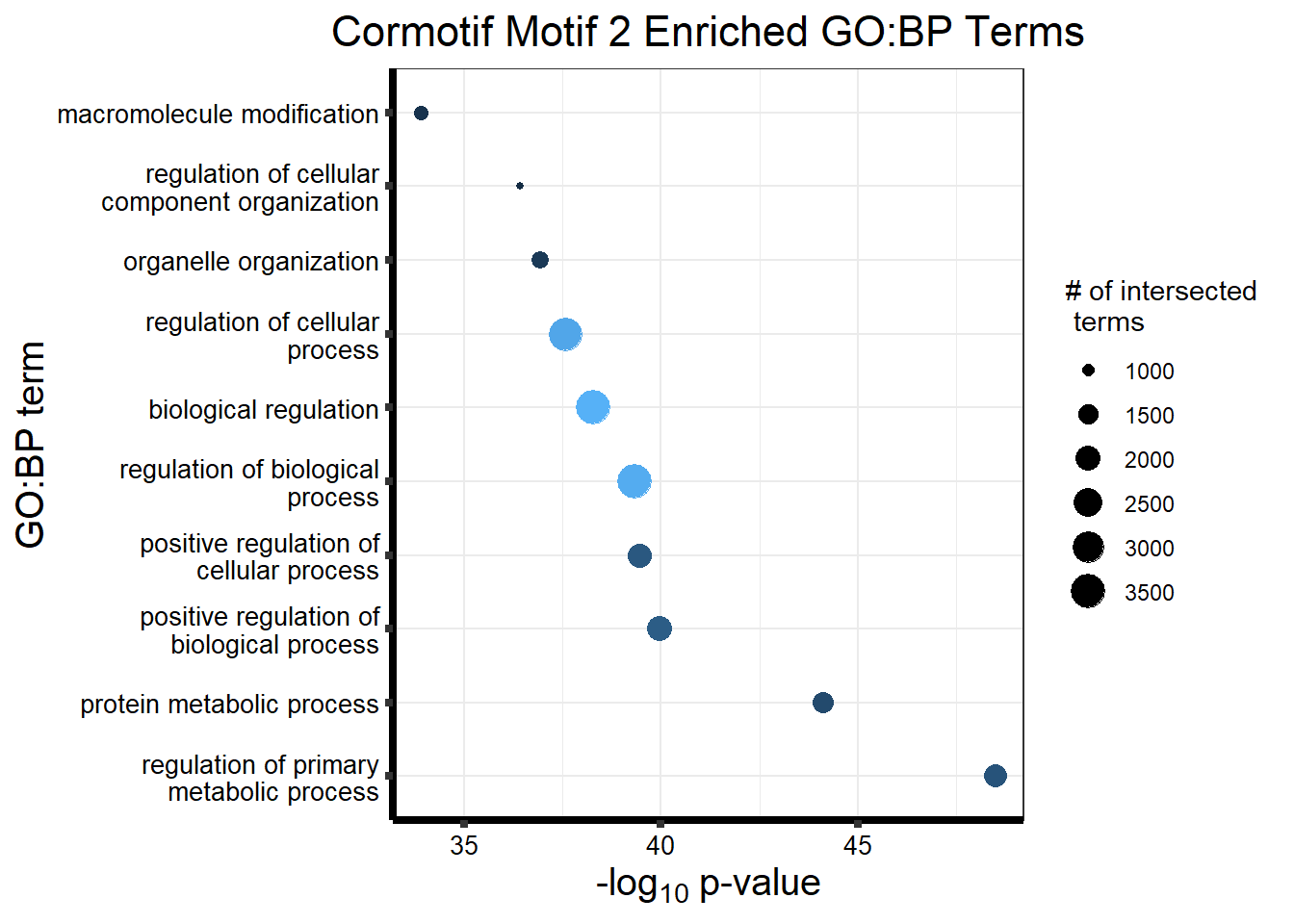
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_motif2_genes_KEGG <- table_motif2_GOKEGG_genes %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_motif2_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("Cormotif Motif 2 DEGs Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))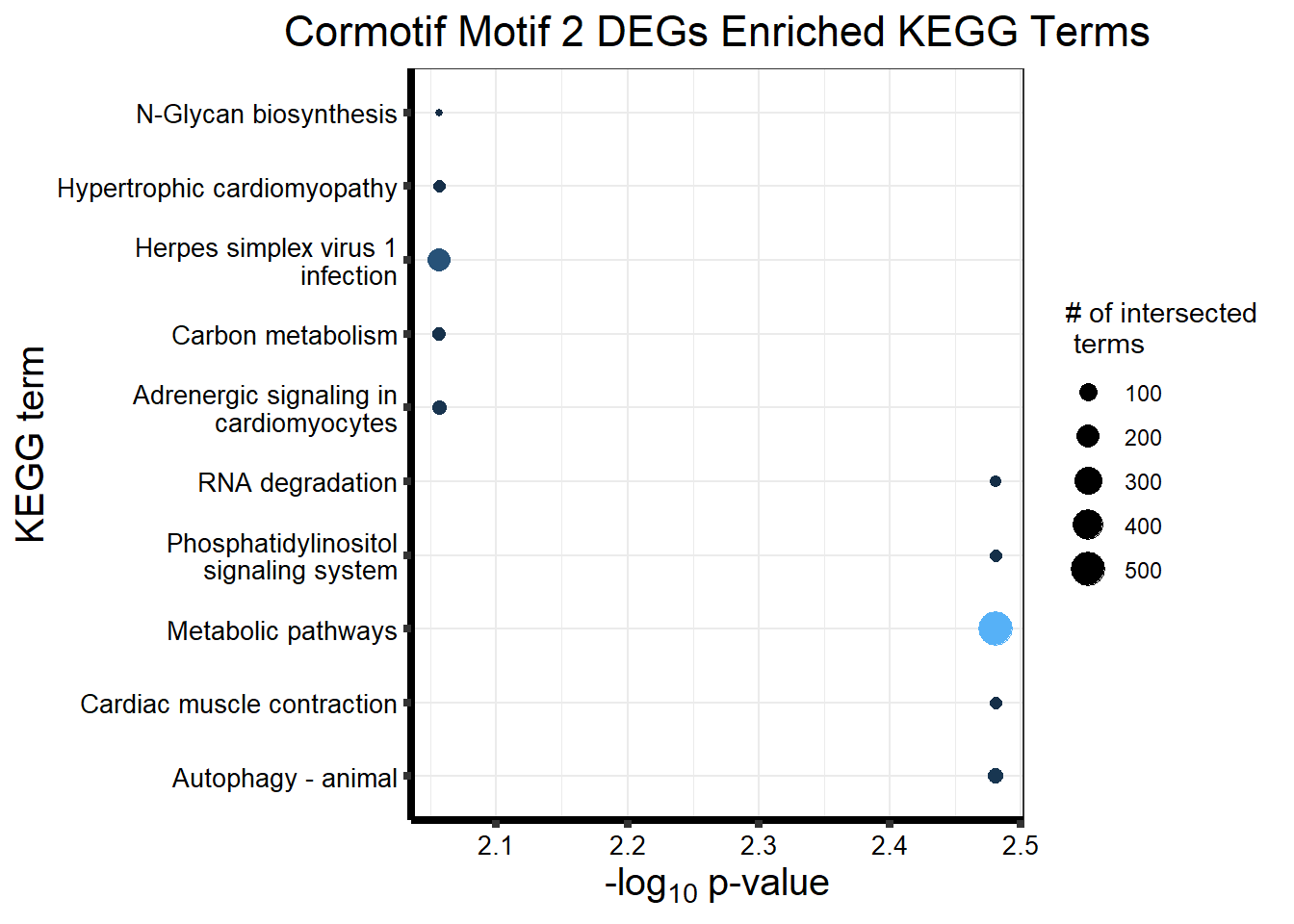
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#####Motif 3 DEGs GO KEGG#####
# motif3_genes_matrix <- as.matrix(final_genes_3)
# colnames(motif3_genes_matrix) <- c("entrezgene_ID")
# saveRDS(motif3_genes_matrix, "data/new/motif3_genes_matrix.RDS")
motif3_genes_matrix <- readRDS("data/new/motif3_genes_matrix.RDS")
length(motif3_genes_matrix)[1] 231#231 genes in this set for motif 3
motif3_mat_GOKEGG <- gost(query = motif3_genes_matrix,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
motif3_GOKEGG_genes <- gostplot(motif3_mat_GOKEGG, capped = FALSE, interactive = TRUE)
motif3_GOKEGG_genestable_motif3_GOKEGG_genes <- motif3_mat_GOKEGG$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_motif3_GOKEGG_genes %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0022402 | cell cycle process | 69 | 1280 | 9.207e-31 |
| GO:BP | GO:0007059 | chromosome segregation | 45 | 427 | 9.207e-31 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 53 | 745 | 1.891e-28 |
| GO:BP | GO:0000278 | mitotic cell cycle | 56 | 892 | 1.301e-27 |
| GO:BP | GO:0007049 | cell cycle | 71 | 1663 | 7.400e-26 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 48 | 720 | 1.962e-24 |
| GO:BP | GO:0051276 | chromosome organization | 42 | 574 | 1.225e-22 |
| GO:BP | GO:0140014 | mitotic nuclear division | 32 | 282 | 1.336e-22 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 33 | 323 | 5.448e-22 |
| GO:BP | GO:0044770 | cell cycle phase transition | 40 | 532 | 5.448e-22 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 27 | 193 | 2.275e-21 |
| GO:BP | GO:0051726 | regulation of cell cycle | 53 | 1087 | 3.696e-21 |
| GO:BP | GO:0006259 | DNA metabolic process | 51 | 1005 | 4.748e-21 |
| GO:BP | GO:0051301 | cell division | 42 | 658 | 1.229e-20 |
| GO:BP | GO:0000819 | sister chromatid segregation | 28 | 234 | 1.908e-20 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 35 | 427 | 1.972e-20 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 35 | 428 | 2.005e-20 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 22 | 121 | 6.255e-20 |
| GO:BP | GO:0000280 | nuclear division | 35 | 452 | 1.086e-19 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 25 | 185 | 1.518e-19 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 36 | 492 | 1.603e-19 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 17 | 59 | 5.873e-19 |
| GO:BP | GO:0006281 | DNA repair | 39 | 621 | 5.873e-19 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 22 | 136 | 6.884e-19 |
| GO:BP | GO:0051310 | metaphase chromosome alignment | 20 | 104 | 1.149e-18 |
| GO:BP | GO:0007051 | spindle organization | 25 | 206 | 1.735e-18 |
| GO:BP | GO:0006260 | DNA replication | 28 | 283 | 1.979e-18 |
| GO:BP | GO:0048285 | organelle fission | 35 | 500 | 1.979e-18 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 17 | 65 | 2.984e-18 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 28 | 289 | 3.157e-18 |
| GO:BP | GO:0051783 | regulation of nuclear division | 22 | 149 | 4.212e-18 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 26 | 248 | 9.826e-18 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 20 | 118 | 1.240e-17 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 22 | 160 | 1.892e-17 |
| GO:BP | GO:0006974 | DNA damage response | 44 | 908 | 1.941e-17 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 30 | 377 | 2.973e-17 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 16 | 62 | 3.970e-17 |
| GO:BP | GO:0050000 | chromosome localization | 20 | 126 | 4.200e-17 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 15 | 51 | 4.914e-17 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 17 | 77 | 4.914e-17 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 15 | 51 | 4.914e-17 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 15 | 51 | 4.914e-17 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 15 | 51 | 4.914e-17 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 18 | 93 | 5.349e-17 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 28 | 327 | 5.797e-17 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 16 | 65 | 7.384e-17 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 15 | 53 | 8.314e-17 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 15 | 53 | 8.314e-17 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 15 | 53 | 8.314e-17 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 18 | 96 | 8.625e-17 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 18 | 97 | 1.030e-16 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 24 | 225 | 1.128e-16 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 18 | 100 | 1.765e-16 |
| GO:BP | GO:0051304 | chromosome separation | 17 | 84 | 1.873e-16 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 15 | 57 | 2.529e-16 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 18 | 107 | 5.966e-16 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 14 | 49 | 8.637e-16 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 14 | 49 | 8.637e-16 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 14 | 49 | 8.637e-16 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 14 | 50 | 1.170e-15 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 19 | 132 | 1.385e-15 |
| GO:BP | GO:0007052 | mitotic spindle organization | 19 | 134 | 1.822e-15 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 14 | 54 | 3.730e-15 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 35 | 670 | 8.784e-15 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 20 | 169 | 9.339e-15 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 20 | 172 | 1.303e-14 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 22 | 242 | 7.483e-14 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 22 | 262 | 3.879e-13 |
| GO:BP | GO:0006996 | organelle organization | 80 | 3594 | 5.571e-13 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 15 | 97 | 1.000e-12 |
| GO:BP | GO:0033043 | regulation of organelle organization | 42 | 1148 | 1.438e-12 |
| GO:BP | GO:0007017 | microtubule-based process | 39 | 999 | 1.774e-12 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 24 | 351 | 1.983e-12 |
| GO:BP | GO:0051225 | spindle assembly | 16 | 136 | 1.028e-11 |
| GO:BP | GO:0033554 | cellular response to stress | 52 | 1830 | 1.603e-11 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 26 | 500 | 8.905e-11 |
| GO:BP | GO:0007080 | mitotic metaphase chromosome alignment | 11 | 63 | 7.329e-10 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 8 | 23 | 1.044e-09 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 15 | 156 | 1.057e-09 |
| GO:BP | GO:0006302 | double-strand break repair | 20 | 319 | 1.275e-09 |
| GO:BP | GO:1903083 | protein localization to condensed chromosome | 7 | 15 | 1.504e-09 |
| GO:BP | GO:0034501 | protein localization to kinetochore | 7 | 15 | 1.504e-09 |
| GO:BP | GO:0051656 | establishment of organelle localization | 24 | 491 | 2.209e-09 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 14 | 139 | 2.494e-09 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 12 | 93 | 3.174e-09 |
| GO:BP | GO:0007010 | cytoskeleton organization | 43 | 1529 | 3.474e-09 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 9 | 41 | 5.251e-09 |
| GO:BP | GO:0006310 | DNA recombination | 20 | 347 | 5.251e-09 |
| GO:BP | GO:0034502 | protein localization to chromosome | 13 | 126 | 8.517e-09 |
| GO:BP | GO:0000725 | recombinational repair | 15 | 193 | 1.905e-08 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 12 | 114 | 3.302e-08 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 21 | 430 | 3.471e-08 |
| GO:BP | GO:0051640 | organelle localization | 25 | 620 | 4.270e-08 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 19 | 352 | 4.305e-08 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 12 | 123 | 7.646e-08 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 12 | 124 | 8.308e-08 |
| GO:BP | GO:0051716 | cellular response to stimulus | 111 | 7376 | 8.331e-08 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 8 | 40 | 1.135e-07 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 11 | 105 | 1.630e-07 |
| GO:BP | GO:0006338 | chromatin remodeling | 26 | 725 | 2.081e-07 |
| GO:BP | GO:0034508 | centromere complex assembly | 7 | 29 | 2.646e-07 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 12 | 139 | 2.890e-07 |
| GO:BP | GO:0051338 | regulation of transferase activity | 21 | 498 | 4.133e-07 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 11 | 118 | 5.351e-07 |
| GO:BP | GO:0006275 | regulation of DNA replication | 11 | 120 | 6.324e-07 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 10 | 94 | 6.652e-07 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 8 | 50 | 6.692e-07 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 93 | 5920 | 6.943e-07 |
| GO:BP | GO:0006325 | chromatin organization | 28 | 884 | 6.943e-07 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 13 | 188 | 9.290e-07 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 15 | 262 | 9.911e-07 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 9 | 74 | 9.931e-07 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 14 | 241 | 2.362e-06 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 8 | 59 | 2.411e-06 |
| GO:BP | GO:0006950 | response to stress | 69 | 3948 | 2.540e-06 |
| GO:BP | GO:0007098 | centrosome cycle | 11 | 140 | 2.834e-06 |
| GO:BP | GO:0050896 | response to stimulus | 122 | 8999 | 4.140e-06 |
| GO:BP | GO:0016043 | cellular component organization | 114 | 8184 | 4.262e-06 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 7 | 44 | 4.888e-06 |
| GO:BP | GO:0048518 | positive regulation of biological process | 94 | 6264 | 5.314e-06 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 7 | 45 | 5.647e-06 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 49 | 2433 | 6.119e-06 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 27 | 928 | 6.206e-06 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 11 | 153 | 6.551e-06 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 8 | 70 | 8.561e-06 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 6 | 30 | 8.563e-06 |
| GO:BP | GO:0090224 | regulation of spindle organization | 7 | 48 | 8.563e-06 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 6 | 32 | 1.257e-05 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 11 | 164 | 1.257e-05 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 12 | 200 | 1.257e-05 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 4 | 8 | 1.328e-05 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 114 | 8393 | 1.734e-05 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 22 | 701 | 2.392e-05 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 85 | 5629 | 2.421e-05 |
| GO:BP | GO:0010212 | response to ionizing radiation | 10 | 141 | 2.423e-05 |
| GO:BP | GO:0009314 | response to radiation | 17 | 437 | 2.455e-05 |
| GO:BP | GO:0051321 | meiotic cell cycle | 14 | 297 | 2.455e-05 |
| GO:BP | GO:0006270 | DNA replication initiation | 6 | 37 | 2.919e-05 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 5 | 21 | 3.089e-05 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 9 | 114 | 3.359e-05 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 19 | 554 | 3.565e-05 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 13 | 266 | 3.896e-05 |
| GO:BP | GO:0051302 | regulation of cell division | 11 | 188 | 4.400e-05 |
| GO:BP | GO:0033260 | nuclear DNA replication | 6 | 41 | 5.242e-05 |
| GO:BP | GO:0070925 | organelle assembly | 27 | 1046 | 5.275e-05 |
| GO:BP | GO:0048519 | negative regulation of biological process | 86 | 5834 | 5.297e-05 |
| GO:BP | GO:0035556 | intracellular signal transduction | 53 | 2965 | 5.841e-05 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 5 | 24 | 5.841e-05 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 5 | 24 | 5.841e-05 |
| GO:BP | GO:0007143 | female meiotic nuclear division | 6 | 42 | 5.841e-05 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 5 | 25 | 7.150e-05 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 5 | 25 | 7.150e-05 |
| GO:BP | GO:0140013 | meiotic nuclear division | 11 | 202 | 8.245e-05 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 7 | 69 | 8.699e-05 |
| GO:BP | GO:0051781 | positive regulation of cell division | 8 | 98 | 9.171e-05 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 7 | 71 | 1.042e-04 |
| GO:BP | GO:1903490 | positive regulation of mitotic cytokinesis | 4 | 13 | 1.091e-04 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 13 | 296 | 1.130e-04 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 6 | 48 | 1.226e-04 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 13 | 299 | 1.244e-04 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 10 | 173 | 1.285e-04 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 15 | 402 | 1.508e-04 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 7 | 76 | 1.575e-04 |
| GO:BP | GO:0031297 | replication fork processing | 6 | 51 | 1.704e-04 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 4 | 15 | 1.924e-04 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 9 | 144 | 1.924e-04 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 4 | 15 | 1.924e-04 |
| GO:BP | GO:0071478 | cellular response to radiation | 10 | 182 | 1.924e-04 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 11 | 224 | 1.996e-04 |
| GO:BP | GO:1902412 | regulation of mitotic cytokinesis | 4 | 16 | 2.507e-04 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 6 | 55 | 2.548e-04 |
| GO:BP | GO:0031507 | heterochromatin formation | 9 | 151 | 2.751e-04 |
| GO:BP | GO:0000910 | cytokinesis | 10 | 192 | 2.961e-04 |
| GO:BP | GO:0050789 | regulation of biological process | 146 | 12336 | 2.961e-04 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 5 | 34 | 3.034e-04 |
| GO:BP | GO:0043549 | regulation of kinase activity | 15 | 431 | 3.158e-04 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 11 | 242 | 3.918e-04 |
| GO:BP | GO:0006301 | postreplication repair | 5 | 36 | 3.979e-04 |
| GO:BP | GO:0050794 | regulation of cellular process | 142 | 11946 | 4.151e-04 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 12 | 291 | 4.276e-04 |
| GO:BP | GO:0019953 | sexual reproduction | 26 | 1122 | 4.382e-04 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 5 | 37 | 4.417e-04 |
| GO:BP | GO:0009966 | regulation of signal transduction | 51 | 3034 | 4.417e-04 |
| GO:BP | GO:0022616 | DNA strand elongation | 5 | 37 | 4.417e-04 |
| GO:BP | GO:0051649 | establishment of localization in cell | 38 | 2010 | 5.228e-04 |
| GO:BP | GO:0000076 | DNA replication checkpoint signaling | 4 | 20 | 5.889e-04 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 4 | 20 | 5.889e-04 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 7 | 96 | 6.342e-04 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 22 | 884 | 6.504e-04 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 9 | 172 | 6.981e-04 |
| GO:BP | GO:0051383 | kinetochore organization | 4 | 21 | 7.069e-04 |
| GO:BP | GO:0007099 | centriole replication | 5 | 43 | 8.925e-04 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 8 | 138 | 8.984e-04 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 5 | 44 | 9.889e-04 |
| GO:BP | GO:0046599 | regulation of centriole replication | 4 | 23 | 1.009e-03 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 6 | 72 | 1.048e-03 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 5 | 45 | 1.087e-03 |
| GO:BP | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | 3 | 9 | 1.106e-03 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 3 | 9 | 1.106e-03 |
| GO:BP | GO:0051298 | centrosome duplication | 6 | 74 | 1.199e-03 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 15 | 492 | 1.252e-03 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 5 | 47 | 1.311e-03 |
| GO:BP | GO:0065007 | biological regulation | 147 | 12743 | 1.313e-03 |
| GO:BP | GO:0065009 | regulation of molecular function | 30 | 1496 | 1.313e-03 |
| GO:BP | GO:0140273 | repair of mitotic kinetochore microtubule attachment defect | 2 | 2 | 1.402e-03 |
| GO:BP | GO:0140274 | repair of kinetochore microtubule attachment defect | 2 | 2 | 1.402e-03 |
| GO:BP | GO:0099607 | lateral attachment of mitotic spindle microtubules to kinetochore | 2 | 2 | 1.402e-03 |
| GO:BP | GO:0098534 | centriole assembly | 5 | 48 | 1.410e-03 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 25 | 1144 | 1.413e-03 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 3 | 10 | 1.466e-03 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 3 | 10 | 1.466e-03 |
| GO:BP | GO:2000104 | negative regulation of DNA-templated DNA replication | 3 | 10 | 1.466e-03 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 3 | 10 | 1.466e-03 |
| GO:BP | GO:0008156 | negative regulation of DNA replication | 4 | 26 | 1.518e-03 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 6 | 80 | 1.725e-03 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 5 | 51 | 1.820e-03 |
| GO:BP | GO:0022414 | reproductive process | 31 | 1608 | 1.902e-03 |
| GO:BP | GO:0051641 | cellular localization | 56 | 3661 | 1.905e-03 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 3 | 11 | 1.938e-03 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 3 | 11 | 1.938e-03 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 15 | 517 | 1.967e-03 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 4 | 28 | 1.975e-03 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 5 | 54 | 2.320e-03 |
| GO:BP | GO:0007056 | spindle assembly involved in female meiosis | 3 | 12 | 2.498e-03 |
| GO:BP | GO:0000022 | mitotic spindle elongation | 3 | 12 | 2.498e-03 |
| GO:BP | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 3 | 12 | 2.498e-03 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 4 | 31 | 2.903e-03 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 59 | 3993 | 2.942e-03 |
| GO:BP | GO:0090232 | positive regulation of spindle checkpoint | 3 | 13 | 3.154e-03 |
| GO:BP | GO:0006264 | mitochondrial DNA replication | 3 | 13 | 3.154e-03 |
| GO:BP | GO:0007144 | female meiosis I | 3 | 13 | 3.154e-03 |
| GO:BP | GO:0023051 | regulation of signaling | 53 | 3478 | 3.206e-03 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 74 | 5390 | 3.323e-03 |
| GO:BP | GO:0010646 | regulation of cell communication | 53 | 3486 | 3.355e-03 |
| GO:BP | GO:0043009 | chordate embryonic development | 17 | 671 | 3.355e-03 |
| GO:BP | GO:0007292 | female gamete generation | 8 | 174 | 3.666e-03 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 5 | 61 | 3.810e-03 |
| GO:BP | GO:0046385 | deoxyribose phosphate biosynthetic process | 3 | 14 | 3.810e-03 |
| GO:BP | GO:0009265 | 2’-deoxyribonucleotide biosynthetic process | 3 | 14 | 3.810e-03 |
| GO:BP | GO:0009263 | deoxyribonucleotide biosynthetic process | 3 | 14 | 3.810e-03 |
| GO:BP | GO:0051231 | spindle elongation | 3 | 14 | 3.810e-03 |
| GO:BP | GO:0051255 | spindle midzone assembly | 3 | 14 | 3.810e-03 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 6 | 95 | 3.924e-03 |
| GO:BP | GO:0006282 | regulation of DNA repair | 9 | 225 | 4.170e-03 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 17 | 692 | 4.586e-03 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 3 | 15 | 4.628e-03 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 3 | 15 | 4.628e-03 |
| GO:BP | GO:0034728 | nucleosome organization | 7 | 138 | 4.674e-03 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 9 | 234 | 5.404e-03 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 11 | 339 | 5.404e-03 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 8 | 186 | 5.404e-03 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 11 | 339 | 5.404e-03 |
| GO:BP | GO:0031399 | regulation of protein modification process | 22 | 1049 | 5.546e-03 |
| GO:BP | GO:0007276 | gamete generation | 19 | 841 | 5.635e-03 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 7 | 143 | 5.635e-03 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 4 | 38 | 5.733e-03 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 4 | 39 | 6.316e-03 |
| GO:BP | GO:0009987 | cellular process | 200 | 20247 | 6.334e-03 |
| GO:BP | GO:0006997 | nucleus organization | 7 | 147 | 6.417e-03 |
| GO:BP | GO:0071932 | replication fork reversal | 2 | 4 | 6.417e-03 |
| GO:BP | GO:0070571 | negative regulation of neuron projection regeneration | 3 | 17 | 6.417e-03 |
| GO:BP | GO:0031508 | pericentric heterochromatin formation | 2 | 4 | 6.417e-03 |
| GO:BP | GO:0007089 | traversing start control point of mitotic cell cycle | 2 | 4 | 6.417e-03 |
| GO:BP | GO:0007057 | spindle assembly involved in female meiosis I | 2 | 4 | 6.417e-03 |
| GO:BP | GO:0031573 | mitotic intra-S DNA damage checkpoint signaling | 3 | 17 | 6.417e-03 |
| GO:BP | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition | 3 | 17 | 6.417e-03 |
| GO:BP | GO:0051382 | kinetochore assembly | 3 | 17 | 6.417e-03 |
| GO:BP | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity | 2 | 4 | 6.417e-03 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 5 | 70 | 6.509e-03 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 4 | 41 | 7.293e-03 |
| GO:BP | GO:1902101 | positive regulation of metaphase/anaphase transition of cell cycle | 3 | 18 | 7.407e-03 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 4 | 42 | 7.943e-03 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 8 | 202 | 8.541e-03 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 3 | 19 | 8.639e-03 |
| GO:BP | GO:0106030 | neuron projection fasciculation | 3 | 20 | 1.002e-02 |
| GO:BP | GO:0007413 | axonal fasciculation | 3 | 20 | 1.002e-02 |
| GO:BP | GO:1902292 | cell cycle DNA replication initiation | 2 | 5 | 1.007e-02 |
| GO:BP | GO:1902315 | nuclear cell cycle DNA replication initiation | 2 | 5 | 1.007e-02 |
| GO:BP | GO:0043137 | DNA replication, removal of RNA primer | 2 | 5 | 1.007e-02 |
| GO:BP | GO:1905784 | regulation of anaphase-promoting complex-dependent catabolic process | 2 | 5 | 1.007e-02 |
| GO:BP | GO:0007165 | signal transduction | 78 | 6002 | 1.007e-02 |
| GO:BP | GO:1902975 | mitotic DNA replication initiation | 2 | 5 | 1.007e-02 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 2 | 5 | 1.007e-02 |
| GO:BP | GO:0006334 | nucleosome assembly | 6 | 118 | 1.063e-02 |
| GO:BP | GO:0006284 | base-excision repair | 4 | 46 | 1.073e-02 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 3 | 21 | 1.120e-02 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 4 | 47 | 1.156e-02 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 3 | 22 | 1.275e-02 |
| GO:BP | GO:0000212 | meiotic spindle organization | 3 | 22 | 1.275e-02 |
| GO:BP | GO:0019222 | regulation of metabolic process | 88 | 7035 | 1.302e-02 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 5 | 83 | 1.307e-02 |
| GO:BP | GO:0033365 | protein localization to organelle | 23 | 1209 | 1.334e-02 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 6 | 124 | 1.334e-02 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 29 | 1680 | 1.399e-02 |
| GO:BP | GO:0090402 | oncogene-induced cell senescence | 2 | 6 | 1.420e-02 |
| GO:BP | GO:0016321 | female meiosis chromosome segregation | 2 | 6 | 1.420e-02 |
| GO:BP | GO:0071139 | resolution of DNA recombination intermediates | 2 | 6 | 1.420e-02 |
| GO:BP | GO:0045162 | clustering of voltage-gated sodium channels | 2 | 6 | 1.420e-02 |
| GO:BP | GO:0032506 | cytokinetic process | 4 | 50 | 1.420e-02 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 13 | 519 | 1.566e-02 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 82 | 6492 | 1.583e-02 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 5 | 89 | 1.719e-02 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 8 | 230 | 1.767e-02 |
| GO:BP | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 3 | 25 | 1.772e-02 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 51 | 3597 | 1.775e-02 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 2 | 7 | 1.905e-02 |
| GO:BP | GO:0032954 | regulation of cytokinetic process | 2 | 7 | 1.905e-02 |
| GO:BP | GO:0044785 | metaphase/anaphase transition of meiotic cell cycle | 2 | 7 | 1.905e-02 |
| GO:BP | GO:1903436 | regulation of mitotic cytokinetic process | 2 | 7 | 1.905e-02 |
| GO:BP | GO:1903438 | positive regulation of mitotic cytokinetic process | 2 | 7 | 1.905e-02 |
| GO:BP | GO:0007154 | cell communication | 82 | 6540 | 1.932e-02 |
| GO:BP | GO:0019985 | translesion synthesis | 3 | 26 | 1.938e-02 |
| GO:BP | GO:0035295 | tube development | 21 | 1101 | 1.981e-02 |
| GO:BP | GO:0008406 | gonad development | 8 | 238 | 2.104e-02 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 14 | 606 | 2.104e-02 |
| GO:BP | GO:0007548 | sex differentiation | 9 | 294 | 2.125e-02 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 8 | 239 | 2.146e-02 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 4 | 57 | 2.146e-02 |
| GO:BP | GO:0031102 | neuron projection regeneration | 4 | 58 | 2.281e-02 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 6 | 141 | 2.345e-02 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 8 | 243 | 2.349e-02 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 8 | 244 | 2.402e-02 |
| GO:BP | GO:2001033 | negative regulation of double-strand break repair via nonhomologous end joining | 2 | 8 | 2.408e-02 |
| GO:BP | GO:0097681 | double-strand break repair via alternative nonhomologous end joining | 2 | 8 | 2.408e-02 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 2 | 8 | 2.408e-02 |
| GO:BP | GO:0046785 | microtubule polymerization | 5 | 98 | 2.433e-02 |
| GO:BP | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 3 | 29 | 2.545e-02 |
| GO:BP | GO:0023052 | signaling | 81 | 6515 | 2.585e-02 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 3 | 30 | 2.785e-02 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 7 | 197 | 2.785e-02 |
| GO:BP | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis | 2 | 9 | 2.950e-02 |
| GO:BP | GO:1904776 | regulation of protein localization to cell cortex | 2 | 9 | 2.950e-02 |
| GO:BP | GO:0051293 | establishment of spindle localization | 4 | 63 | 2.950e-02 |
| GO:BP | GO:0000915 | actomyosin contractile ring assembly | 2 | 9 | 2.950e-02 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 2 | 9 | 2.950e-02 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 2 | 9 | 2.950e-02 |
| GO:BP | GO:0032201 | telomere maintenance via semi-conservative replication | 2 | 9 | 2.950e-02 |
| GO:BP | GO:0046602 | regulation of mitotic centrosome separation | 2 | 9 | 2.950e-02 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 9 | 312 | 2.950e-02 |
| GO:BP | GO:0006468 | protein phosphorylation | 22 | 1227 | 3.045e-02 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 19 | 1000 | 3.106e-02 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 5 | 105 | 3.127e-02 |
| GO:BP | GO:0006298 | mismatch repair | 3 | 32 | 3.216e-02 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 3 | 32 | 3.216e-02 |
| GO:BP | GO:0048856 | anatomical structure development | 75 | 5997 | 3.377e-02 |
| GO:BP | GO:0016310 | phosphorylation | 23 | 1320 | 3.440e-02 |
| GO:BP | GO:0009411 | response to UV | 6 | 155 | 3.456e-02 |
| GO:BP | GO:1901031 | regulation of response to reactive oxygen species | 2 | 10 | 3.559e-02 |
| GO:BP | GO:0044837 | actomyosin contractile ring organization | 2 | 10 | 3.559e-02 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 23 | 1326 | 3.594e-02 |
| GO:BP | GO:0051653 | spindle localization | 4 | 68 | 3.686e-02 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 4 | 68 | 3.686e-02 |
| GO:BP | GO:0030010 | establishment of cell polarity | 6 | 159 | 3.847e-02 |
| GO:BP | GO:0035239 | tube morphogenesis | 17 | 879 | 4.091e-02 |
| GO:BP | GO:0035090 | maintenance of apical/basal cell polarity | 2 | 11 | 4.191e-02 |
| GO:BP | GO:0033504 | floor plate development | 2 | 11 | 4.191e-02 |
| GO:BP | GO:0033314 | mitotic DNA replication checkpoint signaling | 2 | 11 | 4.191e-02 |
| GO:BP | GO:0045199 | maintenance of epithelial cell apical/basal polarity | 2 | 11 | 4.191e-02 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 2 | 11 | 4.191e-02 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 2 | 11 | 4.191e-02 |
| GO:BP | GO:0006915 | apoptotic process | 30 | 1923 | 4.273e-02 |
| GO:BP | GO:0032502 | developmental process | 80 | 6553 | 4.317e-02 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 6 | 164 | 4.347e-02 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 9 | 336 | 4.443e-02 |
| GO:BP | GO:0023057 | negative regulation of signaling | 24 | 1435 | 4.457e-02 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 24 | 1436 | 4.483e-02 |
| GO:BP | GO:0007275 | multicellular organism development | 61 | 4727 | 4.510e-02 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 3 | 37 | 4.549e-02 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 29 | 1855 | 4.705e-02 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 2 | 12 | 4.798e-02 |
| GO:BP | GO:0072710 | response to hydroxyurea | 2 | 12 | 4.798e-02 |
| GO:BP | GO:0046549 | retinal cone cell development | 2 | 12 | 4.798e-02 |
| GO:BP | GO:0045161 | neuronal ion channel clustering | 2 | 12 | 4.798e-02 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 2 | 12 | 4.798e-02 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 2 | 12 | 4.798e-02 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 3 | 38 | 4.801e-02 |
| KEGG | KEGG:04110 | Cell cycle | 18 | 157 | 1.045e-11 |
| KEGG | KEGG:03030 | DNA replication | 10 | 36 | 2.743e-10 |
| KEGG | KEGG:03430 | Mismatch repair | 5 | 23 | 2.299e-04 |
| KEGG | KEGG:04115 | p53 signaling pathway | 7 | 74 | 6.435e-04 |
| KEGG | KEGG:03440 | Homologous recombination | 5 | 41 | 2.625e-03 |
| KEGG | KEGG:03410 | Base excision repair | 5 | 44 | 3.088e-03 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 5 | 54 | 7.063e-03 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 5 | 61 | 1.094e-02 |
# write.csv(table_motif3_GOKEGG_genes, "data/new/table_motif3_GOKEGG_genes.csv")
#GO:BP
table_motif3_genes_GOBP <- table_motif3_GOKEGG_genes %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
# saveRDS(table_motif3_genes_GOBP, "data/table_motif3_genes_GOBP.RDS")
table_motif3_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("Cormotif Motif 3 Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))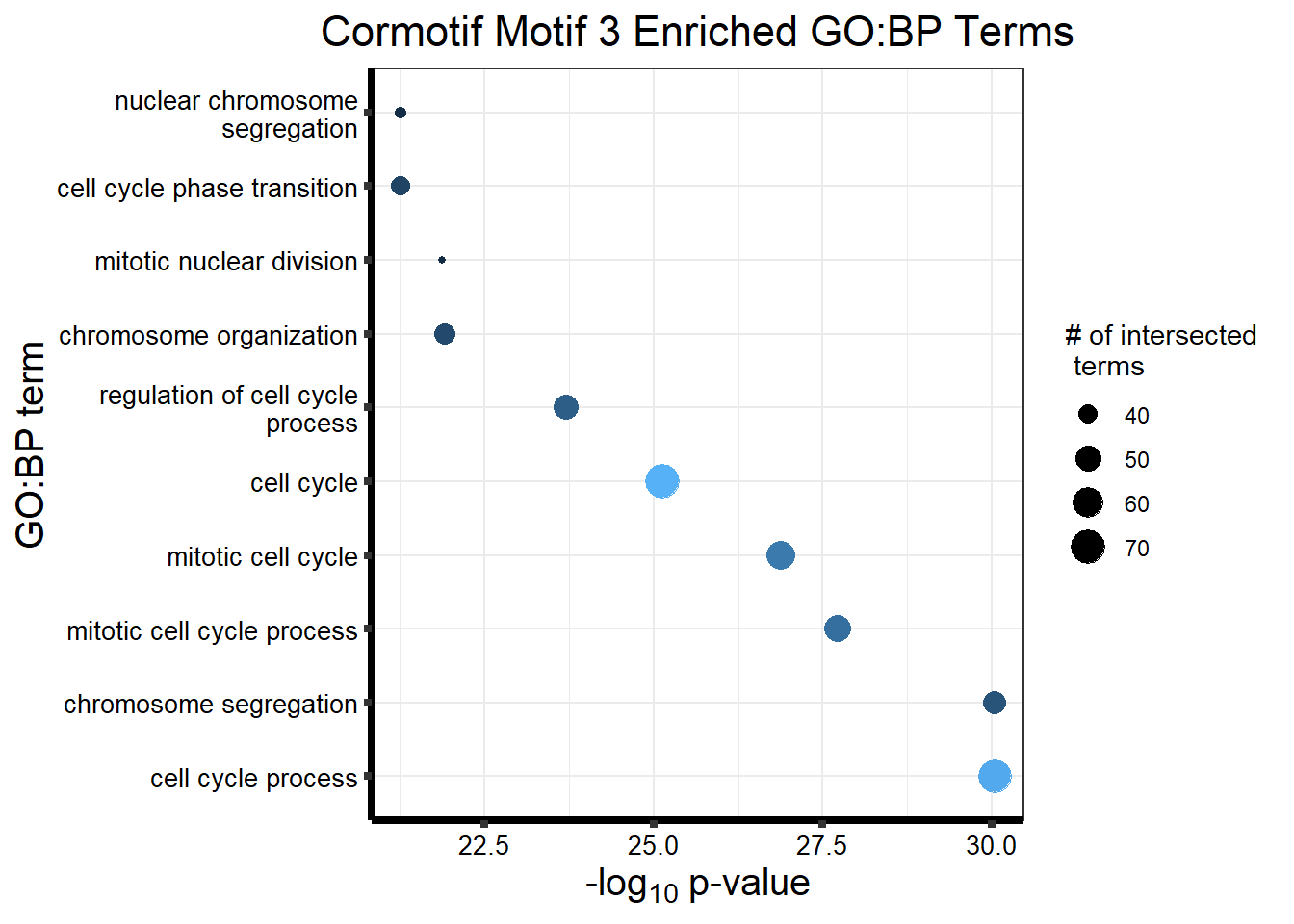
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_motif3_genes_KEGG <- table_motif3_GOKEGG_genes %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_motif3_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("Cormotif Motif 3 DEGs Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))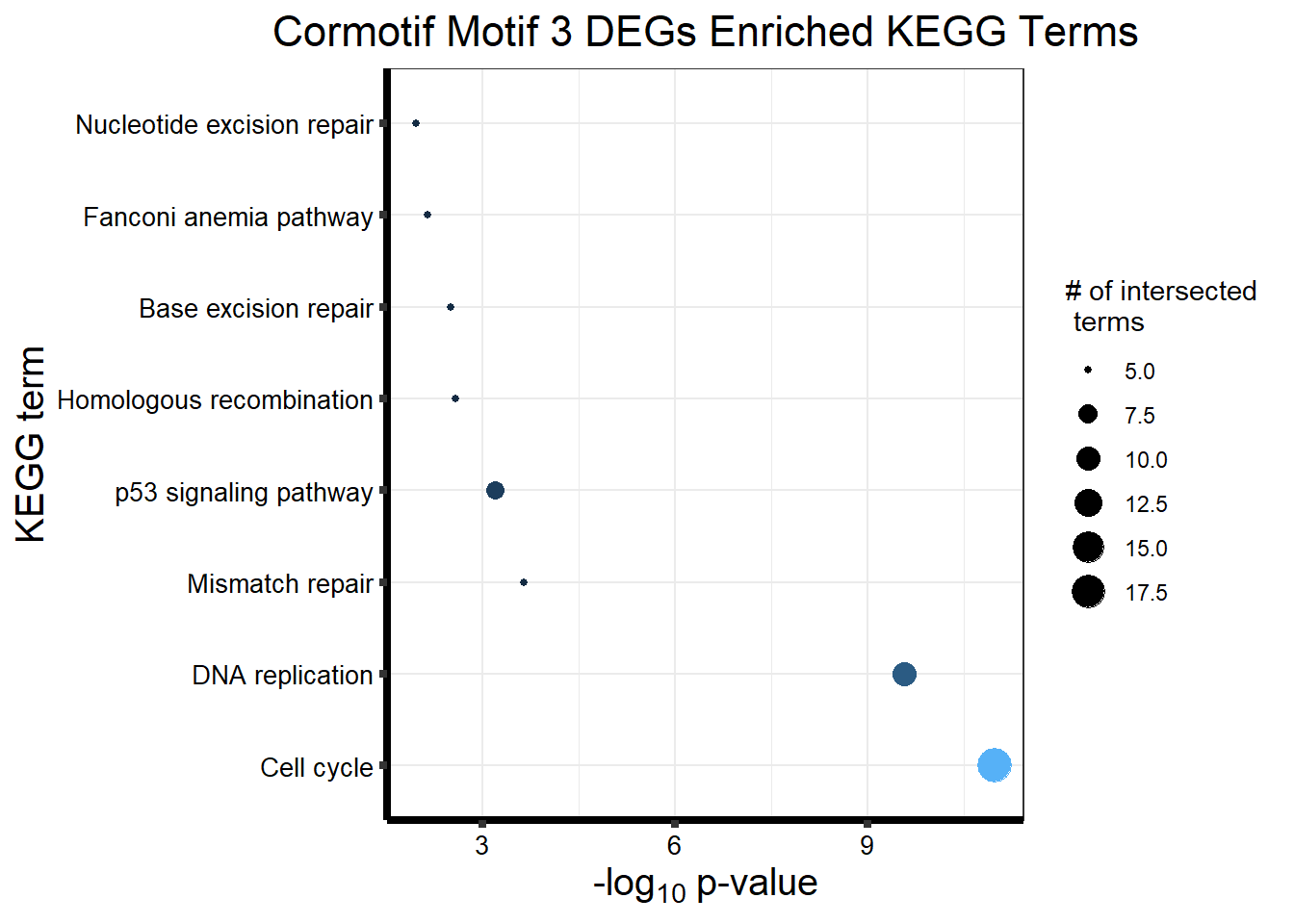
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
###Look at Unassigned Gene Set Cormotif
#now let's pull out the genes that were not assigned to a motif from above
#this one is giving me an issue with wflow_build so I'll fix it later
unassigned_genes_list <- readRDS("data/new/Cormotif/unassigned_genes_Cormotif.RDS")
#with these unassigned genes, I want to plot logFC and GO/KEGG of unassigned genes
#####logFC#####
##no motif genes
unassign_genes_fin <- unassigned_genes_list
length(unassign_genes_fin)[1] 2232#2232 genes
#Combine the toptables I have from pairwise analysis into a single dataframe
combined_toptables_dxr <- bind_rows(
d24_toptable_dxr,
d24r_toptable_dxr,
d144r_toptable_dxr)
#Filter the data based on each motif
# filt_toptable_dxr_fin_unassign <- as.data.frame(combined_toptables_dxr) %>%
# dplyr::filter(Entrez_ID %in% unassign_genes_fin) %>%
# mutate(absFC = abs(logFC)) %>%
# mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
# ggplot(., aes(x = time, y = logFC))+
# geom_boxplot(aes(fill = time))+
# scale_fill_manual(values = time_col)+
# guides(fill = guide_legend(title = "Time"))+
# theme_bw()+
# xlab(" ")+
# ylab("logFC")+
# theme_bw()+
# ggtitle("LogFC of Unassigned Genes (n=2232)")+
# theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
# axis.title = element_text(size = 15, colour = "black"),
# strip.background = element_rect(fill = "#CAD899"),
# axis.text.x = element_text(size = 8, colour = "white", angle = 15),
# strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#
# filt_toptable_dxr_fin_unassign
# filt_toptable_abs_fin_unassign <- as.data.frame(combined_toptables_dxr) %>%
# dplyr::filter(Entrez_ID %in% unassign_genes_fin) %>%
# mutate(absFC = abs(logFC)) %>%
# mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
# ggplot(., aes(x = time, y = absFC))+
# geom_boxplot(aes(fill = time))+
# scale_fill_manual(values = time_col)+
# guides(fill = guide_legend(title = "Time"))+
# theme_bw()+
# xlab(" ")+
# ylab("|logFC|")+
# theme_bw()+
# ggtitle("Abs LogFC of Unassigned Genes (n=2232)")+
# theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
# axis.title = element_text(size = 15, colour = "black"),
# strip.background = element_rect(fill = "#CAD899"),
# axis.text.x = element_text(size = 8, colour = "white", angle = 15),
# strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#
# filt_toptable_abs_fin_unassign
####GO KEGG####
unassigned_genes_matrix <- as.matrix(unassigned_genes_list)
length(unassigned_genes_matrix)[1] 2232#2232 genes in this set
unassign_mat_GOKEGG <- gost(query = unassigned_genes_matrix,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
unassign_GOKEGG_genes <- gostplot(unassign_mat_GOKEGG, capped = FALSE, interactive = TRUE)
unassign_GOKEGG_genestable_unassign_GOKEGG_genes <- unassign_mat_GOKEGG$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_unassign_GOKEGG_genes %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0022402 | cell cycle process | 273 | 1280 | 3.208e-39 |
| GO:BP | GO:0007049 | cell cycle | 320 | 1663 | 2.070e-37 |
| GO:BP | GO:0000278 | mitotic cell cycle | 205 | 892 | 1.338e-33 |
| GO:BP | GO:0006996 | organelle organization | 537 | 3594 | 4.972e-33 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 179 | 745 | 9.607e-32 |
| GO:BP | GO:0051301 | cell division | 156 | 658 | 1.128e-26 |
| GO:BP | GO:0033554 | cellular response to stress | 304 | 1830 | 8.255e-24 |
| GO:BP | GO:0007059 | chromosome segregation | 110 | 427 | 2.368e-21 |
| GO:BP | GO:0051726 | regulation of cell cycle | 202 | 1087 | 1.158e-20 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 149 | 720 | 3.253e-19 |
| GO:BP | GO:0048285 | organelle fission | 117 | 500 | 3.529e-19 |
| GO:BP | GO:0000280 | nuclear division | 109 | 452 | 7.327e-19 |
| GO:BP | GO:0044770 | cell cycle phase transition | 120 | 532 | 2.257e-18 |
| GO:BP | GO:0065007 | biological regulation | 1348 | 12743 | 3.593e-18 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 712 | 5920 | 9.277e-17 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 84 | 323 | 1.799e-16 |
| GO:BP | GO:0050789 | regulation of biological process | 1304 | 12336 | 1.799e-16 |
| GO:BP | GO:0035556 | intracellular signal transduction | 406 | 2965 | 2.038e-16 |
| GO:BP | GO:0140014 | mitotic nuclear division | 77 | 282 | 2.306e-16 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 656 | 5390 | 3.227e-16 |
| GO:BP | GO:0051276 | chromosome organization | 121 | 574 | 3.227e-16 |
| GO:BP | GO:0006974 | DNA damage response | 166 | 908 | 3.356e-16 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 61 | 193 | 3.798e-16 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 99 | 428 | 6.365e-16 |
| GO:BP | GO:0051641 | cellular localization | 477 | 3661 | 7.907e-16 |
| GO:BP | GO:0050794 | regulation of cellular process | 1265 | 11946 | 1.032e-15 |
| GO:BP | GO:0000819 | sister chromatid segregation | 67 | 234 | 2.195e-15 |
| GO:BP | GO:0048518 | positive regulation of biological process | 735 | 6264 | 6.012e-15 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 94 | 427 | 1.162e-13 |
| GO:BP | GO:0032502 | developmental process | 754 | 6553 | 2.467e-13 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 102 | 492 | 4.199e-13 |
| GO:BP | GO:0006260 | DNA replication | 70 | 283 | 1.454e-12 |
| GO:BP | GO:0019538 | protein metabolic process | 568 | 4721 | 2.242e-12 |
| GO:BP | GO:0006259 | DNA metabolic process | 166 | 1005 | 3.752e-12 |
| GO:BP | GO:0070925 | organelle assembly | 168 | 1046 | 3.029e-11 |
| GO:BP | GO:0036211 | protein modification process | 369 | 2846 | 3.029e-11 |
| GO:BP | GO:0023051 | regulation of signaling | 435 | 3478 | 3.149e-11 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 47 | 160 | 4.629e-11 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 361 | 2782 | 4.991e-11 |
| GO:BP | GO:0051179 | localization | 642 | 5555 | 6.201e-11 |
| GO:BP | GO:0010646 | regulation of cell communication | 434 | 3486 | 6.383e-11 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 73 | 327 | 7.886e-11 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 485 | 3990 | 8.037e-11 |
| GO:BP | GO:0007275 | multicellular organism development | 559 | 4727 | 8.630e-11 |
| GO:BP | GO:0008104 | protein localization | 358 | 2770 | 9.831e-11 |
| GO:BP | GO:0051649 | establishment of localization in cell | 276 | 2010 | 1.004e-10 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 50 | 185 | 2.185e-10 |
| GO:BP | GO:0043412 | macromolecule modification | 383 | 3030 | 2.719e-10 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 62 | 262 | 2.791e-10 |
| GO:BP | GO:0033036 | macromolecule localization | 403 | 3228 | 3.432e-10 |
| GO:BP | GO:0033043 | regulation of organelle organization | 176 | 1148 | 3.496e-10 |
| GO:BP | GO:0051716 | cellular response to stimulus | 811 | 7376 | 4.875e-10 |
| GO:BP | GO:0007010 | cytoskeleton organization | 218 | 1529 | 1.009e-09 |
| GO:BP | GO:0048856 | anatomical structure development | 676 | 5997 | 1.290e-09 |
| GO:BP | GO:0046907 | intracellular transport | 200 | 1381 | 1.971e-09 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 58 | 248 | 2.106e-09 |
| GO:BP | GO:0006950 | response to stress | 472 | 3948 | 2.106e-09 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 73 | 351 | 2.140e-09 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 64 | 289 | 2.293e-09 |
| GO:BP | GO:0051310 | metaphase chromosome alignment | 34 | 104 | 2.329e-09 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 39 | 132 | 2.609e-09 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 76 | 377 | 3.744e-09 |
| GO:BP | GO:0019222 | regulation of metabolic process | 771 | 7035 | 5.286e-09 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 36 | 118 | 5.324e-09 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 250 | 1855 | 7.508e-09 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 312 | 2433 | 7.852e-09 |
| GO:BP | GO:0006281 | DNA repair | 107 | 621 | 9.154e-09 |
| GO:BP | GO:0050000 | chromosome localization | 37 | 126 | 9.328e-09 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 441 | 3687 | 1.036e-08 |
| GO:BP | GO:0009966 | regulation of signal transduction | 374 | 3034 | 1.083e-08 |
| GO:BP | GO:0048519 | negative regulation of biological process | 653 | 5834 | 1.178e-08 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 412 | 3409 | 1.309e-08 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 632 | 5629 | 1.667e-08 |
| GO:BP | GO:0007399 | nervous system development | 327 | 2604 | 2.643e-08 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 412 | 3428 | 2.686e-08 |
| GO:BP | GO:0006351 | DNA-templated transcription | 424 | 3549 | 2.983e-08 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 41 | 156 | 3.575e-08 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 27 | 77 | 3.616e-08 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 257 | 1953 | 3.644e-08 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 43 | 169 | 3.911e-08 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 35 | 121 | 4.003e-08 |
| GO:BP | GO:0048731 | system development | 473 | 4053 | 5.369e-08 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 427 | 3597 | 5.535e-08 |
| GO:BP | GO:0006270 | DNA replication initiation | 18 | 37 | 5.838e-08 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 51 | 224 | 6.519e-08 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 51 | 225 | 7.471e-08 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 710 | 6492 | 7.471e-08 |
| GO:BP | GO:0000910 | cytokinesis | 46 | 192 | 7.471e-08 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 321 | 2578 | 8.801e-08 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 79 | 430 | 1.099e-07 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 334 | 2711 | 1.217e-07 |
| GO:BP | GO:0030154 | cell differentiation | 508 | 4437 | 1.474e-07 |
| GO:BP | GO:0048869 | cellular developmental process | 508 | 4438 | 1.508e-07 |
| GO:BP | GO:0051304 | chromosome separation | 27 | 84 | 2.734e-07 |
| GO:BP | GO:0051321 | meiotic cell cycle | 60 | 297 | 2.743e-07 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 36 | 136 | 2.757e-07 |
| GO:BP | GO:0051783 | regulation of nuclear division | 38 | 149 | 3.128e-07 |
| GO:BP | GO:0140013 | meiotic nuclear division | 46 | 202 | 3.830e-07 |
| GO:BP | GO:0015031 | protein transport | 196 | 1444 | 4.337e-07 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 389 | 3288 | 5.444e-07 |
| GO:BP | GO:0060341 | regulation of cellular localization | 147 | 1013 | 6.032e-07 |
| GO:BP | GO:0007052 | mitotic spindle organization | 35 | 134 | 6.251e-07 |
| GO:BP | GO:0071705 | nitrogen compound transport | 247 | 1923 | 6.269e-07 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 177 | 1280 | 6.328e-07 |
| GO:BP | GO:0007051 | spindle organization | 46 | 206 | 6.885e-07 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 298 | 2410 | 6.992e-07 |
| GO:BP | GO:0051234 | establishment of localization | 551 | 4928 | 7.142e-07 |
| GO:BP | GO:0016310 | phosphorylation | 181 | 1320 | 7.667e-07 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 22 | 62 | 8.667e-07 |
| GO:BP | GO:0051640 | organelle localization | 100 | 620 | 8.698e-07 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 297 | 2407 | 8.807e-07 |
| GO:BP | GO:0006468 | protein phosphorylation | 170 | 1227 | 1.038e-06 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 177 | 1296 | 1.449e-06 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 35 | 139 | 1.555e-06 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 327 | 2713 | 1.616e-06 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 105 | 670 | 1.668e-06 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 188 | 1400 | 1.754e-06 |
| GO:BP | GO:0007098 | centrosome cycle | 35 | 140 | 1.814e-06 |
| GO:BP | GO:0051098 | regulation of binding | 46 | 213 | 1.814e-06 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 40 | 173 | 2.002e-06 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 22 | 65 | 2.113e-06 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 35 | 141 | 2.150e-06 |
| GO:BP | GO:0050896 | response to stimulus | 932 | 8999 | 2.380e-06 |
| GO:BP | GO:0023052 | signaling | 699 | 6515 | 2.623e-06 |
| GO:BP | GO:0007154 | cell communication | 701 | 6540 | 2.892e-06 |
| GO:BP | GO:0051383 | kinetochore organization | 12 | 21 | 2.941e-06 |
| GO:BP | GO:0048513 | animal organ development | 363 | 3085 | 2.959e-06 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 32 | 124 | 2.959e-06 |
| GO:BP | GO:0045184 | establishment of protein localization | 244 | 1937 | 3.454e-06 |
| GO:BP | GO:0034508 | centromere complex assembly | 14 | 29 | 3.609e-06 |
| GO:BP | GO:0009790 | embryo development | 157 | 1135 | 3.675e-06 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 200 | 1529 | 4.068e-06 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 20 | 57 | 4.087e-06 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 39 | 172 | 4.662e-06 |
| GO:BP | GO:0065009 | regulation of molecular function | 196 | 1496 | 4.848e-06 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 49 | 242 | 4.874e-06 |
| GO:BP | GO:0007017 | microtubule-based process | 141 | 999 | 5.176e-06 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 36 | 153 | 5.296e-06 |
| GO:BP | GO:0032880 | regulation of protein localization | 130 | 907 | 6.943e-06 |
| GO:BP | GO:0051099 | positive regulation of binding | 28 | 104 | 7.057e-06 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 20 | 59 | 7.449e-06 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 19 | 54 | 7.559e-06 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 18 | 49 | 7.559e-06 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 18 | 49 | 7.559e-06 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 18 | 49 | 7.559e-06 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 31 | 123 | 7.559e-06 |
| GO:BP | GO:0051225 | spindle assembly | 33 | 136 | 7.839e-06 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 21 | 65 | 8.831e-06 |
| GO:BP | GO:0050793 | regulation of developmental process | 295 | 2457 | 1.032e-05 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 18 | 50 | 1.052e-05 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 238 | 1912 | 1.167e-05 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 251 | 2036 | 1.167e-05 |
| GO:BP | GO:0051656 | establishment of organelle localization | 80 | 491 | 1.200e-05 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 28 | 107 | 1.229e-05 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 26 | 95 | 1.257e-05 |
| GO:BP | GO:0030030 | cell projection organization | 208 | 1631 | 1.268e-05 |
| GO:BP | GO:0007127 | meiosis I | 32 | 133 | 1.386e-05 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 18 | 51 | 1.386e-05 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 18 | 51 | 1.386e-05 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 18 | 51 | 1.386e-05 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 18 | 51 | 1.386e-05 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 868 | 8393 | 1.414e-05 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 448 | 3993 | 1.496e-05 |
| GO:BP | GO:0016043 | cellular component organization | 848 | 8184 | 1.617e-05 |
| GO:BP | GO:0010256 | endomembrane system organization | 94 | 613 | 1.778e-05 |
| GO:BP | GO:0007080 | mitotic metaphase chromosome alignment | 20 | 63 | 2.139e-05 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 202 | 1588 | 2.166e-05 |
| GO:BP | GO:0006810 | transport | 487 | 4407 | 2.358e-05 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 79 | 492 | 2.370e-05 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 18 | 53 | 2.517e-05 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 18 | 53 | 2.517e-05 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 18 | 53 | 2.517e-05 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 30 | 124 | 2.611e-05 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 213 | 1697 | 2.634e-05 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 29 | 118 | 2.795e-05 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 213 | 1699 | 2.839e-05 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 56 | 312 | 3.155e-05 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 22 | 76 | 3.215e-05 |
| GO:BP | GO:0035295 | tube development | 148 | 1101 | 3.690e-05 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 13 | 30 | 3.762e-05 |
| GO:BP | GO:0023057 | negative regulation of signaling | 184 | 1435 | 4.059e-05 |
| GO:BP | GO:0009056 | catabolic process | 309 | 2639 | 4.192e-05 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 28 | 114 | 4.192e-05 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 28 | 114 | 4.192e-05 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 184 | 1436 | 4.192e-05 |
| GO:BP | GO:0007165 | signal transduction | 638 | 6002 | 4.574e-05 |
| GO:BP | GO:0009411 | response to UV | 34 | 155 | 5.076e-05 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 25 | 97 | 6.010e-05 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 303 | 2593 | 6.417e-05 |
| GO:BP | GO:0031399 | regulation of protein modification process | 141 | 1049 | 6.436e-05 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 225 | 1835 | 6.456e-05 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 171 | 1326 | 6.733e-05 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 35 | 164 | 6.770e-05 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 99 | 677 | 6.801e-05 |
| GO:BP | GO:0022008 | neurogenesis | 218 | 1771 | 7.121e-05 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 8 | 12 | 7.376e-05 |
| GO:BP | GO:0009888 | tissue development | 245 | 2032 | 7.465e-05 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 26 | 105 | 8.031e-05 |
| GO:BP | GO:0016236 | macroautophagy | 61 | 362 | 8.183e-05 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 128 | 938 | 8.726e-05 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 24 | 93 | 8.880e-05 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 40 | 202 | 8.995e-05 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 180 | 1418 | 9.156e-05 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 25 | 100 | 1.014e-04 |
| GO:BP | GO:0008283 | cell population proliferation | 242 | 2015 | 1.126e-04 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 311 | 2696 | 1.297e-04 |
| GO:BP | GO:0048468 | cell development | 329 | 2876 | 1.333e-04 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 24 | 96 | 1.565e-04 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 24 | 96 | 1.565e-04 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 62 | 378 | 1.592e-04 |
| GO:BP | GO:0000902 | cell morphogenesis | 133 | 996 | 1.637e-04 |
| GO:BP | GO:0006914 | autophagy | 88 | 597 | 1.701e-04 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 88 | 597 | 1.701e-04 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 43 | 230 | 1.725e-04 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 89 | 606 | 1.729e-04 |
| GO:BP | GO:0032879 | regulation of localization | 242 | 2029 | 1.778e-04 |
| GO:BP | GO:0072359 | circulatory system development | 149 | 1145 | 1.778e-04 |
| GO:BP | GO:0008219 | cell death | 238 | 1991 | 1.824e-04 |
| GO:BP | GO:0007033 | vacuole organization | 44 | 239 | 1.988e-04 |
| GO:BP | GO:0051382 | kinetochore assembly | 9 | 17 | 2.078e-04 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 95 | 662 | 2.167e-04 |
| GO:BP | GO:0012501 | programmed cell death | 237 | 1987 | 2.204e-04 |
| GO:BP | GO:0030261 | chromosome condensation | 15 | 45 | 2.252e-04 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 25 | 105 | 2.371e-04 |
| GO:BP | GO:0032456 | endocytic recycling | 22 | 86 | 2.437e-04 |
| GO:BP | GO:0006915 | apoptotic process | 230 | 1923 | 2.498e-04 |
| GO:BP | GO:0007417 | central nervous system development | 139 | 1061 | 2.530e-04 |
| GO:BP | GO:0007420 | brain development | 105 | 754 | 2.649e-04 |
| GO:BP | GO:0007507 | heart development | 88 | 605 | 2.703e-04 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 23 | 93 | 2.734e-04 |
| GO:BP | GO:0061061 | muscle structure development | 98 | 693 | 2.771e-04 |
| GO:BP | GO:0043687 | post-translational protein modification | 135 | 1027 | 2.838e-04 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 47 | 266 | 2.859e-04 |
| GO:BP | GO:0006275 | regulation of DNA replication | 27 | 120 | 3.053e-04 |
| GO:BP | GO:0033260 | nuclear DNA replication | 14 | 41 | 3.177e-04 |
| GO:BP | GO:0006310 | DNA recombination | 57 | 347 | 3.276e-04 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 176 | 1417 | 3.808e-04 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 12 | 32 | 4.614e-04 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 23 | 96 | 4.625e-04 |
| GO:BP | GO:0060284 | regulation of cell development | 114 | 847 | 4.964e-04 |
| GO:BP | GO:0038202 | TORC1 signaling | 24 | 103 | 4.964e-04 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 118 | 884 | 5.111e-04 |
| GO:BP | GO:0051101 | regulation of DNA binding | 19 | 72 | 5.889e-04 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 9 | 19 | 6.012e-04 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 191 | 1576 | 6.897e-04 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 96 | 692 | 7.036e-04 |
| GO:BP | GO:0072073 | kidney epithelium development | 31 | 154 | 7.079e-04 |
| GO:BP | GO:0051960 | regulation of nervous system development | 69 | 457 | 7.103e-04 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 122 | 928 | 7.246e-04 |
| GO:BP | GO:0009267 | cellular response to starvation | 34 | 177 | 8.019e-04 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 76 | 519 | 8.085e-04 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 111 | 830 | 8.110e-04 |
| GO:BP | GO:0010506 | regulation of autophagy | 58 | 367 | 8.286e-04 |
| GO:BP | GO:0060322 | head development | 108 | 806 | 9.534e-04 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 105 | 779 | 9.589e-04 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 114 | 861 | 9.638e-04 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 22 | 94 | 9.666e-04 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 20 | 81 | 9.666e-04 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 14 | 45 | 9.666e-04 |
| GO:BP | GO:0002181 | cytoplasmic translation | 32 | 165 | 1.087e-03 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 329 | 2956 | 1.102e-03 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 140 | 1105 | 1.104e-03 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 126 | 975 | 1.119e-03 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 10 | 25 | 1.141e-03 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 10 | 25 | 1.141e-03 |
| GO:BP | GO:0032501 | multicellular organismal process | 747 | 7322 | 1.141e-03 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 35 | 188 | 1.141e-03 |
| GO:BP | GO:0006325 | chromatin organization | 116 | 884 | 1.163e-03 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 75 | 517 | 1.173e-03 |
| GO:BP | GO:0006446 | regulation of translational initiation | 20 | 83 | 1.352e-03 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 77 | 537 | 1.375e-03 |
| GO:BP | GO:0043009 | chordate embryonic development | 92 | 671 | 1.473e-03 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 53 | 334 | 1.512e-03 |
| GO:BP | GO:0007015 | actin filament organization | 68 | 461 | 1.543e-03 |
| GO:BP | GO:0016050 | vesicle organization | 58 | 376 | 1.543e-03 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 58 | 376 | 1.543e-03 |
| GO:BP | GO:0006412 | translation | 98 | 727 | 1.614e-03 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 15 | 53 | 1.628e-03 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 16 | 59 | 1.636e-03 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 11 | 31 | 1.645e-03 |
| GO:BP | GO:1902969 | mitotic DNA replication | 8 | 17 | 1.664e-03 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 23 | 105 | 1.804e-03 |
| GO:BP | GO:0000725 | recombinational repair | 35 | 193 | 1.889e-03 |
| GO:BP | GO:0001701 | in utero embryonic development | 62 | 413 | 1.896e-03 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 14 | 48 | 1.943e-03 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 14 | 48 | 1.943e-03 |
| GO:BP | GO:0006413 | translational initiation | 26 | 127 | 2.047e-03 |
| GO:BP | GO:0001822 | kidney development | 51 | 322 | 2.073e-03 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 95 | 706 | 2.145e-03 |
| GO:BP | GO:0030029 | actin filament-based process | 108 | 825 | 2.187e-03 |
| GO:BP | GO:0016197 | endosomal transport | 47 | 290 | 2.203e-03 |
| GO:BP | GO:0033365 | protein localization to organelle | 149 | 1209 | 2.213e-03 |
| GO:BP | GO:0072001 | renal system development | 52 | 332 | 2.338e-03 |
| GO:BP | GO:0051302 | regulation of cell division | 34 | 188 | 2.433e-03 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 585 | 5644 | 2.437e-03 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 18 | 74 | 2.577e-03 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 12 | 38 | 2.644e-03 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 97 | 729 | 2.681e-03 |
| GO:BP | GO:0061024 | membrane organization | 107 | 821 | 2.731e-03 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 600 | 5809 | 2.735e-03 |
| GO:BP | GO:0035825 | homologous recombination | 17 | 68 | 2.739e-03 |
| GO:BP | GO:0031647 | regulation of protein stability | 52 | 335 | 2.844e-03 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 188 | 1592 | 2.844e-03 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 197 | 1680 | 2.844e-03 |
| GO:BP | GO:0035970 | peptidyl-threonine dephosphorylation | 7 | 14 | 2.844e-03 |
| GO:BP | GO:0016477 | cell migration | 182 | 1534 | 2.844e-03 |
| GO:BP | GO:1990928 | response to amino acid starvation | 15 | 56 | 2.951e-03 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 24 | 116 | 3.005e-03 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 71 | 499 | 3.005e-03 |
| GO:BP | GO:0009987 | cellular process | 1872 | 20247 | 3.008e-03 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 23 | 109 | 3.008e-03 |
| GO:BP | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 5 | 7 | 3.137e-03 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 71 | 500 | 3.156e-03 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 121 | 956 | 3.277e-03 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 61 | 414 | 3.324e-03 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 28 | 146 | 3.324e-03 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 121 | 957 | 3.391e-03 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 15 | 57 | 3.529e-03 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 15 | 57 | 3.529e-03 |
| GO:BP | GO:0048589 | developmental growth | 89 | 663 | 3.529e-03 |
| GO:BP | GO:0031297 | replication fork processing | 14 | 51 | 3.565e-03 |
| GO:BP | GO:0017148 | negative regulation of translation | 26 | 132 | 3.565e-03 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 41 | 248 | 3.575e-03 |
| GO:BP | GO:2000677 | regulation of transcription regulatory region DNA binding | 11 | 34 | 3.718e-03 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 17 | 70 | 3.732e-03 |
| GO:BP | GO:2000269 | regulation of fibroblast apoptotic process | 8 | 19 | 3.777e-03 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 21 | 97 | 3.828e-03 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 29 | 155 | 3.834e-03 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 107 | 831 | 3.986e-03 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 71 | 505 | 4.064e-03 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 117 | 925 | 4.121e-03 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 196 | 1685 | 4.247e-03 |
| GO:BP | GO:0030031 | cell projection assembly | 83 | 613 | 4.247e-03 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 129 | 1039 | 4.284e-03 |
| GO:BP | GO:0006886 | intracellular protein transport | 91 | 686 | 4.331e-03 |
| GO:BP | GO:0032835 | glomerulus development | 17 | 71 | 4.348e-03 |
| GO:BP | GO:0022414 | reproductive process | 188 | 1608 | 4.383e-03 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 7 | 15 | 4.435e-03 |
| GO:BP | GO:0042594 | response to starvation | 37 | 219 | 4.515e-03 |
| GO:BP | GO:0009314 | response to radiation | 63 | 437 | 4.515e-03 |
| GO:BP | GO:0010468 | regulation of gene expression | 571 | 5536 | 4.601e-03 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 11 | 35 | 4.717e-03 |
| GO:BP | GO:0034504 | protein localization to nucleus | 49 | 318 | 4.730e-03 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 22 | 106 | 4.877e-03 |
| GO:BP | GO:0045995 | regulation of embryonic development | 20 | 92 | 4.877e-03 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 91 | 689 | 4.882e-03 |
| GO:BP | GO:0006302 | double-strand break repair | 49 | 319 | 5.046e-03 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 81 | 599 | 5.054e-03 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 90 | 681 | 5.124e-03 |
| GO:BP | GO:0048699 | generation of neurons | 180 | 1536 | 5.176e-03 |
| GO:BP | GO:0044782 | cilium organization | 62 | 431 | 5.202e-03 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 10 | 30 | 5.207e-03 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 81 | 600 | 5.258e-03 |
| GO:BP | GO:0007034 | vacuolar transport | 31 | 174 | 5.352e-03 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 31 | 174 | 5.352e-03 |
| GO:BP | GO:0019953 | sexual reproduction | 137 | 1122 | 5.352e-03 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 16 | 66 | 5.408e-03 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 9 | 25 | 5.471e-03 |
| GO:BP | GO:0031929 | TOR signaling | 30 | 167 | 5.733e-03 |
| GO:BP | GO:0070848 | response to growth factor | 94 | 721 | 5.843e-03 |
| GO:BP | GO:0051223 | regulation of protein transport | 63 | 442 | 5.843e-03 |
| GO:BP | GO:0043388 | positive regulation of DNA binding | 11 | 36 | 5.893e-03 |
| GO:BP | GO:0007041 | lysosomal transport | 26 | 137 | 5.904e-03 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 89 | 676 | 6.031e-03 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 18 | 80 | 6.031e-03 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 54 | 365 | 6.176e-03 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 21 | 101 | 6.183e-03 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 20 | 94 | 6.203e-03 |
| GO:BP | GO:0071763 | nuclear membrane organization | 13 | 48 | 6.216e-03 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 147 | 1223 | 6.230e-03 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 108 | 853 | 6.276e-03 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 32 | 184 | 6.436e-03 |
| GO:BP | GO:1905821 | positive regulation of chromosome condensation | 5 | 8 | 6.471e-03 |
| GO:BP | GO:0060253 | negative regulation of glial cell proliferation | 7 | 16 | 6.548e-03 |
| GO:BP | GO:0032926 | negative regulation of activin receptor signaling pathway | 7 | 16 | 6.548e-03 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 87 | 660 | 6.565e-03 |
| GO:BP | GO:0051298 | centrosome duplication | 17 | 74 | 6.586e-03 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 10 | 31 | 6.586e-03 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 27 | 146 | 6.727e-03 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 39 | 241 | 6.855e-03 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 9 | 26 | 7.230e-03 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 16 | 68 | 7.279e-03 |
| GO:BP | GO:0022616 | DNA strand elongation | 11 | 37 | 7.279e-03 |
| GO:BP | GO:0043174 | nucleoside salvage | 6 | 12 | 7.351e-03 |
| GO:BP | GO:0006997 | nucleus organization | 27 | 147 | 7.421e-03 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 38 | 234 | 7.460e-03 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 12 | 43 | 7.488e-03 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 32 | 186 | 7.534e-03 |
| GO:BP | GO:0061572 | actin filament bundle organization | 29 | 163 | 7.880e-03 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 124 | 1011 | 8.024e-03 |
| GO:BP | GO:0035239 | tube morphogenesis | 110 | 879 | 8.143e-03 |
| GO:BP | GO:0061036 | positive regulation of cartilage development | 10 | 32 | 8.478e-03 |
| GO:BP | GO:0006417 | regulation of translation | 55 | 380 | 8.832e-03 |
| GO:BP | GO:0032506 | cytokinetic process | 13 | 50 | 8.992e-03 |
| GO:BP | GO:0031032 | actomyosin structure organization | 35 | 212 | 8.992e-03 |
| GO:BP | GO:0034644 | cellular response to UV | 19 | 90 | 9.023e-03 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 89 | 686 | 9.033e-03 |
| GO:BP | GO:0048863 | stem cell differentiation | 39 | 245 | 9.179e-03 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 41 | 262 | 9.488e-03 |
| GO:BP | GO:0048729 | tissue morphogenesis | 81 | 614 | 9.594e-03 |
| GO:BP | GO:0034349 | glial cell apoptotic process | 7 | 17 | 9.594e-03 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 143 | 1199 | 9.702e-03 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 27 | 150 | 9.887e-03 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 50 | 339 | 9.936e-03 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 50 | 339 | 9.936e-03 |
| GO:BP | GO:0061709 | reticulophagy | 8 | 22 | 9.990e-03 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 14 | 57 | 9.990e-03 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 73 | 543 | 1.028e-02 |
| GO:BP | GO:0051338 | regulation of transferase activity | 68 | 498 | 1.028e-02 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 87 | 671 | 1.031e-02 |
| GO:BP | GO:0007040 | lysosome organization | 22 | 113 | 1.049e-02 |
| GO:BP | GO:0080171 | lytic vacuole organization | 22 | 113 | 1.049e-02 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 27 | 151 | 1.075e-02 |
| GO:BP | GO:0030163 | protein catabolic process | 125 | 1030 | 1.091e-02 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 12 | 45 | 1.103e-02 |
| GO:BP | GO:0031503 | protein-containing complex localization | 35 | 215 | 1.112e-02 |
| GO:BP | GO:0090232 | positive regulation of spindle checkpoint | 6 | 13 | 1.160e-02 |
| GO:BP | GO:0043393 | regulation of protein binding | 22 | 114 | 1.173e-02 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 14 | 58 | 1.173e-02 |
| GO:BP | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis | 5 | 9 | 1.184e-02 |
| GO:BP | GO:0048666 | neuron development | 140 | 1177 | 1.184e-02 |
| GO:BP | GO:0000915 | actomyosin contractile ring assembly | 5 | 9 | 1.184e-02 |
| GO:BP | GO:0000301 | retrograde transport, vesicle recycling within Golgi | 5 | 9 | 1.184e-02 |
| GO:BP | GO:0070255 | regulation of mucus secretion | 5 | 9 | 1.184e-02 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 28 | 160 | 1.188e-02 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 30 | 176 | 1.214e-02 |
| GO:BP | GO:0030182 | neuron differentiation | 168 | 1451 | 1.217e-02 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 148 | 1256 | 1.228e-02 |
| GO:BP | GO:0045185 | maintenance of protein location | 15 | 65 | 1.233e-02 |
| GO:BP | GO:0060429 | epithelium development | 146 | 1238 | 1.283e-02 |
| GO:BP | GO:0042692 | muscle cell differentiation | 57 | 405 | 1.293e-02 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 37 | 234 | 1.333e-02 |
| GO:BP | GO:0043149 | stress fiber assembly | 21 | 108 | 1.343e-02 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 11 | 40 | 1.343e-02 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 21 | 108 | 1.343e-02 |
| GO:BP | GO:0006998 | nuclear envelope organization | 14 | 59 | 1.356e-02 |
| GO:BP | GO:0030323 | respiratory tube development | 33 | 202 | 1.411e-02 |
| GO:BP | GO:0048144 | fibroblast proliferation | 21 | 109 | 1.511e-02 |
| GO:BP | GO:0072009 | nephron epithelium development | 23 | 124 | 1.511e-02 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 46 | 312 | 1.522e-02 |
| GO:BP | GO:0007131 | reciprocal meiotic recombination | 14 | 60 | 1.597e-02 |
| GO:BP | GO:0140527 | reciprocal homologous recombination | 14 | 60 | 1.597e-02 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 22 | 117 | 1.597e-02 |
| GO:BP | GO:0007517 | muscle organ development | 52 | 365 | 1.599e-02 |
| GO:BP | GO:0031175 | neuron projection development | 123 | 1023 | 1.599e-02 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 25 | 140 | 1.606e-02 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 19 | 95 | 1.617e-02 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 15 | 67 | 1.655e-02 |
| GO:BP | GO:0034137 | positive regulation of toll-like receptor 2 signaling pathway | 4 | 6 | 1.717e-02 |
| GO:BP | GO:0070257 | positive regulation of mucus secretion | 4 | 6 | 1.717e-02 |
| GO:BP | GO:0010032 | meiotic chromosome condensation | 4 | 6 | 1.717e-02 |
| GO:BP | GO:0051231 | spindle elongation | 6 | 14 | 1.725e-02 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 6 | 14 | 1.725e-02 |
| GO:BP | GO:0009416 | response to light stimulus | 47 | 323 | 1.732e-02 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 8 | 24 | 1.737e-02 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 8 | 24 | 1.737e-02 |
| GO:BP | GO:0034502 | protein localization to chromosome | 23 | 126 | 1.824e-02 |
| GO:BP | GO:0009163 | nucleoside biosynthetic process | 7 | 19 | 1.833e-02 |
| GO:BP | GO:0034404 | nucleobase-containing small molecule biosynthetic process | 7 | 19 | 1.833e-02 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 7 | 19 | 1.833e-02 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 84 | 658 | 1.833e-02 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 14 | 61 | 1.833e-02 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 12 | 48 | 1.854e-02 |
| GO:BP | GO:0001824 | blastocyst development | 22 | 119 | 1.926e-02 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 67 | 503 | 1.926e-02 |
| GO:BP | GO:0006338 | chromatin remodeling | 91 | 725 | 1.938e-02 |
| GO:BP | GO:0071478 | cellular response to radiation | 30 | 182 | 1.943e-02 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 179 | 1578 | 1.944e-02 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 5 | 10 | 1.968e-02 |
| GO:BP | GO:0044837 | actomyosin contractile ring organization | 5 | 10 | 1.968e-02 |
| GO:BP | GO:0009894 | regulation of catabolic process | 124 | 1040 | 1.968e-02 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 5 | 10 | 1.968e-02 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 43 | 291 | 1.970e-02 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 10 | 36 | 1.972e-02 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 50 | 352 | 1.987e-02 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 82 | 642 | 1.987e-02 |
| GO:BP | GO:0050821 | protein stabilization | 35 | 224 | 2.060e-02 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 14 | 62 | 2.089e-02 |
| GO:BP | GO:0030010 | establishment of cell polarity | 27 | 159 | 2.094e-02 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 17 | 83 | 2.100e-02 |
| GO:BP | GO:0021537 | telencephalon development | 42 | 284 | 2.164e-02 |
| GO:BP | GO:0060541 | respiratory system development | 35 | 225 | 2.207e-02 |
| GO:BP | GO:0048870 | cell motility | 200 | 1793 | 2.212e-02 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 8 | 25 | 2.216e-02 |
| GO:BP | GO:0072006 | nephron development | 27 | 160 | 2.276e-02 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 131 | 1113 | 2.292e-02 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 33 | 209 | 2.294e-02 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 18 | 91 | 2.294e-02 |
| GO:BP | GO:0007178 | cell surface receptor protein serine/threonine kinase signaling pathway | 51 | 364 | 2.354e-02 |
| GO:BP | GO:0023056 | positive regulation of signaling | 200 | 1796 | 2.363e-02 |
| GO:BP | GO:0071539 | protein localization to centrosome | 10 | 37 | 2.394e-02 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 10 | 37 | 2.394e-02 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 16 | 77 | 2.400e-02 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 134 | 1144 | 2.400e-02 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 6 | 15 | 2.410e-02 |
| GO:BP | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 6 | 15 | 2.410e-02 |
| GO:BP | GO:2000679 | positive regulation of transcription regulatory region DNA binding | 7 | 20 | 2.410e-02 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 6 | 15 | 2.410e-02 |
| GO:BP | GO:0000076 | DNA replication checkpoint signaling | 7 | 20 | 2.410e-02 |
| GO:BP | GO:0065008 | regulation of biological quality | 313 | 2947 | 2.420e-02 |
| GO:BP | GO:0001568 | blood vessel development | 91 | 732 | 2.423e-02 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 53 | 383 | 2.472e-02 |
| GO:BP | GO:0035850 | epithelial cell differentiation involved in kidney development | 12 | 50 | 2.493e-02 |
| GO:BP | GO:0032924 | activin receptor signaling pathway | 12 | 50 | 2.493e-02 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 12 | 50 | 2.493e-02 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 12 | 50 | 2.493e-02 |
| GO:BP | GO:0060173 | limb development | 31 | 194 | 2.499e-02 |
| GO:BP | GO:0048736 | appendage development | 31 | 194 | 2.499e-02 |
| GO:BP | GO:0003176 | aortic valve development | 11 | 44 | 2.713e-02 |
| GO:BP | GO:1903008 | organelle disassembly | 15 | 71 | 2.723e-02 |
| GO:BP | GO:0001503 | ossification | 58 | 430 | 2.749e-02 |
| GO:BP | GO:1905207 | regulation of cardiocyte differentiation | 8 | 26 | 2.787e-02 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 10 | 38 | 2.865e-02 |
| GO:BP | GO:0043549 | regulation of kinase activity | 58 | 431 | 2.885e-02 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 37 | 247 | 3.045e-02 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 16 | 79 | 3.048e-02 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 5 | 11 | 3.048e-02 |
| GO:BP | GO:0035329 | hippo signaling | 13 | 58 | 3.070e-02 |
| GO:BP | GO:0035148 | tube formation | 25 | 148 | 3.110e-02 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 45 | 317 | 3.173e-02 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 7 | 21 | 3.189e-02 |
| GO:BP | GO:1905684 | regulation of plasma membrane repair | 4 | 7 | 3.198e-02 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 4 | 7 | 3.198e-02 |
| GO:BP | GO:2000270 | negative regulation of fibroblast apoptotic process | 4 | 7 | 3.198e-02 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 11 | 45 | 3.199e-02 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 21 | 117 | 3.206e-02 |
| GO:BP | GO:0031214 | biomineral tissue development | 29 | 181 | 3.227e-02 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 17 | 87 | 3.274e-02 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 80 | 637 | 3.277e-02 |
| GO:BP | GO:0048313 | Golgi inheritance | 6 | 16 | 3.328e-02 |
| GO:BP | GO:0070254 | mucus secretion | 6 | 16 | 3.328e-02 |
| GO:BP | GO:0048308 | organelle inheritance | 6 | 16 | 3.328e-02 |
| GO:BP | GO:0098877 | neurotransmitter receptor transport to plasma membrane | 6 | 16 | 3.328e-02 |
| GO:BP | GO:0030324 | lung development | 31 | 198 | 3.328e-02 |
| GO:BP | GO:0003007 | heart morphogenesis | 39 | 266 | 3.339e-02 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 47 | 336 | 3.345e-02 |
| GO:BP | GO:0007585 | respiratory gaseous exchange by respiratory system | 16 | 80 | 3.345e-02 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 16 | 80 | 3.345e-02 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 16 | 80 | 3.345e-02 |
| GO:BP | GO:0046677 | response to antibiotic | 10 | 39 | 3.365e-02 |
| GO:BP | GO:0014706 | striated muscle tissue development | 55 | 408 | 3.417e-02 |
| GO:BP | GO:0003281 | ventricular septum development | 15 | 73 | 3.427e-02 |
| GO:BP | GO:0061005 | cell differentiation involved in kidney development | 13 | 59 | 3.449e-02 |
| GO:BP | GO:0000045 | autophagosome assembly | 21 | 118 | 3.455e-02 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 14 | 66 | 3.458e-02 |
| GO:BP | GO:0043094 | metabolic compound salvage | 9 | 33 | 3.461e-02 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 9 | 33 | 3.461e-02 |
| GO:BP | GO:0051642 | centrosome localization | 9 | 33 | 3.461e-02 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 24 | 142 | 3.557e-02 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 19 | 103 | 3.561e-02 |
| GO:BP | GO:0014896 | muscle hypertrophy | 17 | 88 | 3.565e-02 |
| GO:BP | GO:0000165 | MAPK cascade | 91 | 744 | 3.573e-02 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 331 | 3157 | 3.573e-02 |
| GO:BP | GO:0031667 | response to nutrient levels | 66 | 510 | 3.621e-02 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 11 | 46 | 3.642e-02 |
| GO:BP | GO:0022411 | cellular component disassembly | 59 | 446 | 3.642e-02 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 11 | 46 | 3.642e-02 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 105 | 879 | 3.674e-02 |
| GO:BP | GO:0003231 | cardiac ventricle development | 22 | 127 | 3.803e-02 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 12 | 53 | 3.836e-02 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 54 | 402 | 3.882e-02 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 10 | 40 | 3.958e-02 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 17 | 89 | 3.958e-02 |
| GO:BP | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 7 | 22 | 3.970e-02 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 7 | 22 | 3.970e-02 |
| GO:BP | GO:0045947 | negative regulation of translational initiation | 7 | 22 | 3.970e-02 |
| GO:BP | GO:0060271 | cilium assembly | 54 | 403 | 4.052e-02 |
| GO:BP | GO:0021915 | neural tube development | 26 | 160 | 4.079e-02 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 25 | 152 | 4.120e-02 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 86 | 701 | 4.127e-02 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 114 | 970 | 4.127e-02 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 351 | 3375 | 4.189e-02 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 8 | 28 | 4.189e-02 |
| GO:BP | GO:0070168 | negative regulation of biomineral tissue development | 8 | 28 | 4.189e-02 |
| GO:BP | GO:0036296 | response to increased oxygen levels | 8 | 28 | 4.189e-02 |
| GO:BP | GO:0015833 | peptide transport | 41 | 288 | 4.206e-02 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 28 | 177 | 4.214e-02 |
| GO:BP | GO:0061512 | protein localization to cilium | 15 | 75 | 4.218e-02 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 197 | 1795 | 4.218e-02 |
| GO:BP | GO:1905636 | positive regulation of RNA polymerase II regulatory region sequence-specific DNA binding | 3 | 4 | 4.218e-02 |
| GO:BP | GO:2000689 | actomyosin contractile ring assembly actin filament organization | 3 | 4 | 4.218e-02 |
| GO:BP | GO:0006404 | RNA import into nucleus | 3 | 4 | 4.218e-02 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 19 | 105 | 4.218e-02 |
| GO:BP | GO:0000022 | mitotic spindle elongation | 5 | 12 | 4.237e-02 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 5 | 12 | 4.237e-02 |
| GO:BP | GO:0099515 | actin filament-based transport | 5 | 12 | 4.237e-02 |
| GO:BP | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 5 | 12 | 4.237e-02 |
| GO:BP | GO:0072178 | nephric duct morphogenesis | 5 | 12 | 4.237e-02 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 5 | 12 | 4.237e-02 |
| GO:BP | GO:0051081 | nuclear membrane disassembly | 5 | 12 | 4.237e-02 |
| GO:BP | GO:0030900 | forebrain development | 56 | 423 | 4.279e-02 |
| GO:BP | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 6 | 17 | 4.279e-02 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 12 | 54 | 4.279e-02 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 21 | 121 | 4.332e-02 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 79 | 637 | 4.338e-02 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 16 | 83 | 4.476e-02 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 79 | 638 | 4.506e-02 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 46 | 334 | 4.507e-02 |
| GO:BP | GO:0051169 | nuclear transport | 46 | 334 | 4.507e-02 |
| GO:BP | GO:0042063 | gliogenesis | 48 | 352 | 4.515e-02 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 10 | 41 | 4.515e-02 |
| GO:BP | GO:0009306 | protein secretion | 51 | 379 | 4.515e-02 |
| GO:BP | GO:0001944 | vasculature development | 92 | 762 | 4.530e-02 |
| GO:BP | GO:0003333 | amino acid transmembrane transport | 19 | 106 | 4.542e-02 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 77 | 620 | 4.609e-02 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 51 | 380 | 4.734e-02 |
| GO:BP | GO:0003013 | circulatory system process | 75 | 602 | 4.758e-02 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 14 | 69 | 4.770e-02 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 14 | 69 | 4.770e-02 |
| GO:BP | GO:1905314 | semi-lunar valve development | 11 | 48 | 4.770e-02 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 9 | 35 | 4.814e-02 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 12 | 55 | 4.848e-02 |
| GO:BP | GO:0003177 | pulmonary valve development | 7 | 23 | 4.848e-02 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 7 | 23 | 4.848e-02 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 13 | 62 | 4.848e-02 |
| GO:BP | GO:0007409 | axonogenesis | 60 | 463 | 4.853e-02 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 74 | 594 | 4.968e-02 |
| KEGG | KEGG:04110 | Cell cycle | 50 | 157 | 3.797e-10 |
| KEGG | KEGG:04218 | Cellular senescence | 35 | 155 | 3.019e-03 |
| KEGG | KEGG:04114 | Oocyte meiosis | 31 | 131 | 3.019e-03 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 34 | 157 | 6.590e-03 |
| KEGG | KEGG:05222 | Small cell lung cancer | 23 | 92 | 8.018e-03 |
| KEGG | KEGG:04115 | p53 signaling pathway | 19 | 74 | 1.773e-02 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 10 | 29 | 3.479e-02 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 55 | 330 | 4.481e-02 |
| KEGG | KEGG:03030 | DNA replication | 11 | 36 | 4.679e-02 |
#GO:BP
table_unassign_genes_GOBP <- table_unassign_GOKEGG_genes %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_unassign_genes_GOBP %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("Cormotif Unassigned Enriched GO:BP Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))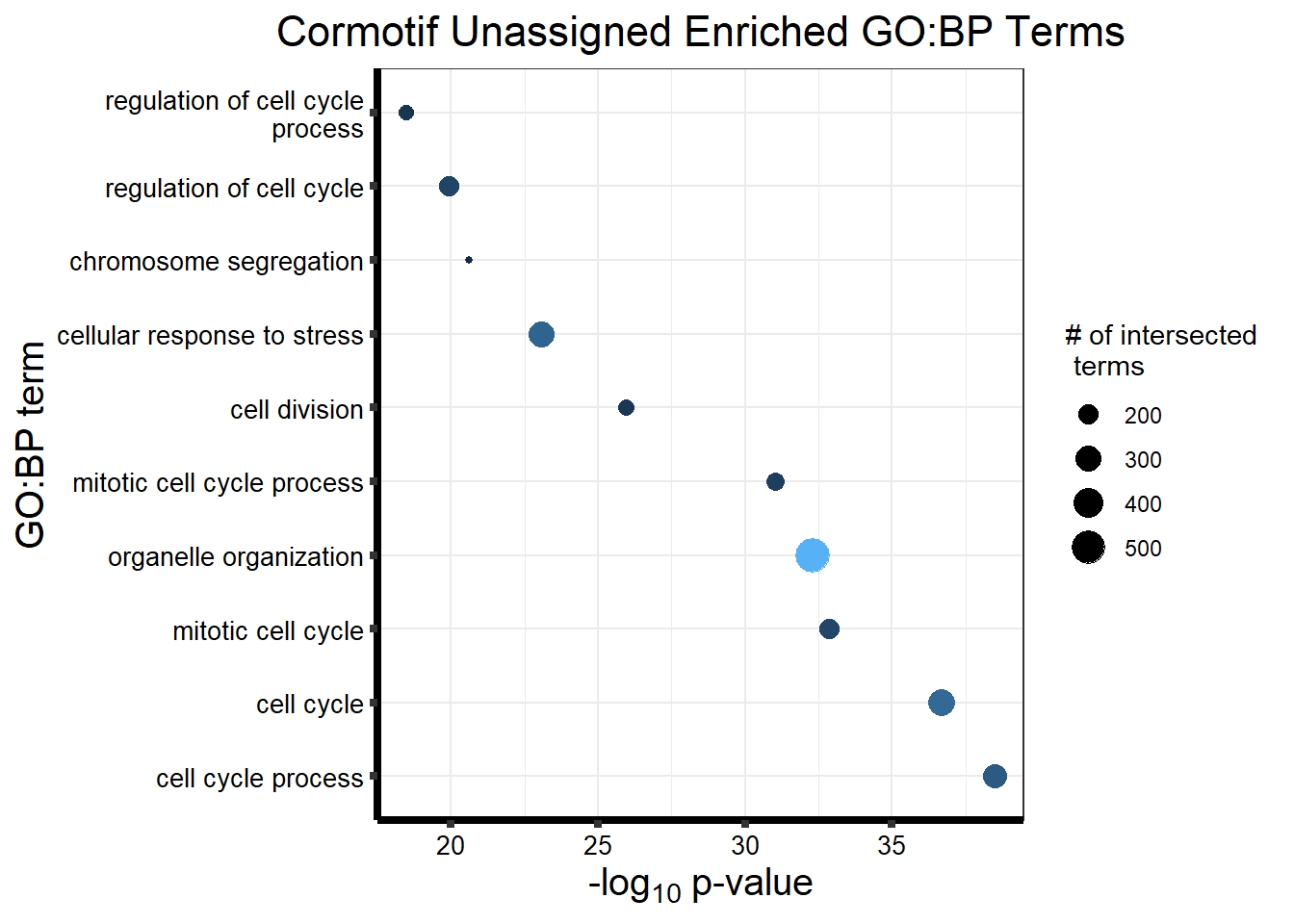
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#KEGG
table_unassign_genes_KEGG <- table_unassign_GOKEGG_genes %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_unassign_genes_KEGG %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("Cormotif Unassigned DEGs Enriched KEGG Terms")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))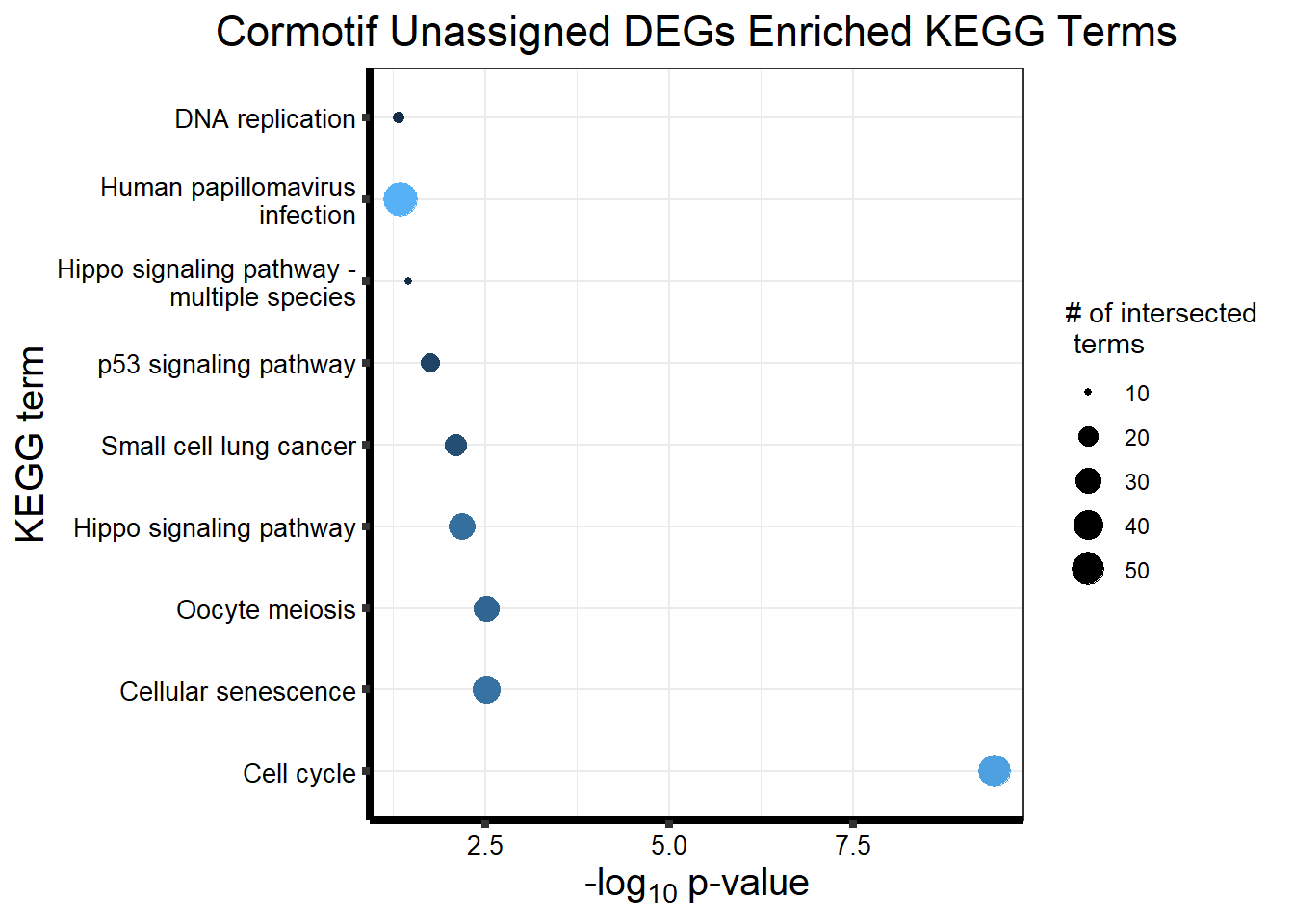
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
##Cormotif RUVs Corrected Norm Counts TMM and log2cpm
# #read in the norm counts df (normalized counts from RUV)
set1 <- readRDS("data/new/RUV/set1_RUVsk1_normcounts.RDS")
NormCounts_RUV_rep <- as.data.frame(set1$normalizedCounts)
#
# #read in my metadata sheet that I'll use as the standard for labelling
# #this one already has the replicate individual removed
Metadata_2 <- readRDS("data/new/Metadata_2_norep.RDS")
#
# #I'll have to remove the replicate at this stage so I can properly run Cormotif
NormCounts_RUV <- NormCounts_RUV_rep %>%
dplyr::select(-(contains("6R")))
#
# #now ensure the column names match the original dge$counts with Sample_name
colnames(NormCounts_RUV) <- Metadata_2$Sample_name
#
# #now, make this into a dge object as I did for DE + Cormotif
dge_RUV <- DGEList(counts = NormCounts_RUV)
#
# #put the Condition names as a factor onto the dge object for further sorting
dge_RUV$samples$group <- factor(Metadata_2[colnames(NormCounts_RUV), "Condition"])
#
# #TMM normalize the normalized counts input with the dge object
dge_RUV_calc <- calcNormFactors(dge_RUV, method = "TMM")Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced
Warning in .calcFactorTMM(obs = x[, i], ref = x[, refColumn], libsize.obs =
lib.size[i], : NaNs produced# #this is producing a lot of NaNs warnings, perhaps because TMM should occur before norm counts?
#
# #try by performing TMM normalization before norm counts in set1
#
counts <- readRDS("data/new/RUV/filt_counts_matrix.RDS")
# #use this counts file to feed into set1 - but instead TMM normalize it first
#
counts_tmm <- calcNormFactors(counts, method = "TMM")
#
# set1_tmm <- RUVSeq::RUVs(x = counts_tmm, k =1, scIdx = scIdx, isLog = FALSE)
# error with tmm normalized counts going into RUVseq
#
# summary(colSums(NormCounts_RUV))
# dge_RUV$samples
# #
ncol(NormCounts_RUV) == nrow(Metadata_2) # Should be TRUE[1] TRUEall(colnames(NormCounts_RUV) == rownames(Metadata_2))[1] TRUE#
#
# #save this file so I can read it in later!
# #saveRDS(dge_RUV_calc, "data/new/Cormotif/dge_matrix_forCormotif.RDS")
#
# #check normalization factors from TMM normalization of LIBRARIES
# dge_RUV_calc$samples
# #it was normalized properly but generated NaNs warnings?
# #still has the same number of columns, properly labelled and in order according to metadata
# #should be okay - let's try running it and see if it works properly
#
# Metadata_2_ordered <- Metadata_2[match(colnames(NormCounts_RUV), Metadata_2$Sample_name), ]
#
# #now cpm these TMM normalized norm counts
# cmf_RUV_cpm <- cpm(dge_RUV_calc, log = TRUE)
#
# #change the column names to match the Metadata sheet I have
# colnames(cmf_RUV_cpm) <- (Metadata_2$Final_sample_name)
#
# cmf_RUV_counts <- dge_RUV_calc
#
# #now make your TMM normalized cpm of norm counts into a dataframe + add Entrez_ID as a col
# cmf_RUV_df <- cmf_RUV_cpm %>%
# as.data.frame() %>%
# rownames_to_column(., var = "Entrez_ID")
#
# #save this to a csv file so I can reorder this matrix easily
# #write.csv(cmf_RUV_cpm, "data/new/Cormotif/RUV/Cormotif_RUVk1_cpm_matrix.csv")
#
# #reorder my test matrix to match the new groupid I've made
# #I want my columns to be in this order:
# #DOX24T 1-6, DOX24R 1-6, DOX144R 1-6, DMSO24T 1-6, DMSO24R 1-6, DMSO144R 1-6
# Cormotif_RUV <- read.csv("data/new/cmf_RUV_final.csv")
# dim(Cormotif_RUV)
# # #14319 genes across 37 cols (1 is Entrez_ID)
# Cormotif_RUV_df <- data.frame(Cormotif_RUV)
# #
# rownames(Cormotif_RUV_df) <- Cormotif_RUV_df$Entrez_ID
# exprs.cmf_RUV <- as.matrix(Cormotif_RUV_df[,2:37])
# dim(exprs.cmf_RUV)
# #
#
# #put together my group id and comparison id to make the correct comparisons between experimental conditions
#
# #groupid tells which experimental conditions are grouped together
# #compid tells which experimental conditions should be compared against one another
# ##ie DOX24T vs DMSO24T matched control
#
# groupid <- c(
# DOX_24T_1 = 1,
# DOX_24T_2 = 1,
# DOX_24T_3 = 1,
# DOX_24T_4 = 1,
# DOX_24T_5 = 1,
# DOX_24T_6 = 1,
# DOX_24R_1 = 2,
# DOX_24R_2 = 2,
# DOX_24R_3 = 2,
# DOX_24R_4 = 2,
# DOX_24R_5 = 2,
# DOX_24R_6 = 2,
# DOX_144R_1 = 3,
# DOX_144R_2 = 3,
# DOX_144R_3 = 3,
# DOX_144R_4 = 3,
# DOX_144R_5 = 3,
# DOX_144R_6 = 3,
# DMSO_24T_1 = 4,
# DMSO_24T_2 = 4,
# DMSO_24T_3 = 4,
# DMSO_24T_4 = 4,
# DMSO_24T_5 = 4,
# DMSO_24T_6 = 4,
# DMSO_24R_1 = 5,
# DMSO_24R_2 = 5,
# DMSO_24R_3 = 5,
# DMSO_24R_4 = 5,
# DMSO_24R_5 = 5,
# DMSO_24R_6 = 5,
# DMSO_144R_1 = 6,
# DMSO_144R_2 = 6,
# DMSO_144R_3 = 6,
# DMSO_144R_4 = 6,
# DMSO_144R_5 = 6,
# DMSO_144R_6 = 6
# )
# #saveRDS(groupid, "data/new/groupidCormotif.RDS")
#
# compid <- data.frame(Cond1 = c(1, 2, 3), Cond2 = c(4, 5, 6))
# #saveRDS(compid, "data/new/compidCormotif.RDS")##RUVs Cormotif TMM norm counts
#raw counts file
counts <- readRDS("data/new/RUV/filt_counts_matrix.RDS")
#create a dgelist object from these counts
dge_test <- DGEList(counts = counts)
dim(dge_test$counts)[1] 14319 42#14319 genes as normal, still contains replicate needed for RUVs
#apply TMM normalization to this DGElist object
dge_test_tmm <- calcNormFactors(object = dge_test, method = "TMM")
#this ran through just fine, must be something to do with applying TMM to normalized counts
dim(dge_test_tmm$counts)[1] 14319 42#14319 genes as normal, still have the replicate so 42 cols
#this still has the entrez_id so I should probably make sure that this is carried down
#pass this TMM normalized DGElist into RUVs correction
colnames(dge_test_tmm) <- Metadata$Final_sample_name
dim(dge_test_tmm)[1] 14319 42#14319 genes as normal
RUV_filt_counts_tmm <- dge_test_tmm$counts %>%
as.data.frame()
Entrez_ID_tmm_RUV <- RUV_filt_counts_tmm$Entrez_ID
# saveRDS(Entrez_ID_tmm_RUV, "data/new/RUV/entrezID_tmm_RUV_list.RDS")
dim(RUV_filt_counts_tmm)[1] 14319 42#14319 genes as normal, no need to add the filtering step as the counts are already filtered
#add in the annotation files
ind_num <- readRDS("data/new/ind_num.RDS")
annot <- read.csv("data/new/Metadata.csv")
#counts need to be integer values and in a numeric matrix
#note: the log transformation needs to be accounted for in the isLog argument in RUVs function.
counts_test_tmm <- as.matrix(RUV_filt_counts_tmm)
#saveRDS(counts_test_tmm, "data/new/RUV/counts_tmm_filtered.RDS")
# Create a DataFrame for the phenoData
phenoData <- DataFrame(annot)
# Now create the RangedSummarizedExperiment necessary for RUVs input
# looks like it did need both the phenodata and the counts.
set_tmm <- SummarizedExperiment(assays = counts_test_tmm, metadata = phenoData)
dim(set_tmm)[1] 14319 42#14319 genes as before
# Generate a background matrix
# The column "Cond" holds the comparisons that you actually want to make. DOX_24, DMSO_24,5FU_24, DOX_3,etc.
scIdx <-RUVSeq::makeGroups(phenoData$Condition)
scIdx [,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 6 12 18 24 30 36 42
[2,] 4 10 16 22 28 34 40
[3,] 2 8 14 20 26 32 38
[4,] 5 11 17 23 29 35 41
[5,] 3 9 15 21 27 33 39
[6,] 1 7 13 19 25 31 37#now run through RUV correction with my tmm normalized data to generate normalized counts
set1_tmm <- RUVSeq::RUVs(x = counts_test_tmm, k =1, scIdx = scIdx, isLog = FALSE)
dim(set1_tmm$normalizedCounts)[1] 14319 42#14319 genes as before, still containing the replicate individual
#saveRDS(set1_tmm, "data/new/RUV/set1_TMMnorm_RUVsk1_normcounts.RDS")
#make a df of the data for Cormotif?
RUV_df1_tmm <- set1_tmm$W %>% as.data.frame()
RUV_df1_tmm$Names <- rownames(RUV_df1_tmm)
#ensure that the names match across samples
RUV_df_rm1_tmm <- RUV_df1_tmm[RUV_df1_tmm$Names %in% annot$Final_sample_name, ]
RUV_1_tmm <- RUV_df_rm1_tmm$W_1
#now that I've passed my TMM normalized counts into the set_tmm, proceed to Cormotif
#####Cormotif Data Setup#####
set1_tmm <- readRDS("data/new/RUV/set1_TMMnorm_RUVsk1_normcounts.RDS")
NormCounts_RUV_rep <- as.data.frame(set1_tmm$normalizedCounts)
#read in my metadata sheet that I'll use as the standard for labelling
#this one already has the replicate individual removed
Metadata_2 <- readRDS("data/new/Metadata_2_norep.RDS")
#I'll have to remove the replicate at this stage so I can properly run Cormotif
NormCounts_RUV <- NormCounts_RUV_rep %>%
dplyr::select(-(contains("6R")))
#14319 genes and 36 columns, the right size matrix for Cormotif
#now ensure the column names match the original dge$counts with Sample_name
colnames(NormCounts_RUV) <- Metadata_2$Sample_name
rownames(NormCounts_RUV) <- Entrez_ID_tmm_RUV
#now cpm the cormotif matrix you're going to use
##this should be tmm normalized log2cpm at the end for going into Cormotif
cmf_test <- cpm(NormCounts_RUV, log = TRUE)
colnames(cmf_test) <- (Metadata_2$Final_sample_name)
cmf_counts <- dge_test_tmm$counts %>%
as.data.frame() %>%
dplyr::select(-(contains("6R")))
cmf_test_df <- cmf_test %>%
as.data.frame() %>%
rownames_to_column(., var = "Entrez_ID")
#write.csv(x = cmf_test, file = "data/new/RUV/cormotif_test_matrix_TMMnorm.csv")
#reorder my test matrix to match the new groupid I've made
#I want my columns to be in this order:
#DOX24T 1-6, DOX24R 1-6, DOX144R 1-6, DMSO24T 1-6, DMSO24R 1-6, DMSO144R 1-6
Cormotif_ruv <- read.csv("data/new/RUV/Cormotif_RUV_final.csv")
dim(Cormotif_ruv)[1] 14319 37#14319 genes across 37 cols (1 is Entrez_ID)
Cormotif_ruv_df <- data.frame(Cormotif_ruv)
rownames(Cormotif_ruv_df) <- Cormotif_ruv_df$Entrez_ID
exprs.cmf_ruv <- as.matrix(Cormotif_ruv_df[,2:37])
dim(exprs.cormotif)[1] 14319 36#14319 genes in 36 columns, set up for plugging into Cormotif
#now I need to set up my groupid and compid accordingly
#put together my group id and comparison id to make the correct comparisons between experimental conditions
#groupid tells which experimental conditions are grouped together
#compid tells which experimental conditions should be compared against one another
##ie DOX24T vs DMSO24T matched control
groupid_csv <- read.csv("data/new/GroupID.csv")
#now I have to make this into a vector (named vector)?
groupid <- c(
DOX_24T_1 = 1,
DOX_24T_2 = 1,
DOX_24T_3 = 1,
DOX_24T_4 = 1,
DOX_24T_5 = 1,
DOX_24T_6 = 1,
DOX_24R_1 = 2,
DOX_24R_2 = 2,
DOX_24R_3 = 2,
DOX_24R_4 = 2,
DOX_24R_5 = 2,
DOX_24R_6 = 2,
DOX_144R_1 = 3,
DOX_144R_2 = 3,
DOX_144R_3 = 3,
DOX_144R_4 = 3,
DOX_144R_5 = 3,
DOX_144R_6 = 3,
DMSO_24T_1 = 4,
DMSO_24T_2 = 4,
DMSO_24T_3 = 4,
DMSO_24T_4 = 4,
DMSO_24T_5 = 4,
DMSO_24T_6 = 4,
DMSO_24R_1 = 5,
DMSO_24R_2 = 5,
DMSO_24R_3 = 5,
DMSO_24R_4 = 5,
DMSO_24R_5 = 5,
DMSO_24R_6 = 5,
DMSO_144R_1 = 6,
DMSO_144R_2 = 6,
DMSO_144R_3 = 6,
DMSO_144R_4 = 6,
DMSO_144R_5 = 6,
DMSO_144R_6 = 6
)
#saveRDS(groupid, "data/new/groupidCormotif.RDS")
compid <- data.frame(Cond1 = c(1, 2, 3), Cond2 = c(4, 5, 6))
#saveRDS(compid, "data/new/compidCormotif.RDS")
compid_cmf <- readRDS("data/new/compidCormotif.RDS")###Run Cormotif RUVs TMM Norm Counts
#set the seed the same as the previous one to ensure consistency
#fit Cormotif model
# set.seed(19191)
#only set the seed ONCE
#
# motif.fitted_ruv <- cormotiffit(
# exprs = exprs.cmf_ruv,
# groupid = groupid_csv,
# compid = compid_cmf,
# K = 1:10,
# max.iter = 1000,
# BIC = TRUE,
# runtype = "logCPM"
# )
# saveRDS(motif.fitted_RUV, "data/new/RUV/motif.fitted_ruv_250624.RDS")
motif.fitted_RUV <- readRDS("data/new/RUV/motif.fitted_ruv_250624.RDS")###Plot TMM Norm Counts RUVs Cormotif
#plot BIC and AIC to see which number of motifs was best for both models
plotIC(motif.fitted_RUV)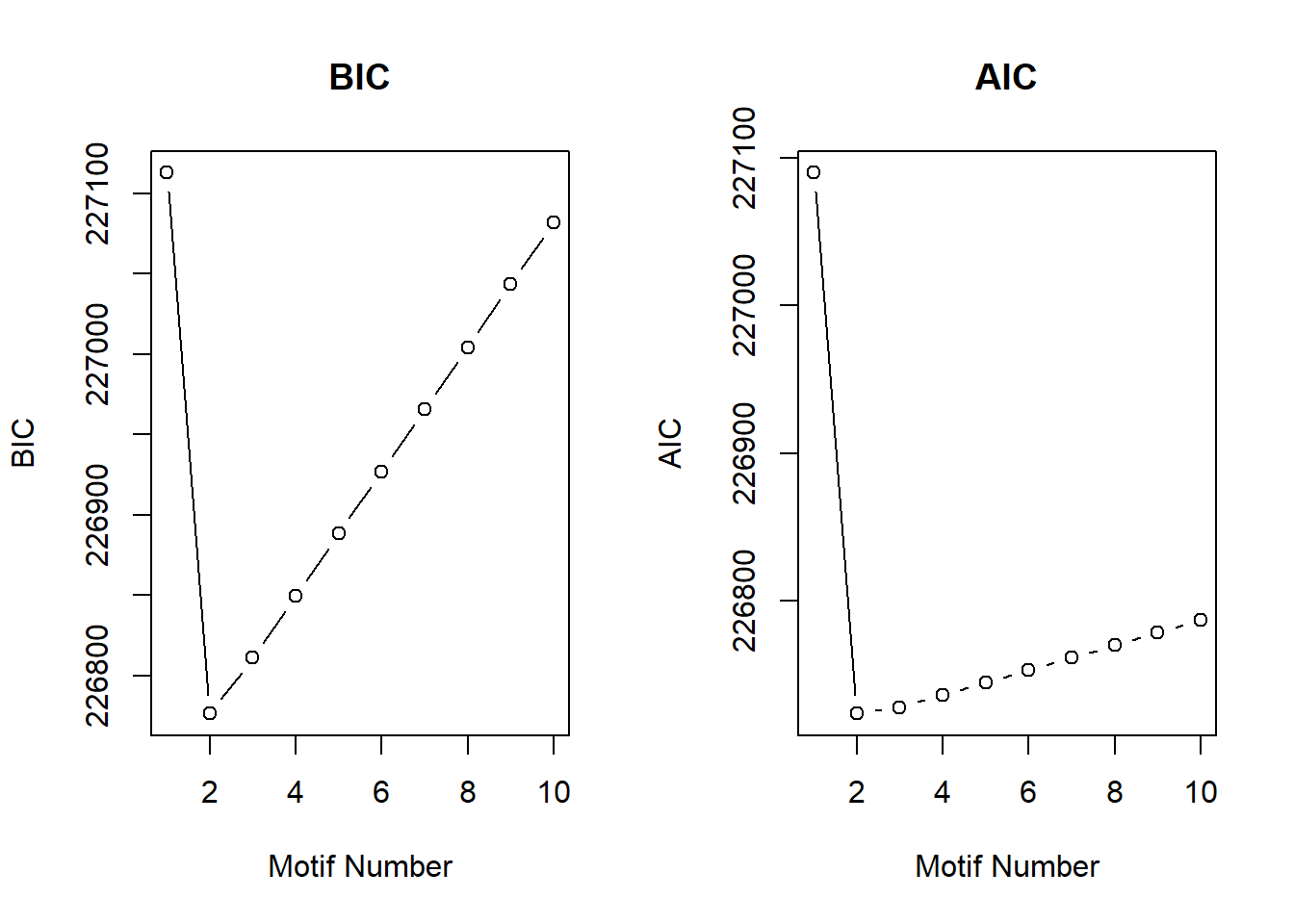
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
motif.fitted_RUV$bic K bic
[1,] 1 227112.8
[2,] 2 226776.5
[3,] 3 226811.1
[4,] 4 226849.7
[5,] 5 226888.4
[6,] 6 226927.1
[7,] 7 226965.9
[8,] 8 227004.3
[9,] 9 227043.6
[10,] 10 227082.0#now plot the motifs themselves
plotMotif(motif.fitted_RUV, title = "Fitted Motifs for DXR RUVs")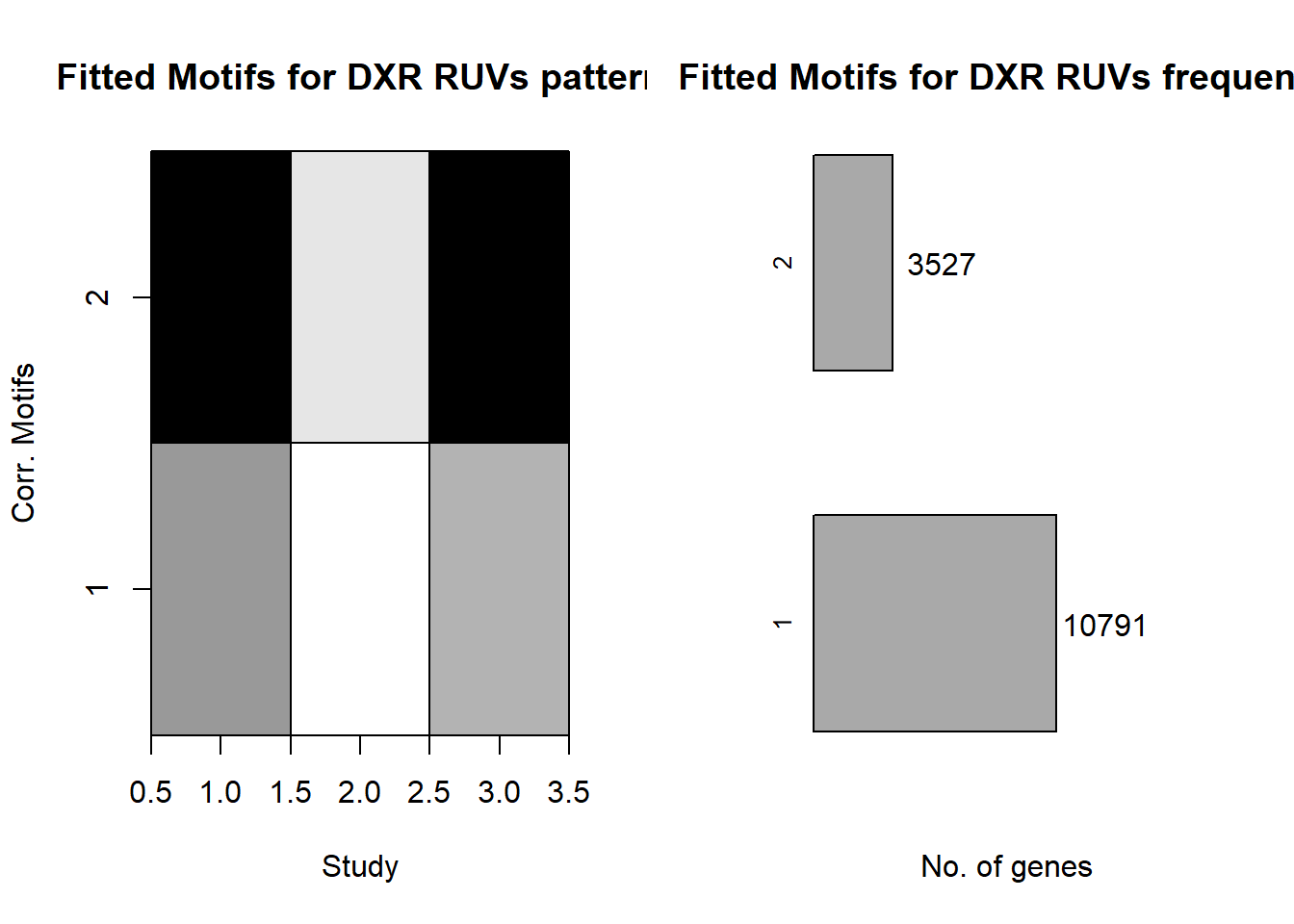
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#plot the probability legend
myColors <- rev(c("#FFFFFF", "#E6E6E6" ,"#CCCCCC", "#B3B3B3", "#999999", "#808080", "#666666","#4C4C4C", "#333333", "#191919","#000000"))
plot.new()
legend('bottomleft',fill=myColors, legend =rev(c("0", "0.1", "0.2", "0.3", "0.4", "0.5", "0.6", "0.7", "0.8","0.9", "1")), box.col="white",title = "Probability\nlegend", horiz=FALSE,title.cex=.8)
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
##Cormotif RUVs no TMM
#plot the AIC and BIC (we're interested in the Bayesian most of all)
# plotIC(motif.fit_RUV)
#
# #visual check of the lowest BIC
# motif.fit_RUV$bic
#
# #plot the motifs
# plotMotif(motif.fit_RUV)
#
# #plot the probability legend
# myColors <- rev(c("#FFFFFF", "#E6E6E6" ,"#CCCCCC", "#B3B3B3", "#999999", "#808080", "#666666","#4C4C4C", "#333333", "#191919","#000000"))
#
# plot.new()
# legend('bottomleft',fill=myColors, legend =rev(c("0", "0.1", "0.2", "0.3", "0.4", "0.5", "0.6", "0.7", "0.8","0.9", "1")), box.col="white",title = "Probability\nlegend", horiz=FALSE,title.cex=.8)##RUVs Cormotif log2 of Norm Counts
# lognormcounts_CMF_rep <- as.matrix(set1$normalizedCounts)
# lognormcounts_CMF <- log2(lognormcounts_CMF_rep + 1)
# #add a pseudocount to ensure any values are not negative or zero
#
# #first want to cut out the replicate samples as not needed for Cormotif
# lognormcounts_CMF <- as.data.frame(lognormcounts_CMF) %>%
# dplyr::select(-(contains("6R"))) %>%
# as.matrix()
#
# #14319 genes in 36 columns, set up for plugging into Cormotif
#
# #now I need to set up my groupid and compid accordingly
#
# #put together my group id and comparison id to make the correct comparisons between experimental conditions
#
# #groupid tells which experimental conditions are grouped together
# #compid tells which experimental conditions should be compared against one another
# ##ie DOX24T vs DMSO24T matched control
#
# groupid_other <- readRDS("data/new/RUV/groupid_other_unrearranged.RDS")
#
# #now put together your compid
# compid1 <- data.frame(Cond1 = c(1, 2, 3), Cond2 = c(4, 5, 6))###Run Cormotif log2 normalized counts RUVs
#set the seed the same as the previous one to ensure consistency
#fit Cormotif model
# set.seed(19191)
#only set the seed ONCE
# motif.fit_RUV_log2 <- cormotiffit(
# exprs = lognormcounts_CMF,
# groupid = groupid_other,
# compid = compid1,
# K = 1:8,
# max.iter = 500,
# BIC = TRUE,
# runtype = "logCPM"
# )
#Warning: Zero sample variances detected, have been offset away from zero#
# saveRDS(motif.fit_RUV_log2, "data/new/RUV/motif.fit_RUVnormcounts_log2NC_250629.RDS")
# motif.fit_RUV_log2 <- readRDS("data/new/RUV/motif.fit_RUVnormcounts_log2NC_250629.RDS")###Plot RUVs Cormotif log2norm counts
# #plot the AIC and BIC (we're interested in the Bayesian most of all)
# plotIC(motif.fit_RUV_log2)
#
# #visual check of the lowest BIC
# motif.fit_RUV_log2$bic
#
# #plot the motifs
# plotMotif(motif.fit_RUV_log2, title = "Log2 RUVs Norm Counts")
#
#plot the probability legend
myColors <- rev(c("#FFFFFF", "#E6E6E6" ,"#CCCCCC", "#B3B3B3", "#999999", "#808080", "#666666","#4C4C4C", "#333333", "#191919","#000000"))
plot.new()
legend('bottomleft',fill=myColors, legend =rev(c("0", "0.1", "0.2", "0.3", "0.4", "0.5", "0.6", "0.7", "0.8","0.9", "1")), box.col="white",title = "Probability\nlegend", horiz=FALSE,title.cex=.8)
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
##Cormotif Log2cpm of Normalized Counts RUVs
#since both the TMM normalized norm counts and the cpm norm counts are giving me issues:
#I am going to try this with just the log2 of the normalized counts from RUVs
lognormcounts_CMF_rep <- as.matrix(set1$normalizedCounts)
lognormcounts_CMF_cpm <- cpm(lognormcounts_CMF_rep + 1, log = TRUE)
#add a pseudocount to ensure any values are not negative or zero
#first want to cut out the replicate samples as not needed for Cormotif
lognormcounts_CMF_cpm <- as.data.frame(lognormcounts_CMF_cpm) %>%
dplyr::select(-(contains("6R"))) %>%
as.matrix()
#14319 genes in 36 cols
#write.csv(x = logNC_CMF_RUV, file = "data/new/RUV/Cormotif_log2normcounts_RUV_matrix.csv")
#14319 genes in 36 columns, set up for plugging into Cormotif
#now I need to set up my groupid and compid accordingly
#put together my group id and comparison id to make the correct comparisons between experimental conditions
#groupid tells which experimental conditions are grouped together
#compid tells which experimental conditions should be compared against one another
##ie DOX24T vs DMSO24T matched control
groupid_other <- readRDS("data/new/RUV/groupid_other_unrearranged.RDS")
#now I have to make this into a vector (named vector)?
compid <- data.frame(Cond1 = c(1, 3, 5), Cond2 = c(2, 4, 6))
compid1 <- data.frame(Cond1 = c(1, 2, 3), Cond2 = c(4, 5, 6))###Run Cormotif log2cpm Norm Counts RUVs
#set the seed the same as the previous one to ensure consistency
#fit Cormotif model
# set.seed(19191)
#only set the seed ONCE
# motif.fit_RUV_logNC <- cormotiffit(
# exprs = lognormcounts_CMF_cpm,
# groupid = groupid_other,
# compid = compid1,
# K = 1:8,
# max.iter = 500,
# BIC = TRUE,
# runtype = "logCPM"
# )
# saveRDS(motif.fit_RUV_logNC, "data/new/RUV/motif.fit_RUVnormcounts_log2cpm_250630.RDS")
motif.fit_RUV_logNC_cpm <- readRDS("data/new/RUV/motif.fit_RUVnormcounts_log2cpm_250630.RDS")###Plot Cormotif log2cpm Norm Counts RUVs
#plot the AIC and BIC (we're interested in the Bayesian most of all)
plotIC(motif.fit_RUV_logNC_cpm)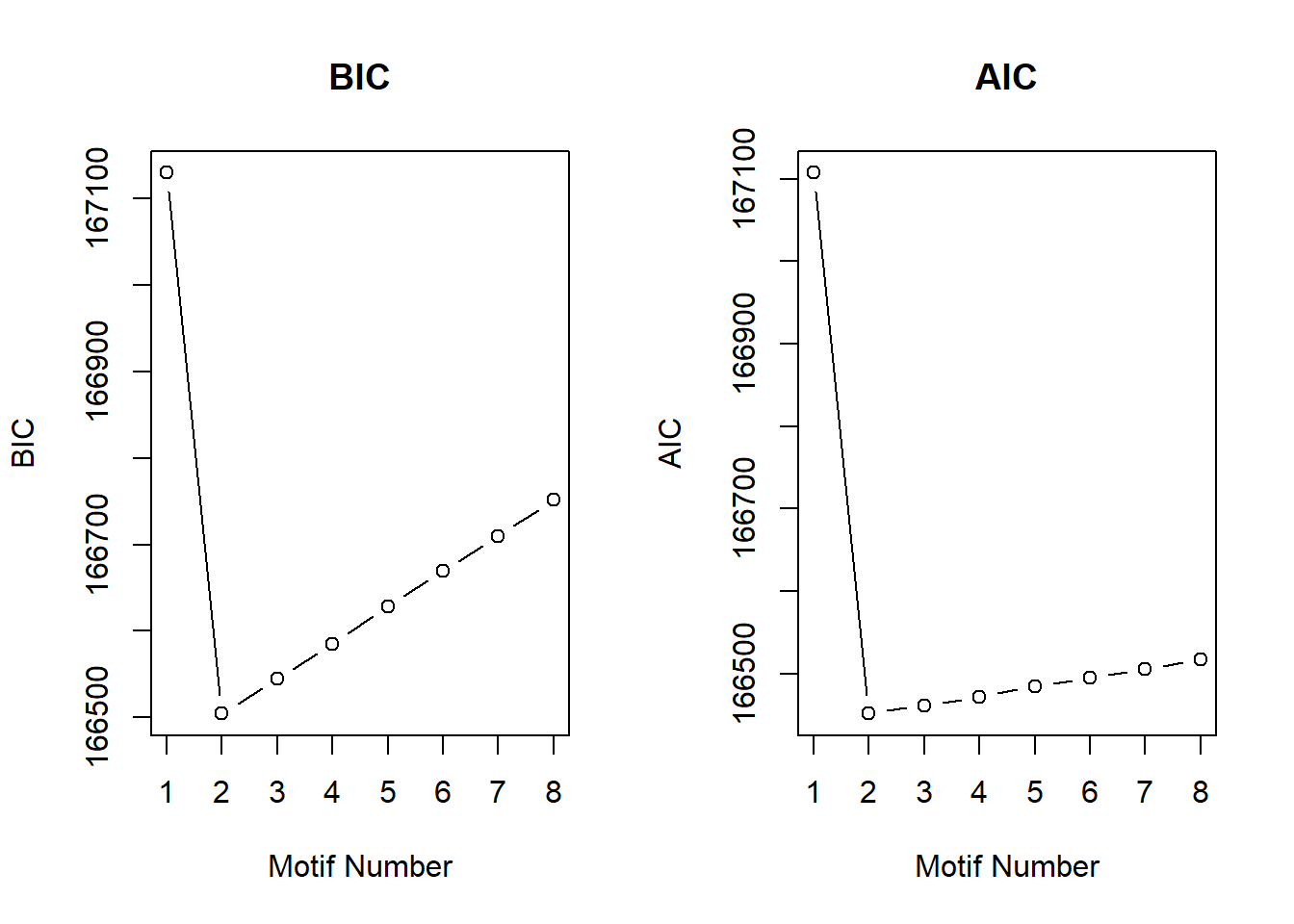
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#visual check of the lowest BIC
motif.fit_RUV_logNC_cpm$bic K bic
[1,] 1 167129.9
[2,] 2 166504.4
[3,] 3 166544.8
[4,] 4 166585.1
[5,] 5 166628.6
[6,] 6 166669.3
[7,] 7 166709.8
[8,] 8 166751.7#plot the motifs
plotMotif(motif.fit_RUV_logNC_cpm)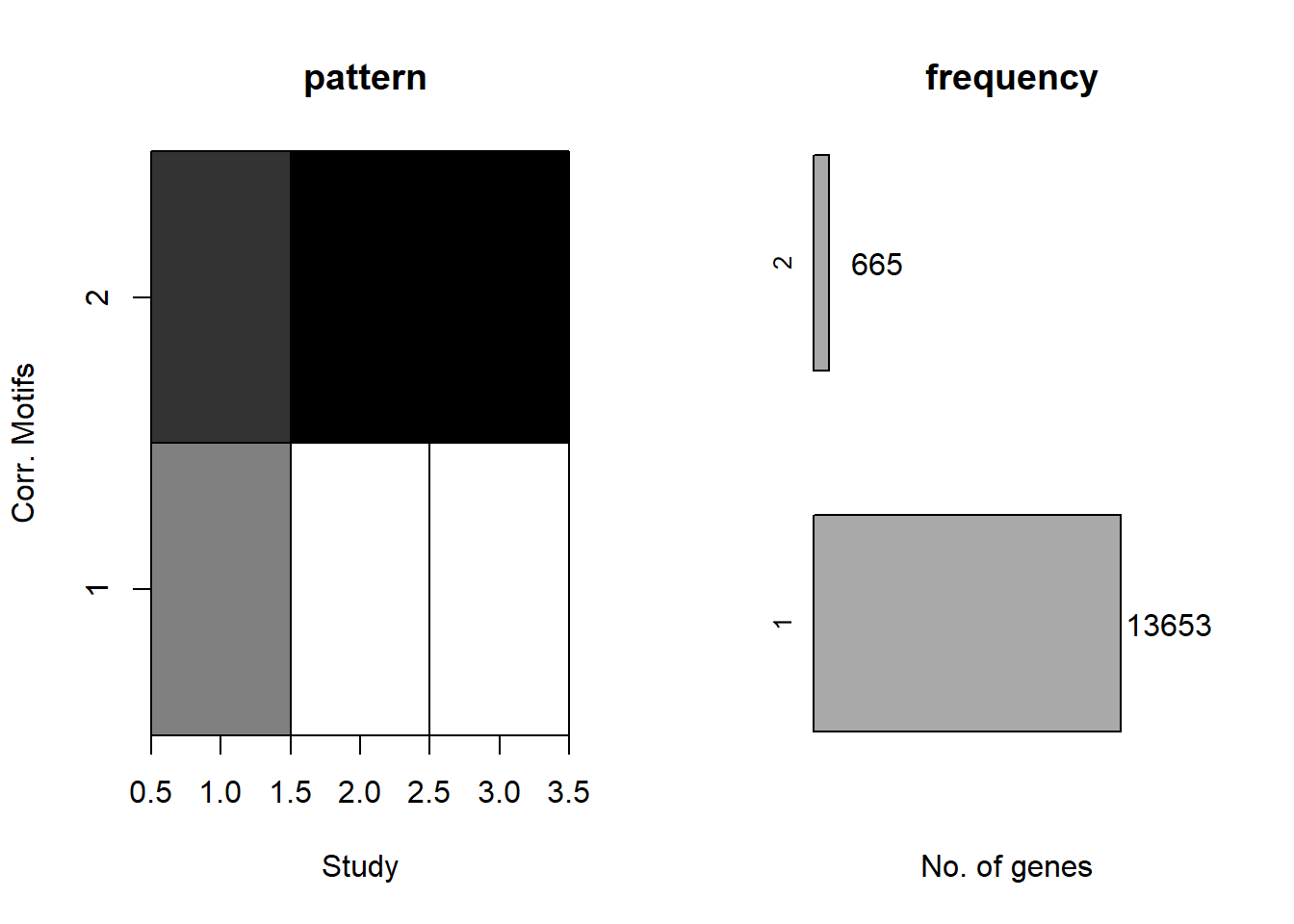
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#plot the probability legend
myColors <- rev(c("#FFFFFF", "#E6E6E6" ,"#CCCCCC", "#B3B3B3", "#999999", "#808080", "#666666","#4C4C4C", "#333333", "#191919","#000000"))
plot.new()
legend('bottomleft',fill=myColors, legend =rev(c("0", "0.1", "0.2", "0.3", "0.4", "0.5", "0.6", "0.7", "0.8","0.9", "1")), box.col="white",title = "Probability\nlegend", horiz=FALSE,title.cex=.8)
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#Log2cpm Norm Counts Cormotif Downstream Analysis
#extract the posterior probability that these DEGs belong to motifs
gene_prob_all_RUV <- motif.fit_RUV_logNC_cpm$bestmotif$p.post
rownames(gene_prob_all_RUV) <- rownames(lognormcounts_CMF_cpm)
#assign each gene to a motif with max post prob
assigned_motifs_RUV <- apply(gene_prob_all_RUV, 1, which.max)
max_probs_RUV <- apply(gene_prob_all_RUV, 1, max)
#combine these into a dataframe - motif assigned genes (p.post)
motif_assignment_df_RUV <- gene_prob_all_RUV %>%
as.data.frame() %>%
rownames_to_column("Gene") %>%
mutate(
Assigned_Motif = assigned_motifs_RUV[Gene],
Max_Probability = max_probs_RUV[Gene]
)
#make some histograms of the unfiltered data from Cormotif p.post
gene_prob_all_RUV %>%
as.data.frame() %>%
ggplot(., aes(x = V1))+
geom_histogram(bins = 50)+
xlim(0,1)+
ggtitle("Cormotif Genes p.post RUV M1")Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).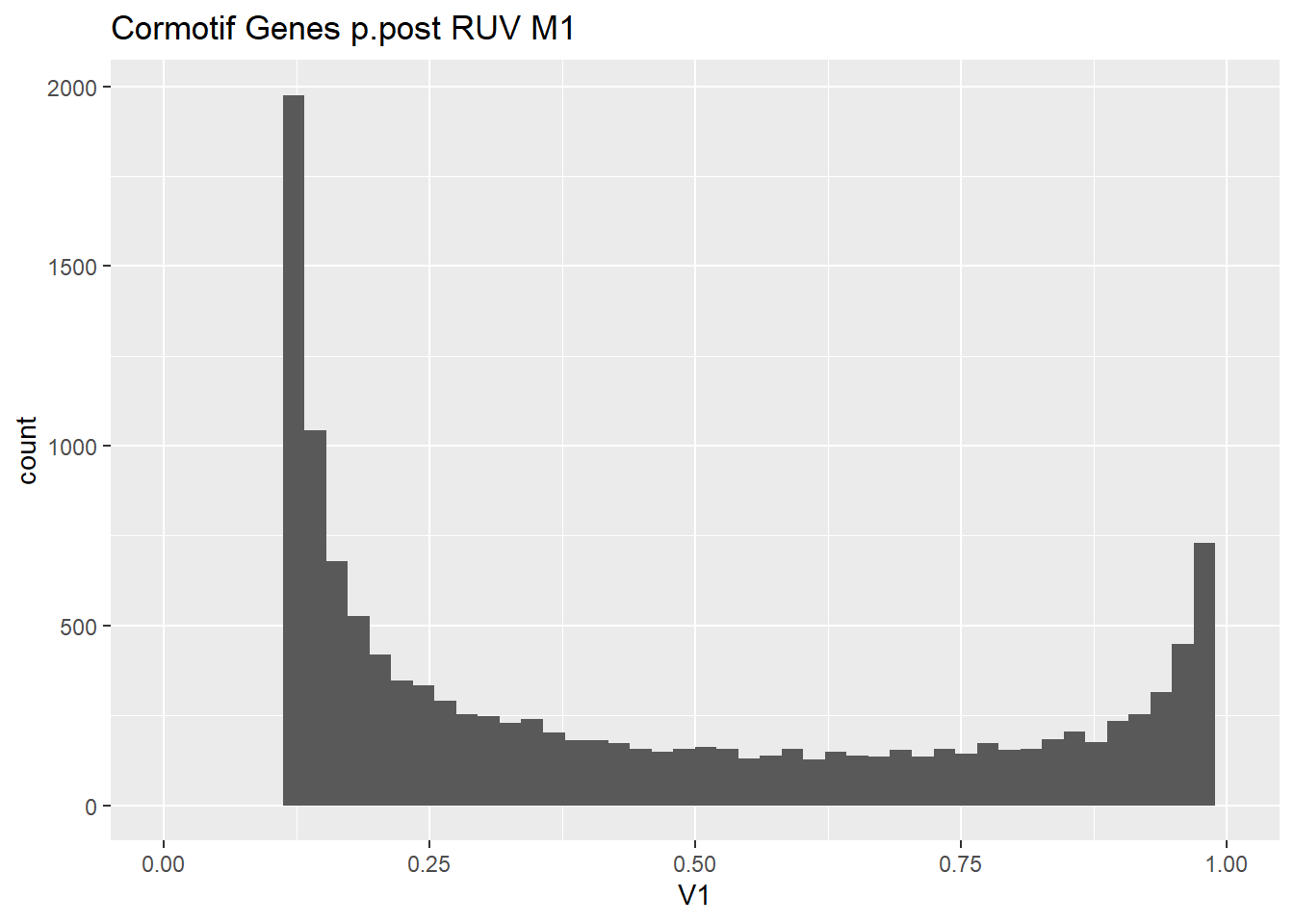
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
gene_prob_all_RUV %>%
as.data.frame() %>%
ggplot(., aes(x = V2))+
geom_histogram(bins = 50)+
xlim(0,1)+
ggtitle("Cormotif Genes p.post RUV M2")Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).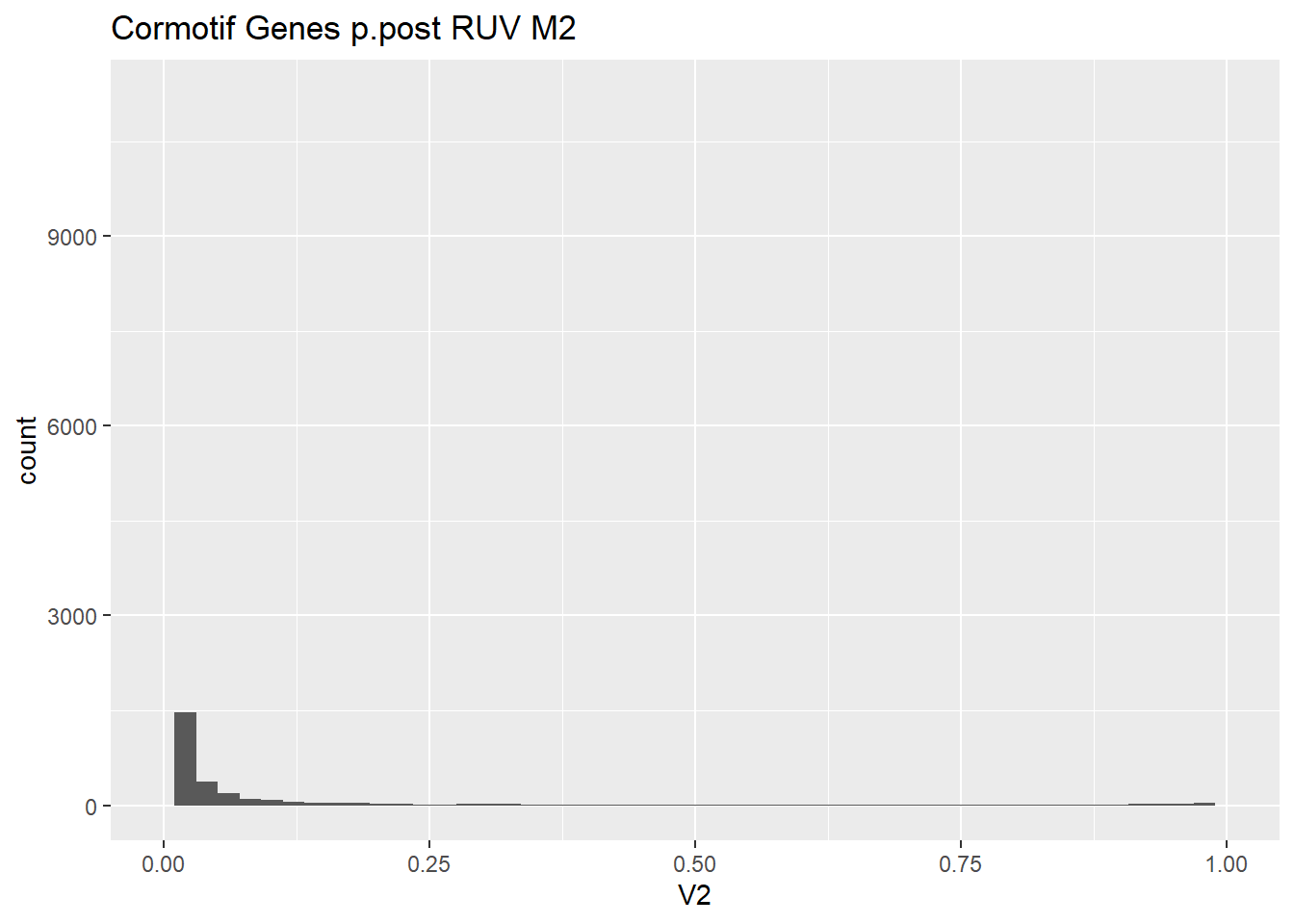
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#extract the cluster likelihood - which DEGs are most likely to be in this cluster
motif_prob_RUV <- motif.fit_RUV_logNC_cpm$bestmotif$clustlike
rownames(motif_prob_RUV) <- rownames(gene_prob_all_RUV)
# write.csv(motif_prob_RUV,"data/new/cormotif_probability_genelist_all_RUV_log2cpm_NormCounts.csv")
#make some histograms to look at the distribution of clustlike genes without filtering
motif_prob_RUV %>%
as.data.frame() %>%
ggplot(., aes(x = V1))+
geom_histogram(bins = 50)+
xlim(0,1)+
ggtitle("Cormotif Genes clustlike Distribution M1")Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).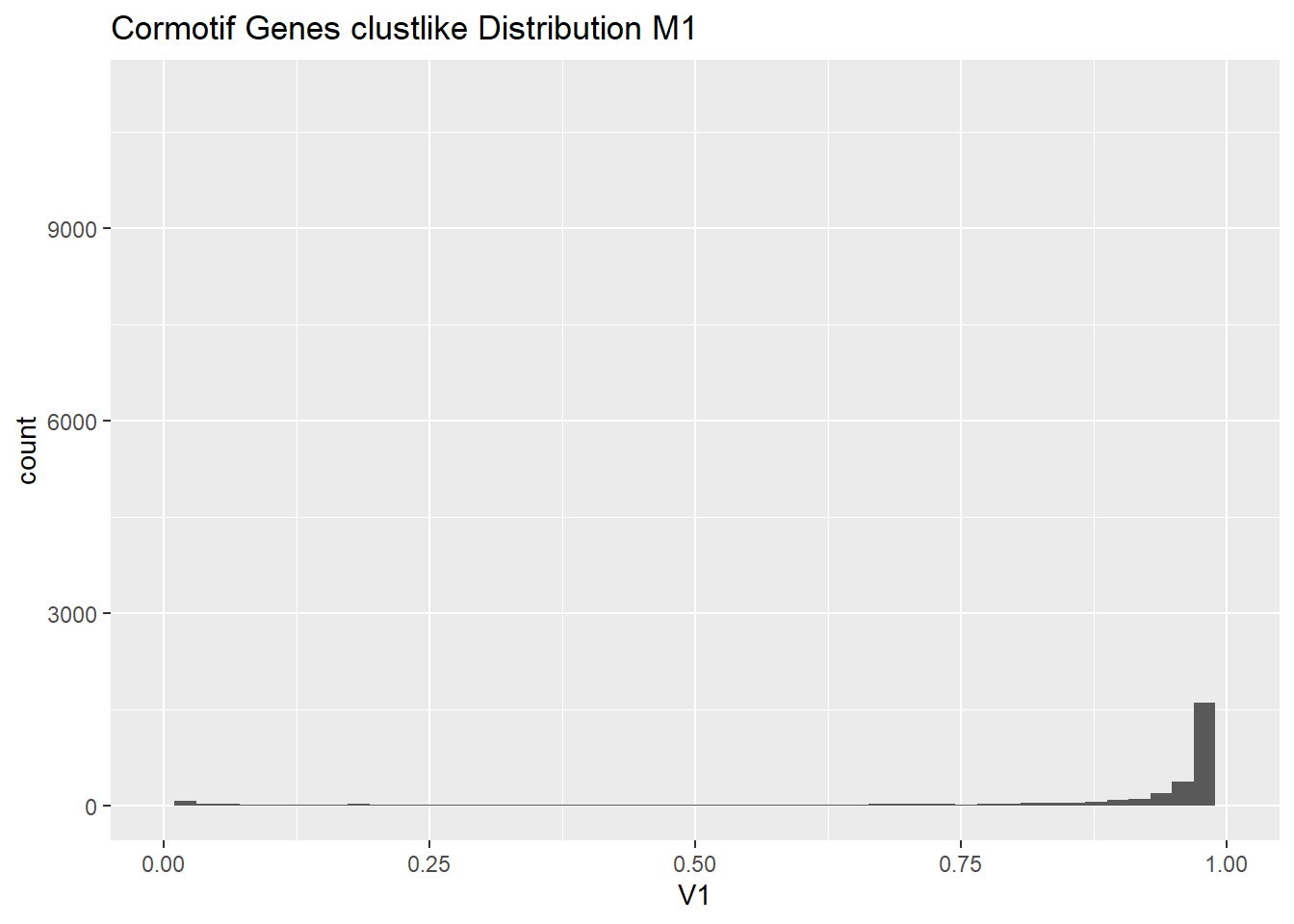
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
motif_prob_RUV %>%
as.data.frame() %>%
ggplot(., aes(x = V2))+
geom_histogram(bins = 50)+
xlim(0,1)+
ggtitle("Cormotif Genes clustlike Distribution M2")Warning: Removed 2 rows containing missing values or values outside the scale range
(`geom_bar()`).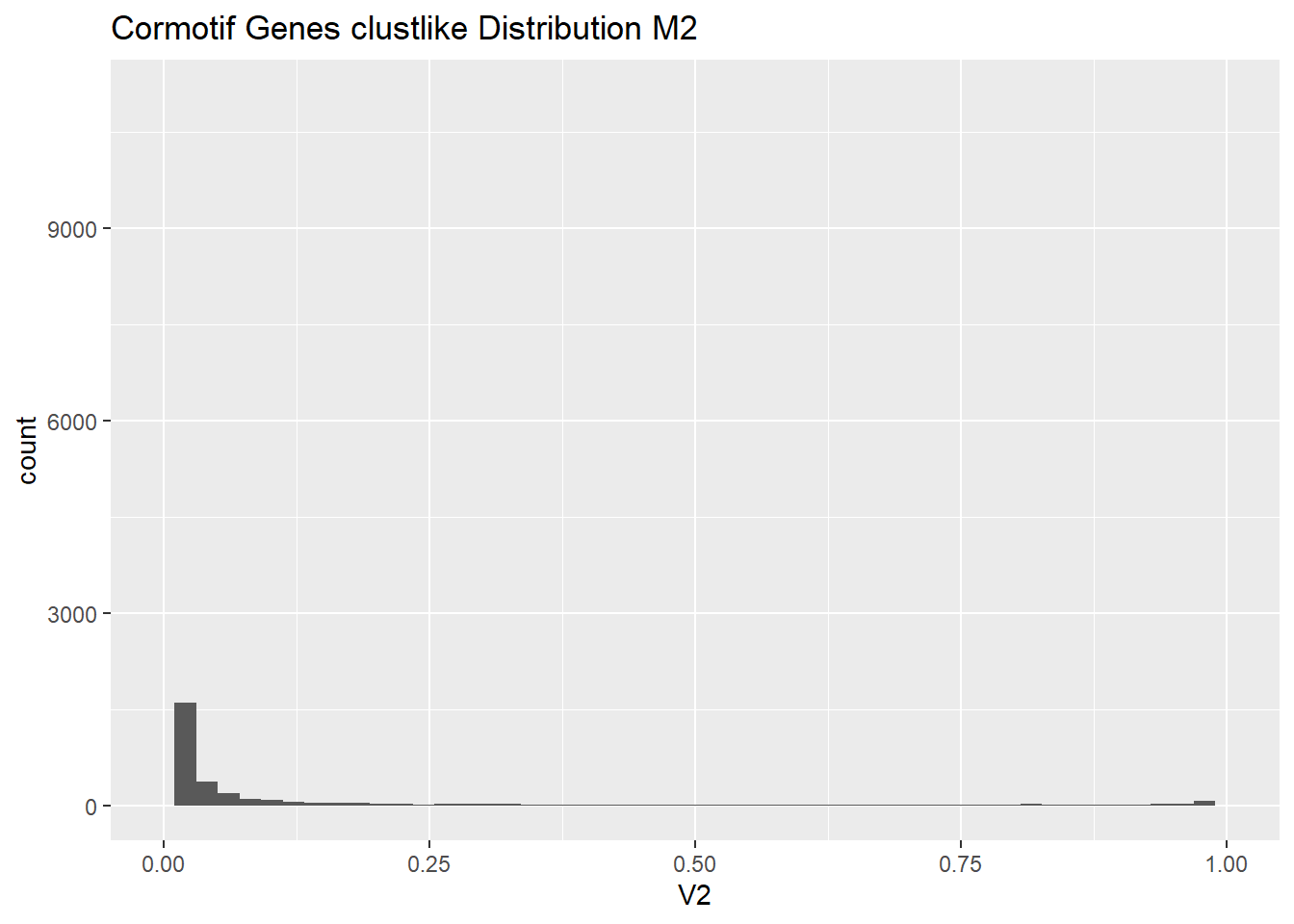
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
##P.post and Clustlike Together log2cpm norm counts Cormotif RUV
#begin with the clustlike cutoffs and then overlay the p.post to check if the genes are present in both
#these filters are the combination of the above analysis to get a gene set that fits in both p.post and clustlike
####motif 1####
#filter 1: clustlike
clust1_genes_RUV <-
motif_prob_RUV %>%
as.data.frame() %>%
filter(V1>0.8 & V2<0.5) %>%
rownames
length(clust1_genes_RUV)[1] 13600#13600 genes
### Filter 2: Gene-level posterior pattern
prob_filtered_genes_1_RUV <- rownames(gene_prob_all_RUV[(gene_prob_all_RUV[,1] >0.05 & gene_prob_all_RUV[,1] <0.5 & gene_prob_all_RUV[,2] <0.5 & gene_prob_all_RUV[,3] <0.5),])
### Final intersection of both filters
final_genes_1_RUV <- intersect(clust1_genes_RUV, prob_filtered_genes_1_RUV)
cat("Number of genes passing both filters:", length(final_genes_1_RUV), "\n")Number of genes passing both filters: 7602 #7602 genes pass both filters here
####motif 2####
#filter 1: clustlike
clust2_genes_RUV <- motif_prob_RUV %>%
as.data.frame() %>%
filter(V2>0.5 & V1<0.5) %>%
rownames
length(clust2_genes_RUV)[1] 517#517 genes
### Filter 2: Gene-level posterior pattern
prob_filtered_genes_2_RUV <- rownames(gene_prob_all_RUV[(gene_prob_all_RUV[,1] >0.3 & gene_prob_all_RUV[,2] >0.5 & gene_prob_all_RUV[,3] >0.5),])
### Final intersection of both filters
final_genes_2_RUV <- intersect(clust2_genes_RUV, prob_filtered_genes_2_RUV)
cat("Number of genes passing both filters:", length(final_genes_2_RUV), "\n")Number of genes passing both filters: 501 #501 genes pass both filters here
#what is the proportion of my genes that are included?
#p.post and clustlike together
7673+501[1] 8174#8174
(8174/14319)*100[1] 57.08499#57.08% of my genes are represented here out of the 14319 original
#what is the proportion of my genes that are included?
#clustlike only
13600+517[1] 14117#14117
(14117/14319)*100[1] 98.58929#98.6% of my genes are included with clustlike only
#want to find out which genes are not assigned to motifs
final_genes_list_RUV <- union(final_genes_1_RUV, final_genes_2_RUV)
length(final_genes_list_RUV)[1] 8103#8174 genes as found above
initial_genes_list_RUV <- rownames(lognormcounts_CMF_cpm)
length(initial_genes_list_RUV)[1] 14319#14319 genes as usual
unassigned_genes_list_RUV <- setdiff(initial_genes_list_RUV, final_genes_list_RUV)
length(unassigned_genes_list_RUV)[1] 6216#6145 genes not assigned
final_genes_list_clustlike_RUV <- union(clust1_genes_RUV, clust2_genes_RUV)
length(final_genes_list_clustlike_RUV)[1] 14117#14117 genes as above with clustlike only
unassigned_genes_list_RUV_clustlike <- setdiff(initial_genes_list_RUV, final_genes_list_clustlike_RUV)
length(unassigned_genes_list_RUV_clustlike)[1] 202#202 genes not assigned
# saveRDS(unassigned_genes_list_RUV, "data/new/RUV/CMF/unassigned_genes_RUVlog2cpm_NormCounts_Cormotif.RDS")
# saveRDS(unassigned_genes_list_RUV_clustlike, "data/new/RUV/CMF/unassigned_genes_clustlike_RUVslog2cpm_NormCounts_Cormotif.RDS")
#now, ensure that none of these genes are shared amongst categories
vennCor_final_RUV <- list(final_genes_1_RUV, final_genes_2_RUV)
ggVennDiagram(
vennCor_final_RUV,
category.names = c("M1", "M2")
) + ggtitle("Cormotif DEGs clustlike + p.post RUVs log2cpm NC")+
theme(
plot.title = element_text(size = 16, face = "bold"), # Increase title size
text = element_text(size = 16) # Increase text size globally
)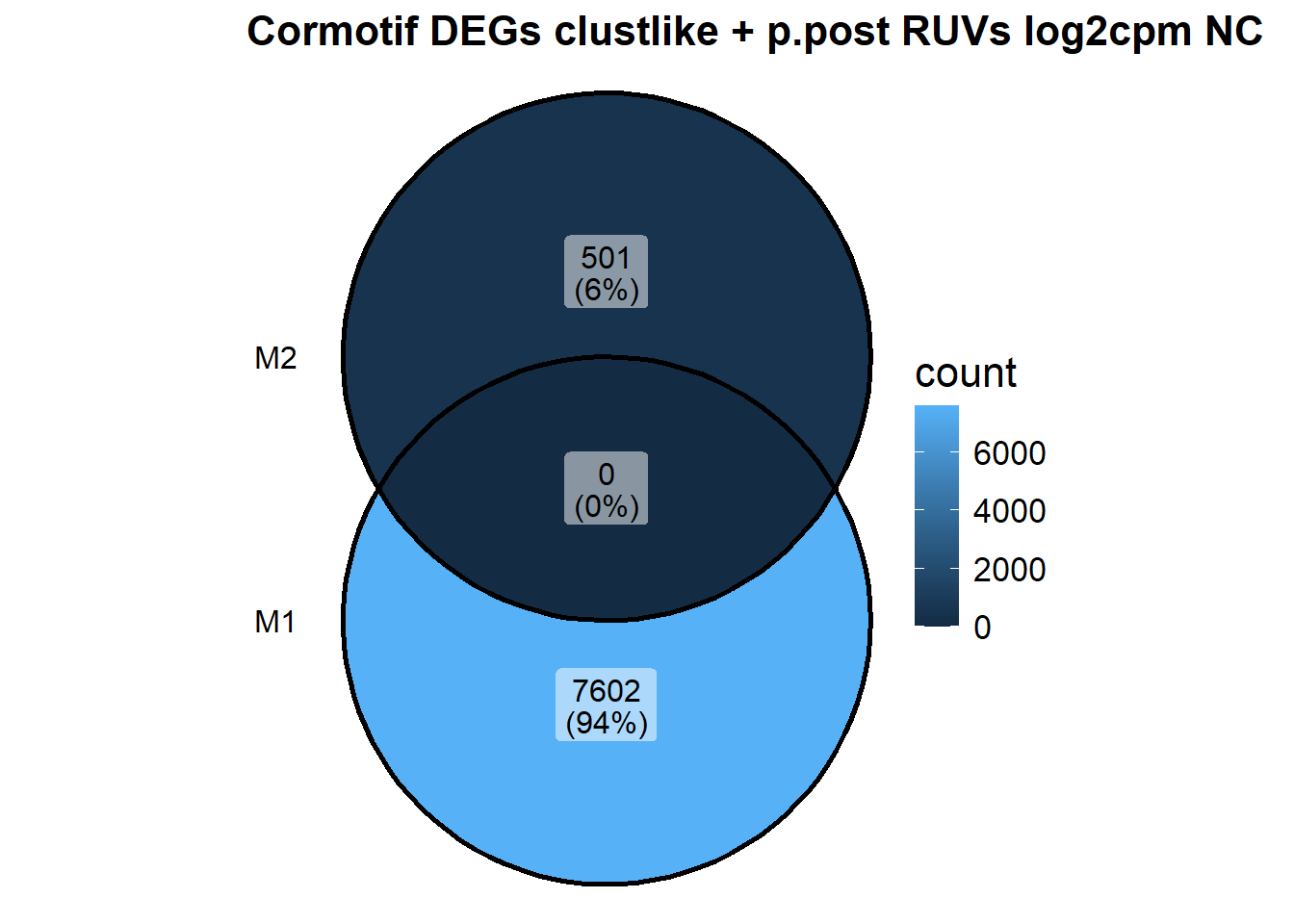
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
vennCor_final_RUV_clust <- list(clust1_genes_RUV, clust2_genes_RUV)
ggVennDiagram(
vennCor_final_RUV_clust,
category.names = c("M1", "M2")
) + ggtitle("Cormotif DEGs clustlike RUVs log2cpm NC")+
theme(
plot.title = element_text(size = 16, face = "bold"), # Increase title size
text = element_text(size = 16) # Increase text size globally
)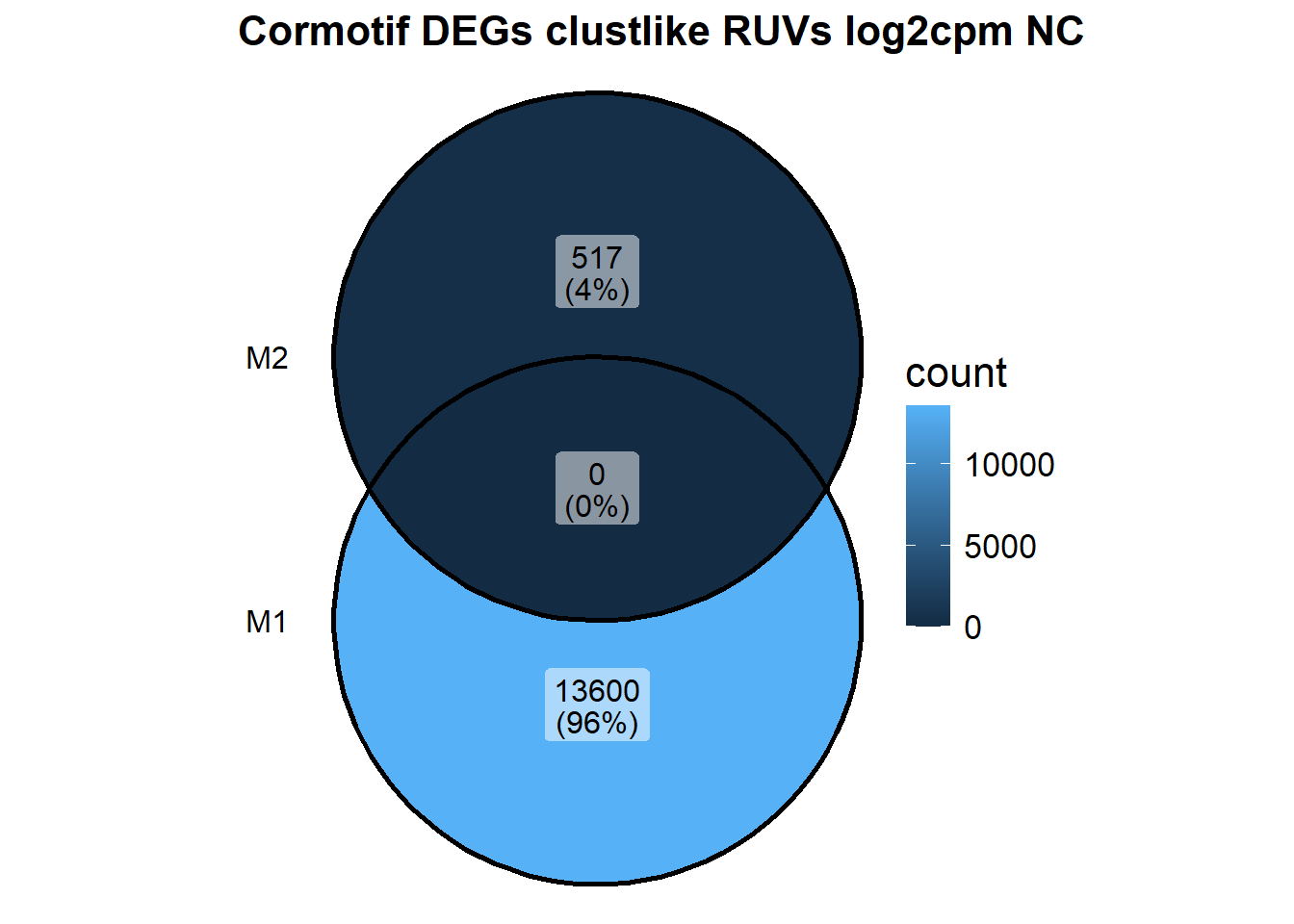
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#no overlapping genes found in these gene sets
clusterdata_dxr_fin_RUV <- data.frame(
Category = c("Motif 1", "Motif 2"),
Value = c(length(final_genes_1_RUV), length(final_genes_2_RUV)
))
clusterdata_dxr_fin_RUV_clust <- data.frame(
Category = c("Motif 1", "Motif 2"),
Value = c(length(clust1_genes_RUV), length(clust2_genes_RUV)
))
piecolors_dxr_fin <- c("Motif 1" = "#007896",
"Motif 2" = "#58508D")
#make a piechart of these distributions
clusterdata_dxr_fin_RUV %>% ggplot(aes(x = "", y = Value, fill = Category))+
geom_bar(width = 1, stat = "identity")+
coord_polar("y", start = 0)+
geom_text(aes(label = Value),
position = position_stack(vjust = 0.5),
size = 4, color = "black")+
labs(title = "Distr of Gene Clusters Cormotif RUVs", x = NULL, y = NULL)+
theme_void()+
scale_fill_manual(values = piecolors_dxr_fin)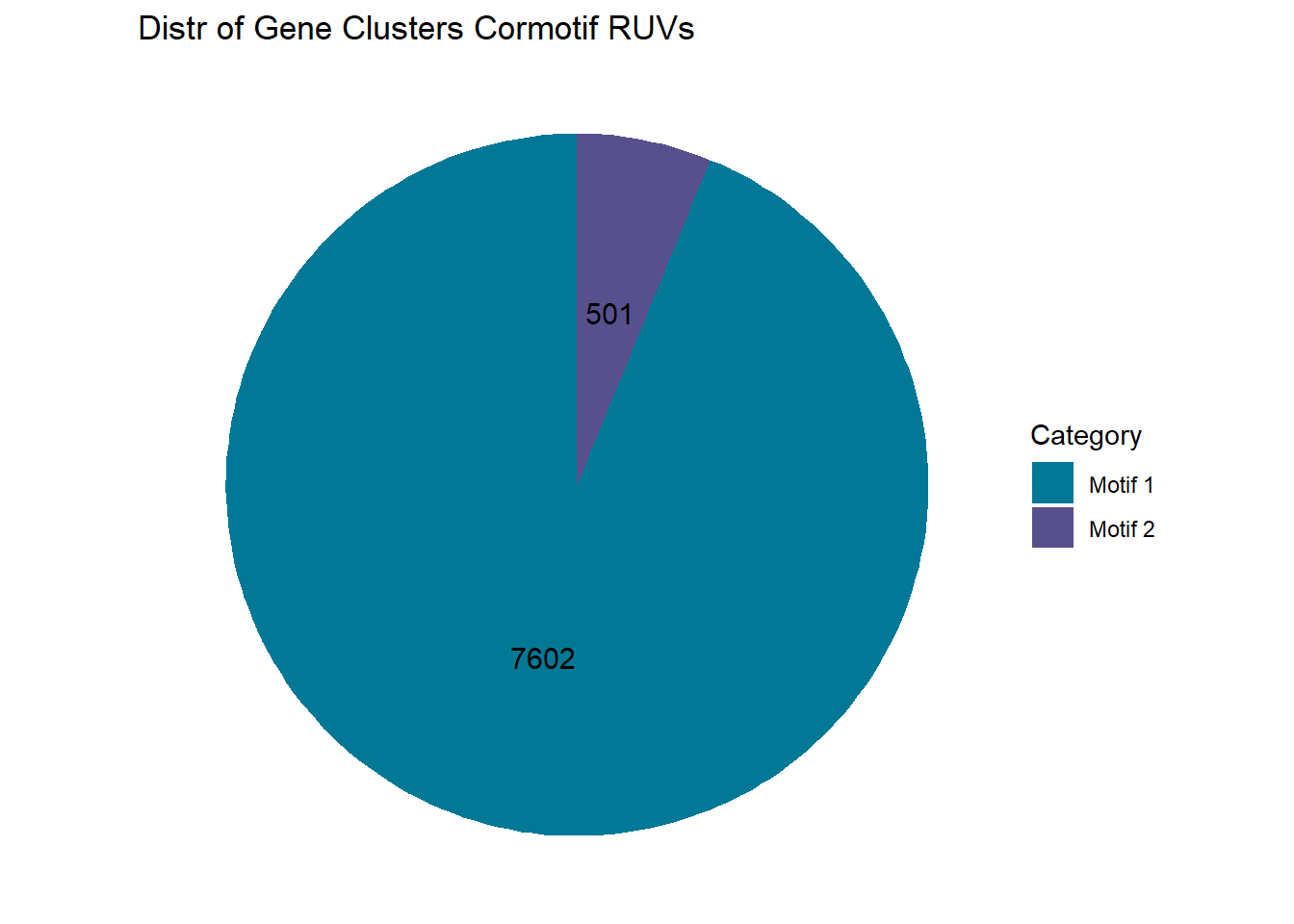
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
clusterdata_dxr_fin_RUV_clust %>% ggplot(aes(x = "", y = Value, fill = Category))+
geom_bar(width = 1, stat = "identity")+
coord_polar("y", start = 0)+
geom_text(aes(label = Value),
position = position_stack(vjust = 0.5),
size = 4, color = "black")+
labs(title = "Distr of Gene Clusters Cormotif RUVs clustlike", x = NULL, y = NULL)+
theme_void()+
scale_fill_manual(values = piecolors_dxr_fin)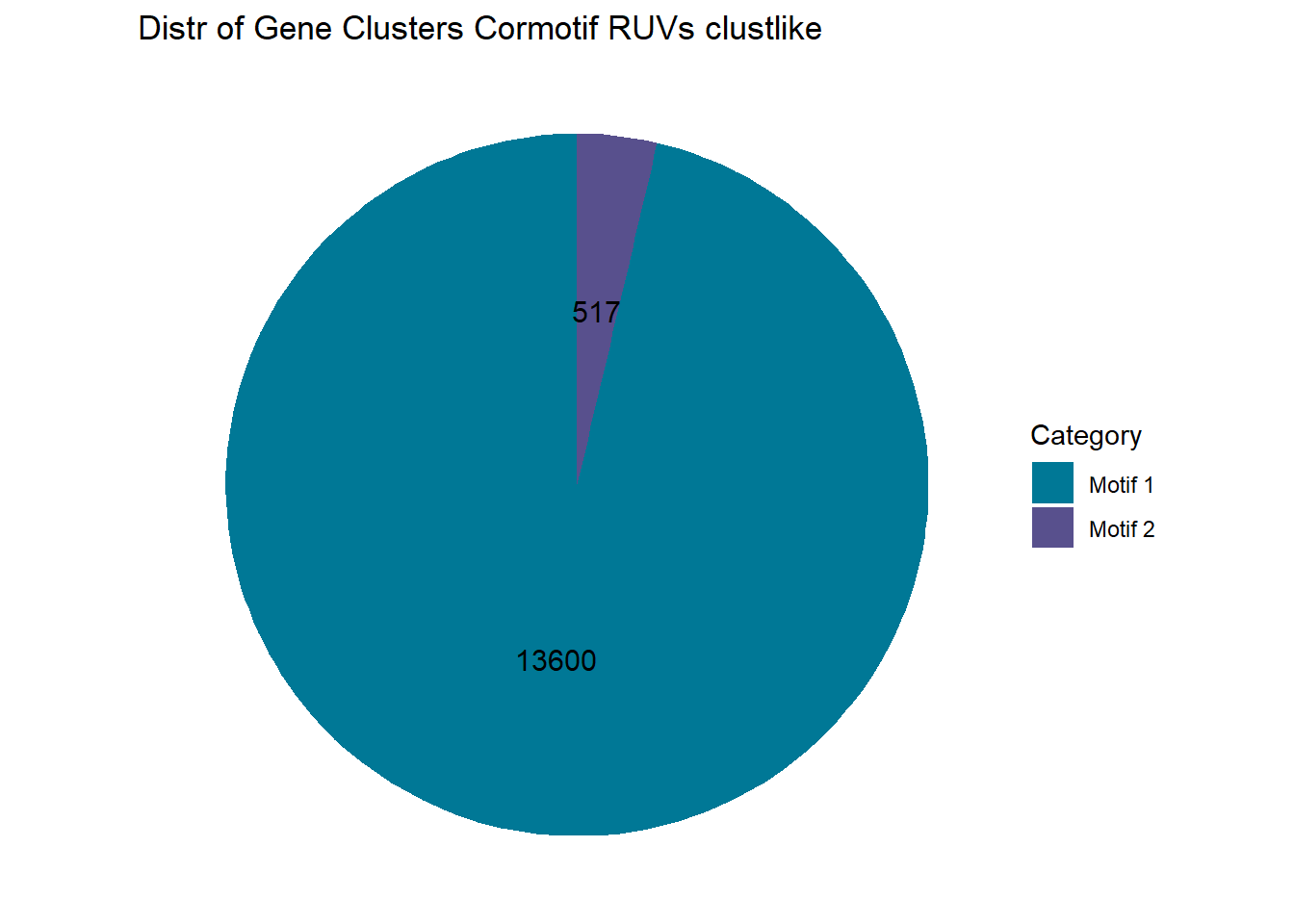
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#now, continue by plotting the logFC of all of these gene sets per motif, as well as look at some random genes for log2cpm to see if the patterns are reflected in the example genes###clustlike RUVs log2cpm Norm Counts RUVs logFC
####clustlike + p.post logFC of initial set####
##motif 1
length(final_genes_1_RUV)[1] 7602##motif 2
length(final_genes_2_RUV)[1] 501#Combine the toptables I have from pairwise analysis into a single dataframe
d24_toptable_dxr_RUV <- Toptable_RUV_24T %>%
rownames_to_column(var = "entrezgene_ID") %>%
mutate(Time = "24T")
d24r_toptable_dxr_RUV <- Toptable_RUV_24R %>%
rownames_to_column(var = "entrezgene_ID") %>%
mutate(Time = "24R")
d144r_toptable_dxr_RUV <- Toptable_RUV_144R %>%
rownames_to_column(var = "entrezgene_ID") %>%
mutate(Time = "144R")
combined_toptables_dxr_RUV <- bind_rows(
d24_toptable_dxr_RUV,
d24r_toptable_dxr_RUV,
d144r_toptable_dxr_RUV)
#Filter the data based on each motif
filt_toptable_dxr_fin_RUV <- combined_toptables_dxr_RUV %>%
dplyr::filter(entrezgene_ID %in% final_genes_1_RUV) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24T", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC Motif 1 RUVs clustlike + p.post (n=7673)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 2
filt_toptable_2_dxr_fin_RUV <- combined_toptables_dxr_RUV %>%
dplyr::filter(entrezgene_ID %in% final_genes_2_RUV) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24T", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC Motif 2 RUVs clustlike + p.post (n=501)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#plots
filt_toptable_dxr_fin_RUV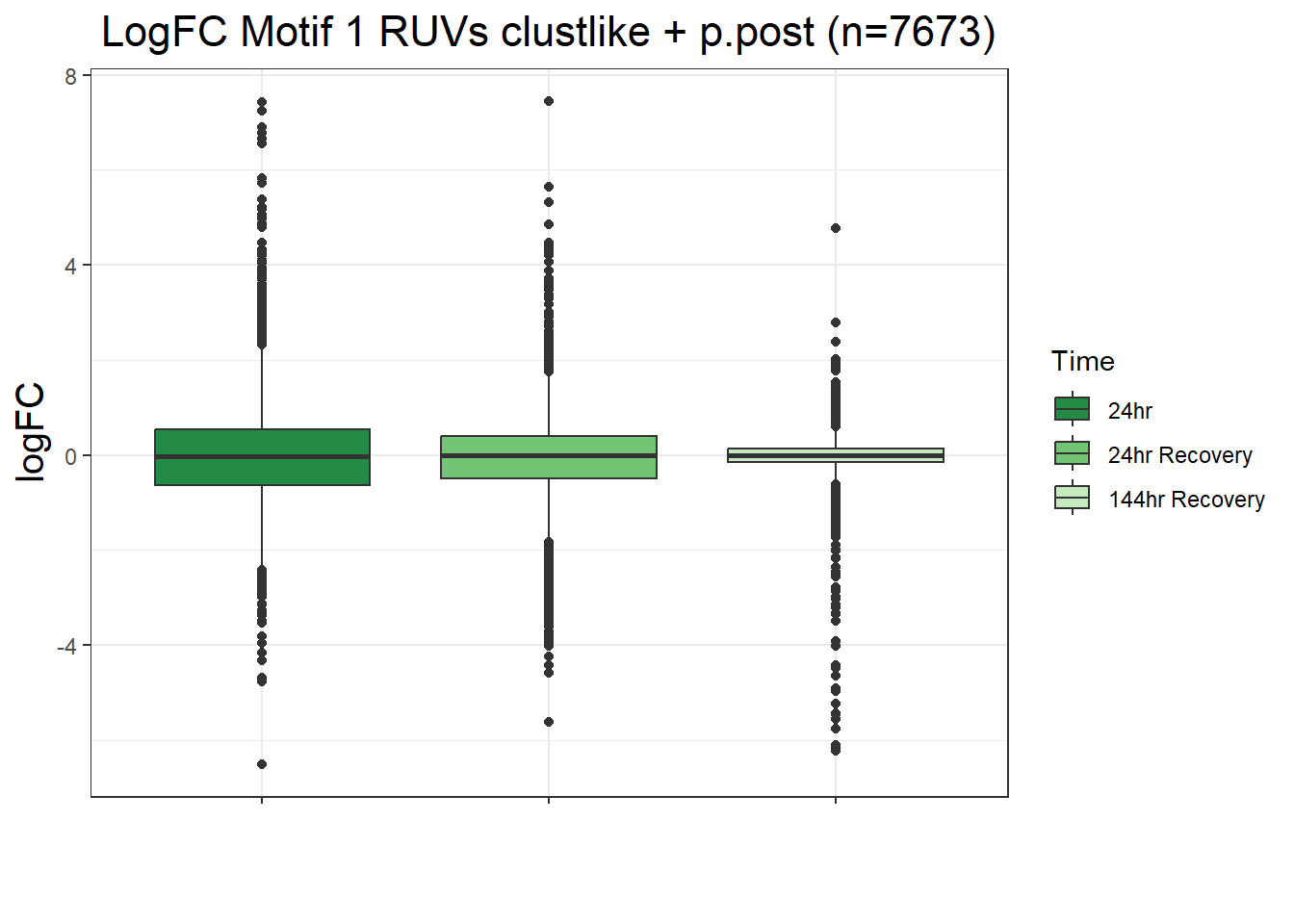
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
filt_toptable_2_dxr_fin_RUV 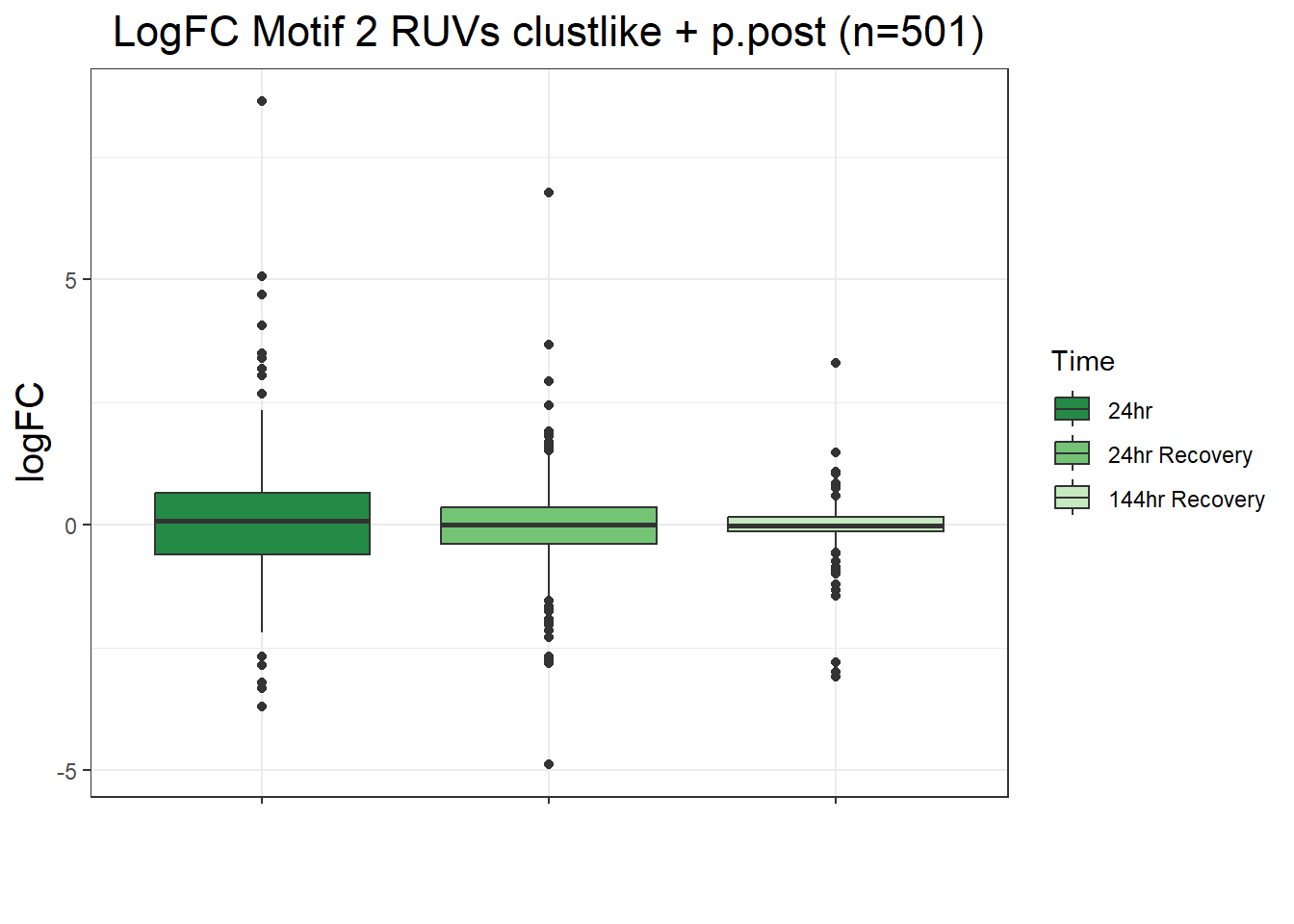
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#now plot the abs logFC for each of these too
filt_toptable_abs_fin_RUV <- combined_toptables_dxr_RUV %>%
dplyr::filter(entrezgene_ID %in% final_genes_1_RUV) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24T", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = absFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("|logFC|")+
theme_bw()+
ggtitle("Abs LogFC Motif 1 RUVs clustlike + p.post (n=7673)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 2
filt_toptable_2_abs_fin_RUV <- combined_toptables_dxr_RUV %>%
dplyr::filter(entrezgene_ID %in% final_genes_2_RUV) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24T", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = absFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("|logFC|")+
theme_bw()+
ggtitle("Abs LogFC Motif 2 RUVs clustlike + p.post (n=501)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 2
filt_toptable_3_abs_fin <- combined_toptables_dxr %>%
dplyr::filter(entrezgene_ID %in% final_genes_3) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = absFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("|logFC|")+
theme_bw()+
ggtitle("Abs LogFC for all genes in Motif 3 (n=231)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#plots
filt_toptable_abs_fin_RUV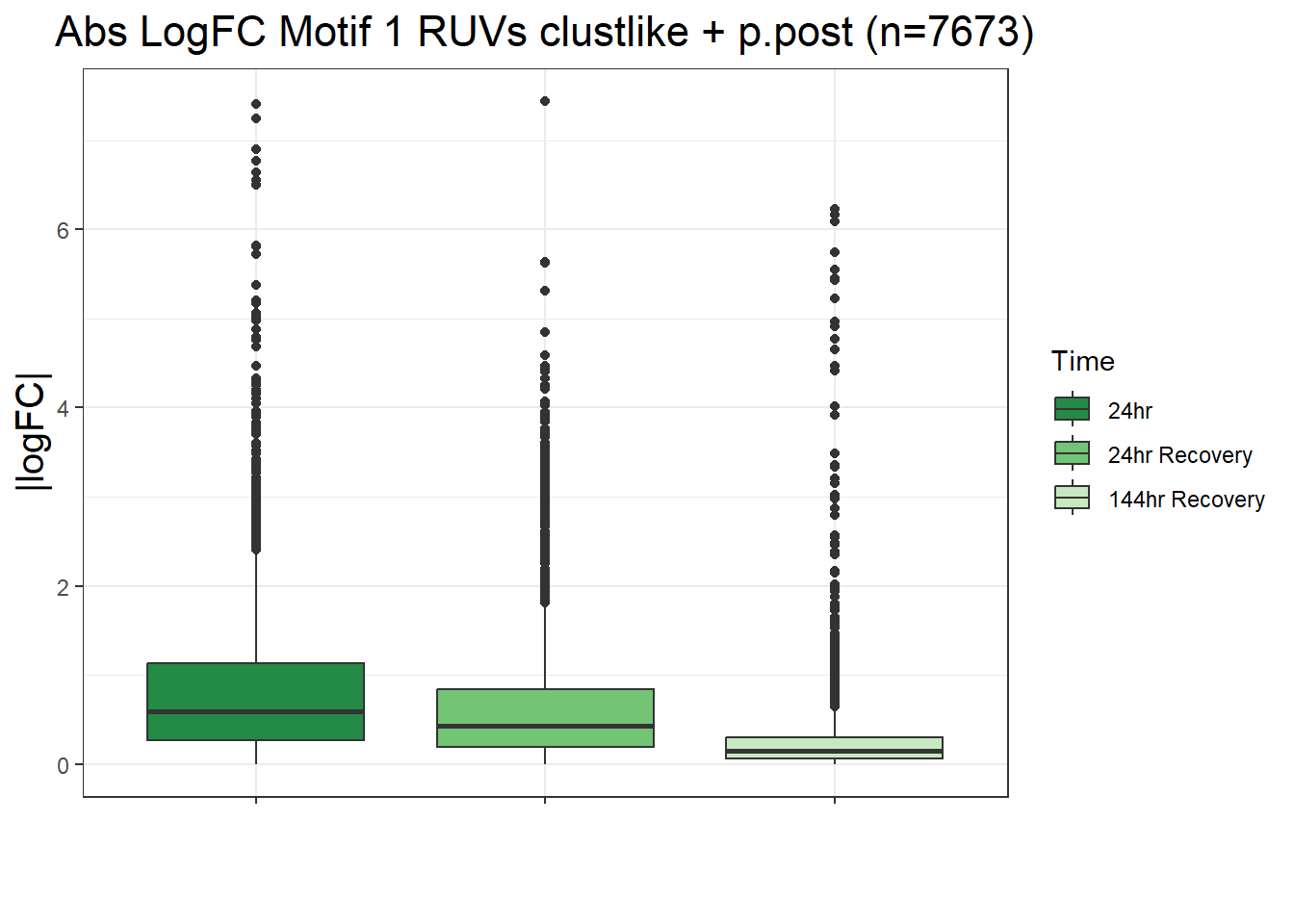
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
filt_toptable_2_abs_fin_RUV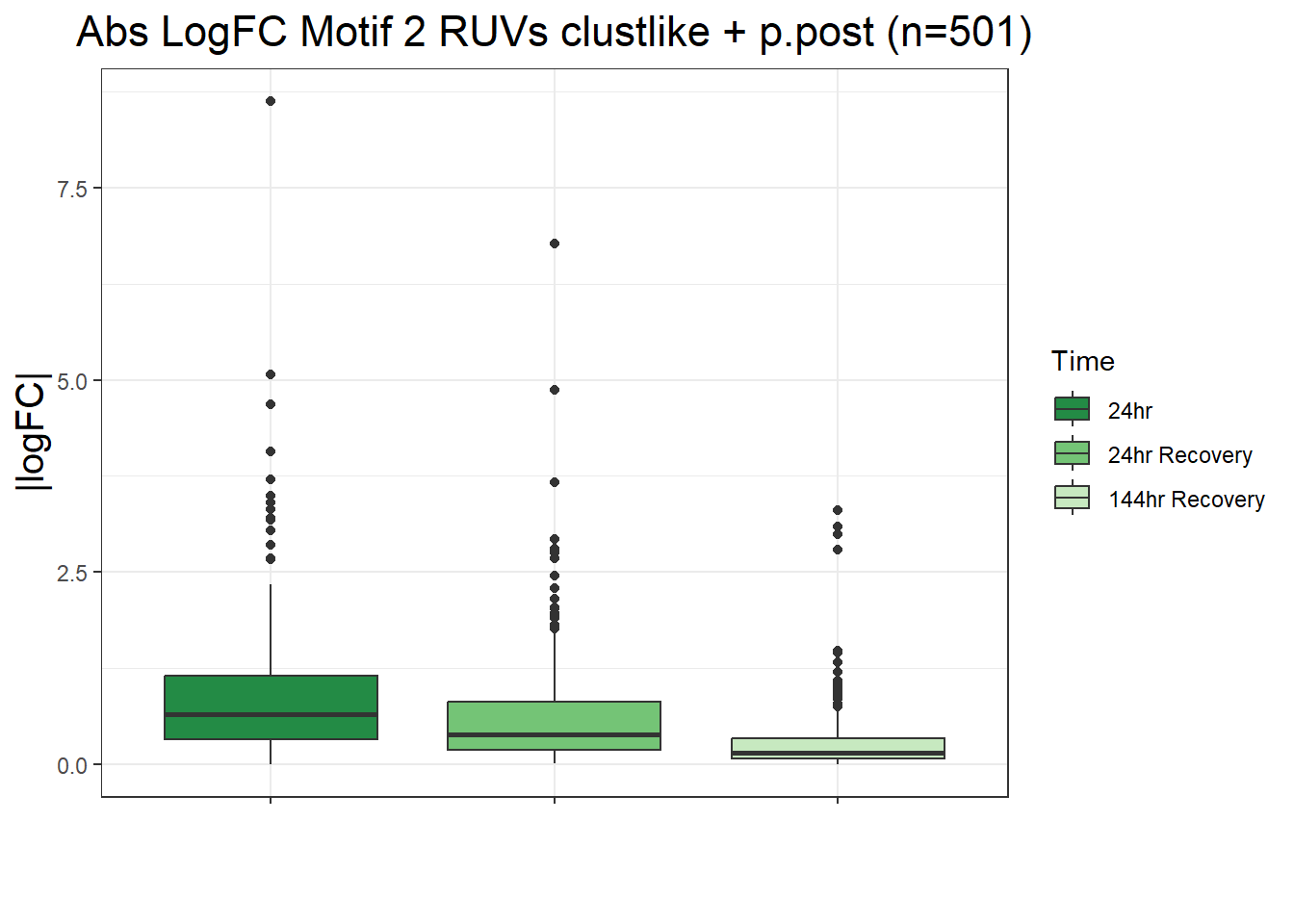
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
####Now do this but have it for the clustlike only genes
##motif 1
length(clust1_genes_RUV)[1] 13600#13600
##motif 2
length(clust2_genes_RUV)[1] 517#517 genes
#Filter the data based on each motif
filt_toptable_dxr_fin_RUV_clust <- combined_toptables_dxr_RUV %>%
dplyr::filter(entrezgene_ID %in% clust1_genes_RUV) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24T", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC Motif 1 RUVs clustlike (n=13600)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 2
filt_toptable_2_dxr_fin_RUV_clust <- combined_toptables_dxr_RUV %>%
dplyr::filter(entrezgene_ID %in% clust2_genes_RUV) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24T", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = logFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("logFC")+
theme_bw()+
ggtitle("LogFC Motif 2 RUVs clustlike (n=517)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#plots
filt_toptable_dxr_fin_RUV_clust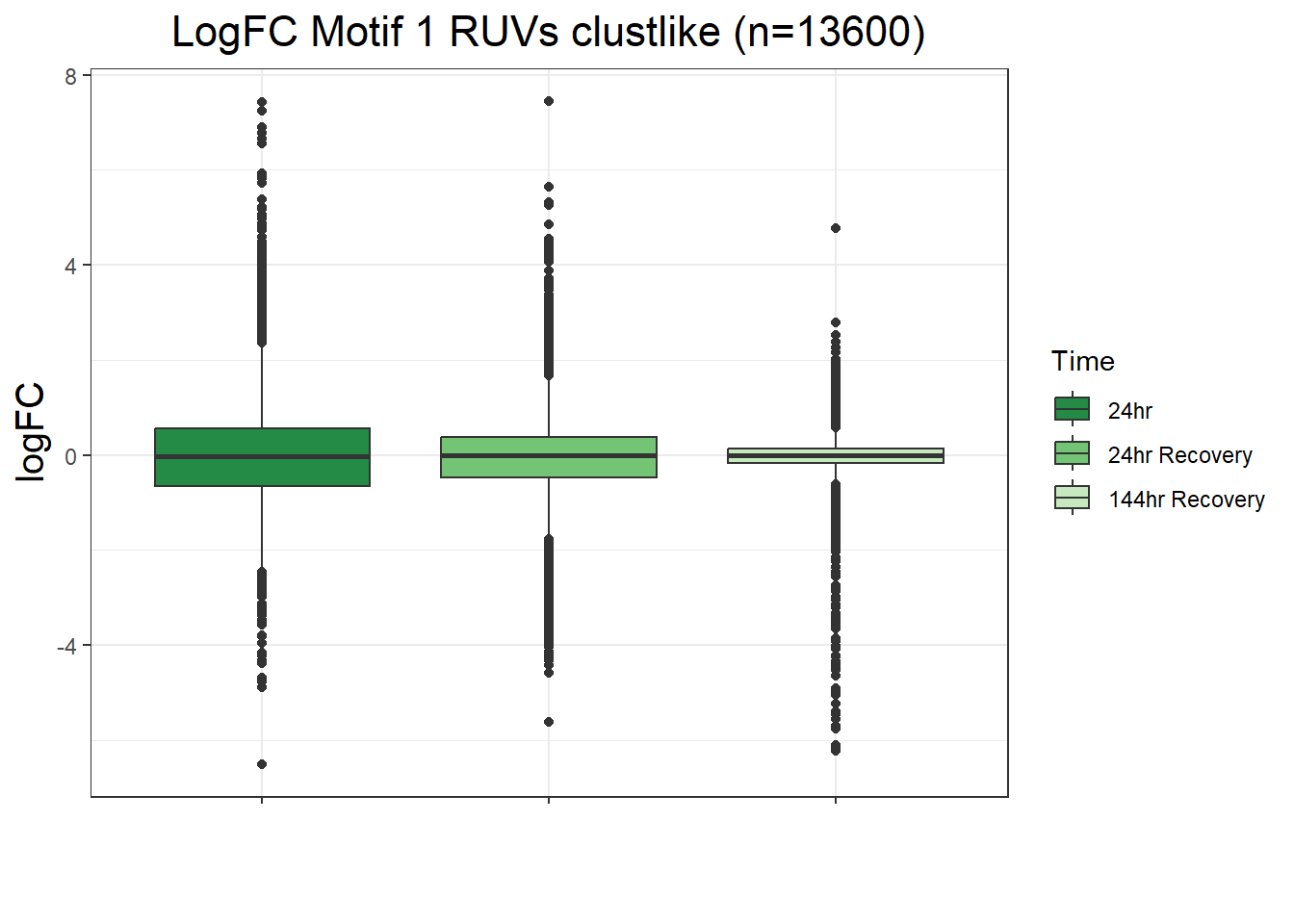
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
filt_toptable_2_dxr_fin_RUV_clust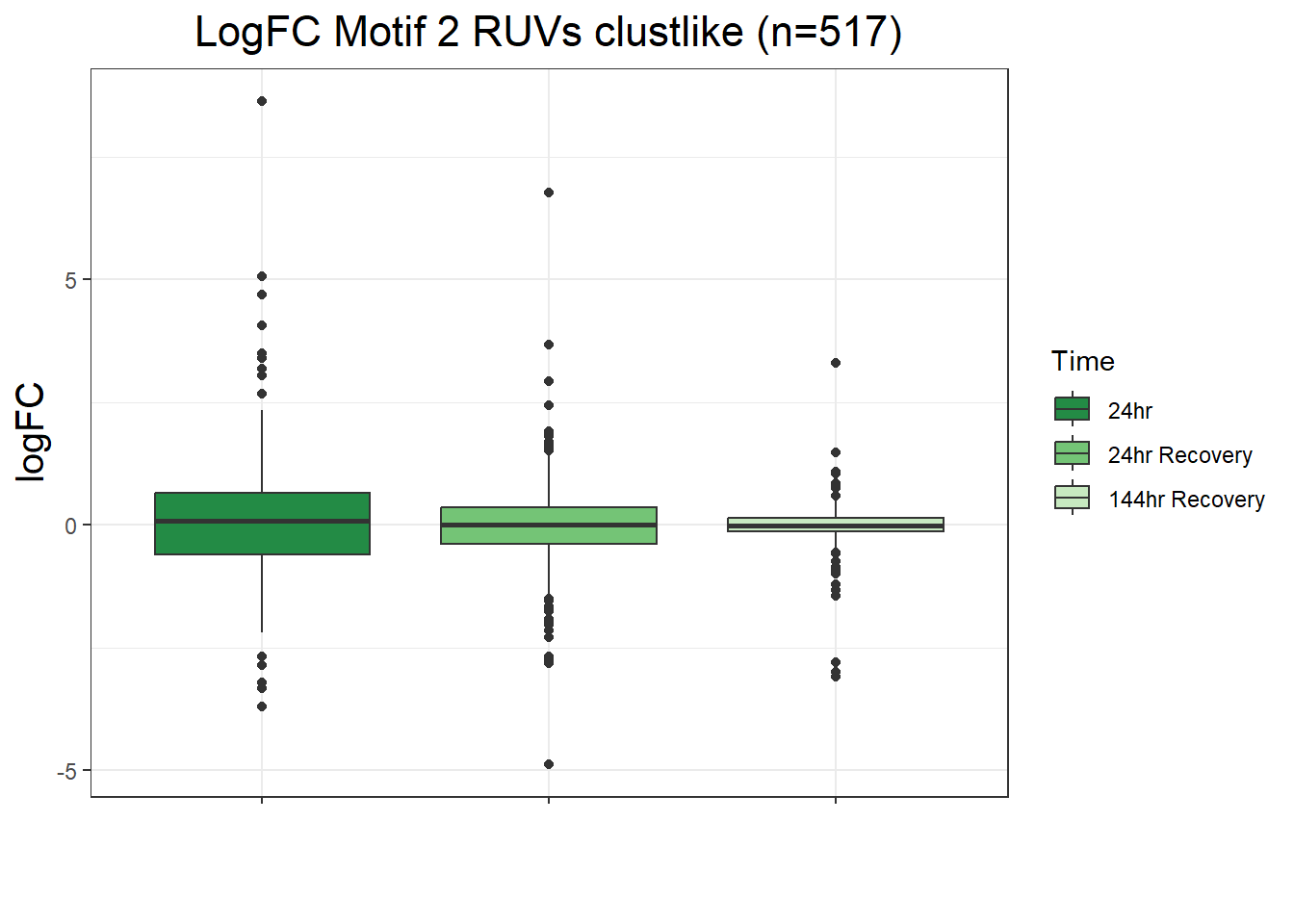
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#now plot the abs logFC for each of these too
filt_toptable_abs_fin_RUV_clust <- combined_toptables_dxr_RUV %>%
dplyr::filter(entrezgene_ID %in% clust1_genes_RUV) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24T", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = absFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("|logFC|")+
theme_bw()+
ggtitle("Abs LogFC Motif 1 RUVs clustlike (n=13600)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#motif 2
filt_toptable_2_abs_fin_RUV_clust <- combined_toptables_dxr_RUV %>%
dplyr::filter(entrezgene_ID %in% clust2_genes_RUV) %>%
mutate(absFC = abs(logFC)) %>%
mutate(time = factor(Time, levels = c("24T", "24R", "144R"), labels = c("24hr", "24hr Recovery", "144hr Recovery"))) %>%
ggplot(., aes(x = time, y = absFC))+
geom_boxplot(aes(fill = time))+
scale_fill_manual(values = time_col)+
guides(fill = guide_legend(title = "Time"))+
theme_bw()+
xlab(" ")+
ylab("|logFC|")+
theme_bw()+
ggtitle("Abs LogFC Motif 2 RUVs clustlike (n=517)")+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
strip.background = element_rect(fill = "#CAD899"),
axis.text.x = element_text(size = 8, colour = "white", angle = 15),
strip.text.x = element_text(size = 12, colour = "black", face = "bold"))
#plots
filt_toptable_abs_fin_RUV_clust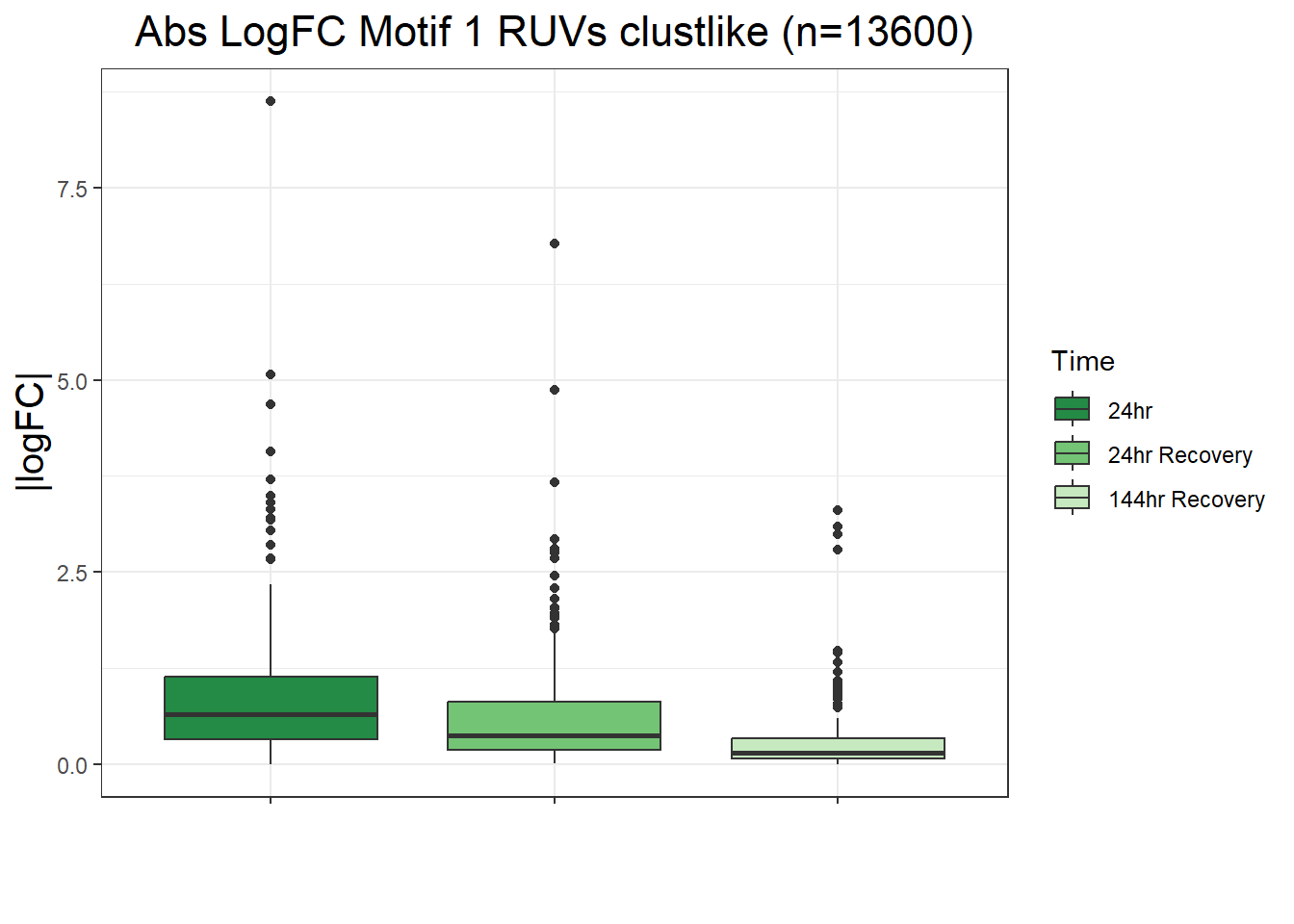
filt_toptable_2_abs_fin_RUV_clust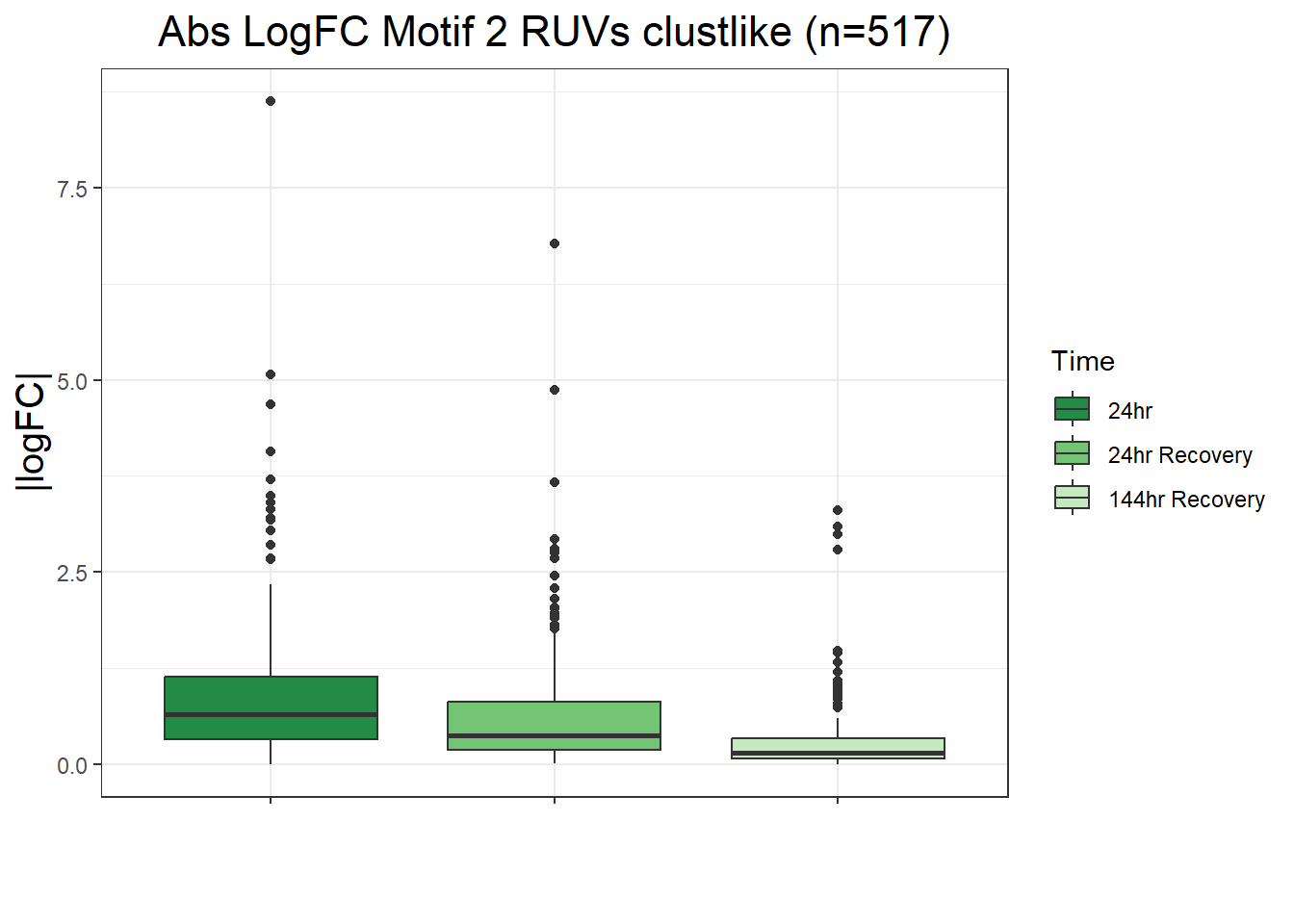
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
###log2cpm Example Genes Cormotif RUVs log2cpm Norm Counts
#use the final genes from cross-verifying with clustlike + p.post
#Load in my count matrices
#final_genes_1_RUV
#final_genes_2_RUV
#clust1_genes_RUV
#clust2_genes_RUV
#these are all the genes that are present in each category, make them into dataframes by filtering out the rest of the genes in my main boxplot df
boxplots_cormotif <- boxplot1
motif1_genes_fin_RUV <- boxplots_cormotif[boxplots_cormotif$Entrez_ID %in% final_genes_1_RUV,]
dim(motif1_genes_fin_RUV)[1] 7602 44#7602 genes in 44 cols
motif2_genes_fin_RUV <- boxplots_cormotif[boxplots_cormotif$Entrez_ID %in% final_genes_2_RUV,]
dim(motif2_genes_fin_RUV)[1] 501 44#501 genes in 44 cols
#pull out some random genes for each using sample
m1_genes_fin_RUV <- motif1_genes_fin_RUV[sample(nrow(motif1_genes_fin_RUV), 3), , drop = FALSE]
#Define gene list(s)
#these are the genes pulled out by m1_genes
initial_test_genes1_fin_RUV <- c(m1_genes_fin_RUV$SYMBOL)
#Add more gene symbols as needed or add more categories
#Now put in the function I want to use to generate boxplots of genes
process_gene_data_1_fin_RUV <- function(gene) {
gene_data <- m1_genes_fin_RUV %>% filter(SYMBOL == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2CPM") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6REP$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#Generate Boxplots from the above function using our gene list above
for (gene in initial_test_genes1_fin_RUV) {
gene_data <- process_gene_data_1_fin_RUV(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2CPM, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2CPM Expression", gene, "Motif 1 RUVs")) +
labs(x = "Treatment", y = "log2CPM") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}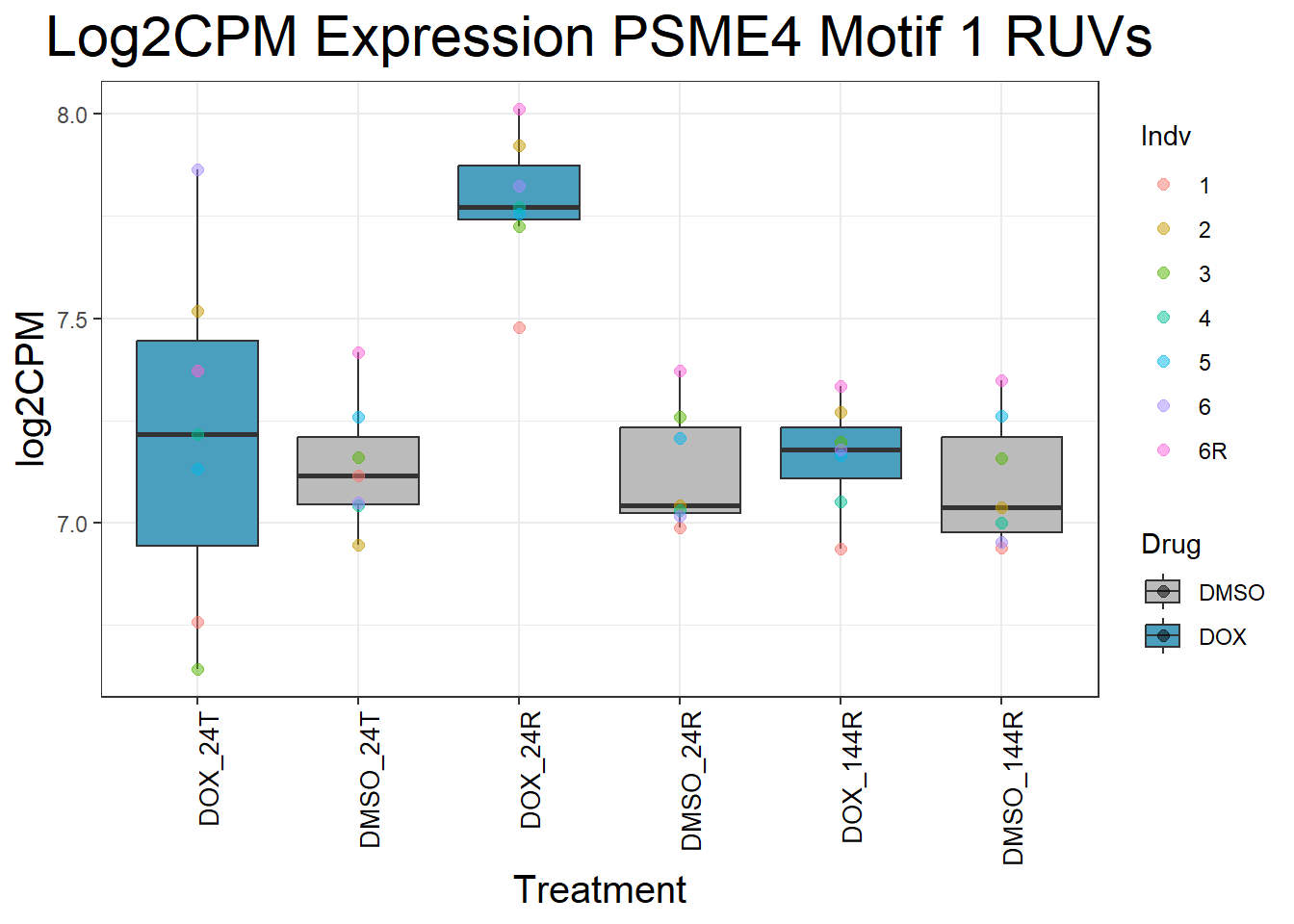
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
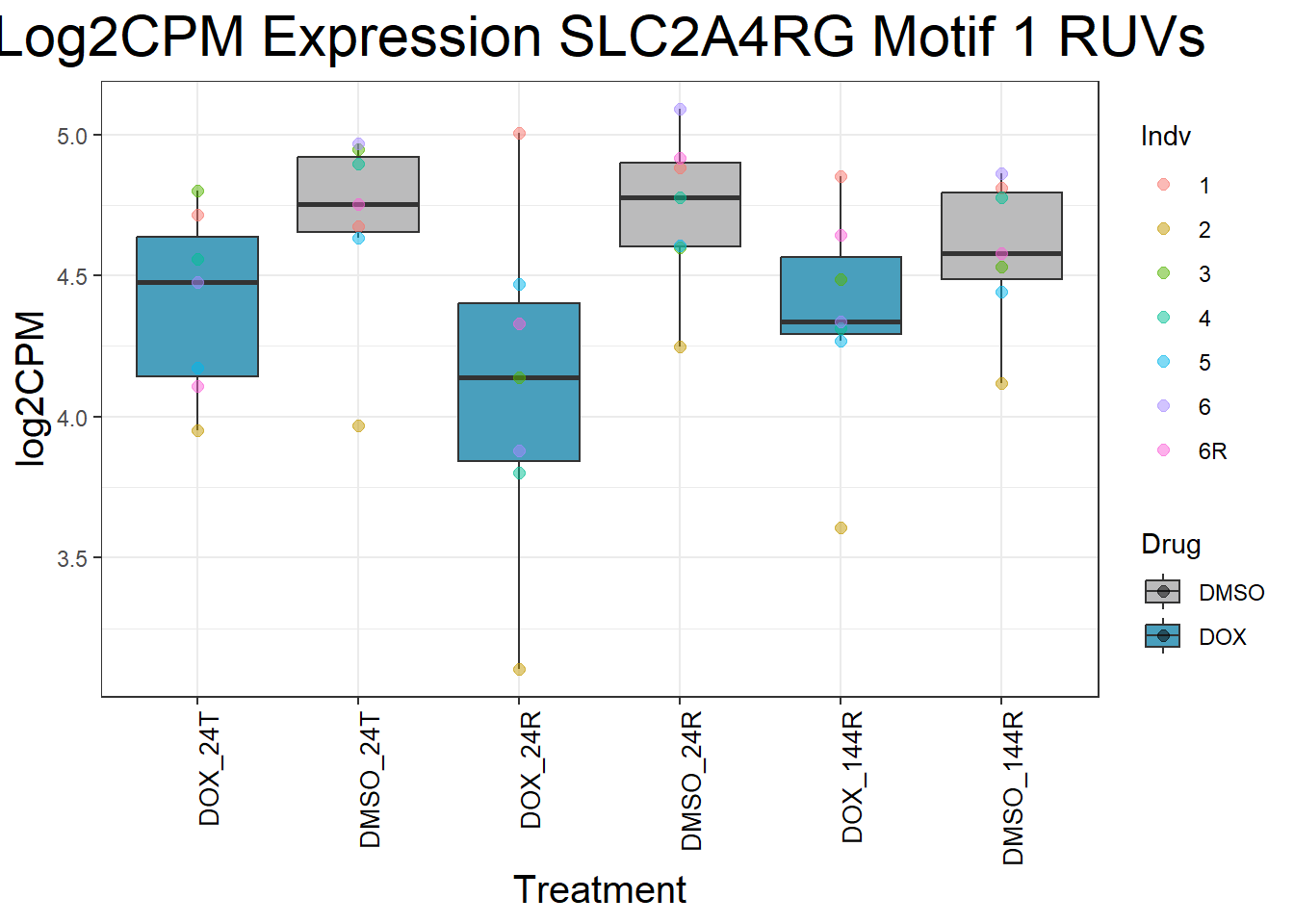
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
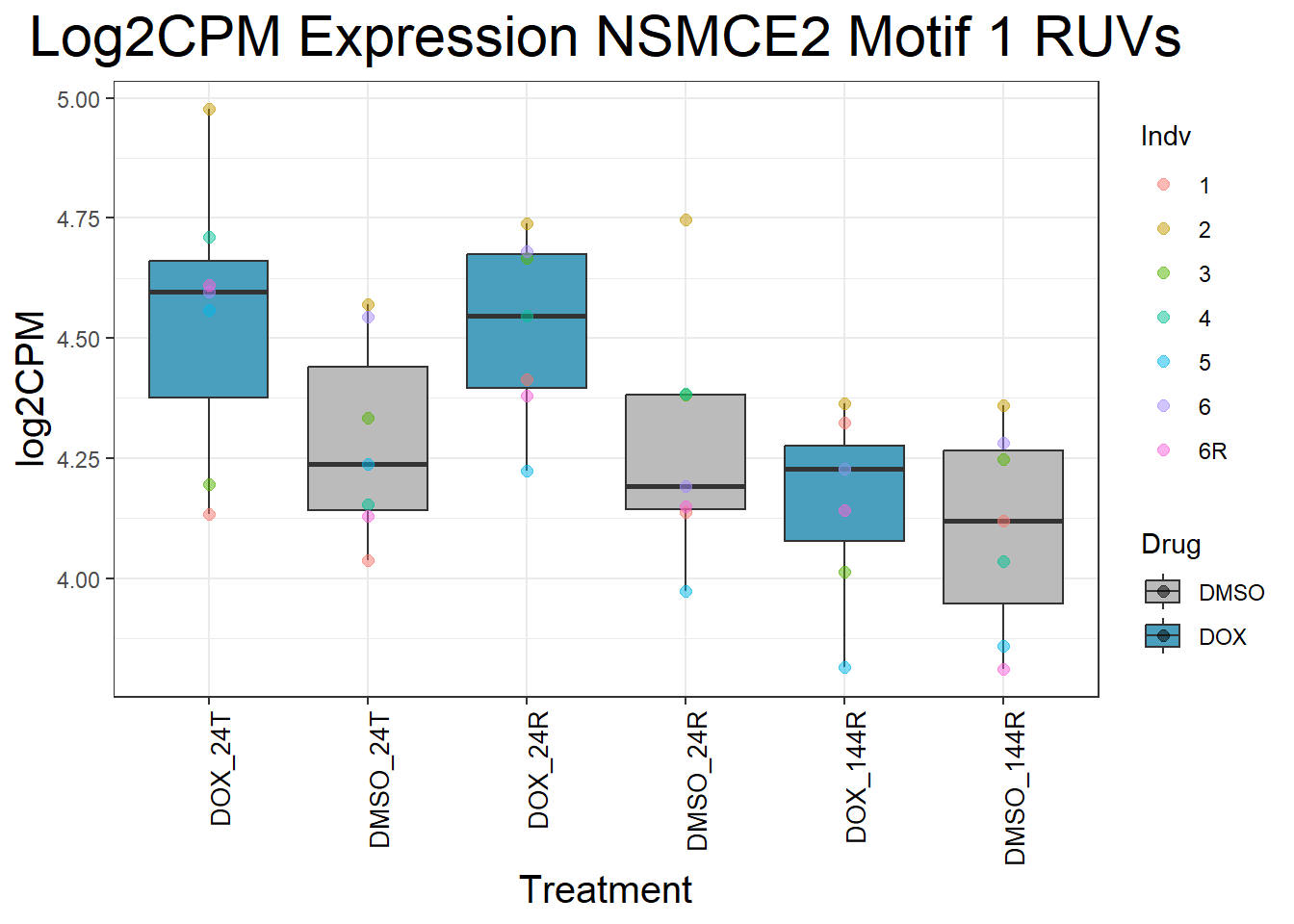
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#####motif 2#####
m2_genes_fin_RUV <- motif2_genes_fin_RUV[sample(nrow(motif2_genes_fin_RUV), 3), , drop = FALSE]
#Define gene list(s)
#these are the genes pulled out by m1_genes
initial_test_genes2_fin_RUV <- c(m2_genes_fin_RUV$SYMBOL)
#Add more gene symbols as needed or add more categories
#Now put in the function I want to use to generate boxplots of genes
process_gene_datam2_fin_RUV <- function(gene) {
gene_data <- motif2_genes_fin_RUV %>% filter(SYMBOL == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2CPM") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6REP$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#this function is saved under process_gene_data so I will save as an R object
#saveRDS(process_gene_data, "data/new/process_gene_data_funct.RDS")
#Generate Boxplots from the above function using our gene list above
for (gene in initial_test_genes2_fin_RUV) {
gene_data <- process_gene_datam2_fin_RUV(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2CPM, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2CPM Expression", gene, "Motif 2 RUVs")) +
labs(x = "Treatment", y = "log2CPM") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}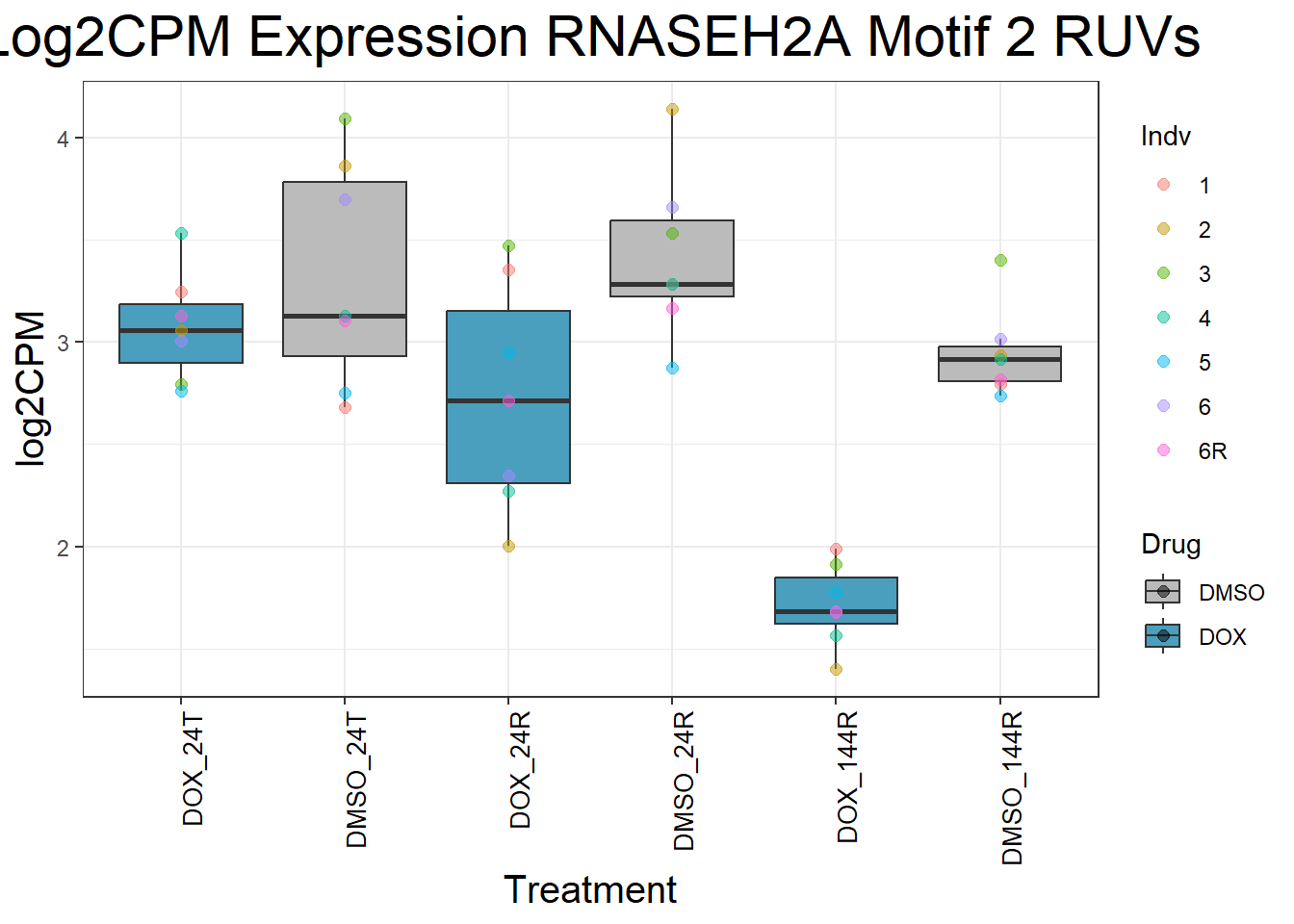
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
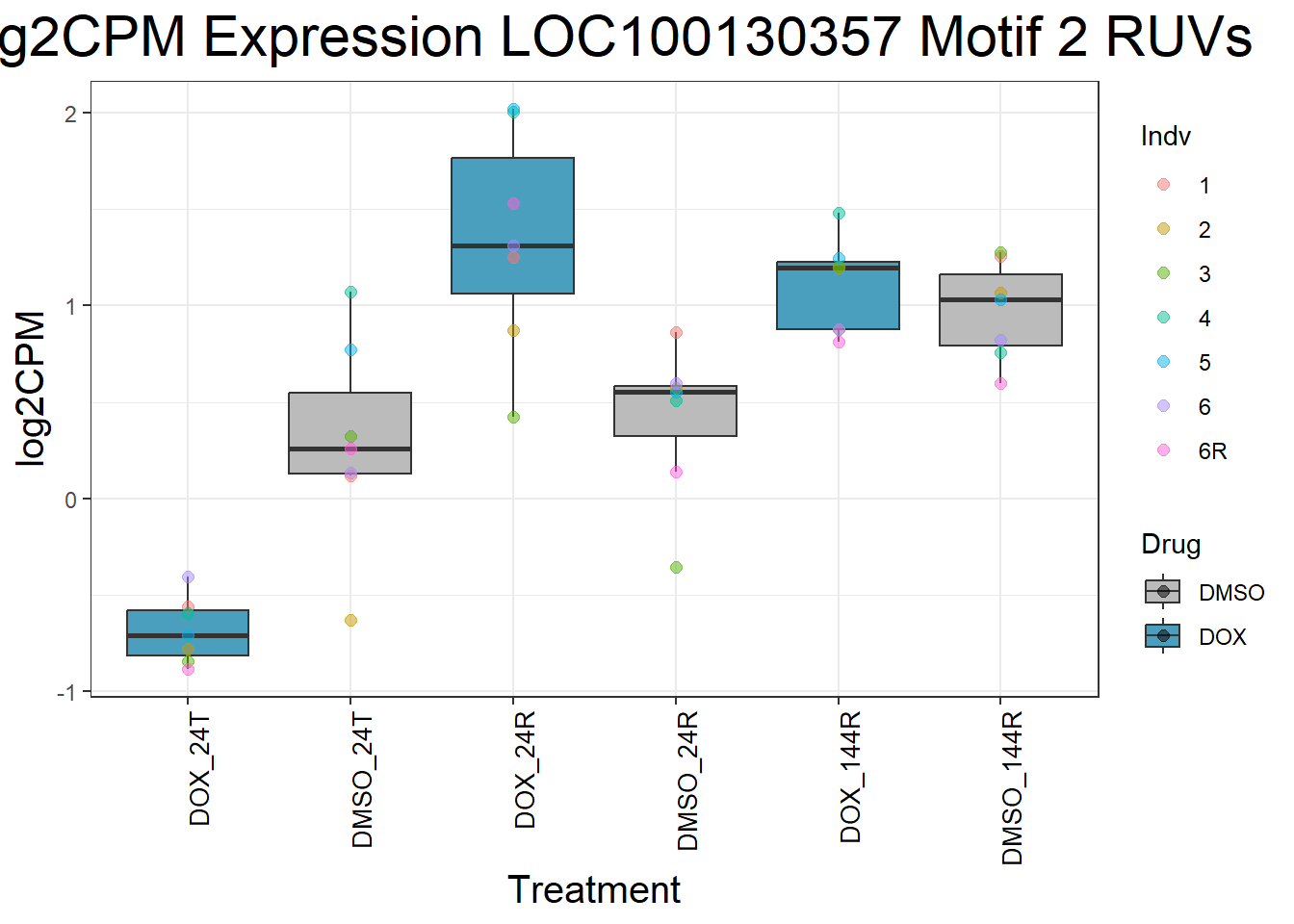
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
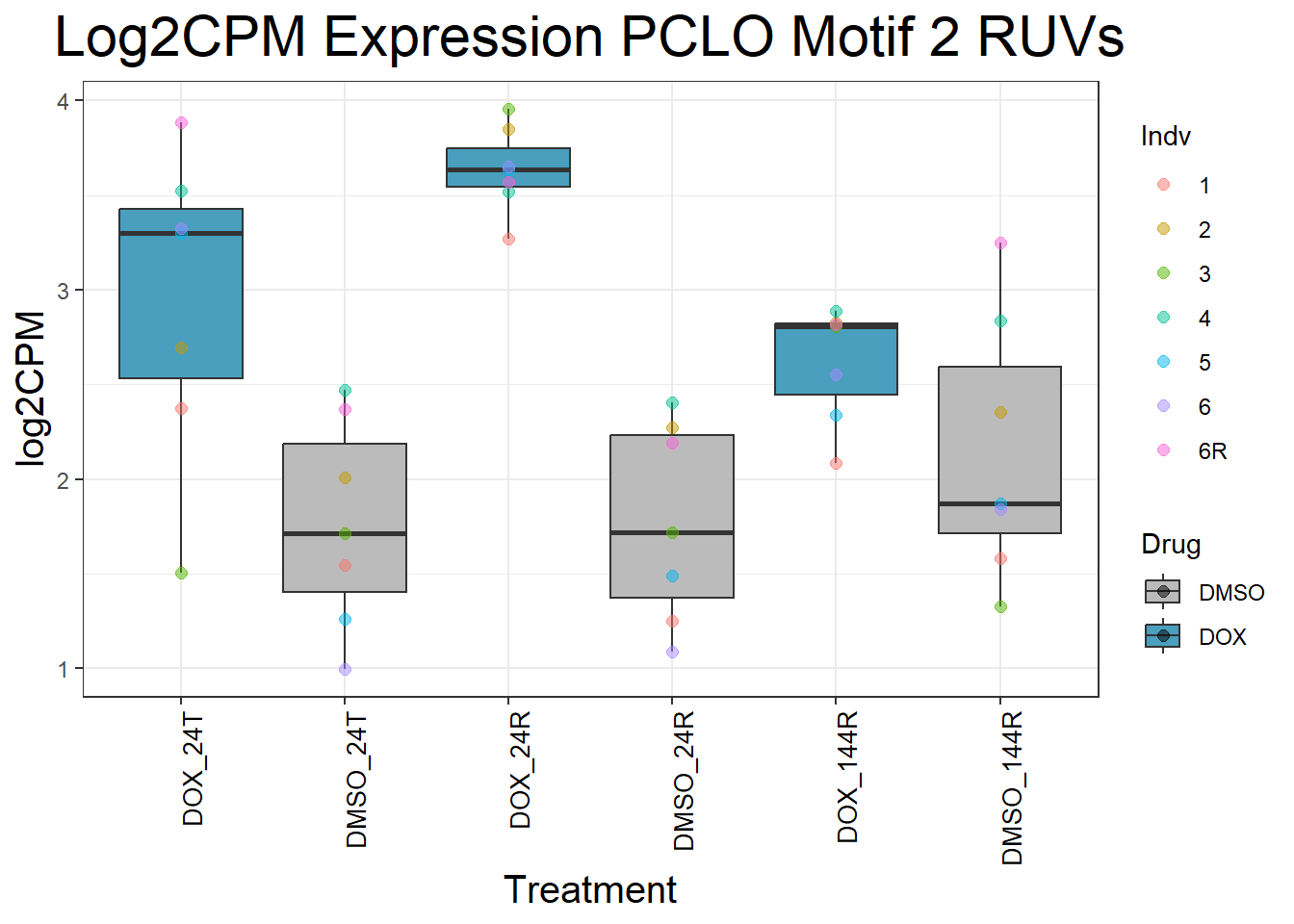
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
##log2cpm Example Genes Cormotif clustlike
#use the final genes from cross-verifying with clustlike + p.post
#Load in my count matrices
#clust1_genes_RUV
#clust2_genes_RUV
#these are all the genes that are present in each category, make them into dataframes by filtering out the rest of the genes in my main boxplot df
boxplots_cormotif <- boxplot1
motif1_genes_fin_RUV_clust <- boxplots_cormotif[boxplots_cormotif$Entrez_ID %in% clust1_genes_RUV,]
dim(motif1_genes_fin_RUV_clust)[1] 13600 44#13600 genes in 44 cols
motif2_genes_fin_RUV_clust <- boxplots_cormotif[boxplots_cormotif$Entrez_ID %in% clust2_genes_RUV,]
dim(motif2_genes_fin_RUV_clust)[1] 517 44#517 genes in 44 cols
#pull out some random genes for each using sample
m1_genes_fin_RUV_clust <- motif1_genes_fin_RUV_clust[sample(nrow(motif1_genes_fin_RUV_clust), 3), , drop = FALSE]
#Define gene list(s)
#these are the genes pulled out by m1_genes
initial_test_genes1_fin_RUV_clust <- c(m1_genes_fin_RUV_clust$SYMBOL)
#Add more gene symbols as needed or add more categories
#Now put in the function I want to use to generate boxplots of genes
process_gene_data_1_fin_RUV_clust <- function(gene) {
gene_data <- m1_genes_fin_RUV_clust %>% filter(SYMBOL == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2CPM") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6REP$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#Generate Boxplots from the above function using our gene list above
for (gene in initial_test_genes1_fin_RUV_clust) {
gene_data <- process_gene_data_1_fin_RUV_clust(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2CPM, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2CPM Expression", gene, "M1 RUVs clustlike")) +
labs(x = "Treatment", y = "log2CPM") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}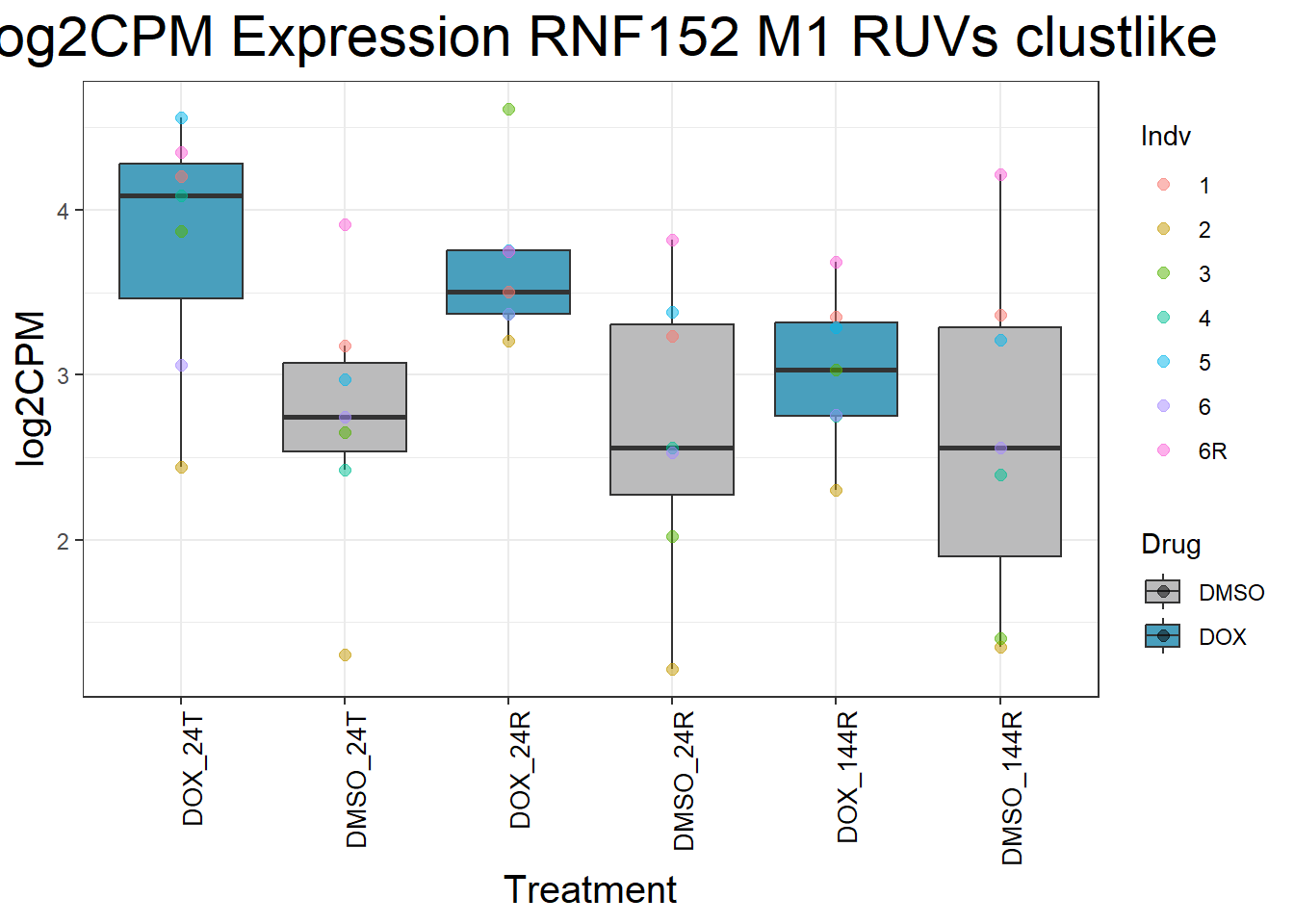
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
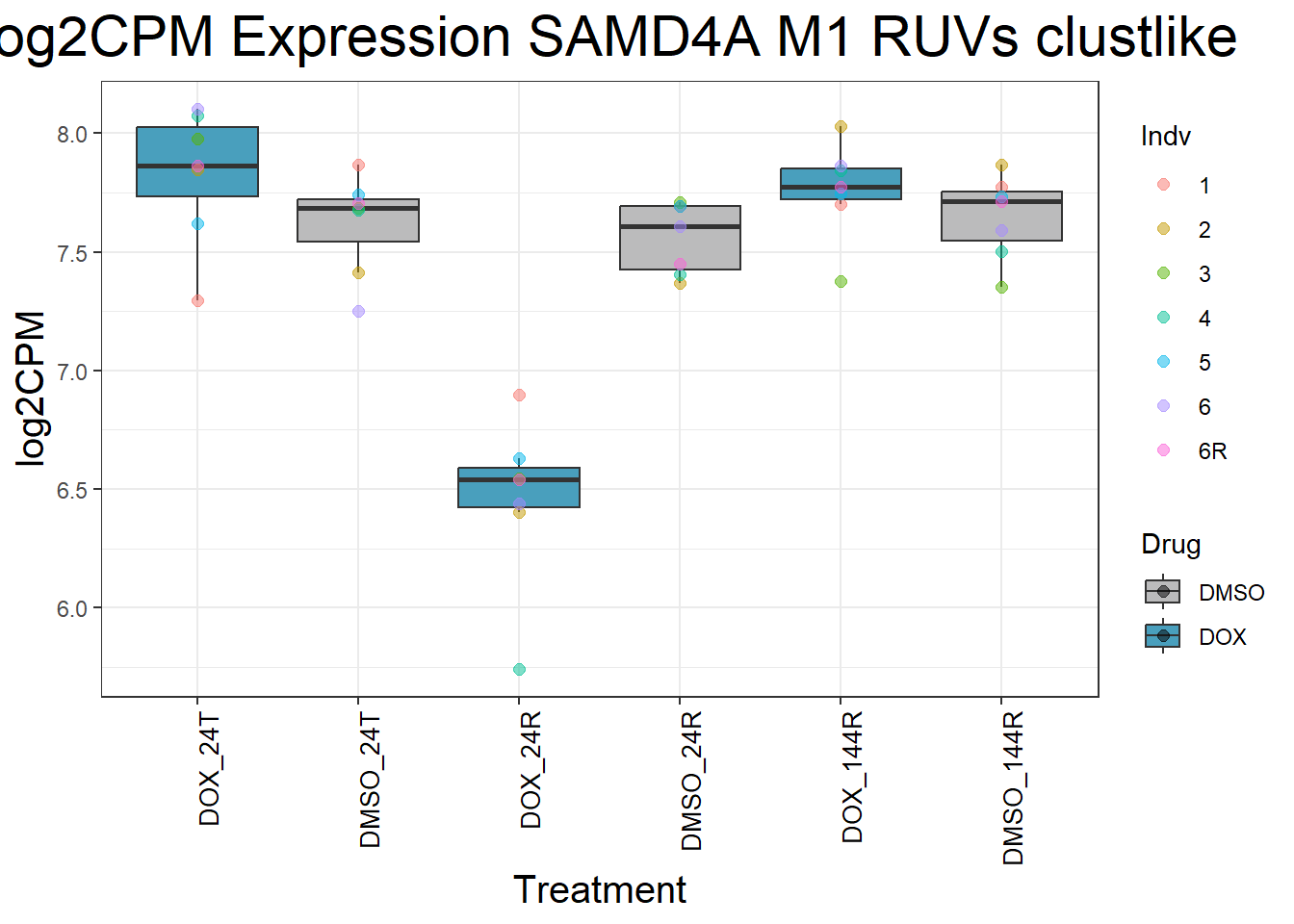
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
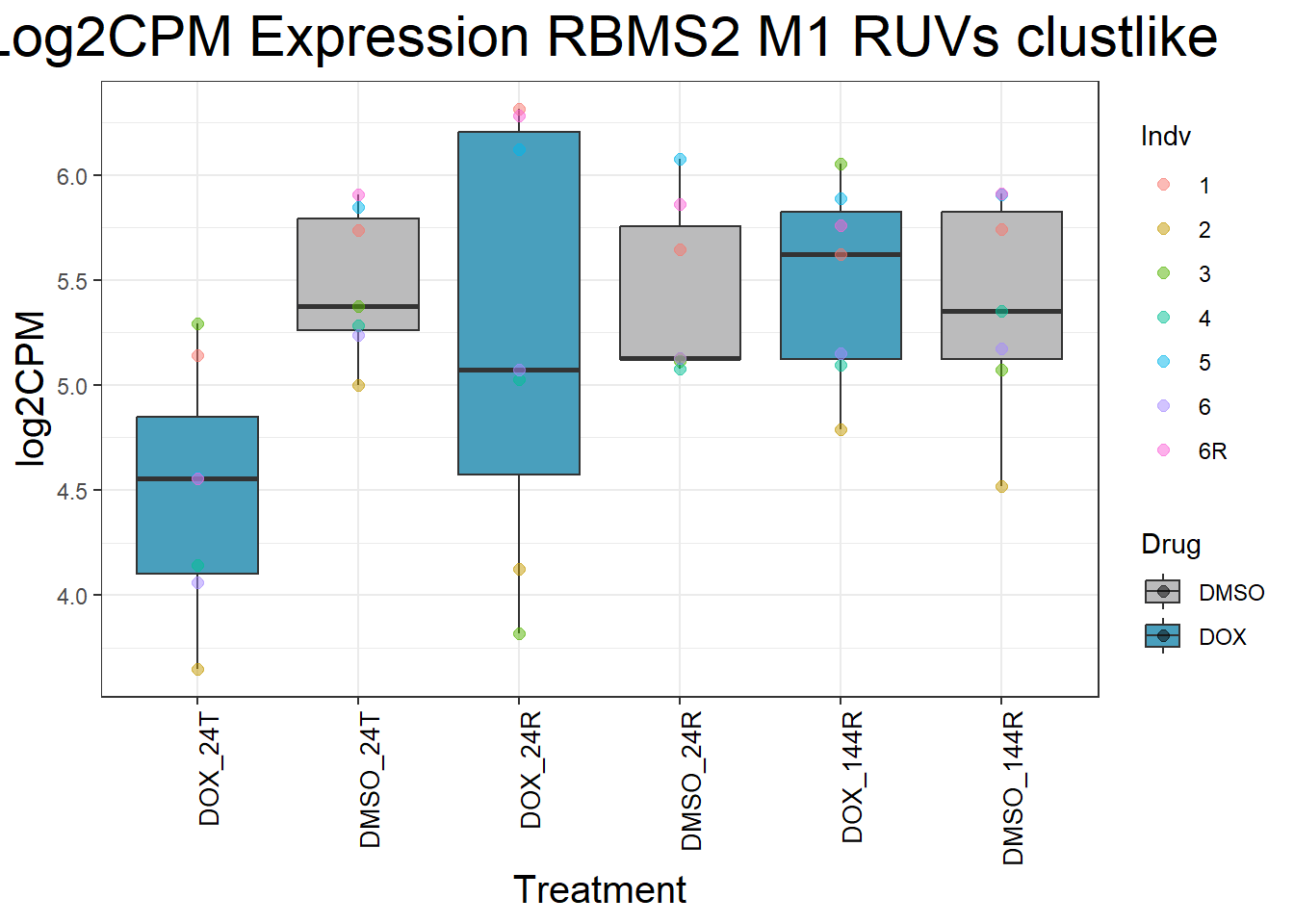
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#####motif 2#####
m2_genes_fin_RUV_clust <- motif2_genes_fin_RUV_clust[sample(nrow(motif2_genes_fin_RUV_clust), 3), , drop = FALSE]
#Define gene list(s)
#these are the genes pulled out by m1_genes
initial_test_genes2_fin_RUV_clust <- c(m2_genes_fin_RUV_clust$SYMBOL)
#Add more gene symbols as needed or add more categories
#Now put in the function I want to use to generate boxplots of genes
process_gene_datam2_fin_RUV_clust <- function(gene) {
gene_data <- motif2_genes_fin_RUV_clust %>% filter(SYMBOL == gene)
long_data <- gene_data %>%
pivot_longer(cols = -c(Entrez_ID, SYMBOL), names_to = "Sample", values_to = "log2CPM") %>%
mutate(
Drug = case_when(
grepl("DOX", Sample) ~ "DOX",
grepl("DMSO", Sample) ~ "DMSO",
TRUE ~ NA_character_
),
Timepoint = case_when(
grepl("_24T_", Sample) ~ "24T",
grepl("_24R_", Sample) ~ "24R",
grepl("_144R_", Sample) ~ "144R",
TRUE ~ NA_character_
),
Indv = case_when(
grepl("Ind1$", Sample) ~ "1",
grepl("Ind2$", Sample) ~ "2",
grepl("Ind3$", Sample) ~ "3",
grepl("Ind4$", Sample) ~ "4",
grepl("Ind5$", Sample) ~ "5",
grepl("Ind6$", Sample) ~ "6",
grepl("Ind6REP$", Sample) ~ "6R",
TRUE ~ NA_character_
),
Condition = paste(Drug, Timepoint, sep = "_")
)
long_data$Condition <- factor(
long_data$Condition,
levels = c(
"DOX_24T",
"DMSO_24T",
"DOX_24R",
"DMSO_24R",
"DOX_144R",
"DMSO_144R"
)
)
return(long_data)
}
#this function is saved under process_gene_data so I will save as an R object
#saveRDS(process_gene_data, "data/new/process_gene_data_funct.RDS")
#Generate Boxplots from the above function using our gene list above
for (gene in initial_test_genes2_fin_RUV_clust) {
gene_data <- process_gene_datam2_fin_RUV_clust(gene)
p <- ggplot(gene_data, aes(x = Condition, y = log2CPM, fill = Drug)) +
geom_boxplot(outlier.shape = NA) +
geom_point(aes(color = Indv), size = 2, alpha = 0.5, position = position_jitter(width = -1, height = 0)) +
scale_fill_manual(values = c("DOX" = "#499FBD", "DMSO" = "#BBBBBC")) +
ggtitle(paste("Log2CPM Expression", gene, "M2 RUVs clustlike")) +
labs(x = "Treatment", y = "log2CPM") +
theme_bw() +
theme(
plot.title = element_text(size = rel(2), hjust = 0.5),
axis.title = element_text(size = 15, color = "black"),
axis.text.x = element_text(size = 10, color = "black", angle = 90, hjust = 1)
)
print(p)
}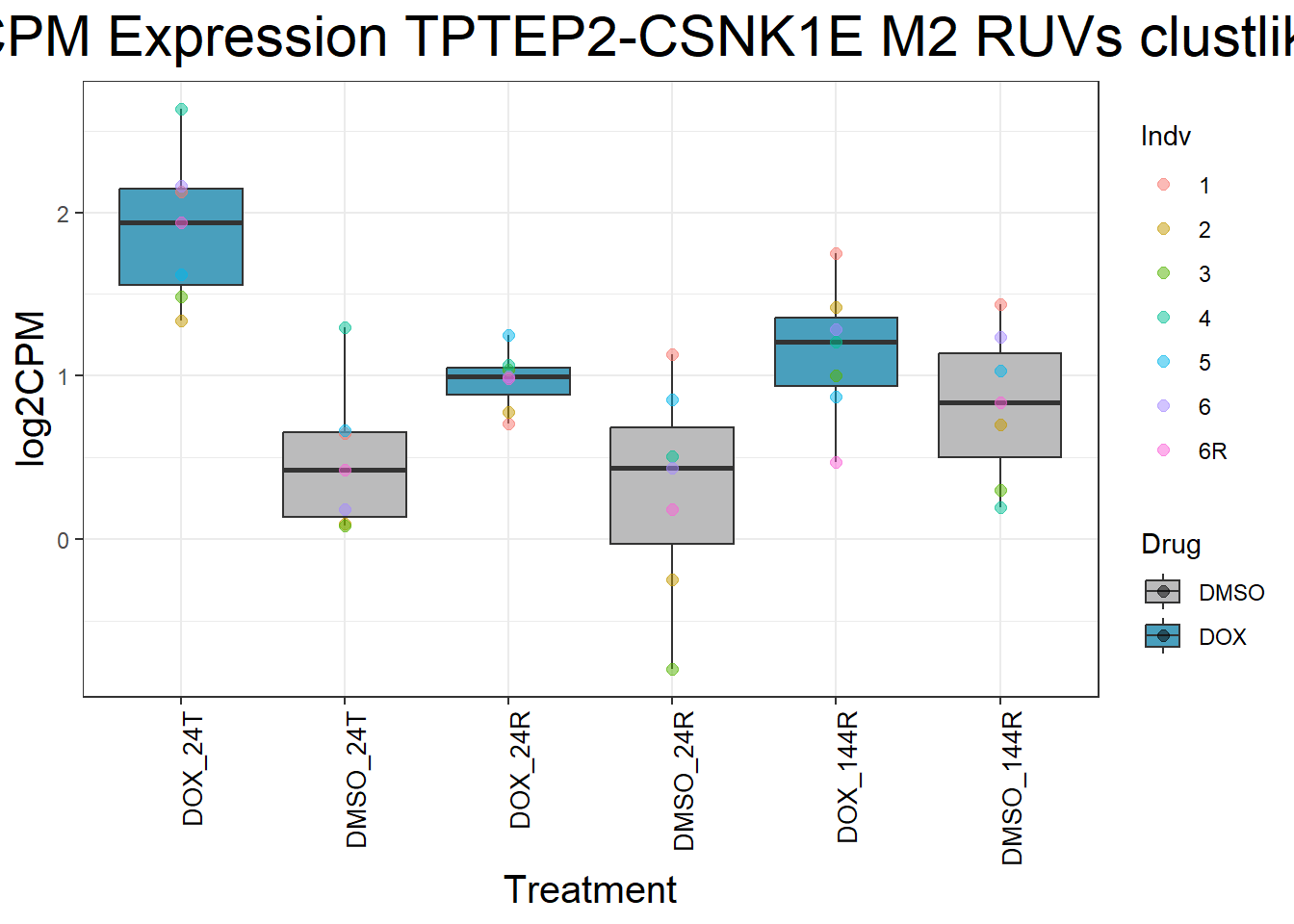
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
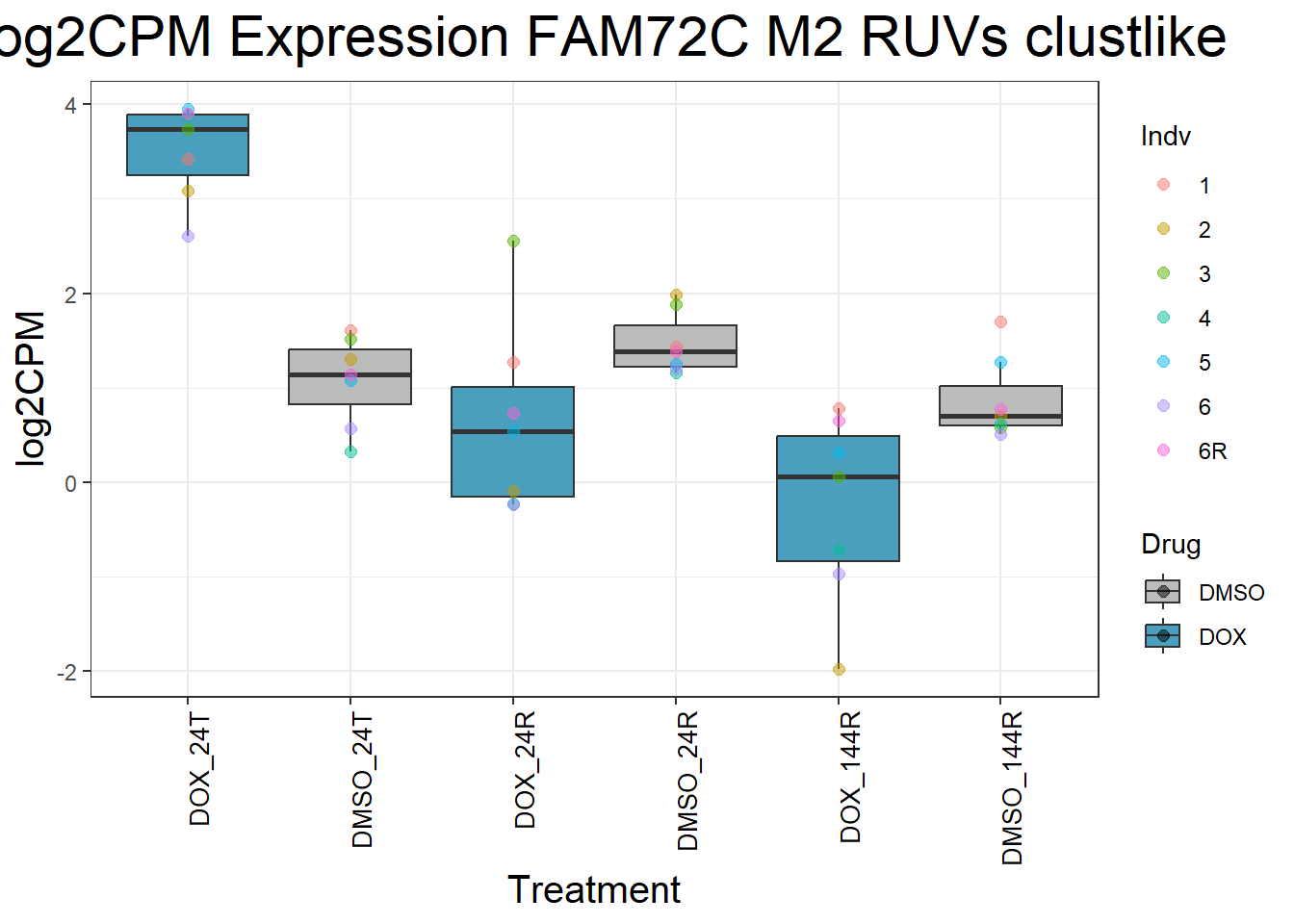
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
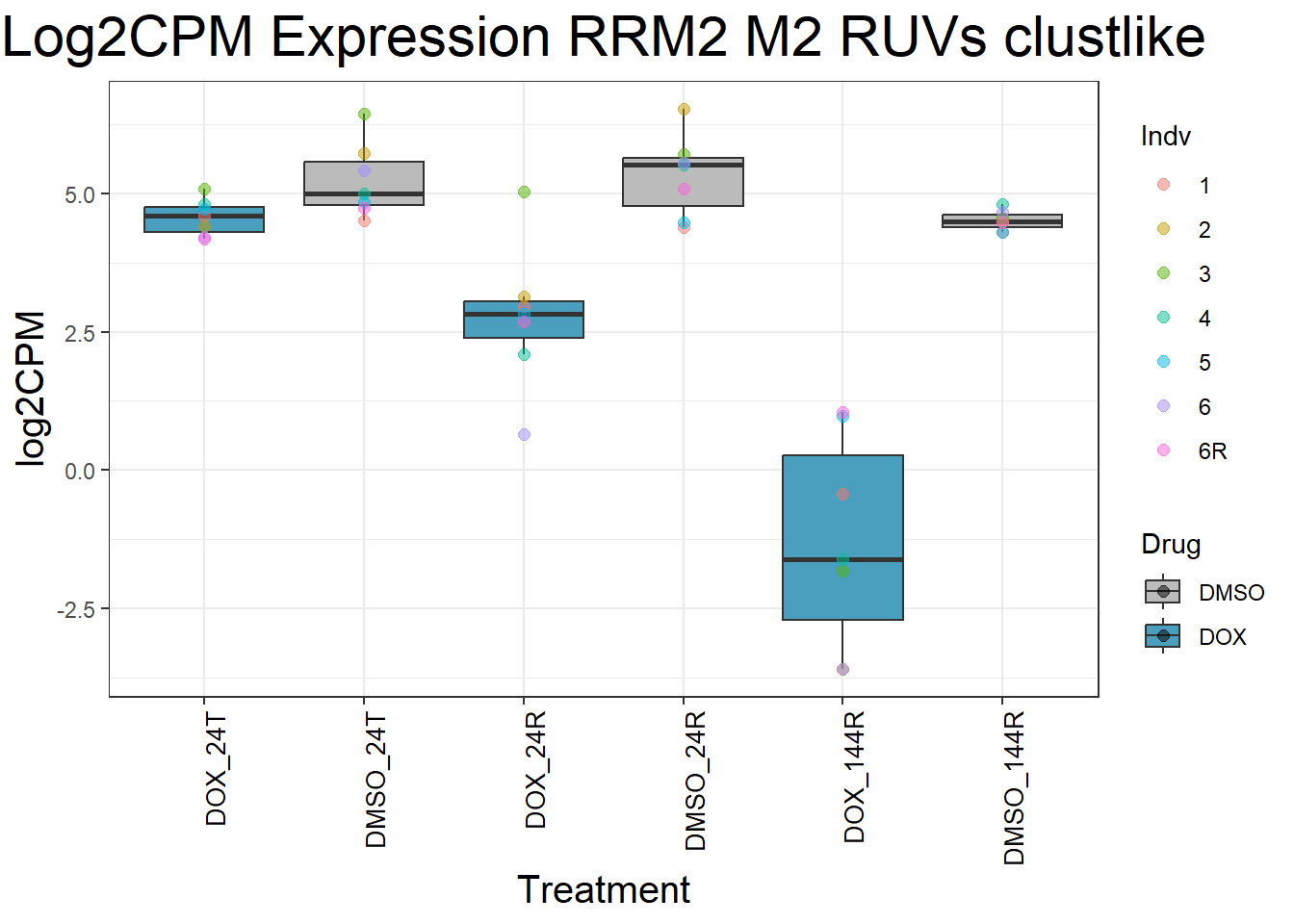
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
###Cormotif RUVs log2cpm Norm Counts GO/KEGG
#now take these final gene sets for each motif - clustlike + p.post consensus genes
# final_genes_1_RUV
# final_genes_2_RUV
#clust1_genes_RUV
#clust2_genes_RUV
library(gprofiler2)
#####Motif 1 Gene Set#####
# motif1_genes_matrix_RUV <- as.matrix(final_genes_1_RUV)
# colnames(motif1_genes_matrix_RUV) <- c("entrezgene_ID")
# saveRDS(motif1_genes_matrix_RUV, "data/new/RUV/CMF/motif1_genes_matrix_RUV.RDS")
motif1_genes_matrix_RUV <- readRDS("data/new/RUV/CMF/motif1_genes_matrix_RUV.RDS")
length(motif1_genes_matrix_RUV)[1] 7602#7602 genes in this set for motif 1
motif1_mat_GOKEGG_RUV <- gost(query = motif1_genes_matrix_RUV,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
motif1_GOKEGG_genes_RUV <- gostplot(motif1_mat_GOKEGG_RUV, capped = FALSE, interactive = TRUE)
motif1_GOKEGG_genes_RUVtable_motif1_GOKEGG_genes_RUV <- motif1_mat_GOKEGG_RUV$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_motif1_GOKEGG_genes_RUV %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0019538 | protein metabolic process | 2065 | 4721 | 4.931e-103 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 2228 | 5390 | 7.135e-83 |
| GO:BP | GO:0006996 | organelle organization | 1583 | 3594 | 3.383e-76 |
| GO:BP | GO:0046907 | intracellular transport | 747 | 1381 | 5.019e-76 |
| GO:BP | GO:0051179 | localization | 2247 | 5555 | 3.318e-73 |
| GO:BP | GO:0051641 | cellular localization | 1585 | 3661 | 7.452e-70 |
| GO:BP | GO:0006810 | transport | 1830 | 4407 | 8.187e-66 |
| GO:BP | GO:0051649 | establishment of localization in cell | 966 | 2010 | 1.457e-64 |
| GO:BP | GO:0043412 | macromolecule modification | 1339 | 3030 | 1.398e-63 |
| GO:BP | GO:0048518 | positive regulation of biological process | 2439 | 6264 | 9.096e-63 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 2324 | 5920 | 2.497e-62 |
| GO:BP | GO:0036211 | protein modification process | 1263 | 2846 | 1.005e-60 |
| GO:BP | GO:0009056 | catabolic process | 1179 | 2639 | 5.706e-58 |
| GO:BP | GO:0008104 | protein localization | 1223 | 2770 | 5.624e-57 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 1227 | 2782 | 6.554e-57 |
| GO:BP | GO:0033036 | macromolecule localization | 1385 | 3228 | 1.242e-56 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 712 | 1417 | 1.769e-55 |
| GO:BP | GO:0065007 | biological regulation | 4404 | 12743 | 4.108e-55 |
| GO:BP | GO:0051234 | establishment of localization | 1960 | 4928 | 2.259e-54 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 1631 | 3990 | 1.070e-51 |
| GO:BP | GO:0071705 | nitrogen compound transport | 895 | 1923 | 1.070e-51 |
| GO:BP | GO:0015031 | protein transport | 711 | 1444 | 3.658e-51 |
| GO:BP | GO:0033554 | cellular response to stress | 856 | 1830 | 2.332e-50 |
| GO:BP | GO:0035556 | intracellular signal transduction | 1268 | 2965 | 3.801e-50 |
| GO:BP | GO:0023051 | regulation of signaling | 1445 | 3478 | 2.022e-49 |
| GO:BP | GO:0009966 | regulation of signal transduction | 1287 | 3034 | 1.001e-48 |
| GO:BP | GO:0050789 | regulation of biological process | 4250 | 12336 | 4.563e-48 |
| GO:BP | GO:0010646 | regulation of cell communication | 1441 | 3486 | 9.475e-48 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 1506 | 3687 | 1.105e-46 |
| GO:BP | GO:0050794 | regulation of cellular process | 4125 | 11946 | 3.132e-46 |
| GO:BP | GO:0030163 | protein catabolic process | 531 | 1030 | 8.599e-45 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 1595 | 3993 | 4.592e-43 |
| GO:BP | GO:0019222 | regulation of metabolic process | 2593 | 7035 | 8.796e-43 |
| GO:BP | GO:0006351 | DNA-templated transcription | 1432 | 3549 | 1.977e-40 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 1385 | 3428 | 3.584e-39 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 1378 | 3409 | 4.491e-39 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 827 | 1855 | 1.476e-38 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 1436 | 3597 | 1.131e-37 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 441 | 853 | 3.256e-37 |
| GO:BP | GO:0045184 | establishment of protein localization | 852 | 1937 | 3.512e-37 |
| GO:BP | GO:0032502 | developmental process | 2402 | 6553 | 4.791e-36 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 486 | 975 | 7.565e-36 |
| GO:BP | GO:0043687 | post-translational protein modification | 506 | 1027 | 1.037e-35 |
| GO:BP | GO:0007275 | multicellular organism development | 1805 | 4727 | 1.310e-35 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 719 | 1592 | 2.561e-35 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 1012 | 2407 | 4.839e-35 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 1013 | 2410 | 4.903e-35 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 849 | 1953 | 8.054e-35 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 407 | 786 | 1.798e-34 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 1014 | 2433 | 2.619e-33 |
| GO:BP | GO:0006886 | intracellular protein transport | 363 | 686 | 4.260e-33 |
| GO:BP | GO:0016567 | protein ubiquitination | 396 | 768 | 5.554e-33 |
| GO:BP | GO:0009894 | regulation of catabolic process | 503 | 1040 | 6.446e-33 |
| GO:BP | GO:0048856 | anatomical structure development | 2204 | 5997 | 9.061e-33 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 1303 | 3288 | 7.825e-32 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 2354 | 6492 | 3.103e-31 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 321 | 597 | 7.677e-31 |
| GO:BP | GO:0006914 | autophagy | 321 | 597 | 7.677e-31 |
| GO:BP | GO:0048731 | system development | 1552 | 4053 | 4.097e-30 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 1086 | 2711 | 9.422e-28 |
| GO:BP | GO:0007005 | mitochondrion organization | 246 | 435 | 1.022e-27 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 594 | 1326 | 1.318e-27 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 723 | 1680 | 1.327e-27 |
| GO:BP | GO:0006412 | translation | 365 | 727 | 2.283e-27 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 802 | 1912 | 1.228e-26 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 845 | 2036 | 1.915e-26 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 288 | 543 | 2.843e-26 |
| GO:BP | GO:0023057 | negative regulation of signaling | 629 | 1435 | 3.007e-26 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 629 | 1436 | 3.722e-26 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 326 | 638 | 4.619e-26 |
| GO:BP | GO:0002181 | cytoplasmic translation | 119 | 165 | 4.898e-26 |
| GO:BP | GO:0006950 | response to stress | 1493 | 3948 | 9.167e-26 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 327 | 643 | 1.090e-25 |
| GO:BP | GO:0048519 | negative regulation of biological process | 2108 | 5834 | 1.108e-25 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 1029 | 2578 | 1.767e-25 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 320 | 628 | 2.548e-25 |
| GO:BP | GO:0006915 | apoptotic process | 800 | 1923 | 2.636e-25 |
| GO:BP | GO:0016236 | macroautophagy | 209 | 362 | 4.216e-25 |
| GO:BP | GO:0051716 | cellular response to stimulus | 2593 | 7376 | 6.940e-25 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 2036 | 5629 | 8.969e-25 |
| GO:BP | GO:0008219 | cell death | 821 | 1991 | 1.215e-24 |
| GO:BP | GO:0012501 | programmed cell death | 819 | 1987 | 1.640e-24 |
| GO:BP | GO:0007399 | nervous system development | 1033 | 2604 | 1.664e-24 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 184 | 312 | 1.567e-23 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 334 | 677 | 2.492e-23 |
| GO:BP | GO:0065008 | regulation of biological quality | 1140 | 2947 | 1.300e-22 |
| GO:BP | GO:0033365 | protein localization to organelle | 531 | 1209 | 3.643e-22 |
| GO:BP | GO:0006508 | proteolysis | 631 | 1486 | 4.204e-22 |
| GO:BP | GO:0044238 | primary metabolic process | 4092 | 12342 | 9.259e-22 |
| GO:BP | GO:0060341 | regulation of cellular localization | 457 | 1013 | 1.063e-21 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 456 | 1011 | 1.248e-21 |
| GO:BP | GO:0032879 | regulation of localization | 820 | 2029 | 1.518e-21 |
| GO:BP | GO:0033043 | regulation of organelle organization | 506 | 1148 | 1.791e-21 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 489 | 1105 | 3.794e-21 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 596 | 1400 | 4.107e-21 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 750 | 1835 | 4.531e-21 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 641 | 1529 | 7.494e-21 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 263 | 519 | 2.842e-20 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 2061 | 5809 | 4.830e-20 |
| GO:BP | GO:0007010 | cytoskeleton organization | 637 | 1529 | 6.275e-20 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 1039 | 2696 | 1.028e-19 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 656 | 1588 | 1.652e-19 |
| GO:BP | GO:0030030 | cell projection organization | 671 | 1631 | 1.778e-19 |
| GO:BP | GO:0032880 | regulation of protein localization | 409 | 907 | 2.724e-19 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 316 | 662 | 3.332e-19 |
| GO:BP | GO:0065009 | regulation of molecular function | 622 | 1496 | 3.408e-19 |
| GO:BP | GO:0034504 | protein localization to nucleus | 177 | 318 | 7.132e-19 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 293 | 606 | 9.092e-19 |
| GO:BP | GO:0006974 | DNA damage response | 407 | 908 | 1.236e-18 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 997 | 2593 | 1.837e-18 |
| GO:BP | GO:0016310 | phosphorylation | 556 | 1320 | 1.980e-18 |
| GO:BP | GO:0070925 | organelle assembly | 456 | 1046 | 3.656e-18 |
| GO:BP | GO:0031399 | regulation of protein modification process | 457 | 1049 | 3.837e-18 |
| GO:BP | GO:0050793 | regulation of developmental process | 949 | 2457 | 3.969e-18 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 546 | 1296 | 4.158e-18 |
| GO:BP | GO:0050896 | response to stimulus | 3046 | 8999 | 4.729e-18 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 540 | 1280 | 4.756e-18 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 687 | 1699 | 7.636e-18 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 1991 | 5644 | 8.673e-18 |
| GO:BP | GO:0030029 | actin filament-based process | 373 | 825 | 8.673e-18 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 686 | 1697 | 8.673e-18 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 154 | 270 | 9.307e-18 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 518 | 1223 | 1.080e-17 |
| GO:BP | GO:0061024 | membrane organization | 371 | 821 | 1.186e-17 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 336 | 729 | 1.629e-17 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 185 | 345 | 1.893e-17 |
| GO:BP | GO:0010506 | regulation of autophagy | 194 | 367 | 2.037e-17 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 180 | 334 | 2.842e-17 |
| GO:BP | GO:0051169 | nuclear transport | 180 | 334 | 2.842e-17 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 126 | 209 | 3.641e-17 |
| GO:BP | GO:0006468 | protein phosphorylation | 517 | 1227 | 3.872e-17 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 424 | 970 | 4.017e-17 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 641 | 1578 | 4.111e-17 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1029 | 2713 | 4.342e-17 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 211 | 414 | 1.327e-16 |
| GO:BP | GO:0016197 | endosomal transport | 160 | 290 | 1.335e-16 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 896 | 2331 | 2.253e-16 |
| GO:BP | GO:0023056 | positive regulation of signaling | 713 | 1796 | 2.342e-16 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 290 | 619 | 3.047e-16 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 596 | 1462 | 3.271e-16 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 611 | 1506 | 3.992e-16 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 605 | 1489 | 4.055e-16 |
| GO:BP | GO:0010468 | regulation of gene expression | 1943 | 5536 | 4.090e-16 |
| GO:BP | GO:0030154 | cell differentiation | 1589 | 4437 | 5.139e-16 |
| GO:BP | GO:0007049 | cell cycle | 665 | 1663 | 5.436e-16 |
| GO:BP | GO:0048869 | cellular developmental process | 1589 | 4438 | 5.606e-16 |
| GO:BP | GO:0044281 | small molecule metabolic process | 704 | 1776 | 5.638e-16 |
| GO:BP | GO:0048468 | cell development | 1075 | 2876 | 9.562e-16 |
| GO:BP | GO:0010256 | endomembrane system organization | 286 | 613 | 9.749e-16 |
| GO:BP | GO:0022008 | neurogenesis | 701 | 1771 | 9.749e-16 |
| GO:BP | GO:0006979 | response to oxidative stress | 203 | 400 | 9.749e-16 |
| GO:BP | GO:0023052 | signaling | 2248 | 6515 | 1.874e-15 |
| GO:BP | GO:0006417 | regulation of translation | 194 | 380 | 2.270e-15 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 377 | 861 | 2.546e-15 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 707 | 1795 | 2.571e-15 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 441 | 1039 | 3.583e-15 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 454 | 1077 | 5.072e-15 |
| GO:BP | GO:0007154 | cell communication | 2250 | 6540 | 8.804e-15 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 311 | 689 | 1.128e-14 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 110 | 184 | 1.231e-14 |
| GO:BP | GO:0042254 | ribosome biogenesis | 169 | 323 | 1.231e-14 |
| GO:BP | GO:0070848 | response to growth factor | 322 | 721 | 1.997e-14 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 166 | 317 | 2.023e-14 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 2825 | 8393 | 2.339e-14 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 146 | 269 | 2.339e-14 |
| GO:BP | GO:0009987 | cellular process | 6272 | 20247 | 2.662e-14 |
| GO:BP | GO:0031647 | regulation of protein stability | 173 | 335 | 2.673e-14 |
| GO:BP | GO:0048513 | animal organ development | 1134 | 3085 | 3.100e-14 |
| GO:BP | GO:0080134 | regulation of response to stress | 559 | 1387 | 4.734e-14 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 416 | 983 | 4.797e-14 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 297 | 658 | 5.181e-14 |
| GO:BP | GO:0010970 | transport along microtubule | 106 | 178 | 5.949e-14 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 462 | 1113 | 5.988e-14 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 142 | 262 | 6.603e-14 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 228 | 478 | 8.520e-14 |
| GO:BP | GO:0031175 | neuron projection development | 429 | 1023 | 9.544e-14 |
| GO:BP | GO:1901698 | response to nitrogen compound | 449 | 1080 | 1.127e-13 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 398 | 938 | 1.241e-13 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 289 | 642 | 1.871e-13 |
| GO:BP | GO:0099111 | microtubule-based transport | 123 | 220 | 2.438e-13 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 131 | 239 | 2.549e-13 |
| GO:BP | GO:0000209 | protein polyubiquitination | 148 | 280 | 2.671e-13 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 142 | 266 | 3.302e-13 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 252 | 546 | 3.433e-13 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 114 | 200 | 3.775e-13 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 304 | 686 | 4.215e-13 |
| GO:BP | GO:0051049 | regulation of transport | 630 | 1607 | 4.354e-13 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 392 | 928 | 4.379e-13 |
| GO:BP | GO:0007015 | actin filament organization | 219 | 461 | 4.985e-13 |
| GO:BP | GO:0048666 | neuron development | 480 | 1177 | 5.539e-13 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 132 | 244 | 7.420e-13 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 375 | 884 | 8.144e-13 |
| GO:BP | GO:0030182 | neuron differentiation | 574 | 1451 | 1.101e-12 |
| GO:BP | GO:0051648 | vesicle localization | 121 | 219 | 1.218e-12 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 94 | 157 | 1.388e-12 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 202 | 421 | 1.554e-12 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 93 | 155 | 1.570e-12 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 234 | 505 | 1.786e-12 |
| GO:BP | GO:0048699 | generation of neurons | 602 | 1536 | 1.873e-12 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 181 | 369 | 2.676e-12 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 465 | 1144 | 2.697e-12 |
| GO:BP | GO:0051604 | protein maturation | 246 | 539 | 3.136e-12 |
| GO:BP | GO:0051640 | organelle localization | 276 | 620 | 3.647e-12 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 228 | 492 | 3.668e-12 |
| GO:BP | GO:0007033 | vacuole organization | 128 | 239 | 4.703e-12 |
| GO:BP | GO:0007165 | signal transduction | 2054 | 6002 | 5.612e-12 |
| GO:BP | GO:0006259 | DNA metabolic process | 414 | 1005 | 6.831e-12 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 232 | 505 | 6.831e-12 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 265 | 594 | 8.360e-12 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 229 | 499 | 1.120e-11 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 114 | 208 | 1.315e-11 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 819 | 2192 | 1.315e-11 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 148 | 291 | 1.399e-11 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 174 | 357 | 1.582e-11 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 130 | 247 | 1.582e-11 |
| GO:BP | GO:0009060 | aerobic respiration | 110 | 199 | 1.590e-11 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 293 | 673 | 1.590e-11 |
| GO:BP | GO:0007017 | microtubule-based process | 410 | 999 | 1.648e-11 |
| GO:BP | GO:0000278 | mitotic cell cycle | 372 | 892 | 1.648e-11 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 283 | 646 | 1.667e-11 |
| GO:BP | GO:0032501 | multicellular organismal process | 2463 | 7322 | 1.779e-11 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 109 | 197 | 1.824e-11 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 175 | 361 | 2.372e-11 |
| GO:BP | GO:0000902 | cell morphogenesis | 408 | 996 | 2.558e-11 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 529 | 1343 | 2.637e-11 |
| GO:BP | GO:0006325 | chromatin organization | 368 | 884 | 2.889e-11 |
| GO:BP | GO:0072359 | circulatory system development | 460 | 1145 | 2.926e-11 |
| GO:BP | GO:0006897 | endocytosis | 305 | 709 | 2.926e-11 |
| GO:BP | GO:0045333 | cellular respiration | 127 | 242 | 3.656e-11 |
| GO:BP | GO:0016032 | viral process | 198 | 423 | 4.617e-11 |
| GO:BP | GO:0050821 | protein stabilization | 119 | 224 | 6.437e-11 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 181 | 380 | 6.562e-11 |
| GO:BP | GO:0022402 | cell cycle process | 505 | 1280 | 6.648e-11 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 237 | 528 | 6.803e-11 |
| GO:BP | GO:0036503 | ERAD pathway | 69 | 109 | 7.681e-11 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 1021 | 2819 | 7.681e-11 |
| GO:BP | GO:0016043 | cellular component organization | 2724 | 8184 | 7.724e-11 |
| GO:BP | GO:0006986 | response to unfolded protein | 83 | 140 | 7.855e-11 |
| GO:BP | GO:0006401 | RNA catabolic process | 155 | 315 | 1.076e-10 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 154 | 314 | 1.754e-10 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 209 | 458 | 2.145e-10 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 231 | 517 | 2.230e-10 |
| GO:BP | GO:0141188 | nucleic acid catabolic process | 165 | 343 | 2.245e-10 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 278 | 645 | 2.295e-10 |
| GO:BP | GO:0042592 | homeostatic process | 657 | 1736 | 2.649e-10 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 238 | 537 | 2.852e-10 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1061 | 2956 | 3.159e-10 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 173 | 365 | 3.228e-10 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 241 | 546 | 3.448e-10 |
| GO:BP | GO:0034330 | cell junction organization | 345 | 834 | 3.710e-10 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 297 | 701 | 4.568e-10 |
| GO:BP | GO:0007178 | cell surface receptor protein serine/threonine kinase signaling pathway | 172 | 364 | 4.992e-10 |
| GO:BP | GO:0051656 | establishment of organelle localization | 220 | 491 | 4.992e-10 |
| GO:BP | GO:0035295 | tube development | 438 | 1101 | 5.283e-10 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 130 | 257 | 5.333e-10 |
| GO:BP | GO:0030031 | cell projection assembly | 264 | 613 | 8.415e-10 |
| GO:BP | GO:0006606 | protein import into nucleus | 92 | 166 | 9.478e-10 |
| GO:BP | GO:0006457 | protein folding | 114 | 219 | 9.478e-10 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 151 | 312 | 9.496e-10 |
| GO:BP | GO:0016477 | cell migration | 585 | 1534 | 9.654e-10 |
| GO:BP | GO:0098657 | import into cell | 374 | 922 | 9.894e-10 |
| GO:BP | GO:0006839 | mitochondrial transport | 97 | 178 | 9.894e-10 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 196 | 430 | 1.031e-09 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 202 | 446 | 1.033e-09 |
| GO:BP | GO:0051170 | import into nucleus | 94 | 171 | 1.037e-09 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 165 | 349 | 1.181e-09 |
| GO:BP | GO:0016482 | cytosolic transport | 81 | 141 | 1.246e-09 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 128 | 255 | 1.429e-09 |
| GO:BP | GO:0030334 | regulation of cell migration | 386 | 960 | 1.782e-09 |
| GO:BP | GO:0008152 | metabolic process | 4514 | 14136 | 1.831e-09 |
| GO:BP | GO:0016072 | rRNA metabolic process | 132 | 266 | 1.929e-09 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 177 | 383 | 2.337e-09 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 340 | 831 | 2.383e-09 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 68 | 113 | 2.720e-09 |
| GO:BP | GO:0006403 | RNA localization | 105 | 200 | 2.831e-09 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 257 | 600 | 2.835e-09 |
| GO:BP | GO:0001701 | in utero embryonic development | 188 | 413 | 2.873e-09 |
| GO:BP | GO:0051168 | nuclear export | 92 | 169 | 3.274e-09 |
| GO:BP | GO:2000145 | regulation of cell motility | 406 | 1022 | 3.539e-09 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 365 | 905 | 3.806e-09 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 486 | 1256 | 4.655e-09 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 164 | 352 | 5.291e-09 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 118 | 234 | 5.422e-09 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 281 | 670 | 5.422e-09 |
| GO:BP | GO:0016050 | vesicle organization | 173 | 376 | 5.673e-09 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 80 | 142 | 5.786e-09 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 232 | 535 | 5.828e-09 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 148 | 311 | 6.334e-09 |
| GO:BP | GO:0032456 | endocytic recycling | 55 | 86 | 6.410e-09 |
| GO:BP | GO:0007507 | heart development | 257 | 605 | 7.361e-09 |
| GO:BP | GO:0006338 | chromatin remodeling | 300 | 725 | 7.367e-09 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 81 | 145 | 7.702e-09 |
| GO:BP | GO:0015986 | proton motive force-driven ATP synthesis | 51 | 78 | 8.965e-09 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 62 | 102 | 9.756e-09 |
| GO:BP | GO:0061061 | muscle structure development | 288 | 693 | 9.847e-09 |
| GO:BP | GO:0040012 | regulation of locomotion | 419 | 1067 | 1.009e-08 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 155 | 331 | 1.012e-08 |
| GO:BP | GO:0009142 | nucleoside triphosphate biosynthetic process | 73 | 127 | 1.012e-08 |
| GO:BP | GO:0006281 | DNA repair | 262 | 621 | 1.082e-08 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 241 | 563 | 1.105e-08 |
| GO:BP | GO:0009165 | nucleotide biosynthetic process | 115 | 229 | 1.210e-08 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 336 | 830 | 1.222e-08 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 380 | 956 | 1.240e-08 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 237 | 553 | 1.342e-08 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 118 | 237 | 1.390e-08 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 106 | 207 | 1.390e-08 |
| GO:BP | GO:0008088 | axo-dendritic transport | 53 | 83 | 1.411e-08 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 380 | 957 | 1.420e-08 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 63 | 105 | 1.435e-08 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 351 | 874 | 1.449e-08 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 130 | 268 | 1.604e-08 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 62 | 103 | 1.624e-08 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 147 | 312 | 1.628e-08 |
| GO:BP | GO:0006364 | rRNA processing | 113 | 225 | 1.635e-08 |
| GO:BP | GO:0007169 | cell surface receptor protein tyrosine kinase signaling pathway | 264 | 629 | 1.635e-08 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 463 | 1199 | 1.639e-08 |
| GO:BP | GO:0009790 | embryo development | 441 | 1135 | 1.677e-08 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 96 | 183 | 1.677e-08 |
| GO:BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | 69 | 119 | 1.719e-08 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 159 | 344 | 1.850e-08 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 93 | 176 | 1.875e-08 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 317 | 779 | 1.880e-08 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 60 | 99 | 2.037e-08 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 282 | 681 | 2.148e-08 |
| GO:BP | GO:0060322 | head development | 326 | 806 | 2.357e-08 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 280 | 676 | 2.384e-08 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 76 | 136 | 2.423e-08 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 266 | 637 | 2.468e-08 |
| GO:BP | GO:0000165 | MAPK cascade | 304 | 744 | 2.507e-08 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 116 | 234 | 2.533e-08 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 125 | 257 | 2.622e-08 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 62 | 104 | 2.653e-08 |
| GO:BP | GO:0009725 | response to hormone | 361 | 907 | 2.653e-08 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 589 | 1576 | 2.875e-08 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 304 | 745 | 2.918e-08 |
| GO:BP | GO:0035239 | tube morphogenesis | 351 | 879 | 2.928e-08 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 80 | 146 | 3.011e-08 |
| GO:BP | GO:0007420 | brain development | 307 | 754 | 3.145e-08 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 147 | 315 | 3.447e-08 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 170 | 376 | 3.573e-08 |
| GO:BP | GO:0006164 | purine nucleotide biosynthetic process | 99 | 193 | 4.177e-08 |
| GO:BP | GO:0051726 | regulation of cell cycle | 422 | 1087 | 4.271e-08 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 265 | 638 | 4.735e-08 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 454 | 1182 | 5.177e-08 |
| GO:BP | GO:0007034 | vacuolar transport | 91 | 174 | 5.343e-08 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 1176 | 3375 | 5.976e-08 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 85 | 160 | 6.748e-08 |
| GO:BP | GO:0043009 | chordate embryonic development | 276 | 671 | 6.759e-08 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 272 | 660 | 7.064e-08 |
| GO:BP | GO:0001944 | vasculature development | 308 | 762 | 7.124e-08 |
| GO:BP | GO:0030162 | regulation of proteolysis | 180 | 406 | 7.326e-08 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 283 | 692 | 8.255e-08 |
| GO:BP | GO:0006402 | mRNA catabolic process | 125 | 261 | 8.255e-08 |
| GO:BP | GO:0061572 | actin filament bundle organization | 86 | 163 | 8.299e-08 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 60 | 102 | 9.644e-08 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 85 | 161 | 9.758e-08 |
| GO:BP | GO:0009152 | purine ribonucleotide biosynthetic process | 85 | 161 | 9.758e-08 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 115 | 236 | 9.984e-08 |
| GO:BP | GO:0051028 | mRNA transport | 72 | 130 | 1.044e-07 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 211 | 493 | 1.205e-07 |
| GO:BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | 65 | 114 | 1.238e-07 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 166 | 371 | 1.333e-07 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 120 | 250 | 1.408e-07 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 91 | 177 | 1.557e-07 |
| GO:BP | GO:0009411 | response to UV | 82 | 155 | 1.591e-07 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 213 | 500 | 1.597e-07 |
| GO:BP | GO:0050658 | RNA transport | 84 | 160 | 1.660e-07 |
| GO:BP | GO:0050657 | nucleic acid transport | 84 | 160 | 1.660e-07 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 68 | 122 | 1.938e-07 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 530 | 1418 | 1.950e-07 |
| GO:BP | GO:0051301 | cell division | 269 | 658 | 2.031e-07 |
| GO:BP | GO:0051236 | establishment of RNA localization | 85 | 163 | 2.031e-07 |
| GO:BP | GO:0048870 | cell motility | 655 | 1793 | 2.033e-07 |
| GO:BP | GO:0007040 | lysosome organization | 64 | 113 | 2.249e-07 |
| GO:BP | GO:0080171 | lytic vacuole organization | 64 | 113 | 2.249e-07 |
| GO:BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | 64 | 113 | 2.249e-07 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 100 | 201 | 2.550e-07 |
| GO:BP | GO:0007041 | lysosomal transport | 74 | 137 | 2.625e-07 |
| GO:BP | GO:0044042 | glucan metabolic process | 52 | 86 | 2.717e-07 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 151 | 334 | 2.722e-07 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 99 | 199 | 3.000e-07 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 78 | 147 | 3.000e-07 |
| GO:BP | GO:0005977 | glycogen metabolic process | 51 | 84 | 3.062e-07 |
| GO:BP | GO:0007020 | microtubule nucleation | 33 | 46 | 3.149e-07 |
| GO:BP | GO:0001568 | blood vessel development | 294 | 732 | 3.183e-07 |
| GO:BP | GO:0000423 | mitophagy | 45 | 71 | 3.354e-07 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 100 | 202 | 3.454e-07 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 98 | 197 | 3.504e-07 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 90 | 177 | 3.555e-07 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 57 | 98 | 3.849e-07 |
| GO:BP | GO:0042776 | proton motive force-driven mitochondrial ATP synthesis | 43 | 67 | 4.038e-07 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 611 | 1669 | 4.621e-07 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 121 | 257 | 4.651e-07 |
| GO:BP | GO:0007029 | endoplasmic reticulum organization | 59 | 103 | 4.659e-07 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 167 | 380 | 5.212e-07 |
| GO:BP | GO:0040007 | growth | 361 | 929 | 5.399e-07 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 128 | 276 | 5.539e-07 |
| GO:BP | GO:0007417 | central nervous system development | 406 | 1061 | 5.738e-07 |
| GO:BP | GO:0009888 | tissue development | 730 | 2032 | 5.939e-07 |
| GO:BP | GO:0017148 | negative regulation of translation | 71 | 132 | 6.228e-07 |
| GO:BP | GO:0043603 | amide metabolic process | 194 | 455 | 6.473e-07 |
| GO:BP | GO:1905037 | autophagosome organization | 68 | 125 | 6.714e-07 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 88 | 174 | 6.894e-07 |
| GO:BP | GO:0000045 | autophagosome assembly | 65 | 118 | 7.091e-07 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 291 | 729 | 7.237e-07 |
| GO:BP | GO:0006282 | regulation of DNA repair | 108 | 225 | 7.344e-07 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 79 | 152 | 7.603e-07 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 221 | 531 | 7.630e-07 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 64 | 116 | 8.250e-07 |
| GO:BP | GO:0019080 | viral gene expression | 61 | 109 | 8.497e-07 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 163 | 372 | 9.451e-07 |
| GO:BP | GO:0043487 | regulation of RNA stability | 95 | 193 | 1.067e-06 |
| GO:BP | GO:0051258 | protein polymerization | 130 | 284 | 1.085e-06 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 49 | 82 | 1.139e-06 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 64 | 117 | 1.262e-06 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 144 | 322 | 1.266e-06 |
| GO:BP | GO:0051223 | regulation of protein transport | 188 | 442 | 1.268e-06 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 90 | 181 | 1.303e-06 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 36 | 54 | 1.332e-06 |
| GO:BP | GO:0051338 | regulation of transferase activity | 208 | 498 | 1.369e-06 |
| GO:BP | GO:0070482 | response to oxygen levels | 152 | 344 | 1.386e-06 |
| GO:BP | GO:0040011 | locomotion | 469 | 1255 | 1.392e-06 |
| GO:BP | GO:0043434 | response to peptide hormone | 187 | 440 | 1.454e-06 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 63 | 115 | 1.458e-06 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 42 | 67 | 1.493e-06 |
| GO:BP | GO:0030010 | establishment of cell polarity | 81 | 159 | 1.543e-06 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 83 | 164 | 1.549e-06 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 70 | 132 | 1.556e-06 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 41 | 65 | 1.651e-06 |
| GO:BP | GO:0006629 | lipid metabolic process | 514 | 1391 | 1.661e-06 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 76 | 147 | 1.751e-06 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 146 | 329 | 1.779e-06 |
| GO:BP | GO:0042255 | ribosome assembly | 40 | 63 | 1.813e-06 |
| GO:BP | GO:0031929 | TOR signaling | 84 | 167 | 1.813e-06 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 45 | 74 | 1.844e-06 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 45 | 74 | 1.844e-06 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 147 | 332 | 1.885e-06 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 208 | 500 | 1.896e-06 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 120 | 260 | 1.920e-06 |
| GO:BP | GO:0007249 | canonical NF-kappaB signal transduction | 138 | 308 | 1.973e-06 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 52 | 90 | 1.982e-06 |
| GO:BP | GO:0034644 | cellular response to UV | 52 | 90 | 1.982e-06 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 605 | 1667 | 2.032e-06 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 227 | 554 | 2.080e-06 |
| GO:BP | GO:0048729 | tissue morphogenesis | 248 | 614 | 2.251e-06 |
| GO:BP | GO:0045727 | positive regulation of translation | 72 | 138 | 2.314e-06 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 256 | 637 | 2.316e-06 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 1089 | 3157 | 2.362e-06 |
| GO:BP | GO:0042594 | response to starvation | 104 | 219 | 2.365e-06 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 109 | 232 | 2.365e-06 |
| GO:BP | GO:0051668 | localization within membrane | 355 | 924 | 2.514e-06 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 50 | 86 | 2.616e-06 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 91 | 186 | 2.670e-06 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 75 | 146 | 2.856e-06 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 182 | 430 | 2.872e-06 |
| GO:BP | GO:0032868 | response to insulin | 123 | 270 | 3.144e-06 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 55 | 98 | 3.168e-06 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 153 | 351 | 3.302e-06 |
| GO:BP | GO:0050808 | synapse organization | 229 | 563 | 3.542e-06 |
| GO:BP | GO:0006413 | translational initiation | 67 | 127 | 3.592e-06 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 119 | 260 | 3.633e-06 |
| GO:BP | GO:0048589 | developmental growth | 264 | 663 | 3.638e-06 |
| GO:BP | GO:0009314 | response to radiation | 184 | 437 | 3.734e-06 |
| GO:BP | GO:0009267 | cellular response to starvation | 87 | 177 | 3.792e-06 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 104 | 221 | 4.043e-06 |
| GO:BP | GO:0035329 | hippo signaling | 37 | 58 | 4.390e-06 |
| GO:BP | GO:0098930 | axonal transport | 42 | 69 | 4.519e-06 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 227 | 559 | 4.585e-06 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 141 | 320 | 4.589e-06 |
| GO:BP | GO:0014706 | striated muscle tissue development | 173 | 408 | 4.923e-06 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 207 | 503 | 4.960e-06 |
| GO:BP | GO:0034329 | cell junction assembly | 207 | 503 | 4.960e-06 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 120 | 264 | 4.980e-06 |
| GO:BP | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 133 | 299 | 5.216e-06 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 130 | 291 | 5.291e-06 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 57 | 104 | 5.419e-06 |
| GO:BP | GO:0001666 | response to hypoxia | 134 | 302 | 5.563e-06 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 90 | 186 | 5.603e-06 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 51 | 90 | 5.703e-06 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 88 | 181 | 5.794e-06 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 59 | 109 | 5.908e-06 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 47 | 81 | 6.404e-06 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 101 | 215 | 6.484e-06 |
| GO:BP | GO:0021537 | telencephalon development | 127 | 284 | 6.669e-06 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 99 | 210 | 6.856e-06 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 65 | 124 | 7.078e-06 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 161 | 377 | 7.278e-06 |
| GO:BP | GO:0051098 | regulation of binding | 100 | 213 | 7.607e-06 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 42 | 70 | 7.611e-06 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 318 | 825 | 7.654e-06 |
| GO:BP | GO:0046034 | ATP metabolic process | 106 | 229 | 8.141e-06 |
| GO:BP | GO:1902600 | proton transmembrane transport | 91 | 190 | 8.368e-06 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 54 | 98 | 8.568e-06 |
| GO:BP | GO:0046785 | microtubule polymerization | 54 | 98 | 8.568e-06 |
| GO:BP | GO:0032535 | regulation of cellular component size | 153 | 356 | 8.800e-06 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 94 | 198 | 8.884e-06 |
| GO:BP | GO:0021543 | pallium development | 94 | 198 | 8.884e-06 |
| GO:BP | GO:0019827 | stem cell population maintenance | 87 | 180 | 9.029e-06 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 113 | 248 | 9.096e-06 |
| GO:BP | GO:0019058 | viral life cycle | 136 | 310 | 9.539e-06 |
| GO:BP | GO:0006903 | vesicle targeting | 40 | 66 | 9.717e-06 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 235 | 587 | 9.893e-06 |
| GO:BP | GO:0051646 | mitochondrion localization | 34 | 53 | 1.055e-05 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 71 | 140 | 1.055e-05 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 96 | 204 | 1.098e-05 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 82 | 168 | 1.137e-05 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 52 | 94 | 1.159e-05 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 64 | 123 | 1.180e-05 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 57 | 106 | 1.189e-05 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 70 | 138 | 1.236e-05 |
| GO:BP | GO:0060537 | muscle tissue development | 179 | 430 | 1.236e-05 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 38 | 62 | 1.236e-05 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 95 | 202 | 1.281e-05 |
| GO:BP | GO:0051896 | regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 118 | 263 | 1.327e-05 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 118 | 263 | 1.327e-05 |
| GO:BP | GO:0022900 | electron transport chain | 65 | 126 | 1.420e-05 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 41 | 69 | 1.434e-05 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 30 | 45 | 1.435e-05 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 114 | 253 | 1.561e-05 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 80 | 164 | 1.565e-05 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 134 | 307 | 1.565e-05 |
| GO:BP | GO:0045048 | protein insertion into ER membrane | 24 | 33 | 1.659e-05 |
| GO:BP | GO:0019725 | cellular homeostasis | 319 | 835 | 1.784e-05 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 96 | 206 | 1.839e-05 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 137 | 316 | 1.853e-05 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 239 | 603 | 1.887e-05 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 105 | 230 | 1.958e-05 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 250 | 635 | 1.973e-05 |
| GO:BP | GO:0055085 | transmembrane transport | 567 | 1578 | 1.977e-05 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 159 | 377 | 1.988e-05 |
| GO:BP | GO:1901875 | positive regulation of post-translational protein modification | 67 | 132 | 2.001e-05 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 97 | 209 | 2.001e-05 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 82 | 170 | 2.001e-05 |
| GO:BP | GO:0032981 | mitochondrial respiratory chain complex I assembly | 38 | 63 | 2.097e-05 |
| GO:BP | GO:0010257 | NADH dehydrogenase complex assembly | 38 | 63 | 2.097e-05 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 224 | 561 | 2.161e-05 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 60 | 115 | 2.227e-05 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 116 | 260 | 2.231e-05 |
| GO:BP | GO:0001525 | angiogenesis | 219 | 547 | 2.251e-05 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 70 | 140 | 2.336e-05 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 50 | 91 | 2.353e-05 |
| GO:BP | GO:0006405 | RNA export from nucleus | 50 | 91 | 2.353e-05 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 57 | 108 | 2.501e-05 |
| GO:BP | GO:0043149 | stress fiber assembly | 57 | 108 | 2.501e-05 |
| GO:BP | GO:0008283 | cell population proliferation | 709 | 2015 | 2.510e-05 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 31 | 48 | 2.532e-05 |
| GO:BP | GO:0051050 | positive regulation of transport | 322 | 847 | 2.538e-05 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 52 | 96 | 2.567e-05 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 59 | 113 | 2.589e-05 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 182 | 443 | 2.602e-05 |
| GO:BP | GO:0098727 | maintenance of cell number | 87 | 184 | 2.655e-05 |
| GO:BP | GO:0051457 | maintenance of protein location in nucleus | 19 | 24 | 2.677e-05 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 88 | 187 | 2.960e-05 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 35 | 57 | 2.997e-05 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 74 | 151 | 3.029e-05 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 39 | 66 | 3.040e-05 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 60 | 116 | 3.132e-05 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 28 | 42 | 3.168e-05 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 68 | 136 | 3.192e-05 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 68 | 136 | 3.192e-05 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 53 | 99 | 3.202e-05 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 66 | 131 | 3.202e-05 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 53 | 99 | 3.202e-05 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 21 | 28 | 3.371e-05 |
| GO:BP | GO:0036010 | protein localization to endosome | 21 | 28 | 3.371e-05 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 21 | 28 | 3.371e-05 |
| GO:BP | GO:0099518 | vesicle cytoskeletal trafficking | 38 | 64 | 3.441e-05 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 236 | 599 | 3.461e-05 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 127 | 292 | 3.469e-05 |
| GO:BP | GO:0006516 | glycoprotein catabolic process | 26 | 38 | 3.473e-05 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 67 | 134 | 3.727e-05 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 163 | 392 | 3.791e-05 |
| GO:BP | GO:0043122 | regulation of canonical NF-kappaB signal transduction | 125 | 287 | 3.799e-05 |
| GO:BP | GO:0061564 | axon development | 211 | 528 | 3.859e-05 |
| GO:BP | GO:0009416 | response to light stimulus | 138 | 323 | 4.093e-05 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 56 | 107 | 4.145e-05 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 79 | 165 | 4.188e-05 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 127 | 293 | 4.232e-05 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 117 | 266 | 4.430e-05 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 82 | 173 | 4.461e-05 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 48 | 88 | 4.740e-05 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 96 | 210 | 4.761e-05 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 83 | 176 | 4.974e-05 |
| GO:BP | GO:0071806 | protein transmembrane transport | 42 | 74 | 5.051e-05 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 109 | 245 | 5.051e-05 |
| GO:BP | GO:0098732 | macromolecule deacylation | 35 | 58 | 5.052e-05 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 50 | 93 | 5.116e-05 |
| GO:BP | GO:0043542 | endothelial cell migration | 100 | 221 | 5.164e-05 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 132 | 308 | 5.439e-05 |
| GO:BP | GO:0045185 | maintenance of protein location | 38 | 65 | 5.624e-05 |
| GO:BP | GO:0016049 | cell growth | 195 | 485 | 5.709e-05 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 28 | 43 | 6.066e-05 |
| GO:BP | GO:0030900 | forebrain development | 173 | 423 | 6.083e-05 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 60 | 118 | 6.083e-05 |
| GO:BP | GO:0007409 | axonogenesis | 187 | 463 | 6.420e-05 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 37 | 63 | 6.456e-05 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 71 | 146 | 6.498e-05 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 27 | 41 | 6.524e-05 |
| GO:BP | GO:0000266 | mitochondrial fission | 27 | 41 | 6.524e-05 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 43 | 77 | 6.717e-05 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 89 | 193 | 6.830e-05 |
| GO:BP | GO:0010586 | miRNA metabolic process | 55 | 106 | 6.904e-05 |
| GO:BP | GO:0006901 | vesicle coating | 26 | 39 | 6.981e-05 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 48 | 89 | 6.981e-05 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 138 | 326 | 7.062e-05 |
| GO:BP | GO:0007517 | muscle organ development | 152 | 365 | 7.114e-05 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 130 | 304 | 7.158e-05 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 130 | 304 | 7.158e-05 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 90 | 196 | 7.404e-05 |
| GO:BP | GO:0030097 | hemopoiesis | 362 | 976 | 7.685e-05 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 165 | 402 | 7.737e-05 |
| GO:BP | GO:0042692 | muscle cell differentiation | 166 | 405 | 7.970e-05 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 91 | 199 | 8.056e-05 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 104 | 234 | 8.546e-05 |
| GO:BP | GO:0006997 | nucleus organization | 71 | 147 | 8.586e-05 |
| GO:BP | GO:0043549 | regulation of kinase activity | 175 | 431 | 8.698e-05 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 98 | 218 | 8.738e-05 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 38 | 66 | 8.944e-05 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 30 | 48 | 9.023e-05 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 30 | 48 | 9.023e-05 |
| GO:BP | GO:0021987 | cerebral cortex development | 65 | 132 | 9.416e-05 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 57 | 112 | 9.809e-05 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 75 | 158 | 1.035e-04 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 40 | 71 | 1.050e-04 |
| GO:BP | GO:0046777 | protein autophosphorylation | 83 | 179 | 1.061e-04 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 33 | 55 | 1.091e-04 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 64 | 130 | 1.110e-04 |
| GO:BP | GO:0006446 | regulation of translational initiation | 45 | 83 | 1.116e-04 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 112 | 257 | 1.124e-04 |
| GO:BP | GO:0006611 | protein export from nucleus | 36 | 62 | 1.192e-04 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 79 | 169 | 1.203e-04 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 67 | 138 | 1.230e-04 |
| GO:BP | GO:0000723 | telomere maintenance | 77 | 164 | 1.277e-04 |
| GO:BP | GO:0007032 | endosome organization | 51 | 98 | 1.314e-04 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 70 | 146 | 1.337e-04 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 35 | 60 | 1.374e-04 |
| GO:BP | GO:0031032 | actomyosin structure organization | 95 | 212 | 1.383e-04 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 78 | 167 | 1.410e-04 |
| GO:BP | GO:0060271 | cilium assembly | 164 | 403 | 1.419e-04 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 130 | 308 | 1.503e-04 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 155 | 378 | 1.520e-04 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 17 | 22 | 1.559e-04 |
| GO:BP | GO:0007035 | vacuolar acidification | 24 | 36 | 1.565e-04 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 162 | 398 | 1.573e-04 |
| GO:BP | GO:0090398 | cellular senescence | 52 | 101 | 1.577e-04 |
| GO:BP | GO:0006399 | tRNA metabolic process | 94 | 210 | 1.606e-04 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 40 | 72 | 1.607e-04 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 40 | 72 | 1.607e-04 |
| GO:BP | GO:0051654 | establishment of mitochondrion localization | 23 | 34 | 1.654e-04 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 95 | 213 | 1.720e-04 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 22 | 32 | 1.735e-04 |
| GO:BP | GO:1903533 | regulation of protein targeting | 42 | 77 | 1.781e-04 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 29 | 47 | 1.781e-04 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 49 | 94 | 1.804e-04 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 342 | 925 | 1.811e-04 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 103 | 235 | 1.859e-04 |
| GO:BP | GO:0099173 | postsynapse organization | 107 | 246 | 1.903e-04 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 66 | 137 | 1.911e-04 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 71 | 150 | 1.947e-04 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 79 | 171 | 1.976e-04 |
| GO:BP | GO:0044782 | cilium organization | 173 | 431 | 2.036e-04 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 145 | 352 | 2.142e-04 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 326 | 879 | 2.149e-04 |
| GO:BP | GO:0044770 | cell cycle phase transition | 208 | 532 | 2.156e-04 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 58 | 117 | 2.156e-04 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 54 | 107 | 2.204e-04 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 27 | 43 | 2.218e-04 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 65 | 135 | 2.239e-04 |
| GO:BP | GO:0022411 | cellular component disassembly | 178 | 446 | 2.250e-04 |
| GO:BP | GO:0035148 | tube formation | 70 | 148 | 2.275e-04 |
| GO:BP | GO:0003281 | ventricular septum development | 40 | 73 | 2.423e-04 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 40 | 73 | 2.423e-04 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 66 | 138 | 2.514e-04 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 79 | 172 | 2.514e-04 |
| GO:BP | GO:0061462 | protein localization to lysosome | 34 | 59 | 2.514e-04 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 140 | 339 | 2.521e-04 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 140 | 339 | 2.521e-04 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 49 | 95 | 2.535e-04 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 126 | 300 | 2.572e-04 |
| GO:BP | GO:0031503 | protein-containing complex localization | 95 | 215 | 2.618e-04 |
| GO:BP | GO:0035601 | protein deacylation | 30 | 50 | 2.658e-04 |
| GO:BP | GO:0070085 | glycosylation | 106 | 245 | 2.658e-04 |
| GO:BP | GO:0006486 | protein glycosylation | 99 | 226 | 2.698e-04 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 99 | 226 | 2.698e-04 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 60 | 123 | 2.813e-04 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 36 | 64 | 2.921e-04 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 70 | 149 | 2.946e-04 |
| GO:BP | GO:1901524 | regulation of mitophagy | 24 | 37 | 2.958e-04 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 52 | 103 | 3.009e-04 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 29 | 48 | 3.009e-04 |
| GO:BP | GO:0072583 | clathrin-dependent endocytosis | 29 | 48 | 3.009e-04 |
| GO:BP | GO:0038202 | TORC1 signaling | 52 | 103 | 3.009e-04 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 52 | 103 | 3.009e-04 |
| GO:BP | GO:0010968 | regulation of microtubule nucleation | 15 | 19 | 3.247e-04 |
| GO:BP | GO:0016925 | protein sumoylation | 38 | 69 | 3.275e-04 |
| GO:BP | GO:0001558 | regulation of cell growth | 163 | 406 | 3.373e-04 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 28 | 46 | 3.418e-04 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 64 | 134 | 3.437e-04 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 108 | 252 | 3.557e-04 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 40 | 74 | 3.578e-04 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 67 | 142 | 3.642e-04 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 21 | 31 | 3.670e-04 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 138 | 336 | 3.852e-04 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 334 | 909 | 3.853e-04 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 27 | 44 | 3.855e-04 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 34 | 60 | 3.922e-04 |
| GO:BP | GO:0030200 | heparan sulfate proteoglycan catabolic process | 9 | 9 | 3.930e-04 |
| GO:BP | GO:0160063 | multi-pass transmembrane protein insertion into ER membrane | 9 | 9 | 3.930e-04 |
| GO:BP | GO:0070841 | inclusion body assembly | 18 | 25 | 3.967e-04 |
| GO:BP | GO:0043393 | regulation of protein binding | 56 | 114 | 3.974e-04 |
| GO:BP | GO:0071478 | cellular response to radiation | 82 | 182 | 3.978e-04 |
| GO:BP | GO:0072665 | protein localization to vacuole | 46 | 89 | 4.082e-04 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 52 | 104 | 4.125e-04 |
| GO:BP | GO:0141193 | nuclear receptor-mediated signaling pathway | 80 | 177 | 4.310e-04 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 30 | 51 | 4.354e-04 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 30 | 51 | 4.354e-04 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 59 | 122 | 4.368e-04 |
| GO:BP | GO:0060485 | mesenchyme development | 132 | 320 | 4.393e-04 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 132 | 320 | 4.393e-04 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 36 | 65 | 4.408e-04 |
| GO:BP | GO:0034605 | cellular response to heat | 36 | 65 | 4.408e-04 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 250 | 661 | 4.487e-04 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 197 | 506 | 4.487e-04 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 128 | 309 | 4.540e-04 |
| GO:BP | GO:0051960 | regulation of nervous system development | 180 | 457 | 4.567e-04 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 51 | 102 | 4.824e-04 |
| GO:BP | GO:0061157 | mRNA destabilization | 51 | 102 | 4.824e-04 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 38 | 70 | 4.837e-04 |
| GO:BP | GO:0043401 | steroid hormone receptor signaling pathway | 65 | 138 | 4.940e-04 |
| GO:BP | GO:0006082 | organic acid metabolic process | 335 | 915 | 5.043e-04 |
| GO:BP | GO:0060284 | regulation of cell development | 312 | 847 | 5.504e-04 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 52 | 105 | 5.616e-04 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 69 | 149 | 5.670e-04 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 28 | 47 | 5.688e-04 |
| GO:BP | GO:0043038 | amino acid activation | 28 | 47 | 5.688e-04 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 75 | 165 | 5.823e-04 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 23 | 36 | 5.951e-04 |
| GO:BP | GO:0010761 | fibroblast migration | 31 | 54 | 6.051e-04 |
| GO:BP | GO:0000910 | cytokinesis | 85 | 192 | 6.068e-04 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 55 | 113 | 6.244e-04 |
| GO:BP | GO:0021915 | neural tube development | 73 | 160 | 6.267e-04 |
| GO:BP | GO:0003279 | cardiac septum development | 53 | 108 | 6.438e-04 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 51 | 103 | 6.598e-04 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 43 | 83 | 6.612e-04 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 36 | 66 | 6.642e-04 |
| GO:BP | GO:0030167 | proteoglycan catabolic process | 13 | 16 | 6.650e-04 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 188 | 483 | 6.662e-04 |
| GO:BP | GO:0014812 | muscle cell migration | 45 | 88 | 6.693e-04 |
| GO:BP | GO:0009615 | response to virus | 164 | 414 | 6.916e-04 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 30 | 52 | 6.948e-04 |
| GO:BP | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 30 | 52 | 6.948e-04 |
| GO:BP | GO:0031648 | protein destabilization | 30 | 52 | 6.948e-04 |
| GO:BP | GO:0035751 | regulation of lysosomal lumen pH | 21 | 32 | 7.089e-04 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 61 | 129 | 7.155e-04 |
| GO:BP | GO:0140352 | export from cell | 338 | 928 | 7.242e-04 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 72 | 158 | 7.262e-04 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 389 | 1082 | 7.262e-04 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 26 | 43 | 7.290e-04 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 26 | 43 | 7.290e-04 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 26 | 43 | 7.290e-04 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 26 | 43 | 7.290e-04 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 586 | 1685 | 7.335e-04 |
| GO:BP | GO:0050779 | RNA destabilization | 52 | 106 | 7.463e-04 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 50 | 101 | 7.652e-04 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 35 | 64 | 7.658e-04 |
| GO:BP | GO:0046903 | secretion | 359 | 992 | 7.658e-04 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 48 | 96 | 7.744e-04 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 48 | 96 | 7.744e-04 |
| GO:BP | GO:1901652 | response to peptide | 349 | 962 | 7.744e-04 |
| GO:BP | GO:0003205 | cardiac chamber development | 76 | 169 | 7.745e-04 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 46 | 91 | 7.777e-04 |
| GO:BP | GO:0170036 | import into the mitochondrion | 29 | 50 | 7.908e-04 |
| GO:BP | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 32 | 57 | 7.937e-04 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 32 | 57 | 7.937e-04 |
| GO:BP | GO:0061245 | establishment or maintenance of bipolar cell polarity | 32 | 57 | 7.937e-04 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 32 | 57 | 7.937e-04 |
| GO:BP | GO:0047497 | mitochondrion transport along microtubule | 19 | 28 | 7.964e-04 |
| GO:BP | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated | 19 | 28 | 7.964e-04 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 19 | 28 | 7.964e-04 |
| GO:BP | GO:0035330 | regulation of hippo signaling | 25 | 41 | 8.153e-04 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 37 | 69 | 8.156e-04 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 15 | 20 | 8.257e-04 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 18 | 26 | 8.336e-04 |
| GO:BP | GO:0045947 | negative regulation of translational initiation | 16 | 22 | 8.555e-04 |
| GO:BP | GO:0007042 | lysosomal lumen acidification | 17 | 24 | 8.555e-04 |
| GO:BP | GO:0075522 | IRES-dependent viral translational initiation | 10 | 11 | 8.578e-04 |
| GO:BP | GO:0051099 | positive regulation of binding | 51 | 104 | 8.626e-04 |
| GO:BP | GO:0007254 | JNK cascade | 78 | 175 | 8.810e-04 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 41 | 79 | 8.860e-04 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 28 | 48 | 9.015e-04 |
| GO:BP | GO:0060606 | tube closure | 45 | 89 | 9.044e-04 |
| GO:BP | GO:0032543 | mitochondrial translation | 61 | 130 | 9.046e-04 |
| GO:BP | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | 31 | 55 | 9.149e-04 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 54 | 112 | 9.446e-04 |
| GO:BP | GO:0006520 | amino acid metabolic process | 122 | 297 | 9.789e-04 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 52 | 107 | 9.821e-04 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 57 | 120 | 1.013e-03 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 108 | 258 | 1.015e-03 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 113 | 272 | 1.021e-03 |
| GO:BP | GO:0031667 | response to nutrient levels | 196 | 510 | 1.024e-03 |
| GO:BP | GO:0040008 | regulation of growth | 228 | 604 | 1.034e-03 |
| GO:BP | GO:0061614 | miRNA transcription | 40 | 77 | 1.037e-03 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 48 | 97 | 1.037e-03 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 40 | 77 | 1.037e-03 |
| GO:BP | GO:0032259 | methylation | 100 | 236 | 1.049e-03 |
| GO:BP | GO:0009791 | post-embryonic development | 46 | 92 | 1.050e-03 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 46 | 92 | 1.050e-03 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 44 | 87 | 1.052e-03 |
| GO:BP | GO:0048545 | response to steroid hormone | 137 | 340 | 1.052e-03 |
| GO:BP | GO:1990089 | response to nerve growth factor | 30 | 53 | 1.052e-03 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 30 | 53 | 1.052e-03 |
| GO:BP | GO:0043123 | positive regulation of canonical NF-kappaB signal transduction | 92 | 214 | 1.052e-03 |
| GO:BP | GO:0072175 | epithelial tube formation | 63 | 136 | 1.083e-03 |
| GO:BP | GO:0050807 | regulation of synapse organization | 126 | 309 | 1.083e-03 |
| GO:BP | GO:0007051 | spindle organization | 89 | 206 | 1.097e-03 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 75 | 168 | 1.097e-03 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 66 | 144 | 1.100e-03 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 66 | 144 | 1.100e-03 |
| GO:BP | GO:0035264 | multicellular organism growth | 72 | 160 | 1.105e-03 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 51 | 105 | 1.136e-03 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 37 | 70 | 1.165e-03 |
| GO:BP | GO:0048863 | stem cell differentiation | 103 | 245 | 1.172e-03 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 270 | 730 | 1.183e-03 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 67 | 147 | 1.194e-03 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 47 | 95 | 1.203e-03 |
| GO:BP | GO:0008089 | anterograde axonal transport | 29 | 51 | 1.216e-03 |
| GO:BP | GO:0097581 | lamellipodium organization | 45 | 90 | 1.221e-03 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 45 | 90 | 1.221e-03 |
| GO:BP | GO:0048041 | focal adhesion assembly | 43 | 85 | 1.229e-03 |
| GO:BP | GO:0034333 | adherens junction assembly | 11 | 13 | 1.258e-03 |
| GO:BP | GO:1904152 | regulation of retrograde protein transport, ER to cytosol | 11 | 13 | 1.258e-03 |
| GO:BP | GO:0048199 | vesicle targeting, to, from or within Golgi | 21 | 33 | 1.258e-03 |
| GO:BP | GO:0003174 | mitral valve development | 11 | 13 | 1.258e-03 |
| GO:BP | GO:0034097 | response to cytokine | 342 | 947 | 1.270e-03 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 52 | 108 | 1.274e-03 |
| GO:BP | GO:0030518 | nuclear receptor-mediated steroid hormone signaling pathway | 57 | 121 | 1.288e-03 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 97 | 229 | 1.304e-03 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 322 | 887 | 1.304e-03 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 128 | 316 | 1.312e-03 |
| GO:BP | GO:0072657 | protein localization to membrane | 302 | 827 | 1.319e-03 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 50 | 103 | 1.321e-03 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 25 | 42 | 1.325e-03 |
| GO:BP | GO:0006476 | protein deacetylation | 25 | 42 | 1.325e-03 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 142 | 356 | 1.327e-03 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 60 | 129 | 1.327e-03 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 429 | 1212 | 1.329e-03 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 31 | 56 | 1.357e-03 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 48 | 98 | 1.357e-03 |
| GO:BP | GO:1901799 | negative regulation of proteasomal protein catabolic process | 31 | 56 | 1.357e-03 |
| GO:BP | GO:0001822 | kidney development | 130 | 322 | 1.365e-03 |
| GO:BP | GO:0007030 | Golgi organization | 69 | 153 | 1.366e-03 |
| GO:BP | GO:0051085 | chaperone cofactor-dependent protein refolding | 20 | 31 | 1.366e-03 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 103 | 246 | 1.366e-03 |
| GO:BP | GO:0051205 | protein insertion into membrane | 28 | 49 | 1.378e-03 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 28 | 49 | 1.378e-03 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 28 | 49 | 1.378e-03 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 46 | 93 | 1.382e-03 |
| GO:BP | GO:0043112 | receptor metabolic process | 38 | 73 | 1.382e-03 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 266 | 720 | 1.392e-03 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 58 | 124 | 1.396e-03 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 212 | 560 | 1.398e-03 |
| GO:BP | GO:0001843 | neural tube closure | 44 | 88 | 1.398e-03 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 212 | 560 | 1.398e-03 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 83 | 191 | 1.399e-03 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 42 | 83 | 1.405e-03 |
| GO:BP | GO:0007018 | microtubule-based movement | 184 | 478 | 1.405e-03 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 70 | 156 | 1.453e-03 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 84 | 194 | 1.476e-03 |
| GO:BP | GO:0090140 | regulation of mitochondrial fission | 19 | 29 | 1.478e-03 |
| GO:BP | GO:0003013 | circulatory system process | 226 | 602 | 1.505e-03 |
| GO:BP | GO:0001841 | neural tube formation | 49 | 101 | 1.519e-03 |
| GO:BP | GO:0009408 | response to heat | 49 | 101 | 1.519e-03 |
| GO:BP | GO:0003231 | cardiac ventricle development | 59 | 127 | 1.525e-03 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 12 | 15 | 1.525e-03 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 65 | 143 | 1.573e-03 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 27 | 47 | 1.580e-03 |
| GO:BP | GO:0048011 | neurotrophin TRK receptor signaling pathway | 18 | 27 | 1.594e-03 |
| GO:BP | GO:0042026 | protein refolding | 18 | 27 | 1.594e-03 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 37 | 71 | 1.608e-03 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 57 | 122 | 1.620e-03 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 43 | 86 | 1.635e-03 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 39 | 76 | 1.636e-03 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 39 | 76 | 1.636e-03 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 176 | 456 | 1.638e-03 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 23 | 38 | 1.681e-03 |
| GO:BP | GO:0061339 | establishment or maintenance of monopolar cell polarity | 17 | 25 | 1.694e-03 |
| GO:BP | GO:0006998 | nuclear envelope organization | 32 | 59 | 1.701e-03 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 92 | 217 | 1.769e-03 |
| GO:BP | GO:0035357 | peroxisome proliferator activated receptor signaling pathway | 16 | 23 | 1.773e-03 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 48 | 99 | 1.773e-03 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 16 | 23 | 1.773e-03 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 53 | 112 | 1.810e-03 |
| GO:BP | GO:0033059 | cellular pigmentation | 34 | 64 | 1.810e-03 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 166 | 428 | 1.876e-03 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 36 | 69 | 1.885e-03 |
| GO:BP | GO:0051402 | neuron apoptotic process | 120 | 296 | 1.919e-03 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 38 | 74 | 1.923e-03 |
| GO:BP | GO:0031623 | receptor internalization | 62 | 136 | 1.966e-03 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 65 | 144 | 1.967e-03 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 31 | 57 | 1.987e-03 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 31 | 57 | 1.987e-03 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 31 | 57 | 1.987e-03 |
| GO:BP | GO:0045216 | cell-cell junction organization | 91 | 215 | 2.025e-03 |
| GO:BP | GO:0048511 | rhythmic process | 123 | 305 | 2.058e-03 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 231 | 620 | 2.077e-03 |
| GO:BP | GO:0048489 | synaptic vesicle transport | 25 | 43 | 2.092e-03 |
| GO:BP | GO:1990000 | amyloid fibril formation | 25 | 43 | 2.092e-03 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 28 | 50 | 2.097e-03 |
| GO:BP | GO:0019079 | viral genome replication | 60 | 131 | 2.103e-03 |
| GO:BP | GO:0034063 | stress granule assembly | 21 | 34 | 2.141e-03 |
| GO:BP | GO:0051647 | nucleus localization | 21 | 34 | 2.141e-03 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 102 | 246 | 2.145e-03 |
| GO:BP | GO:0032940 | secretion by cell | 309 | 854 | 2.152e-03 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 108 | 263 | 2.198e-03 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 35 | 67 | 2.200e-03 |
| GO:BP | GO:0036446 | myofibroblast differentiation | 9 | 10 | 2.202e-03 |
| GO:BP | GO:0030050 | vesicle transport along actin filament | 9 | 10 | 2.202e-03 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 41 | 82 | 2.238e-03 |
| GO:BP | GO:0034502 | protein localization to chromosome | 58 | 126 | 2.247e-03 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 48 | 100 | 2.332e-03 |
| GO:BP | GO:1903955 | positive regulation of protein targeting to mitochondrion | 20 | 32 | 2.404e-03 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 20 | 32 | 2.404e-03 |
| GO:BP | GO:0006513 | protein monoubiquitination | 27 | 48 | 2.433e-03 |
| GO:BP | GO:0001881 | receptor recycling | 27 | 48 | 2.433e-03 |
| GO:BP | GO:0001101 | response to acid chemical | 65 | 145 | 2.443e-03 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 32 | 60 | 2.476e-03 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 69 | 156 | 2.572e-03 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 40 | 80 | 2.645e-03 |
| GO:BP | GO:0061512 | protein localization to cilium | 38 | 75 | 2.664e-03 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 19 | 30 | 2.690e-03 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 90 | 214 | 2.773e-03 |
| GO:BP | GO:0032438 | melanosome organization | 23 | 39 | 2.773e-03 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 67 | 151 | 2.806e-03 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 26 | 46 | 2.840e-03 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 58 | 127 | 2.868e-03 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 71 | 162 | 2.908e-03 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 31 | 58 | 2.915e-03 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 119 | 296 | 2.915e-03 |
| GO:BP | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining | 18 | 28 | 2.980e-03 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 120 | 299 | 2.980e-03 |
| GO:BP | GO:0050847 | progesterone receptor signaling pathway | 10 | 12 | 3.099e-03 |
| GO:BP | GO:0099515 | actin filament-based transport | 10 | 12 | 3.099e-03 |
| GO:BP | GO:0003183 | mitral valve morphogenesis | 10 | 12 | 3.099e-03 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 10 | 12 | 3.099e-03 |
| GO:BP | GO:0140627 | ubiquitin-dependent protein catabolic process via the C-end degron rule pathway | 10 | 12 | 3.099e-03 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 35 | 68 | 3.103e-03 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 35 | 68 | 3.103e-03 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 28 | 51 | 3.165e-03 |
| GO:BP | GO:0051259 | protein complex oligomerization | 110 | 271 | 3.165e-03 |
| GO:BP | GO:0051276 | chromosome organization | 214 | 574 | 3.253e-03 |
| GO:BP | GO:0070198 | protein localization to chromosome, telomeric region | 17 | 26 | 3.260e-03 |
| GO:BP | GO:0005980 | glycogen catabolic process | 17 | 26 | 3.260e-03 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 74 | 171 | 3.419e-03 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 258 | 706 | 3.483e-03 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 42 | 86 | 3.483e-03 |
| GO:BP | GO:0003007 | heart morphogenesis | 108 | 266 | 3.513e-03 |
| GO:BP | GO:0061162 | establishment of monopolar cell polarity | 16 | 24 | 3.537e-03 |
| GO:BP | GO:0000027 | ribosomal large subunit assembly | 16 | 24 | 3.537e-03 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 186 | 492 | 3.537e-03 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 16 | 24 | 3.537e-03 |
| GO:BP | GO:1902653 | secondary alcohol biosynthetic process | 32 | 61 | 3.557e-03 |
| GO:BP | GO:0006695 | cholesterol biosynthetic process | 32 | 61 | 3.557e-03 |
| GO:BP | GO:0051100 | negative regulation of binding | 47 | 99 | 3.573e-03 |
| GO:BP | GO:0043045 | epigenetic programming of gene expression | 21 | 35 | 3.609e-03 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 116 | 289 | 3.632e-03 |
| GO:BP | GO:0072001 | renal system development | 131 | 332 | 3.642e-03 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 11 | 14 | 3.652e-03 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 27 | 49 | 3.678e-03 |
| GO:BP | GO:0014020 | primary neural tube formation | 45 | 94 | 3.750e-03 |
| GO:BP | GO:0035089 | establishment of apical/basal cell polarity | 15 | 22 | 3.766e-03 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 246 | 671 | 3.772e-03 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 62 | 139 | 3.816e-03 |
| GO:BP | GO:0043200 | response to amino acid | 59 | 131 | 3.860e-03 |
| GO:BP | GO:0032200 | telomere organization | 81 | 191 | 3.895e-03 |
| GO:BP | GO:0043543 | protein acylation | 43 | 89 | 3.911e-03 |
| GO:BP | GO:1901663 | quinone biosynthetic process | 12 | 16 | 3.916e-03 |
| GO:BP | GO:0006517 | protein deglycosylation | 14 | 20 | 3.916e-03 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 77 | 180 | 3.916e-03 |
| GO:BP | GO:0034497 | protein localization to phagophore assembly site | 12 | 16 | 3.916e-03 |
| GO:BP | GO:0006744 | ubiquinone biosynthetic process | 12 | 16 | 3.916e-03 |
| GO:BP | GO:0051788 | response to misfolded protein | 14 | 20 | 3.916e-03 |
| GO:BP | GO:0044849 | estrous cycle | 12 | 16 | 3.916e-03 |
| GO:BP | GO:0045088 | regulation of innate immune response | 171 | 449 | 3.937e-03 |
| GO:BP | GO:0071218 | cellular response to misfolded protein | 13 | 18 | 3.991e-03 |
| GO:BP | GO:0043604 | amide biosynthetic process | 74 | 172 | 4.091e-03 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 87 | 208 | 4.102e-03 |
| GO:BP | GO:0033002 | muscle cell proliferation | 87 | 208 | 4.102e-03 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 39 | 79 | 4.152e-03 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 37 | 74 | 4.238e-03 |
| GO:BP | GO:0050919 | negative chemotaxis | 26 | 47 | 4.266e-03 |
| GO:BP | GO:0016126 | sterol biosynthetic process | 35 | 69 | 4.266e-03 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 64 | 145 | 4.307e-03 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 49 | 105 | 4.320e-03 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 118 | 296 | 4.320e-03 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 99 | 242 | 4.322e-03 |
| GO:BP | GO:0048753 | pigment granule organization | 23 | 40 | 4.334e-03 |
| GO:BP | GO:0006302 | double-strand break repair | 126 | 319 | 4.334e-03 |
| GO:BP | GO:0007416 | synapse assembly | 105 | 259 | 4.334e-03 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 23 | 40 | 4.334e-03 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 119 | 299 | 4.383e-03 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 42 | 87 | 4.542e-03 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 42 | 87 | 4.542e-03 |
| GO:BP | GO:0048878 | chemical homeostasis | 368 | 1043 | 4.568e-03 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 19 | 31 | 4.592e-03 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 19 | 31 | 4.592e-03 |
| GO:BP | GO:0009299 | mRNA transcription | 28 | 52 | 4.592e-03 |
| GO:BP | GO:0009266 | response to temperature stimulus | 73 | 170 | 4.680e-03 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 30 | 57 | 4.841e-03 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 38 | 77 | 4.860e-03 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 32 | 62 | 4.969e-03 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 34 | 67 | 5.002e-03 |
| GO:BP | GO:0039531 | regulation of cytoplasmic pattern recognition receptor signaling pathway | 54 | 119 | 5.209e-03 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 18 | 29 | 5.214e-03 |
| GO:BP | GO:0051235 | maintenance of location | 90 | 218 | 5.368e-03 |
| GO:BP | GO:0003158 | endothelium development | 61 | 138 | 5.451e-03 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 49 | 106 | 5.556e-03 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 58 | 130 | 5.562e-03 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 39 | 80 | 5.564e-03 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 39 | 80 | 5.564e-03 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 29 | 55 | 5.709e-03 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 29 | 55 | 5.709e-03 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 24 | 43 | 5.802e-03 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 24 | 43 | 5.802e-03 |
| GO:BP | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion | 21 | 36 | 5.819e-03 |
| GO:BP | GO:0009251 | glucan catabolic process | 17 | 27 | 5.838e-03 |
| GO:BP | GO:0009648 | photoperiodism | 17 | 27 | 5.838e-03 |
| GO:BP | GO:1904153 | negative regulation of retrograde protein transport, ER to cytosol | 8 | 9 | 5.838e-03 |
| GO:BP | GO:0034214 | protein hexamerization | 8 | 9 | 5.838e-03 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 74 | 174 | 5.908e-03 |
| GO:BP | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway | 74 | 174 | 5.908e-03 |
| GO:BP | GO:0002682 | regulation of immune system process | 541 | 1581 | 6.027e-03 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 45 | 96 | 6.301e-03 |
| GO:BP | GO:0007097 | nuclear migration | 16 | 25 | 6.523e-03 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 97 | 239 | 6.523e-03 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 16 | 25 | 6.523e-03 |
| GO:BP | GO:0030859 | polarized epithelial cell differentiation | 16 | 25 | 6.523e-03 |
| GO:BP | GO:1901659 | glycosyl compound biosynthetic process | 16 | 25 | 6.523e-03 |
| GO:BP | GO:0030148 | sphingolipid biosynthetic process | 51 | 112 | 6.538e-03 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 366 | 1042 | 6.600e-03 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 43 | 91 | 6.666e-03 |
| GO:BP | GO:0016358 | dendrite development | 98 | 242 | 6.679e-03 |
| GO:BP | GO:0008361 | regulation of cell size | 77 | 183 | 6.684e-03 |
| GO:BP | GO:0034405 | response to fluid shear stress | 20 | 34 | 6.684e-03 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 200 | 540 | 6.838e-03 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 34 | 68 | 6.866e-03 |
| GO:BP | GO:0022029 | telencephalon cell migration | 32 | 63 | 6.922e-03 |
| GO:BP | GO:0051897 | positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 78 | 186 | 6.944e-03 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 55 | 123 | 6.980e-03 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 49 | 107 | 7.018e-03 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 62 | 142 | 7.050e-03 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 62 | 142 | 7.050e-03 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 304 | 854 | 7.241e-03 |
| GO:BP | GO:0042063 | gliogenesis | 136 | 352 | 7.298e-03 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 102 | 254 | 7.304e-03 |
| GO:BP | GO:0030641 | regulation of cellular pH | 44 | 94 | 7.337e-03 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 25 | 46 | 7.384e-03 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 25 | 46 | 7.384e-03 |
| GO:BP | GO:0001825 | blastocyst formation | 25 | 46 | 7.384e-03 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 80 | 192 | 7.436e-03 |
| GO:BP | GO:0010720 | positive regulation of cell development | 172 | 458 | 7.593e-03 |
| GO:BP | GO:0046710 | GDP metabolic process | 9 | 11 | 7.602e-03 |
| GO:BP | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway | 9 | 11 | 7.602e-03 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 37 | 76 | 7.611e-03 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 37 | 76 | 7.611e-03 |
| GO:BP | GO:0099022 | vesicle tethering | 19 | 32 | 7.633e-03 |
| GO:BP | GO:0071514 | genomic imprinting | 14 | 21 | 7.748e-03 |
| GO:BP | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 22 | 39 | 7.748e-03 |
| GO:BP | GO:0048009 | insulin-like growth factor receptor signaling pathway | 27 | 51 | 7.748e-03 |
| GO:BP | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 27 | 51 | 7.748e-03 |
| GO:BP | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | 14 | 21 | 7.748e-03 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 22 | 39 | 7.748e-03 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 22 | 39 | 7.748e-03 |
| GO:BP | GO:1901661 | quinone metabolic process | 14 | 21 | 7.748e-03 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 22 | 39 | 7.748e-03 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 64 | 148 | 7.754e-03 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 35 | 71 | 7.788e-03 |
| GO:BP | GO:0030032 | lamellipodium assembly | 35 | 71 | 7.788e-03 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 77 | 184 | 7.830e-03 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 57 | 129 | 7.830e-03 |
| GO:BP | GO:0042221 | response to chemical | 1276 | 3909 | 7.830e-03 |
| GO:BP | GO:0009306 | protein secretion | 145 | 379 | 7.830e-03 |
| GO:BP | GO:0021885 | forebrain cell migration | 33 | 66 | 7.934e-03 |
| GO:BP | GO:1990928 | response to amino acid starvation | 29 | 56 | 7.934e-03 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 54 | 121 | 7.969e-03 |
| GO:BP | GO:0051898 | negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 31 | 61 | 7.990e-03 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 61 | 140 | 8.007e-03 |
| GO:BP | GO:0007098 | centrosome cycle | 61 | 140 | 8.007e-03 |
| GO:BP | GO:0048675 | axon extension | 51 | 113 | 8.036e-03 |
| GO:BP | GO:0006970 | response to osmotic stress | 40 | 84 | 8.055e-03 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 65 | 151 | 8.081e-03 |
| GO:BP | GO:1903232 | melanosome assembly | 13 | 19 | 8.188e-03 |
| GO:BP | GO:0046931 | pore complex assembly | 13 | 19 | 8.188e-03 |
| GO:BP | GO:0045945 | positive regulation of transcription by RNA polymerase III | 13 | 19 | 8.188e-03 |
| GO:BP | GO:0097284 | hepatocyte apoptotic process | 13 | 19 | 8.188e-03 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 118 | 301 | 8.288e-03 |
| GO:BP | GO:0036295 | cellular response to increased oxygen levels | 10 | 13 | 8.378e-03 |
| GO:BP | GO:0010421 | hydrogen peroxide-mediated programmed cell death | 10 | 13 | 8.378e-03 |
| GO:BP | GO:0061795 | Golgi lumen acidification | 10 | 13 | 8.378e-03 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 43 | 92 | 8.385e-03 |
| GO:BP | GO:0051051 | negative regulation of transport | 156 | 412 | 8.425e-03 |
| GO:BP | GO:0034205 | amyloid-beta formation | 24 | 44 | 8.429e-03 |
| GO:BP | GO:0010907 | positive regulation of glucose metabolic process | 24 | 44 | 8.429e-03 |
| GO:BP | GO:0090114 | COPII-coated vesicle budding | 24 | 44 | 8.429e-03 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 55 | 124 | 8.429e-03 |
| GO:BP | GO:0046112 | nucleobase biosynthetic process | 12 | 17 | 8.489e-03 |
| GO:BP | GO:0044790 | suppression of viral release by host | 12 | 17 | 8.489e-03 |
| GO:BP | GO:0036508 | protein alpha-1,2-demannosylation | 12 | 17 | 8.489e-03 |
| GO:BP | GO:0036507 | protein demannosylation | 12 | 17 | 8.489e-03 |
| GO:BP | GO:0090136 | epithelial cell-cell adhesion | 12 | 17 | 8.489e-03 |
| GO:BP | GO:0070071 | proton-transporting two-sector ATPase complex assembly | 12 | 17 | 8.489e-03 |
| GO:BP | GO:0090083 | regulation of inclusion body assembly | 12 | 17 | 8.489e-03 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 18 | 30 | 8.493e-03 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 18 | 30 | 8.493e-03 |
| GO:BP | GO:0061684 | chaperone-mediated autophagy | 11 | 15 | 8.572e-03 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 11 | 15 | 8.572e-03 |
| GO:BP | GO:0006999 | nuclear pore organization | 11 | 15 | 8.572e-03 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 11 | 15 | 8.572e-03 |
| GO:BP | GO:0001503 | ossification | 162 | 430 | 8.618e-03 |
| GO:BP | GO:1904760 | regulation of myofibroblast differentiation | 6 | 6 | 8.707e-03 |
| GO:BP | GO:0010935 | regulation of macrophage cytokine production | 21 | 37 | 8.707e-03 |
| GO:BP | GO:0010934 | macrophage cytokine production | 21 | 37 | 8.707e-03 |
| GO:BP | GO:0010616 | negative regulation of cardiac muscle adaptation | 6 | 6 | 8.707e-03 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 67 | 157 | 8.707e-03 |
| GO:BP | GO:1903243 | negative regulation of cardiac muscle hypertrophy in response to stress | 6 | 6 | 8.707e-03 |
| GO:BP | GO:0048205 | COPI coating of Golgi vesicle | 6 | 6 | 8.707e-03 |
| GO:BP | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region | 6 | 6 | 8.707e-03 |
| GO:BP | GO:0046826 | negative regulation of protein export from nucleus | 6 | 6 | 8.707e-03 |
| GO:BP | GO:0048200 | Golgi transport vesicle coating | 6 | 6 | 8.707e-03 |
| GO:BP | GO:0035964 | COPI-coated vesicle budding | 6 | 6 | 8.707e-03 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 21 | 37 | 8.707e-03 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 26 | 49 | 8.793e-03 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 34 | 69 | 8.867e-03 |
| GO:BP | GO:1990138 | neuron projection extension | 72 | 171 | 8.896e-03 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 77 | 185 | 8.992e-03 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 44 | 95 | 8.992e-03 |
| GO:BP | GO:0016125 | sterol metabolic process | 68 | 160 | 9.027e-03 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 32 | 64 | 9.045e-03 |
| GO:BP | GO:1901342 | regulation of vasculature development | 117 | 299 | 9.064e-03 |
| GO:BP | GO:0072577 | endothelial cell apoptotic process | 30 | 59 | 9.115e-03 |
| GO:BP | GO:0009451 | RNA modification | 73 | 174 | 9.224e-03 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 190 | 514 | 9.410e-03 |
| GO:BP | GO:0010212 | response to ionizing radiation | 61 | 141 | 9.483e-03 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 65 | 152 | 9.492e-03 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 65 | 152 | 9.492e-03 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 17 | 28 | 9.562e-03 |
| GO:BP | GO:0051453 | regulation of intracellular pH | 42 | 90 | 9.574e-03 |
| GO:BP | GO:0009611 | response to wounding | 208 | 568 | 9.576e-03 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 97 | 242 | 9.634e-03 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 23 | 42 | 9.638e-03 |
| GO:BP | GO:1990776 | response to angiotensin | 23 | 42 | 9.638e-03 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 51 | 114 | 9.716e-03 |
| GO:BP | GO:0051651 | maintenance of location in cell | 85 | 208 | 9.716e-03 |
| GO:BP | GO:0045926 | negative regulation of growth | 92 | 228 | 9.978e-03 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 20 | 35 | 1.007e-02 |
| GO:BP | GO:0030100 | regulation of endocytosis | 116 | 297 | 1.015e-02 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 71 | 169 | 1.015e-02 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 25 | 47 | 1.028e-02 |
| GO:BP | GO:0050792 | regulation of viral process | 67 | 158 | 1.033e-02 |
| GO:BP | GO:0051225 | spindle assembly | 59 | 136 | 1.035e-02 |
| GO:BP | GO:0044091 | membrane biogenesis | 33 | 67 | 1.036e-02 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 216 | 593 | 1.046e-02 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 72 | 172 | 1.051e-02 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 72 | 172 | 1.051e-02 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 27 | 52 | 1.058e-02 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 145 | 382 | 1.061e-02 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 95 | 237 | 1.062e-02 |
| GO:BP | GO:0034332 | adherens junction organization | 29 | 57 | 1.065e-02 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 29 | 57 | 1.065e-02 |
| GO:BP | GO:0030041 | actin filament polymerization | 68 | 161 | 1.071e-02 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 147 | 388 | 1.072e-02 |
| GO:BP | GO:1900078 | positive regulation of cellular response to insulin stimulus | 16 | 26 | 1.077e-02 |
| GO:BP | GO:0019081 | viral translation | 16 | 26 | 1.077e-02 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 96 | 240 | 1.080e-02 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 64 | 150 | 1.081e-02 |
| GO:BP | GO:0001764 | neuron migration | 78 | 189 | 1.082e-02 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 73 | 175 | 1.082e-02 |
| GO:BP | GO:0003170 | heart valve development | 36 | 75 | 1.121e-02 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 22 | 40 | 1.121e-02 |
| GO:BP | GO:0048588 | developmental cell growth | 91 | 226 | 1.121e-02 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 189 | 513 | 1.123e-02 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 54 | 123 | 1.168e-02 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 34 | 70 | 1.171e-02 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 34 | 70 | 1.171e-02 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 62 | 145 | 1.191e-02 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 62 | 145 | 1.191e-02 |
| GO:BP | GO:0018279 | protein N-linked glycosylation via asparagine | 24 | 45 | 1.198e-02 |
| GO:BP | GO:0035196 | miRNA processing | 24 | 45 | 1.198e-02 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 32 | 65 | 1.211e-02 |
| GO:BP | GO:1901673 | regulation of mitotic spindle assembly | 15 | 24 | 1.212e-02 |
| GO:BP | GO:0016311 | dephosphorylation | 88 | 218 | 1.212e-02 |
| GO:BP | GO:0071359 | cellular response to dsRNA | 15 | 24 | 1.212e-02 |
| GO:BP | GO:1904893 | negative regulation of receptor signaling pathway via STAT | 15 | 24 | 1.212e-02 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 45 | 99 | 1.222e-02 |
| GO:BP | GO:0044788 | modulation by host of viral process | 26 | 50 | 1.233e-02 |
| GO:BP | GO:0021795 | cerebral cortex cell migration | 26 | 50 | 1.233e-02 |
| GO:BP | GO:0032506 | cytokinetic process | 26 | 50 | 1.233e-02 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 30 | 60 | 1.233e-02 |
| GO:BP | GO:0045444 | fat cell differentiation | 96 | 241 | 1.238e-02 |
| GO:BP | GO:0043489 | RNA stabilization | 37 | 78 | 1.244e-02 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 114 | 293 | 1.258e-02 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 105 | 267 | 1.261e-02 |
| GO:BP | GO:0055006 | cardiac cell development | 40 | 86 | 1.294e-02 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 21 | 38 | 1.306e-02 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 35 | 73 | 1.311e-02 |
| GO:BP | GO:0106027 | neuron projection organization | 43 | 94 | 1.315e-02 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 18 | 31 | 1.337e-02 |
| GO:BP | GO:0019082 | viral protein processing | 18 | 31 | 1.337e-02 |
| GO:BP | GO:0034067 | protein localization to Golgi apparatus | 18 | 31 | 1.337e-02 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 57 | 132 | 1.366e-02 |
| GO:BP | GO:0097485 | neuron projection guidance | 95 | 239 | 1.402e-02 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 31 | 63 | 1.419e-02 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 31 | 63 | 1.419e-02 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 31 | 63 | 1.419e-02 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 31 | 63 | 1.419e-02 |
| GO:BP | GO:0003012 | muscle system process | 163 | 438 | 1.419e-02 |
| GO:BP | GO:0035383 | thioester metabolic process | 44 | 97 | 1.421e-02 |
| GO:BP | GO:0006637 | acyl-CoA metabolic process | 44 | 97 | 1.421e-02 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 7 | 8 | 1.438e-02 |
| GO:BP | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway | 7 | 8 | 1.438e-02 |
| GO:BP | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation | 7 | 8 | 1.438e-02 |
| GO:BP | GO:0048227 | plasma membrane to endosome transport | 7 | 8 | 1.438e-02 |
| GO:BP | GO:0061724 | lipophagy | 7 | 8 | 1.438e-02 |
| GO:BP | GO:0070973 | protein localization to endoplasmic reticulum exit site | 7 | 8 | 1.438e-02 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 25 | 48 | 1.443e-02 |
| GO:BP | GO:0071763 | nuclear membrane organization | 25 | 48 | 1.443e-02 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 25 | 48 | 1.443e-02 |
| GO:BP | GO:0019083 | viral transcription | 27 | 53 | 1.456e-02 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 27 | 53 | 1.456e-02 |
| GO:BP | GO:0006684 | sphingomyelin metabolic process | 13 | 20 | 1.500e-02 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 48 | 108 | 1.500e-02 |
| GO:BP | GO:0090630 | activation of GTPase activity | 20 | 36 | 1.517e-02 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 20 | 36 | 1.517e-02 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 20 | 36 | 1.517e-02 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 926 | 2813 | 1.518e-02 |
| GO:BP | GO:0000041 | transition metal ion transport | 45 | 100 | 1.518e-02 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 45 | 100 | 1.518e-02 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 17 | 29 | 1.533e-02 |
| GO:BP | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 17 | 29 | 1.533e-02 |
| GO:BP | GO:0001649 | osteoblast differentiation | 93 | 234 | 1.534e-02 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 52 | 119 | 1.542e-02 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 69 | 166 | 1.544e-02 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 376 | 1087 | 1.571e-02 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 32 | 66 | 1.597e-02 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 37 | 79 | 1.600e-02 |
| GO:BP | GO:0097494 | regulation of vesicle size | 12 | 18 | 1.633e-02 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 12 | 18 | 1.633e-02 |
| GO:BP | GO:2000641 | regulation of early endosome to late endosome transport | 12 | 18 | 1.633e-02 |
| GO:BP | GO:0006743 | ubiquinone metabolic process | 12 | 18 | 1.633e-02 |
| GO:BP | GO:0032239 | regulation of nucleobase-containing compound transport | 12 | 18 | 1.633e-02 |
| GO:BP | GO:0019896 | axonal transport of mitochondrion | 12 | 18 | 1.633e-02 |
| GO:BP | GO:0000028 | ribosomal small subunit assembly | 12 | 18 | 1.633e-02 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 22 | 41 | 1.633e-02 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 30 | 61 | 1.649e-02 |
| GO:BP | GO:0019318 | hexose metabolic process | 90 | 226 | 1.663e-02 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 62 | 147 | 1.686e-02 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 24 | 46 | 1.688e-02 |
| GO:BP | GO:0006473 | protein acetylation | 28 | 56 | 1.689e-02 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 35 | 74 | 1.689e-02 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 35 | 74 | 1.689e-02 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 26 | 51 | 1.702e-02 |
| GO:BP | GO:0030048 | actin filament-based movement | 58 | 136 | 1.704e-02 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 110 | 284 | 1.719e-02 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 8 | 10 | 1.721e-02 |
| GO:BP | GO:1902513 | regulation of organelle transport along microtubule | 8 | 10 | 1.721e-02 |
| GO:BP | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway | 8 | 10 | 1.721e-02 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 47 | 106 | 1.721e-02 |
| GO:BP | GO:1904667 | negative regulation of ubiquitin protein ligase activity | 8 | 10 | 1.721e-02 |
| GO:BP | GO:0060052 | neurofilament cytoskeleton organization | 8 | 10 | 1.721e-02 |
| GO:BP | GO:1904814 | regulation of protein localization to chromosome, telomeric region | 8 | 10 | 1.721e-02 |
| GO:BP | GO:1901298 | regulation of hydrogen peroxide-mediated programmed cell death | 8 | 10 | 1.721e-02 |
| GO:BP | GO:0048194 | Golgi vesicle budding | 8 | 10 | 1.721e-02 |
| GO:BP | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum | 8 | 10 | 1.721e-02 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 11 | 16 | 1.736e-02 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 19 | 34 | 1.736e-02 |
| GO:BP | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 11 | 16 | 1.736e-02 |
| GO:BP | GO:0043201 | response to L-leucine | 11 | 16 | 1.736e-02 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 16 | 27 | 1.736e-02 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 19 | 34 | 1.736e-02 |
| GO:BP | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining | 11 | 16 | 1.736e-02 |
| GO:BP | GO:0030033 | microvillus assembly | 11 | 16 | 1.736e-02 |
| GO:BP | GO:0018198 | peptidyl-cysteine modification | 19 | 34 | 1.736e-02 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 181 | 494 | 1.736e-02 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 33 | 69 | 1.758e-02 |
| GO:BP | GO:0007411 | axon guidance | 94 | 238 | 1.758e-02 |
| GO:BP | GO:0060429 | epithelium development | 424 | 1238 | 1.781e-02 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 64 | 153 | 1.790e-02 |
| GO:BP | GO:0097468 | programmed cell death in response to reactive oxygen species | 10 | 14 | 1.806e-02 |
| GO:BP | GO:0060816 | random inactivation of X chromosome | 10 | 14 | 1.806e-02 |
| GO:BP | GO:0007008 | outer mitochondrial membrane organization | 10 | 14 | 1.806e-02 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 10 | 14 | 1.806e-02 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 10 | 14 | 1.806e-02 |
| GO:BP | GO:0001921 | positive regulation of receptor recycling | 10 | 14 | 1.806e-02 |
| GO:BP | GO:0045040 | protein insertion into mitochondrial outer membrane | 10 | 14 | 1.806e-02 |
| GO:BP | GO:0035358 | regulation of peroxisome proliferator activated receptor signaling pathway | 10 | 14 | 1.806e-02 |
| GO:BP | GO:0006006 | glucose metabolic process | 75 | 184 | 1.808e-02 |
| GO:BP | GO:0034139 | regulation of toll-like receptor 3 signaling pathway | 9 | 12 | 1.810e-02 |
| GO:BP | GO:1905671 | regulation of lysosome organization | 9 | 12 | 1.810e-02 |
| GO:BP | GO:0010172 | embryonic body morphogenesis | 9 | 12 | 1.810e-02 |
| GO:BP | GO:0061723 | glycophagy | 9 | 12 | 1.810e-02 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 31 | 64 | 1.828e-02 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 98 | 250 | 1.860e-02 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 21 | 39 | 1.869e-02 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 27 | 54 | 1.949e-02 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 23 | 44 | 1.949e-02 |
| GO:BP | GO:0044273 | sulfur compound catabolic process | 23 | 44 | 1.949e-02 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 34 | 72 | 1.949e-02 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 27 | 54 | 1.949e-02 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 102 | 262 | 1.965e-02 |
| GO:BP | GO:0030521 | androgen receptor signaling pathway | 25 | 49 | 1.965e-02 |
| GO:BP | GO:0042073 | intraciliary transport | 25 | 49 | 1.965e-02 |
| GO:BP | GO:0051882 | mitochondrial depolarization | 15 | 25 | 1.976e-02 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 15 | 25 | 1.976e-02 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 15 | 25 | 1.976e-02 |
| GO:BP | GO:0070861 | regulation of protein exit from endoplasmic reticulum | 15 | 25 | 1.976e-02 |
| GO:BP | GO:0060155 | platelet dense granule organization | 15 | 25 | 1.976e-02 |
| GO:BP | GO:0007059 | chromosome segregation | 158 | 427 | 2.005e-02 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 18 | 32 | 2.009e-02 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 63 | 151 | 2.032e-02 |
| GO:BP | GO:0070050 | neuron cellular homeostasis | 32 | 67 | 2.048e-02 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 97 | 248 | 2.085e-02 |
| GO:BP | GO:0008360 | regulation of cell shape | 59 | 140 | 2.085e-02 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 89 | 225 | 2.102e-02 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 89 | 225 | 2.102e-02 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 44 | 99 | 2.139e-02 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 35 | 75 | 2.139e-02 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 30 | 62 | 2.143e-02 |
| GO:BP | GO:0008347 | glial cell migration | 30 | 62 | 2.143e-02 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 30 | 62 | 2.143e-02 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 30 | 62 | 2.143e-02 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 70 | 171 | 2.143e-02 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 60 | 143 | 2.159e-02 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 279 | 794 | 2.161e-02 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 38 | 83 | 2.165e-02 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 38 | 83 | 2.165e-02 |
| GO:BP | GO:0061448 | connective tissue development | 112 | 292 | 2.176e-02 |
| GO:BP | GO:0071539 | protein localization to centrosome | 20 | 37 | 2.180e-02 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 28 | 57 | 2.216e-02 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 28 | 57 | 2.216e-02 |
| GO:BP | GO:0051452 | intracellular pH reduction | 28 | 57 | 2.216e-02 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 28 | 57 | 2.216e-02 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 28 | 57 | 2.216e-02 |
| GO:BP | GO:0008015 | blood circulation | 188 | 518 | 2.219e-02 |
| GO:BP | GO:0000963 | mitochondrial RNA processing | 14 | 23 | 2.244e-02 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 14 | 23 | 2.244e-02 |
| GO:BP | GO:0046823 | negative regulation of nucleocytoplasmic transport | 14 | 23 | 2.244e-02 |
| GO:BP | GO:2000114 | regulation of establishment of cell polarity | 14 | 23 | 2.244e-02 |
| GO:BP | GO:0033962 | P-body assembly | 14 | 23 | 2.244e-02 |
| GO:BP | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | 33 | 70 | 2.252e-02 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 22 | 42 | 2.258e-02 |
| GO:BP | GO:0042307 | positive regulation of protein import into nucleus | 22 | 42 | 2.258e-02 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 22 | 42 | 2.258e-02 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 22 | 42 | 2.258e-02 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 26 | 52 | 2.260e-02 |
| GO:BP | GO:0006812 | monoatomic cation transport | 368 | 1069 | 2.281e-02 |
| GO:BP | GO:0047496 | vesicle transport along microtubule | 24 | 47 | 2.281e-02 |
| GO:BP | GO:0021762 | substantia nigra development | 24 | 47 | 2.281e-02 |
| GO:BP | GO:0061337 | cardiac conduction | 42 | 94 | 2.296e-02 |
| GO:BP | GO:0007155 | cell adhesion | 516 | 1530 | 2.303e-02 |
| GO:BP | GO:1903077 | negative regulation of protein localization to plasma membrane | 17 | 30 | 2.313e-02 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 39 | 86 | 2.316e-02 |
| GO:BP | GO:0048864 | stem cell development | 39 | 86 | 2.316e-02 |
| GO:BP | GO:0008643 | carbohydrate transport | 58 | 138 | 2.346e-02 |
| GO:BP | GO:2001197 | basement membrane assembly involved in embryonic body morphogenesis | 5 | 5 | 2.346e-02 |
| GO:BP | GO:0038027 | apolipoprotein A-I-mediated signaling pathway | 5 | 5 | 2.346e-02 |
| GO:BP | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis | 5 | 5 | 2.346e-02 |
| GO:BP | GO:2000435 | negative regulation of protein neddylation | 5 | 5 | 2.346e-02 |
| GO:BP | GO:0043335 | protein unfolding | 5 | 5 | 2.346e-02 |
| GO:BP | GO:0070370 | cellular heat acclimation | 5 | 5 | 2.346e-02 |
| GO:BP | GO:0010286 | heat acclimation | 5 | 5 | 2.346e-02 |
| GO:BP | GO:1904504 | positive regulation of lipophagy | 5 | 5 | 2.346e-02 |
| GO:BP | GO:1905167 | positive regulation of lysosomal protein catabolic process | 5 | 5 | 2.346e-02 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 58 | 138 | 2.346e-02 |
| GO:BP | GO:1904261 | positive regulation of basement membrane assembly involved in embryonic body morphogenesis | 5 | 5 | 2.346e-02 |
| GO:BP | GO:1904502 | regulation of lipophagy | 5 | 5 | 2.346e-02 |
| GO:BP | GO:0002520 | immune system development | 81 | 203 | 2.346e-02 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 31 | 65 | 2.347e-02 |
| GO:BP | GO:0001890 | placenta development | 63 | 152 | 2.347e-02 |
| GO:BP | GO:0006885 | regulation of pH | 46 | 105 | 2.354e-02 |
| GO:BP | GO:0043414 | macromolecule methylation | 54 | 127 | 2.375e-02 |
| GO:BP | GO:1904659 | D-glucose transmembrane transport | 50 | 116 | 2.385e-02 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 59 | 141 | 2.424e-02 |
| GO:BP | GO:0021766 | hippocampus development | 43 | 97 | 2.424e-02 |
| GO:BP | GO:0002218 | activation of innate immune response | 116 | 305 | 2.480e-02 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 274 | 781 | 2.490e-02 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 51 | 119 | 2.502e-02 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 51 | 119 | 2.502e-02 |
| GO:BP | GO:1902414 | protein localization to cell junction | 51 | 119 | 2.502e-02 |
| GO:BP | GO:0019068 | virion assembly | 19 | 35 | 2.502e-02 |
| GO:BP | GO:0010288 | response to lead ion | 13 | 21 | 2.502e-02 |
| GO:BP | GO:0016556 | mRNA modification | 13 | 21 | 2.502e-02 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 19 | 35 | 2.502e-02 |
| GO:BP | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 19 | 35 | 2.502e-02 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 19 | 35 | 2.502e-02 |
| GO:BP | GO:0048147 | negative regulation of fibroblast proliferation | 19 | 35 | 2.502e-02 |
| GO:BP | GO:0044321 | response to leptin | 13 | 21 | 2.502e-02 |
| GO:BP | GO:2001135 | regulation of endocytic recycling | 13 | 21 | 2.502e-02 |
| GO:BP | GO:0060216 | definitive hemopoiesis | 13 | 21 | 2.502e-02 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 144 | 388 | 2.528e-02 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 27 | 55 | 2.545e-02 |
| GO:BP | GO:0140374 | antiviral innate immune response | 27 | 55 | 2.545e-02 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 27 | 55 | 2.545e-02 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 148 | 400 | 2.548e-02 |
| GO:BP | GO:0071346 | cellular response to type II interferon | 44 | 100 | 2.556e-02 |
| GO:BP | GO:1904646 | cellular response to amyloid-beta | 21 | 40 | 2.607e-02 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 21 | 40 | 2.607e-02 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 21 | 40 | 2.607e-02 |
| GO:BP | GO:0090148 | membrane fission | 23 | 45 | 2.639e-02 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 23 | 45 | 2.639e-02 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 16 | 28 | 2.642e-02 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 16 | 28 | 2.642e-02 |
| GO:BP | GO:0036296 | response to increased oxygen levels | 16 | 28 | 2.642e-02 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 16 | 28 | 2.642e-02 |
| GO:BP | GO:0030516 | regulation of axon extension | 38 | 84 | 2.647e-02 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 57 | 136 | 2.647e-02 |
| GO:BP | GO:0030832 | regulation of actin filament length | 62 | 150 | 2.649e-02 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 228 | 642 | 2.741e-02 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 139 | 374 | 2.745e-02 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 58 | 139 | 2.747e-02 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 360 | 1048 | 2.796e-02 |
| GO:BP | GO:0006144 | purine nucleobase metabolic process | 12 | 19 | 2.812e-02 |
| GO:BP | GO:0034404 | nucleobase-containing small molecule biosynthetic process | 12 | 19 | 2.812e-02 |
| GO:BP | GO:0090670 | RNA localization to Cajal body | 12 | 19 | 2.812e-02 |
| GO:BP | GO:0090672 | telomerase RNA localization | 12 | 19 | 2.812e-02 |
| GO:BP | GO:0090685 | RNA localization to nucleus | 12 | 19 | 2.812e-02 |
| GO:BP | GO:0009163 | nucleoside biosynthetic process | 12 | 19 | 2.812e-02 |
| GO:BP | GO:1902992 | negative regulation of amyloid precursor protein catabolic process | 12 | 19 | 2.812e-02 |
| GO:BP | GO:0090671 | telomerase RNA localization to Cajal body | 12 | 19 | 2.812e-02 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 28 | 58 | 2.866e-02 |
| GO:BP | GO:0008366 | axon ensheathment | 65 | 159 | 2.894e-02 |
| GO:BP | GO:0007272 | ensheathment of neurons | 65 | 159 | 2.894e-02 |
| GO:BP | GO:0021591 | ventricular system development | 18 | 33 | 2.917e-02 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 26 | 53 | 2.990e-02 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 31 | 66 | 3.027e-02 |
| GO:BP | GO:0021782 | glial cell development | 56 | 134 | 3.037e-02 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 56 | 134 | 3.037e-02 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 40 | 90 | 3.041e-02 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 67 | 165 | 3.043e-02 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 20 | 38 | 3.043e-02 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 24 | 48 | 3.043e-02 |
| GO:BP | GO:0007549 | sex-chromosome dosage compensation | 15 | 26 | 3.043e-02 |
| GO:BP | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 15 | 26 | 3.043e-02 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 271 | 775 | 3.043e-02 |
| GO:BP | GO:0048208 | COPII vesicle coating | 15 | 26 | 3.043e-02 |
| GO:BP | GO:0006515 | protein quality control for misfolded or incompletely synthesized proteins | 15 | 26 | 3.043e-02 |
| GO:BP | GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 15 | 26 | 3.043e-02 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 271 | 775 | 3.043e-02 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 24 | 48 | 3.043e-02 |
| GO:BP | GO:0101023 | vascular endothelial cell proliferation | 15 | 26 | 3.043e-02 |
| GO:BP | GO:1905562 | regulation of vascular endothelial cell proliferation | 15 | 26 | 3.043e-02 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 20 | 38 | 3.043e-02 |
| GO:BP | GO:0034453 | microtubule anchoring | 15 | 26 | 3.043e-02 |
| GO:BP | GO:0034331 | cell junction maintenance | 24 | 48 | 3.043e-02 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 20 | 38 | 3.043e-02 |
| GO:BP | GO:0048638 | regulation of developmental growth | 117 | 310 | 3.045e-02 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 74 | 185 | 3.050e-02 |
| GO:BP | GO:0042698 | ovulation cycle | 34 | 74 | 3.050e-02 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 37 | 82 | 3.050e-02 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 22 | 43 | 3.063e-02 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 22 | 43 | 3.063e-02 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 44 | 101 | 3.083e-02 |
| GO:BP | GO:0060996 | dendritic spine development | 44 | 101 | 3.083e-02 |
| GO:BP | GO:0006895 | Golgi to endosome transport | 11 | 17 | 3.108e-02 |
| GO:BP | GO:0045198 | establishment of epithelial cell apical/basal polarity | 11 | 17 | 3.108e-02 |
| GO:BP | GO:0007063 | regulation of sister chromatid cohesion | 11 | 17 | 3.108e-02 |
| GO:BP | GO:0034349 | glial cell apoptotic process | 11 | 17 | 3.108e-02 |
| GO:BP | GO:0030575 | nuclear body organization | 11 | 17 | 3.108e-02 |
| GO:BP | GO:0071709 | membrane assembly | 29 | 61 | 3.159e-02 |
| GO:BP | GO:0030901 | midbrain development | 41 | 93 | 3.184e-02 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 53 | 126 | 3.184e-02 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 41 | 93 | 3.184e-02 |
| GO:BP | GO:0043473 | pigmentation | 49 | 115 | 3.238e-02 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 49 | 115 | 3.238e-02 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 32 | 69 | 3.263e-02 |
| GO:BP | GO:0048255 | mRNA stabilization | 32 | 69 | 3.263e-02 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 32 | 69 | 3.263e-02 |
| GO:BP | GO:0007369 | gastrulation | 78 | 197 | 3.274e-02 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 87 | 223 | 3.289e-02 |
| GO:BP | GO:0042060 | wound healing | 158 | 433 | 3.348e-02 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 42 | 96 | 3.373e-02 |
| GO:BP | GO:0046685 | response to arsenic-containing substance | 17 | 31 | 3.373e-02 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 42 | 96 | 3.373e-02 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 17 | 31 | 3.373e-02 |
| GO:BP | GO:0061308 | cardiac neural crest cell development involved in heart development | 10 | 15 | 3.390e-02 |
| GO:BP | GO:0061502 | early endosome to recycling endosome transport | 6 | 7 | 3.390e-02 |
| GO:BP | GO:0050904 | diapedesis | 6 | 7 | 3.390e-02 |
| GO:BP | GO:0046831 | regulation of RNA export from nucleus | 10 | 15 | 3.390e-02 |
| GO:BP | GO:0042780 | tRNA 3’-end processing | 6 | 7 | 3.390e-02 |
| GO:BP | GO:0030091 | protein repair | 6 | 7 | 3.390e-02 |
| GO:BP | GO:0106057 | negative regulation of calcineurin-mediated signaling | 10 | 15 | 3.390e-02 |
| GO:BP | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 10 | 15 | 3.390e-02 |
| GO:BP | GO:1905279 | regulation of retrograde transport, endosome to Golgi | 6 | 7 | 3.390e-02 |
| GO:BP | GO:0061307 | cardiac neural crest cell differentiation involved in heart development | 10 | 15 | 3.390e-02 |
| GO:BP | GO:0048298 | positive regulation of isotype switching to IgA isotypes | 6 | 7 | 3.390e-02 |
| GO:BP | GO:0048296 | regulation of isotype switching to IgA isotypes | 6 | 7 | 3.390e-02 |
| GO:BP | GO:0033240 | positive regulation of amine metabolic process | 6 | 7 | 3.390e-02 |
| GO:BP | GO:0010715 | regulation of extracellular matrix disassembly | 10 | 15 | 3.390e-02 |
| GO:BP | GO:0072126 | positive regulation of glomerular mesangial cell proliferation | 6 | 7 | 3.390e-02 |
| GO:BP | GO:0033211 | adiponectin-activated signaling pathway | 6 | 7 | 3.390e-02 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 55 | 132 | 3.390e-02 |
| GO:BP | GO:0006672 | ceramide metabolic process | 46 | 107 | 3.390e-02 |
| GO:BP | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide | 6 | 7 | 3.390e-02 |
| GO:BP | GO:1901299 | negative regulation of hydrogen peroxide-mediated programmed cell death | 6 | 7 | 3.390e-02 |
| GO:BP | GO:0034334 | adherens junction maintenance | 6 | 7 | 3.390e-02 |
| GO:BP | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 25 | 51 | 3.416e-02 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 25 | 51 | 3.416e-02 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 25 | 51 | 3.416e-02 |
| GO:BP | GO:0045824 | negative regulation of innate immune response | 39 | 88 | 3.427e-02 |
| GO:BP | GO:0014896 | muscle hypertrophy | 39 | 88 | 3.427e-02 |
| GO:BP | GO:0007219 | Notch signaling pathway | 74 | 186 | 3.451e-02 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 14 | 24 | 3.453e-02 |
| GO:BP | GO:0036119 | response to platelet-derived growth factor | 14 | 24 | 3.453e-02 |
| GO:BP | GO:0051131 | chaperone-mediated protein complex assembly | 14 | 24 | 3.453e-02 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 14 | 24 | 3.453e-02 |
| GO:BP | GO:0046628 | positive regulation of insulin receptor signaling pathway | 14 | 24 | 3.453e-02 |
| GO:BP | GO:0051560 | mitochondrial calcium ion homeostasis | 14 | 24 | 3.453e-02 |
| GO:BP | GO:0002090 | regulation of receptor internalization | 33 | 72 | 3.477e-02 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 19 | 36 | 3.477e-02 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 375 | 1099 | 3.477e-02 |
| GO:BP | GO:0006414 | translational elongation | 36 | 80 | 3.477e-02 |
| GO:BP | GO:0006099 | tricarboxylic acid cycle | 19 | 36 | 3.477e-02 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 19 | 36 | 3.477e-02 |
| GO:BP | GO:0014032 | neural crest cell development | 36 | 80 | 3.477e-02 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 33 | 72 | 3.477e-02 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 23 | 46 | 3.486e-02 |
| GO:BP | GO:0035337 | fatty-acyl-CoA metabolic process | 21 | 41 | 3.524e-02 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 21 | 41 | 3.524e-02 |
| GO:BP | GO:0071233 | cellular response to L-leucine | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0038203 | TORC2 signaling | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0042761 | very long-chain fatty acid biosynthetic process | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0043558 | regulation of translational initiation in response to stress | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0060612 | adipose tissue development | 28 | 59 | 3.605e-02 |
| GO:BP | GO:0071679 | commissural neuron axon guidance | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0010666 | positive regulation of cardiac muscle cell apoptotic process | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0010766 | negative regulation of sodium ion transport | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0140588 | chromatin looping | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0017062 | respiratory chain complex III assembly | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0098935 | dendritic transport | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0010225 | response to UV-C | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 28 | 59 | 3.605e-02 |
| GO:BP | GO:1904380 | endoplasmic reticulum mannose trimming | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0034551 | mitochondrial respiratory chain complex III assembly | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0010663 | positive regulation of striated muscle cell apoptotic process | 9 | 13 | 3.605e-02 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 63 | 155 | 3.605e-02 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 63 | 155 | 3.605e-02 |
| GO:BP | GO:0031347 | regulation of defense response | 281 | 809 | 3.626e-02 |
| GO:BP | GO:0097061 | dendritic spine organization | 37 | 83 | 3.682e-02 |
| GO:BP | GO:0043954 | cellular component maintenance | 34 | 75 | 3.718e-02 |
| GO:BP | GO:0006930 | substrate-dependent cell migration, cell extension | 7 | 9 | 3.718e-02 |
| GO:BP | GO:0014745 | negative regulation of muscle adaptation | 7 | 9 | 3.718e-02 |
| GO:BP | GO:1901724 | positive regulation of cell proliferation involved in kidney development | 7 | 9 | 3.718e-02 |
| GO:BP | GO:1990481 | mRNA pseudouridine synthesis | 7 | 9 | 3.718e-02 |
| GO:BP | GO:1990314 | cellular response to insulin-like growth factor stimulus | 7 | 9 | 3.718e-02 |
| GO:BP | GO:0051561 | positive regulation of mitochondrial calcium ion concentration | 7 | 9 | 3.718e-02 |
| GO:BP | GO:0015886 | heme transport | 7 | 9 | 3.718e-02 |
| GO:BP | GO:0010998 | regulation of translational initiation by eIF2 alpha phosphorylation | 7 | 9 | 3.718e-02 |
| GO:BP | GO:0002191 | cap-dependent translational initiation | 7 | 9 | 3.718e-02 |
| GO:BP | GO:0007220 | Notch receptor processing | 7 | 9 | 3.718e-02 |
| GO:BP | GO:1903332 | regulation of protein folding | 7 | 9 | 3.718e-02 |
| GO:BP | GO:0009113 | purine nucleobase biosynthetic process | 7 | 9 | 3.718e-02 |
| GO:BP | GO:2000678 | negative regulation of transcription regulatory region DNA binding | 7 | 9 | 3.718e-02 |
| GO:BP | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation | 7 | 9 | 3.718e-02 |
| GO:BP | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway | 7 | 9 | 3.718e-02 |
| GO:BP | GO:0032201 | telomere maintenance via semi-conservative replication | 7 | 9 | 3.718e-02 |
| GO:BP | GO:0090085 | regulation of protein deubiquitination | 8 | 11 | 3.726e-02 |
| GO:BP | GO:0046129 | purine ribonucleoside biosynthetic process | 8 | 11 | 3.726e-02 |
| GO:BP | GO:0042451 | purine nucleoside biosynthetic process | 8 | 11 | 3.726e-02 |
| GO:BP | GO:0042455 | ribonucleoside biosynthetic process | 8 | 11 | 3.726e-02 |
| GO:BP | GO:0010807 | regulation of synaptic vesicle priming | 8 | 11 | 3.726e-02 |
| GO:BP | GO:1905383 | protein localization to presynapse | 8 | 11 | 3.726e-02 |
| GO:BP | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus | 8 | 11 | 3.726e-02 |
| GO:BP | GO:0006107 | oxaloacetate metabolic process | 8 | 11 | 3.726e-02 |
| GO:BP | GO:0099532 | synaptic vesicle endosomal processing | 8 | 11 | 3.726e-02 |
| GO:BP | GO:0070372 | regulation of ERK1 and ERK2 cascade | 104 | 274 | 3.726e-02 |
| GO:BP | GO:0006686 | sphingomyelin biosynthetic process | 8 | 11 | 3.726e-02 |
| GO:BP | GO:0023061 | signal release | 180 | 501 | 3.726e-02 |
| GO:BP | GO:0021954 | central nervous system neuron development | 41 | 94 | 3.734e-02 |
| GO:BP | GO:2000351 | regulation of endothelial cell apoptotic process | 26 | 54 | 3.734e-02 |
| GO:BP | GO:0060840 | artery development | 45 | 105 | 3.753e-02 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 16 | 29 | 3.772e-02 |
| GO:BP | GO:0035265 | organ growth | 66 | 164 | 3.772e-02 |
| GO:BP | GO:0072012 | glomerulus vasculature development | 16 | 29 | 3.772e-02 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 66 | 164 | 3.772e-02 |
| GO:BP | GO:0035872 | nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | 16 | 29 | 3.772e-02 |
| GO:BP | GO:0045116 | protein neddylation | 16 | 29 | 3.772e-02 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 16 | 29 | 3.772e-02 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 16 | 29 | 3.772e-02 |
| GO:BP | GO:0006811 | monoatomic ion transport | 433 | 1282 | 3.791e-02 |
| GO:BP | GO:0099536 | synaptic signaling | 280 | 807 | 3.813e-02 |
| GO:BP | GO:0009743 | response to carbohydrate | 94 | 245 | 3.830e-02 |
| GO:BP | GO:0009746 | response to hexose | 83 | 213 | 3.835e-02 |
| GO:BP | GO:0001824 | blastocyst development | 50 | 119 | 3.835e-02 |
| GO:BP | GO:0022406 | membrane docking | 38 | 86 | 3.839e-02 |
| GO:BP | GO:1900273 | positive regulation of long-term synaptic potentiation | 13 | 22 | 3.861e-02 |
| GO:BP | GO:0036120 | cellular response to platelet-derived growth factor stimulus | 13 | 22 | 3.861e-02 |
| GO:BP | GO:0032482 | Rab protein signal transduction | 13 | 22 | 3.861e-02 |
| GO:BP | GO:0061709 | reticulophagy | 13 | 22 | 3.861e-02 |
| GO:BP | GO:0150105 | protein localization to cell-cell junction | 13 | 22 | 3.861e-02 |
| GO:BP | GO:0016082 | synaptic vesicle priming | 13 | 22 | 3.861e-02 |
| GO:BP | GO:0009605 | response to external stimulus | 792 | 2415 | 3.862e-02 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 24 | 49 | 3.863e-02 |
| GO:BP | GO:0045010 | actin nucleation | 29 | 62 | 3.868e-02 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 61 | 150 | 3.868e-02 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 29 | 62 | 3.868e-02 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 32 | 70 | 3.929e-02 |
| GO:BP | GO:0048857 | neural nucleus development | 32 | 70 | 3.929e-02 |
| GO:BP | GO:0032376 | positive regulation of cholesterol transport | 18 | 34 | 3.943e-02 |
| GO:BP | GO:0098884 | postsynaptic neurotransmitter receptor internalization | 18 | 34 | 3.943e-02 |
| GO:BP | GO:0140058 | neuron projection arborization | 18 | 34 | 3.943e-02 |
| GO:BP | GO:0140239 | postsynaptic endocytosis | 18 | 34 | 3.943e-02 |
| GO:BP | GO:0032373 | positive regulation of sterol transport | 18 | 34 | 3.943e-02 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 18 | 34 | 3.943e-02 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 18 | 34 | 3.943e-02 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 51 | 122 | 3.943e-02 |
| GO:BP | GO:0009112 | nucleobase metabolic process | 18 | 34 | 3.943e-02 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 295 | 854 | 3.946e-02 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 62 | 153 | 3.946e-02 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 22 | 44 | 3.947e-02 |
| GO:BP | GO:0042398 | modified amino acid biosynthetic process | 22 | 44 | 3.947e-02 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 22 | 44 | 3.947e-02 |
| GO:BP | GO:0030258 | lipid modification | 77 | 196 | 3.963e-02 |
| GO:BP | GO:1905476 | negative regulation of protein localization to membrane | 20 | 39 | 3.988e-02 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 20 | 39 | 3.988e-02 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 20 | 39 | 3.988e-02 |
| GO:BP | GO:0099068 | postsynapse assembly | 39 | 89 | 4.015e-02 |
| GO:BP | GO:0061515 | myeloid cell development | 39 | 89 | 4.015e-02 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 39 | 89 | 4.015e-02 |
| GO:BP | GO:0008033 | tRNA processing | 57 | 139 | 4.023e-02 |
| GO:BP | GO:0030323 | respiratory tube development | 79 | 202 | 4.075e-02 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 146 | 400 | 4.111e-02 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 48 | 114 | 4.189e-02 |
| GO:BP | GO:0009749 | response to glucose | 81 | 208 | 4.191e-02 |
| GO:BP | GO:0021761 | limbic system development | 53 | 128 | 4.197e-02 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 30 | 65 | 4.197e-02 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 53 | 128 | 4.197e-02 |
| GO:BP | GO:0072348 | sulfur compound transport | 30 | 65 | 4.197e-02 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 302 | 877 | 4.281e-02 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 15 | 27 | 4.362e-02 |
| GO:BP | GO:0060343 | trabecula formation | 15 | 27 | 4.362e-02 |
| GO:BP | GO:0006929 | substrate-dependent cell migration | 15 | 27 | 4.362e-02 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 15 | 27 | 4.362e-02 |
| GO:BP | GO:0031468 | nuclear membrane reassembly | 15 | 27 | 4.362e-02 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 15 | 27 | 4.362e-02 |
| GO:BP | GO:0034284 | response to monosaccharide | 85 | 220 | 4.388e-02 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 37 | 84 | 4.398e-02 |
| GO:BP | GO:2000786 | positive regulation of autophagosome assembly | 12 | 20 | 4.405e-02 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 12 | 20 | 4.405e-02 |
| GO:BP | GO:0046782 | regulation of viral transcription | 12 | 20 | 4.405e-02 |
| GO:BP | GO:0090141 | positive regulation of mitochondrial fission | 12 | 20 | 4.405e-02 |
| GO:BP | GO:0051851 | host-mediated perturbation of symbiont process | 45 | 106 | 4.426e-02 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 55 | 134 | 4.446e-02 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 61 | 151 | 4.446e-02 |
| GO:BP | GO:0007052 | mitotic spindle organization | 55 | 134 | 4.446e-02 |
| GO:BP | GO:0035296 | regulation of tube diameter | 61 | 151 | 4.446e-02 |
| GO:BP | GO:0097746 | blood vessel diameter maintenance | 61 | 151 | 4.446e-02 |
| GO:BP | GO:0080164 | regulation of nitric oxide metabolic process | 28 | 60 | 4.482e-02 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 23 | 47 | 4.485e-02 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 23 | 47 | 4.485e-02 |
| GO:BP | GO:0051865 | protein autoubiquitination | 34 | 76 | 4.485e-02 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 31 | 68 | 4.531e-02 |
| GO:BP | GO:1904376 | negative regulation of protein localization to cell periphery | 17 | 32 | 4.612e-02 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 17 | 32 | 4.612e-02 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 894 | 2745 | 4.616e-02 |
| GO:BP | GO:0034341 | response to type II interferon | 51 | 123 | 4.627e-02 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 21 | 42 | 4.627e-02 |
| GO:BP | GO:0042552 | myelination | 63 | 157 | 4.644e-02 |
| GO:BP | GO:0045324 | late endosome to vacuole transport | 19 | 37 | 4.683e-02 |
| GO:BP | GO:1901655 | cellular response to ketone | 47 | 112 | 4.775e-02 |
| GO:BP | GO:0038066 | p38MAPK cascade | 26 | 55 | 4.775e-02 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 52 | 126 | 4.775e-02 |
| GO:BP | GO:0033209 | tumor necrosis factor-mediated signaling pathway | 47 | 112 | 4.775e-02 |
| GO:BP | GO:0006836 | neurotransmitter transport | 82 | 212 | 4.775e-02 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 35 | 79 | 4.780e-02 |
| GO:BP | GO:0002200 | somatic diversification of immune receptors | 35 | 79 | 4.780e-02 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 29 | 63 | 4.886e-02 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 29 | 63 | 4.886e-02 |
| GO:BP | GO:0006518 | peptide metabolic process | 29 | 63 | 4.886e-02 |
| KEGG | KEGG:04144 | Endocytosis | 147 | 248 | 1.209e-10 |
| KEGG | KEGG:05012 | Parkinson disease | 151 | 265 | 2.204e-09 |
| KEGG | KEGG:04142 | Lysosome | 84 | 131 | 1.592e-08 |
| KEGG | KEGG:05016 | Huntington disease | 166 | 305 | 1.592e-08 |
| KEGG | KEGG:03010 | Ribosome | 94 | 153 | 2.724e-08 |
| KEGG | KEGG:05010 | Alzheimer disease | 199 | 382 | 2.724e-08 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 237 | 474 | 7.445e-08 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 186 | 363 | 3.970e-07 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 98 | 168 | 3.977e-07 |
| KEGG | KEGG:05132 | Salmonella infection | 135 | 249 | 4.527e-07 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 110 | 202 | 6.832e-06 |
| KEGG | KEGG:04714 | Thermogenesis | 123 | 232 | 8.747e-06 |
| KEGG | KEGG:05171 | Coronavirus disease - COVID-19 | 122 | 231 | 1.184e-05 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 77 | 134 | 2.231e-05 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 116 | 221 | 2.963e-05 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 235 | 506 | 8.223e-05 |
| KEGG | KEGG:05020 | Prion disease | 136 | 271 | 8.270e-05 |
| KEGG | KEGG:04140 | Autophagy - animal | 89 | 165 | 1.006e-04 |
| KEGG | KEGG:05212 | Pancreatic cancer | 47 | 76 | 1.523e-04 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 75 | 137 | 2.464e-04 |
| KEGG | KEGG:05210 | Colorectal cancer | 51 | 86 | 3.132e-04 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 82 | 154 | 3.617e-04 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 56 | 98 | 5.136e-04 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 49 | 84 | 7.274e-04 |
| KEGG | KEGG:04148 | Efferocytosis | 81 | 156 | 1.070e-03 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 74 | 140 | 1.070e-03 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 64 | 118 | 1.070e-03 |
| KEGG | KEGG:04510 | Focal adhesion | 101 | 202 | 1.070e-03 |
| KEGG | KEGG:04137 | Mitophagy - animal | 57 | 103 | 1.194e-03 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 80 | 155 | 1.366e-03 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 40 | 68 | 2.122e-03 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 73 | 141 | 2.213e-03 |
| KEGG | KEGG:04218 | Cellular senescence | 79 | 155 | 2.324e-03 |
| KEGG | KEGG:04360 | Axon guidance | 90 | 181 | 2.608e-03 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 100 | 205 | 2.912e-03 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 41 | 72 | 3.992e-03 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 40 | 70 | 4.128e-03 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 20 | 29 | 4.181e-03 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 44 | 79 | 4.451e-03 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 43 | 77 | 4.529e-03 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 57 | 108 | 4.529e-03 |
| KEGG | KEGG:01522 | Endocrine resistance | 51 | 95 | 5.003e-03 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 106 | 223 | 5.282e-03 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 30 | 50 | 5.911e-03 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 42 | 76 | 6.379e-03 |
| KEGG | KEGG:05131 | Shigellosis | 115 | 246 | 6.497e-03 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 61 | 120 | 8.903e-03 |
| KEGG | KEGG:04520 | Adherens junction | 49 | 93 | 9.398e-03 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 45 | 84 | 9.398e-03 |
| KEGG | KEGG:05213 | Endometrial cancer | 33 | 58 | 1.050e-02 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 47 | 89 | 1.055e-02 |
| KEGG | KEGG:04146 | Peroxisome | 44 | 83 | 1.306e-02 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 26 | 44 | 1.488e-02 |
| KEGG | KEGG:01100 | Metabolic pathways | 615 | 1537 | 1.944e-02 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 49 | 96 | 1.994e-02 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 134 | 301 | 2.192e-02 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 59 | 120 | 2.354e-02 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 91 | 197 | 2.616e-02 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 37 | 70 | 2.638e-02 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 54 | 109 | 2.638e-02 |
| KEGG | KEGG:00604 | Glycosphingolipid biosynthesis - ganglio series | 11 | 15 | 2.638e-02 |
| KEGG | KEGG:04072 | Phospholipase D signaling pathway | 70 | 147 | 2.690e-02 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 50 | 100 | 2.703e-02 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 96 | 210 | 2.703e-02 |
| KEGG | KEGG:03060 | Protein export | 19 | 31 | 2.703e-02 |
| KEGG | KEGG:05214 | Glioma | 39 | 75 | 2.766e-02 |
| KEGG | KEGG:00531 | Glycosaminoglycan degradation | 13 | 19 | 2.766e-02 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 25 | 44 | 2.879e-02 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 48 | 96 | 2.919e-02 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 33 | 62 | 3.147e-02 |
| KEGG | KEGG:03020 | RNA polymerase | 20 | 34 | 3.716e-02 |
| KEGG | KEGG:04210 | Apoptosis | 64 | 135 | 3.716e-02 |
| KEGG | KEGG:04540 | Gap junction | 44 | 88 | 3.831e-02 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 52 | 107 | 3.973e-02 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 143 | 330 | 3.973e-02 |
| KEGG | KEGG:04550 | Signaling pathways regulating pluripotency of stem cells | 67 | 143 | 4.127e-02 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 32 | 61 | 4.242e-02 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 44 | 89 | 4.601e-02 |
| KEGG | KEGG:04931 | Insulin resistance | 52 | 108 | 4.710e-02 |
# write.csv(table_motif1_GOKEGG_genes, "data/new/table_motif1_GOKEGG_genes.csv")
#GO:BP
table_motif1_genes_GOBP_RUV <- table_motif1_GOKEGG_genes_RUV %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
# saveRDS(table_motif1_genes_GOBP, "data/table_motif1_genes_GOBP.RDS")
table_motif1_genes_GOBP_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("M1 Enriched GO:BP Terms RUVs")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))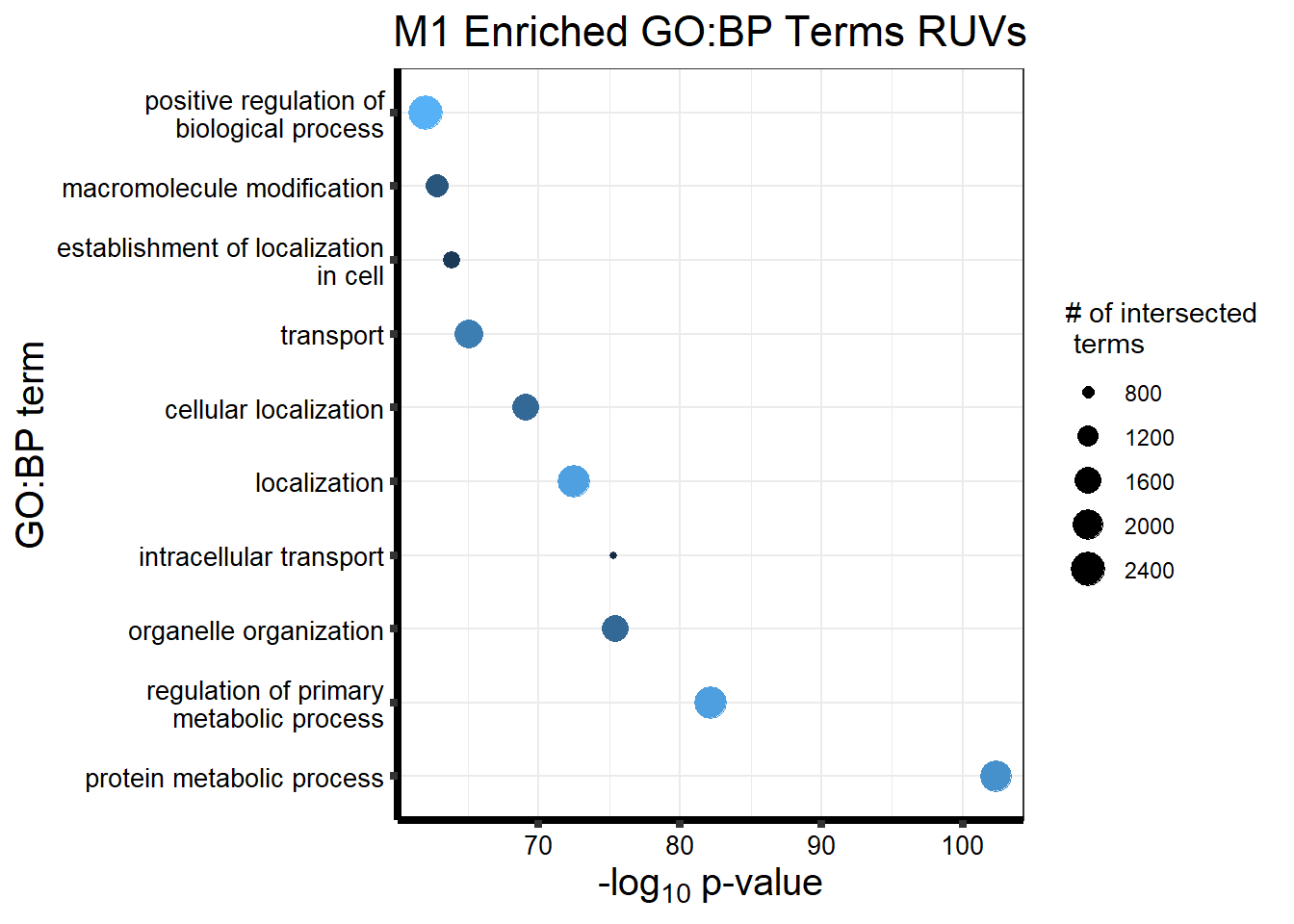
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#KEGG
table_motif1_genes_KEGG_RUV <- table_motif1_GOKEGG_genes_RUV %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_motif1_genes_KEGG_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("M1 DEGs Enriched KEGG Terms RUVs")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))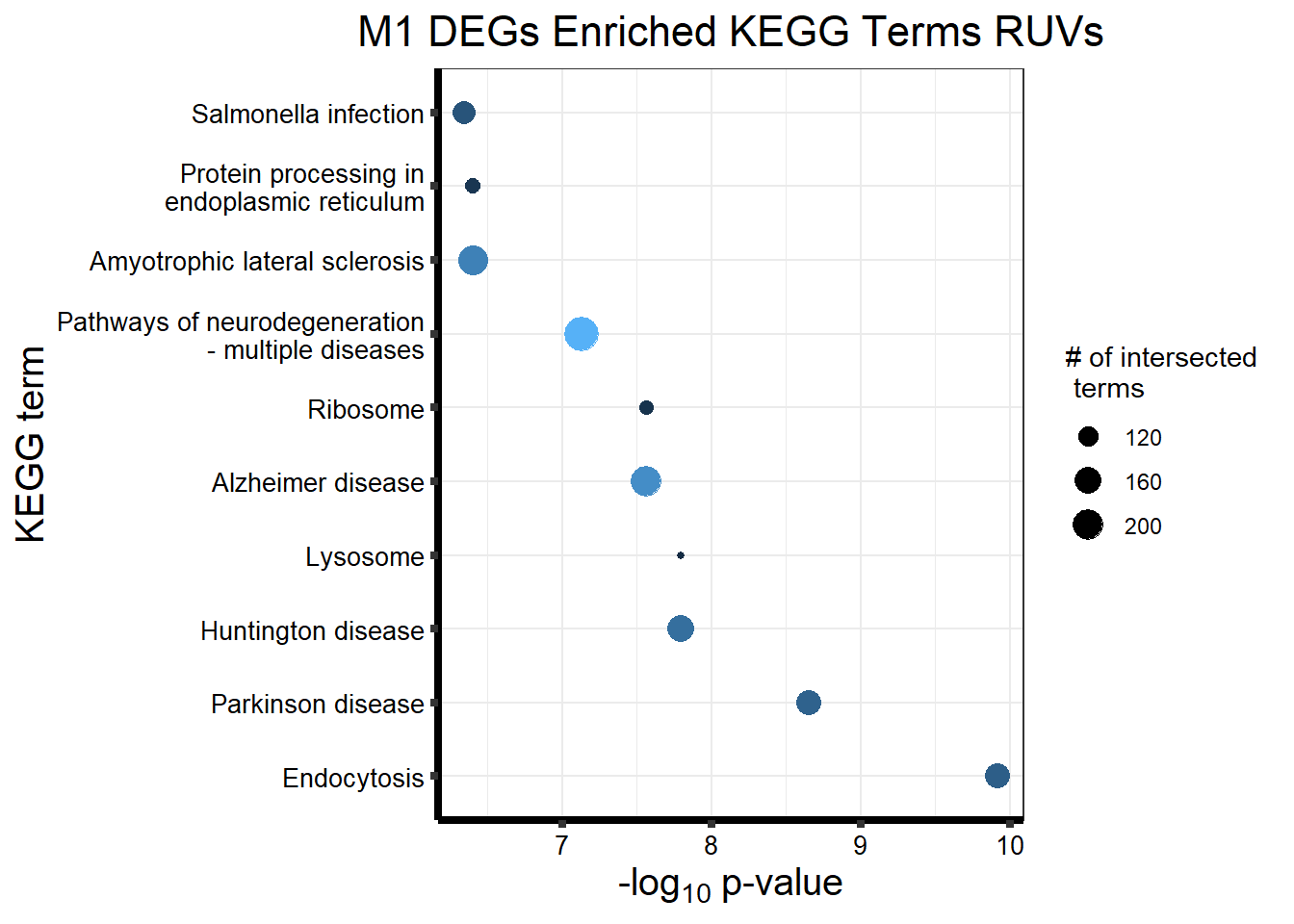
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#####Motif 2 Genes#####
# motif2_genes_matrix_RUV <- as.matrix(final_genes_2_RUV)
# colnames(motif2_genes_matrix_RUV) <- c("entrezgene_ID")
# saveRDS(motif2_genes_matrix_RUV, "data/new/RUV/CMF/motif2_genes_matrix_RUV.RDS")
motif2_genes_matrix_RUV <- readRDS("data/new/RUV/CMF/motif2_genes_matrix_RUV.RDS")
length(motif2_genes_matrix_RUV)[1] 501#501 genes in this set for motif 2
motif2_mat_GOKEGG_RUV <- gost(query = motif2_genes_matrix_RUV,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
motif2_GOKEGG_genes_RUV <- gostplot(motif2_mat_GOKEGG_RUV, capped = FALSE, interactive = TRUE)
motif2_GOKEGG_genes_RUVtable_motif2_GOKEGG_genes_RUV <- motif2_mat_GOKEGG_RUV$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_motif2_GOKEGG_genes_RUV %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0022402 | cell cycle process | 175 | 1280 | 5.510e-90 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 134 | 745 | 6.757e-82 |
| GO:BP | GO:0007049 | cell cycle | 185 | 1663 | 7.203e-81 |
| GO:BP | GO:0000278 | mitotic cell cycle | 141 | 892 | 4.021e-79 |
| GO:BP | GO:0007059 | chromosome segregation | 97 | 427 | 1.026e-67 |
| GO:BP | GO:0000280 | nuclear division | 95 | 452 | 6.218e-63 |
| GO:BP | GO:0051301 | cell division | 109 | 658 | 1.017e-61 |
| GO:BP | GO:0048285 | organelle fission | 96 | 500 | 6.643e-60 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 111 | 720 | 1.112e-59 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 80 | 323 | 1.652e-58 |
| GO:BP | GO:0140014 | mitotic nuclear division | 73 | 282 | 1.257e-54 |
| GO:BP | GO:0051726 | regulation of cell cycle | 126 | 1087 | 4.813e-54 |
| GO:BP | GO:0051276 | chromosome organization | 95 | 574 | 2.601e-53 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 62 | 193 | 1.623e-52 |
| GO:BP | GO:0000819 | sister chromatid segregation | 66 | 234 | 6.445e-52 |
| GO:BP | GO:0044770 | cell cycle phase transition | 89 | 532 | 3.637e-50 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 78 | 428 | 2.033e-46 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 75 | 427 | 2.437e-43 |
| GO:BP | GO:0006260 | DNA replication | 63 | 283 | 1.360e-42 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 78 | 492 | 7.753e-42 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 50 | 185 | 6.541e-38 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 59 | 289 | 1.551e-37 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 62 | 327 | 1.551e-37 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 43 | 132 | 2.536e-36 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 46 | 160 | 3.571e-36 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 54 | 248 | 8.462e-36 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 35 | 77 | 1.831e-35 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 63 | 377 | 7.789e-35 |
| GO:BP | GO:0006259 | DNA metabolic process | 98 | 1005 | 1.265e-34 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 40 | 121 | 4.455e-34 |
| GO:BP | GO:0051304 | chromosome separation | 35 | 84 | 7.309e-34 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 41 | 136 | 3.947e-33 |
| GO:BP | GO:0051310 | metaphase chromosome alignment | 37 | 104 | 7.726e-33 |
| GO:BP | GO:0051783 | regulation of nuclear division | 42 | 149 | 1.230e-32 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 52 | 262 | 2.154e-32 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 43 | 169 | 2.160e-31 |
| GO:BP | GO:0050000 | chromosome localization | 38 | 126 | 1.096e-30 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 37 | 118 | 1.452e-30 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 42 | 172 | 7.227e-30 |
| GO:BP | GO:0007051 | spindle organization | 45 | 206 | 7.918e-30 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 29 | 62 | 7.918e-30 |
| GO:BP | GO:0051321 | meiotic cell cycle | 52 | 297 | 1.216e-29 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 46 | 225 | 3.511e-29 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 29 | 65 | 4.249e-29 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 55 | 351 | 6.912e-29 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 34 | 107 | 2.629e-28 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 45 | 224 | 3.283e-28 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 26 | 51 | 5.082e-28 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 26 | 51 | 5.082e-28 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 26 | 51 | 5.082e-28 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 26 | 51 | 5.082e-28 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 27 | 57 | 5.296e-28 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 28 | 65 | 1.310e-27 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 27 | 59 | 1.690e-27 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 26 | 53 | 1.714e-27 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 26 | 53 | 1.714e-27 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 26 | 53 | 1.714e-27 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 32 | 97 | 2.779e-27 |
| GO:BP | GO:0006281 | DNA repair | 69 | 621 | 2.866e-27 |
| GO:BP | GO:0007052 | mitotic spindle organization | 36 | 134 | 3.114e-27 |
| GO:BP | GO:0006974 | DNA damage response | 83 | 908 | 3.686e-27 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 25 | 49 | 5.060e-27 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 25 | 49 | 5.060e-27 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 25 | 49 | 5.060e-27 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 32 | 100 | 7.532e-27 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 25 | 50 | 9.617e-27 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 31 | 93 | 1.253e-26 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 31 | 96 | 3.802e-26 |
| GO:BP | GO:0140013 | meiotic nuclear division | 41 | 202 | 5.875e-26 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 25 | 54 | 1.114e-25 |
| GO:BP | GO:0006996 | organelle organization | 175 | 3594 | 2.359e-25 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 43 | 242 | 8.496e-25 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 68 | 670 | 1.344e-24 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 34 | 156 | 1.686e-22 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 32 | 139 | 6.208e-22 |
| GO:BP | GO:0007017 | microtubule-based process | 79 | 999 | 9.475e-22 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 27 | 97 | 8.380e-21 |
| GO:BP | GO:0006270 | DNA replication initiation | 19 | 37 | 1.540e-20 |
| GO:BP | GO:0051225 | spindle assembly | 29 | 136 | 7.605e-19 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 26 | 105 | 1.315e-18 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 29 | 141 | 2.161e-18 |
| GO:BP | GO:0007080 | mitotic metaphase chromosome alignment | 21 | 63 | 6.248e-18 |
| GO:BP | GO:0051656 | establishment of organelle localization | 50 | 491 | 7.124e-18 |
| GO:BP | GO:0006302 | double-strand break repair | 40 | 319 | 2.430e-17 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 24 | 96 | 2.855e-17 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 26 | 118 | 2.929e-17 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 16 | 32 | 5.150e-17 |
| GO:BP | GO:0051640 | organelle localization | 55 | 620 | 5.237e-17 |
| GO:BP | GO:0006310 | DNA recombination | 41 | 347 | 7.288e-17 |
| GO:BP | GO:0000725 | recombinational repair | 31 | 193 | 1.886e-16 |
| GO:BP | GO:0033043 | regulation of organelle organization | 76 | 1148 | 2.120e-16 |
| GO:BP | GO:0033554 | cellular response to stress | 100 | 1830 | 3.261e-16 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 15 | 30 | 5.967e-16 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 30 | 188 | 7.674e-16 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 17 | 45 | 1.441e-15 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 43 | 430 | 4.952e-15 |
| GO:BP | GO:0033260 | nuclear DNA replication | 16 | 41 | 6.607e-15 |
| GO:BP | GO:0007127 | meiosis I | 25 | 133 | 6.919e-15 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 13 | 23 | 1.005e-14 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 24 | 123 | 1.129e-14 |
| GO:BP | GO:0034508 | centromere complex assembly | 14 | 29 | 1.289e-14 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 23 | 114 | 2.159e-14 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 21 | 93 | 3.628e-14 |
| GO:BP | GO:0051716 | cellular response to stimulus | 245 | 7376 | 4.997e-14 |
| GO:BP | GO:0007010 | cytoskeleton organization | 85 | 1529 | 5.517e-14 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 10 | 12 | 6.432e-14 |
| GO:BP | GO:0006275 | regulation of DNA replication | 23 | 120 | 6.738e-14 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 15 | 40 | 1.089e-13 |
| GO:BP | GO:0051383 | kinetochore organization | 12 | 21 | 1.118e-13 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 23 | 124 | 1.389e-13 |
| GO:BP | GO:0019953 | sexual reproduction | 69 | 1122 | 3.050e-13 |
| GO:BP | GO:0022414 | reproductive process | 86 | 1608 | 3.220e-13 |
| GO:BP | GO:0035556 | intracellular signal transduction | 128 | 2965 | 3.789e-13 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 36 | 352 | 7.247e-13 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 18 | 76 | 1.561e-12 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 12 | 25 | 1.716e-12 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 15 | 48 | 2.327e-12 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 10 | 15 | 2.457e-12 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 10 | 15 | 2.457e-12 |
| GO:BP | GO:0000910 | cytokinesis | 26 | 192 | 4.917e-12 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 19 | 95 | 7.826e-12 |
| GO:BP | GO:1902969 | mitotic DNA replication | 10 | 17 | 1.492e-11 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 41 | 500 | 1.706e-11 |
| GO:BP | GO:0050896 | response to stimulus | 272 | 8999 | 5.867e-11 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 11 | 25 | 6.242e-11 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 18 | 94 | 7.121e-11 |
| GO:BP | GO:0051231 | spindle elongation | 9 | 14 | 7.127e-11 |
| GO:BP | GO:0007143 | female meiotic nuclear division | 13 | 42 | 1.185e-10 |
| GO:BP | GO:0007098 | centrosome cycle | 21 | 140 | 1.430e-10 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 105 | 2433 | 1.934e-10 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 19 | 114 | 2.213e-10 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 10 | 21 | 2.311e-10 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 8 | 11 | 2.702e-10 |
| GO:BP | GO:0030261 | chromosome condensation | 13 | 45 | 3.049e-10 |
| GO:BP | GO:0000022 | mitotic spindle elongation | 8 | 12 | 7.775e-10 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 21 | 153 | 7.775e-10 |
| GO:BP | GO:0051302 | regulation of cell division | 23 | 188 | 9.490e-10 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 19 | 124 | 9.706e-10 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 10 | 24 | 1.142e-09 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 10 | 24 | 1.142e-09 |
| GO:BP | GO:0034502 | protein localization to chromosome | 19 | 126 | 1.267e-09 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 12 | 41 | 1.547e-09 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 15 | 74 | 1.883e-09 |
| GO:BP | GO:0016043 | cellular component organization | 248 | 8184 | 1.900e-09 |
| GO:BP | GO:0008283 | cell population proliferation | 89 | 2015 | 3.354e-09 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 23 | 202 | 3.872e-09 |
| GO:BP | GO:0051255 | spindle midzone assembly | 8 | 14 | 4.198e-09 |
| GO:BP | GO:0050789 | regulation of biological process | 336 | 12336 | 4.430e-09 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 25 | 241 | 4.452e-09 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 26 | 262 | 5.037e-09 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 13 | 57 | 7.139e-09 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 14 | 69 | 7.654e-09 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 7 | 10 | 7.846e-09 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 7 | 10 | 7.846e-09 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 8 | 15 | 8.209e-09 |
| GO:BP | GO:1903083 | protein localization to condensed chromosome | 8 | 15 | 8.209e-09 |
| GO:BP | GO:0034501 | protein localization to kinetochore | 8 | 15 | 8.209e-09 |
| GO:BP | GO:0006468 | protein phosphorylation | 63 | 1227 | 8.209e-09 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 24 | 230 | 8.660e-09 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 14 | 71 | 1.088e-08 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 12 | 50 | 1.753e-08 |
| GO:BP | GO:0070925 | organelle assembly | 56 | 1046 | 1.915e-08 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 6 | 7 | 2.072e-08 |
| GO:BP | GO:0016310 | phosphorylation | 65 | 1320 | 2.183e-08 |
| GO:BP | GO:0051382 | kinetochore assembly | 8 | 17 | 2.857e-08 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 248 | 8393 | 2.872e-08 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 20 | 173 | 4.228e-08 |
| GO:BP | GO:0051338 | regulation of transferase activity | 35 | 498 | 4.450e-08 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 7 | 12 | 4.542e-08 |
| GO:BP | GO:0050794 | regulation of cellular process | 324 | 11946 | 4.811e-08 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 12 | 55 | 5.401e-08 |
| GO:BP | GO:0051653 | spindle localization | 13 | 68 | 6.372e-08 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 13 | 68 | 6.372e-08 |
| GO:BP | GO:0035825 | homologous recombination | 13 | 68 | 6.372e-08 |
| GO:BP | GO:1905821 | positive regulation of chromosome condensation | 6 | 8 | 7.575e-08 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 8 | 19 | 8.011e-08 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 13 | 70 | 9.108e-08 |
| GO:BP | GO:0090232 | positive regulation of spindle checkpoint | 7 | 13 | 9.166e-08 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 12 | 59 | 1.216e-07 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 9 | 28 | 1.319e-07 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 186 | 5920 | 2.542e-07 |
| GO:BP | GO:0051293 | establishment of spindle localization | 12 | 63 | 2.633e-07 |
| GO:BP | GO:0000212 | meiotic spindle organization | 8 | 22 | 3.076e-07 |
| GO:BP | GO:0065007 | biological regulation | 337 | 12743 | 3.148e-07 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 7 | 15 | 3.183e-07 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 9 | 31 | 3.515e-07 |
| GO:BP | GO:2000241 | regulation of reproductive process | 20 | 203 | 6.005e-07 |
| GO:BP | GO:0051781 | positive regulation of cell division | 14 | 98 | 7.307e-07 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 40 | 701 | 9.799e-07 |
| GO:BP | GO:0009314 | response to radiation | 30 | 437 | 1.003e-06 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 18 | 172 | 1.166e-06 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 32 | 492 | 1.230e-06 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 10 | 47 | 1.391e-06 |
| GO:BP | GO:0051447 | negative regulation of meiotic cell cycle | 7 | 18 | 1.418e-06 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 14 | 105 | 1.722e-06 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 9 | 37 | 1.831e-06 |
| GO:BP | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 6 | 12 | 2.039e-06 |
| GO:BP | GO:0007056 | spindle assembly involved in female meiosis | 6 | 12 | 2.039e-06 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 6 | 12 | 2.039e-06 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 7 | 19 | 2.136e-06 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 47 | 928 | 2.161e-06 |
| GO:BP | GO:0031297 | replication fork processing | 10 | 51 | 3.032e-06 |
| GO:BP | GO:0000076 | DNA replication checkpoint signaling | 7 | 20 | 3.177e-06 |
| GO:BP | GO:0007144 | female meiosis I | 6 | 13 | 3.608e-06 |
| GO:BP | GO:1903490 | positive regulation of mitotic cytokinesis | 6 | 13 | 3.608e-06 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 12 | 80 | 3.746e-06 |
| GO:BP | GO:0048518 | positive regulation of biological process | 189 | 6264 | 3.876e-06 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 11 | 66 | 4.014e-06 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 11 | 67 | 4.686e-06 |
| GO:BP | GO:0007057 | spindle assembly involved in female meiosis I | 4 | 4 | 4.942e-06 |
| GO:BP | GO:0048856 | anatomical structure development | 182 | 5997 | 5.192e-06 |
| GO:BP | GO:0006950 | response to stress | 131 | 3948 | 5.764e-06 |
| GO:BP | GO:0046385 | deoxyribose phosphate biosynthetic process | 6 | 14 | 5.882e-06 |
| GO:BP | GO:0009263 | deoxyribonucleotide biosynthetic process | 6 | 14 | 5.882e-06 |
| GO:BP | GO:0009265 | 2’-deoxyribonucleotide biosynthetic process | 6 | 14 | 5.882e-06 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 6 | 14 | 5.882e-06 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 9 | 43 | 6.669e-06 |
| GO:BP | GO:0007275 | multicellular organism development | 150 | 4727 | 8.169e-06 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 9 | 44 | 8.169e-06 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 15 | 139 | 8.891e-06 |
| GO:BP | GO:0032502 | developmental process | 194 | 6553 | 9.316e-06 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 5 | 9 | 1.193e-05 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 11 | 74 | 1.244e-05 |
| GO:BP | GO:0007131 | reciprocal meiotic recombination | 10 | 60 | 1.346e-05 |
| GO:BP | GO:0140527 | reciprocal homologous recombination | 10 | 60 | 1.346e-05 |
| GO:BP | GO:0045840 | positive regulation of mitotic nuclear division | 9 | 47 | 1.431e-05 |
| GO:BP | GO:1902412 | regulation of mitotic cytokinesis | 6 | 16 | 1.438e-05 |
| GO:BP | GO:0090224 | regulation of spindle organization | 9 | 48 | 1.712e-05 |
| GO:BP | GO:0007154 | cell communication | 192 | 6540 | 2.052e-05 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 26 | 402 | 2.076e-05 |
| GO:BP | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition | 6 | 17 | 2.143e-05 |
| GO:BP | GO:1902975 | mitotic DNA replication initiation | 4 | 5 | 2.160e-05 |
| GO:BP | GO:0022616 | DNA strand elongation | 8 | 37 | 2.160e-05 |
| GO:BP | GO:1902315 | nuclear cell cycle DNA replication initiation | 4 | 5 | 2.160e-05 |
| GO:BP | GO:1902292 | cell cycle DNA replication initiation | 4 | 5 | 2.160e-05 |
| GO:BP | GO:0110030 | regulation of G2/MI transition of meiotic cell cycle | 4 | 5 | 2.160e-05 |
| GO:BP | GO:0051307 | meiotic chromosome separation | 5 | 10 | 2.185e-05 |
| GO:BP | GO:0045835 | negative regulation of meiotic nuclear division | 5 | 10 | 2.185e-05 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 10 | 64 | 2.354e-05 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 67 | 1685 | 2.848e-05 |
| GO:BP | GO:0007292 | female gamete generation | 16 | 174 | 2.944e-05 |
| GO:BP | GO:1902101 | positive regulation of metaphase/anaphase transition of cell cycle | 6 | 18 | 3.011e-05 |
| GO:BP | GO:0048519 | negative regulation of biological process | 174 | 5834 | 3.436e-05 |
| GO:BP | GO:0007165 | signal transduction | 178 | 6002 | 3.444e-05 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 19 | 242 | 3.444e-05 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 20 | 266 | 3.604e-05 |
| GO:BP | GO:0006338 | chromatin remodeling | 37 | 725 | 3.647e-05 |
| GO:BP | GO:0023052 | signaling | 190 | 6515 | 3.697e-05 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 5 | 11 | 3.760e-05 |
| GO:BP | GO:0065009 | regulation of molecular function | 61 | 1496 | 3.847e-05 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 21 | 296 | 4.956e-05 |
| GO:BP | GO:0071478 | cellular response to radiation | 16 | 182 | 5.085e-05 |
| GO:BP | GO:0010212 | response to ionizing radiation | 14 | 141 | 5.095e-05 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 6 | 20 | 5.771e-05 |
| GO:BP | GO:0010032 | meiotic chromosome condensation | 4 | 6 | 5.894e-05 |
| GO:BP | GO:0051649 | establishment of localization in cell | 75 | 2010 | 6.215e-05 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 15 | 164 | 6.239e-05 |
| GO:BP | GO:0043549 | regulation of kinase activity | 26 | 431 | 6.661e-05 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 7 | 31 | 6.851e-05 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 8 | 44 | 7.850e-05 |
| GO:BP | GO:0051298 | centrosome duplication | 10 | 74 | 8.404e-05 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 167 | 5629 | 8.496e-05 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 14 | 150 | 1.015e-04 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 85 | 2407 | 1.062e-04 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 85 | 2410 | 1.110e-04 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 43 | 954 | 1.205e-04 |
| GO:BP | GO:1901994 | negative regulation of meiotic cell cycle phase transition | 4 | 7 | 1.286e-04 |
| GO:BP | GO:0044785 | metaphase/anaphase transition of meiotic cell cycle | 4 | 7 | 1.286e-04 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 15 | 176 | 1.417e-04 |
| GO:BP | GO:0001833 | inner cell mass cell proliferation | 5 | 14 | 1.423e-04 |
| GO:BP | GO:0009411 | response to UV | 14 | 155 | 1.441e-04 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 7 | 35 | 1.546e-04 |
| GO:BP | GO:0007276 | gamete generation | 39 | 841 | 1.674e-04 |
| GO:BP | GO:0110032 | positive regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 1.722e-04 |
| GO:BP | GO:0033316 | meiotic spindle assembly checkpoint signaling | 3 | 3 | 1.722e-04 |
| GO:BP | GO:0071163 | DNA replication preinitiation complex assembly | 3 | 3 | 1.722e-04 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 13 | 138 | 1.869e-04 |
| GO:BP | GO:0030010 | establishment of cell polarity | 14 | 159 | 1.882e-04 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 34 | 692 | 1.891e-04 |
| GO:BP | GO:0032506 | cytokinetic process | 8 | 50 | 1.968e-04 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 7 | 37 | 2.191e-04 |
| GO:BP | GO:0006325 | chromatin organization | 40 | 884 | 2.191e-04 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 48 | 1144 | 2.191e-04 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 4 | 8 | 2.368e-04 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 4 | 8 | 2.368e-04 |
| GO:BP | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process | 4 | 8 | 2.368e-04 |
| GO:BP | GO:0048731 | system development | 126 | 4053 | 2.376e-04 |
| GO:BP | GO:0008156 | negative regulation of DNA replication | 6 | 26 | 2.703e-04 |
| GO:BP | GO:0051299 | centrosome separation | 5 | 16 | 2.796e-04 |
| GO:BP | GO:0048513 | animal organ development | 101 | 3085 | 2.835e-04 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 21 | 339 | 3.393e-04 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 21 | 339 | 3.393e-04 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 43 | 1000 | 3.415e-04 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 36 | 779 | 3.698e-04 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 46 | 1105 | 3.890e-04 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 19 | 291 | 3.979e-04 |
| GO:BP | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | 4 | 9 | 3.979e-04 |
| GO:BP | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis | 4 | 9 | 3.979e-04 |
| GO:BP | GO:0000915 | actomyosin contractile ring assembly | 4 | 9 | 3.979e-04 |
| GO:BP | GO:1904776 | regulation of protein localization to cell cortex | 4 | 9 | 3.979e-04 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 4 | 9 | 3.979e-04 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 9 | 72 | 4.117e-04 |
| GO:BP | GO:0009394 | 2’-deoxyribonucleotide metabolic process | 7 | 41 | 4.142e-04 |
| GO:BP | GO:0048144 | fibroblast proliferation | 11 | 109 | 4.207e-04 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 70 | 1953 | 4.373e-04 |
| GO:BP | GO:0031399 | regulation of protein modification process | 44 | 1049 | 4.742e-04 |
| GO:BP | GO:0009262 | deoxyribonucleotide metabolic process | 7 | 42 | 4.782e-04 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 7 | 42 | 4.782e-04 |
| GO:BP | GO:0019692 | deoxyribose phosphate metabolic process | 7 | 42 | 4.782e-04 |
| GO:BP | GO:0043009 | chordate embryonic development | 32 | 671 | 5.496e-04 |
| GO:BP | GO:0007099 | centriole replication | 7 | 43 | 5.551e-04 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 7 | 43 | 5.551e-04 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 37 | 830 | 5.759e-04 |
| GO:BP | GO:1901995 | positive regulation of meiotic cell cycle phase transition | 3 | 4 | 5.937e-04 |
| GO:BP | GO:0044779 | meiotic spindle checkpoint signaling | 3 | 4 | 5.937e-04 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 28 | 554 | 6.074e-04 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 4 | 10 | 6.129e-04 |
| GO:BP | GO:0033313 | meiotic cell cycle checkpoint signaling | 4 | 10 | 6.129e-04 |
| GO:BP | GO:2000104 | negative regulation of DNA-templated DNA replication | 4 | 10 | 6.129e-04 |
| GO:BP | GO:0044837 | actomyosin contractile ring organization | 4 | 10 | 6.129e-04 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 8 | 61 | 7.623e-04 |
| GO:BP | GO:0033314 | mitotic DNA replication checkpoint signaling | 4 | 11 | 9.377e-04 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 4 | 11 | 9.377e-04 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 38 | 884 | 9.456e-04 |
| GO:BP | GO:0098534 | centriole assembly | 7 | 48 | 1.108e-03 |
| GO:BP | GO:0001832 | blastocyst growth | 5 | 22 | 1.340e-03 |
| GO:BP | GO:1905784 | regulation of anaphase-promoting complex-dependent catabolic process | 3 | 5 | 1.398e-03 |
| GO:BP | GO:1905133 | negative regulation of meiotic chromosome separation | 3 | 5 | 1.398e-03 |
| GO:BP | GO:1902103 | negative regulation of metaphase/anaphase transition of meiotic cell cycle | 3 | 5 | 1.398e-03 |
| GO:BP | GO:0007019 | microtubule depolymerization | 7 | 50 | 1.425e-03 |
| GO:BP | GO:0009966 | regulation of signal transduction | 96 | 3034 | 1.566e-03 |
| GO:BP | GO:0006301 | postreplication repair | 6 | 36 | 1.647e-03 |
| GO:BP | GO:0071695 | anatomical structure maturation | 17 | 277 | 1.988e-03 |
| GO:BP | GO:0009162 | deoxyribonucleoside monophosphate metabolic process | 5 | 24 | 2.036e-03 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 6 | 38 | 2.229e-03 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 7 | 54 | 2.294e-03 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 7 | 54 | 2.294e-03 |
| GO:BP | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 5 | 25 | 2.470e-03 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 6 | 39 | 2.556e-03 |
| GO:BP | GO:2000105 | positive regulation of DNA-templated DNA replication | 4 | 14 | 2.557e-03 |
| GO:BP | GO:0007100 | mitotic centrosome separation | 4 | 14 | 2.557e-03 |
| GO:BP | GO:0006335 | DNA replication-dependent chromatin assembly | 3 | 6 | 2.585e-03 |
| GO:BP | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process | 3 | 6 | 2.585e-03 |
| GO:BP | GO:0046073 | dTMP metabolic process | 3 | 6 | 2.585e-03 |
| GO:BP | GO:1905132 | regulation of meiotic chromosome separation | 3 | 6 | 2.585e-03 |
| GO:BP | GO:0090402 | oncogene-induced cell senescence | 3 | 6 | 2.585e-03 |
| GO:BP | GO:0016321 | female meiosis chromosome segregation | 3 | 6 | 2.585e-03 |
| GO:BP | GO:0042148 | DNA strand invasion | 3 | 6 | 2.585e-03 |
| GO:BP | GO:1902102 | regulation of metaphase/anaphase transition of meiotic cell cycle | 3 | 6 | 2.585e-03 |
| GO:BP | GO:0090166 | Golgi disassembly | 3 | 6 | 2.585e-03 |
| GO:BP | GO:0023051 | regulation of signaling | 106 | 3478 | 2.593e-03 |
| GO:BP | GO:0010646 | regulation of cell communication | 106 | 3486 | 2.823e-03 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 15 | 234 | 2.846e-03 |
| GO:BP | GO:0019985 | translesion synthesis | 5 | 26 | 2.867e-03 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 6 | 41 | 3.241e-03 |
| GO:BP | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process | 4 | 15 | 3.282e-03 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 7 | 58 | 3.410e-03 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 12 | 164 | 3.564e-03 |
| GO:BP | GO:0001824 | blastocyst development | 10 | 119 | 3.764e-03 |
| GO:BP | GO:0036211 | protein modification process | 89 | 2846 | 4.033e-03 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 11 | 143 | 4.096e-03 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 38 | 956 | 4.096e-03 |
| GO:BP | GO:0072528 | pyrimidine-containing compound biosynthetic process | 6 | 43 | 4.130e-03 |
| GO:BP | GO:0009124 | nucleoside monophosphate biosynthetic process | 6 | 43 | 4.130e-03 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 38 | 957 | 4.145e-03 |
| GO:BP | GO:0048313 | Golgi inheritance | 4 | 16 | 4.171e-03 |
| GO:BP | GO:0048308 | organelle inheritance | 4 | 16 | 4.171e-03 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 63 | 1855 | 4.227e-03 |
| GO:BP | GO:0030154 | cell differentiation | 128 | 4437 | 4.784e-03 |
| GO:BP | GO:0048869 | cellular developmental process | 128 | 4438 | 4.817e-03 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 7 | 62 | 4.965e-03 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 7 | 62 | 4.965e-03 |
| GO:BP | GO:0006997 | nucleus organization | 11 | 147 | 5.021e-03 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 40 | 1038 | 5.148e-03 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 6 | 45 | 5.161e-03 |
| GO:BP | GO:1905325 | regulation of meiosis I spindle assembly checkpoint | 2 | 2 | 5.599e-03 |
| GO:BP | GO:0110029 | negative regulation of meiosis I | 2 | 2 | 5.599e-03 |
| GO:BP | GO:0099607 | lateral attachment of mitotic spindle microtubules to kinetochore | 2 | 2 | 5.599e-03 |
| GO:BP | GO:1902298 | cell cycle DNA replication maintenance of fidelity | 2 | 2 | 5.599e-03 |
| GO:BP | GO:1990426 | mitotic recombination-dependent replication fork processing | 2 | 2 | 5.599e-03 |
| GO:BP | GO:0014846 | esophagus smooth muscle contraction | 2 | 2 | 5.599e-03 |
| GO:BP | GO:1990505 | mitotic DNA replication maintenance of fidelity | 2 | 2 | 5.599e-03 |
| GO:BP | GO:0031619 | homologous chromosome orientation in meiotic metaphase I | 2 | 2 | 5.599e-03 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 8 | 83 | 5.599e-03 |
| GO:BP | GO:0140274 | repair of kinetochore microtubule attachment defect | 2 | 2 | 5.599e-03 |
| GO:BP | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway | 2 | 2 | 5.599e-03 |
| GO:BP | GO:1905318 | meiosis I spindle assembly checkpoint signaling | 2 | 2 | 5.599e-03 |
| GO:BP | GO:0140273 | repair of mitotic kinetochore microtubule attachment defect | 2 | 2 | 5.599e-03 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 13 | 200 | 5.845e-03 |
| GO:BP | GO:0006939 | smooth muscle contraction | 9 | 105 | 5.954e-03 |
| GO:BP | GO:0031507 | heterochromatin formation | 11 | 151 | 6.014e-03 |
| GO:BP | GO:0010458 | exit from mitosis | 5 | 31 | 6.107e-03 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 150 | 5390 | 6.107e-03 |
| GO:BP | GO:0007077 | mitotic nuclear membrane disassembly | 3 | 8 | 6.221e-03 |
| GO:BP | GO:0007096 | regulation of exit from mitosis | 4 | 18 | 6.292e-03 |
| GO:BP | GO:0051641 | cellular localization | 108 | 3661 | 6.743e-03 |
| GO:BP | GO:0048608 | reproductive structure development | 17 | 314 | 7.129e-03 |
| GO:BP | GO:0009790 | embryo development | 42 | 1135 | 7.895e-03 |
| GO:BP | GO:0051642 | centrosome localization | 5 | 33 | 8.048e-03 |
| GO:BP | GO:0006221 | pyrimidine nucleotide biosynthetic process | 5 | 33 | 8.048e-03 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 5 | 33 | 8.048e-03 |
| GO:BP | GO:0061458 | reproductive system development | 17 | 318 | 8.087e-03 |
| GO:BP | GO:0031100 | animal organ regeneration | 7 | 68 | 8.087e-03 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 3 | 9 | 8.931e-03 |
| GO:BP | GO:0046602 | regulation of mitotic centrosome separation | 3 | 9 | 8.931e-03 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 8 | 90 | 9.099e-03 |
| GO:BP | GO:0034644 | cellular response to UV | 8 | 90 | 9.099e-03 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 5 | 34 | 9.108e-03 |
| GO:BP | GO:0021700 | developmental maturation | 18 | 352 | 9.400e-03 |
| GO:BP | GO:0001701 | in utero embryonic development | 20 | 413 | 9.716e-03 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 5 | 35 | 1.034e-02 |
| GO:BP | GO:0010332 | response to gamma radiation | 6 | 52 | 1.034e-02 |
| GO:BP | GO:0007018 | microtubule-based movement | 22 | 478 | 1.060e-02 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 4 | 21 | 1.113e-02 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 14 | 243 | 1.113e-02 |
| GO:BP | GO:0001556 | oocyte maturation | 5 | 36 | 1.168e-02 |
| GO:BP | GO:0110025 | DNA strand resection involved in replication fork processing | 3 | 10 | 1.219e-02 |
| GO:BP | GO:0007399 | nervous system development | 80 | 2604 | 1.237e-02 |
| GO:BP | GO:0007129 | homologous chromosome pairing at meiosis | 6 | 54 | 1.241e-02 |
| GO:BP | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process | 4 | 22 | 1.316e-02 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 4 | 22 | 1.316e-02 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 7 | 75 | 1.384e-02 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 10 | 144 | 1.420e-02 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 5 | 38 | 1.468e-02 |
| GO:BP | GO:0071312 | cellular response to alkaloid | 5 | 38 | 1.468e-02 |
| GO:BP | GO:0032501 | multicellular organismal process | 192 | 7322 | 1.475e-02 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 6 | 56 | 1.475e-02 |
| GO:BP | GO:0034421 | post-translational protein acetylation | 2 | 3 | 1.493e-02 |
| GO:BP | GO:0006231 | dTMP biosynthetic process | 2 | 3 | 1.493e-02 |
| GO:BP | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process | 2 | 3 | 1.493e-02 |
| GO:BP | GO:0046599 | regulation of centriole replication | 4 | 23 | 1.526e-02 |
| GO:BP | GO:0009219 | pyrimidine deoxyribonucleotide metabolic process | 4 | 23 | 1.526e-02 |
| GO:BP | GO:0006282 | regulation of DNA repair | 13 | 225 | 1.553e-02 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 3 | 11 | 1.566e-02 |
| GO:BP | GO:0009987 | cellular process | 456 | 20247 | 1.566e-02 |
| GO:BP | GO:0009650 | UV protection | 3 | 11 | 1.566e-02 |
| GO:BP | GO:0009176 | pyrimidine deoxyribonucleoside monophosphate metabolic process | 3 | 11 | 1.566e-02 |
| GO:BP | GO:1990918 | double-strand break repair involved in meiotic recombination | 3 | 11 | 1.566e-02 |
| GO:BP | GO:0070601 | centromeric sister chromatid cohesion | 3 | 11 | 1.566e-02 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 8 | 100 | 1.671e-02 |
| GO:BP | GO:0090398 | cellular senescence | 8 | 101 | 1.777e-02 |
| GO:BP | GO:0009123 | nucleoside monophosphate metabolic process | 7 | 79 | 1.790e-02 |
| GO:BP | GO:0006998 | nuclear envelope organization | 6 | 59 | 1.867e-02 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 26 | 637 | 1.998e-02 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 3 | 12 | 2.001e-02 |
| GO:BP | GO:0051081 | nuclear membrane disassembly | 3 | 12 | 2.001e-02 |
| GO:BP | GO:0072710 | response to hydroxyurea | 3 | 12 | 2.001e-02 |
| GO:BP | GO:0008584 | male gonad development | 10 | 152 | 2.001e-02 |
| GO:BP | GO:0014831 | gastro-intestinal system smooth muscle contraction | 3 | 12 | 2.001e-02 |
| GO:BP | GO:0071168 | protein localization to chromatin | 6 | 60 | 2.001e-02 |
| GO:BP | GO:0071493 | cellular response to UV-B | 3 | 12 | 2.001e-02 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 10 | 153 | 2.087e-02 |
| GO:BP | GO:0009416 | response to light stimulus | 16 | 323 | 2.226e-02 |
| GO:BP | GO:0043412 | macromolecule modification | 89 | 3030 | 2.267e-02 |
| GO:BP | GO:0007548 | sex differentiation | 15 | 294 | 2.270e-02 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 4 | 26 | 2.313e-02 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 4 | 26 | 2.313e-02 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 5 | 43 | 2.388e-02 |
| GO:BP | GO:0008406 | gonad development | 13 | 238 | 2.403e-02 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 21 | 483 | 2.444e-02 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 13 | 239 | 2.481e-02 |
| GO:BP | GO:1903251 | multi-ciliated epithelial cell differentiation | 3 | 13 | 2.486e-02 |
| GO:BP | GO:0048469 | cell maturation | 12 | 211 | 2.486e-02 |
| GO:BP | GO:0030397 | membrane disassembly | 3 | 13 | 2.486e-02 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 22 | 517 | 2.508e-02 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 15 | 299 | 2.602e-02 |
| GO:BP | GO:0051296 | establishment of meiotic spindle orientation | 2 | 4 | 2.648e-02 |
| GO:BP | GO:1901563 | response to camptothecin | 2 | 4 | 2.648e-02 |
| GO:BP | GO:0072757 | cellular response to camptothecin | 2 | 4 | 2.648e-02 |
| GO:BP | GO:0043060 | meiotic metaphase I homologous chromosome alignment | 2 | 4 | 2.648e-02 |
| GO:BP | GO:1905448 | positive regulation of mitochondrial ATP synthesis coupled electron transport | 2 | 4 | 2.648e-02 |
| GO:BP | GO:0009212 | pyrimidine deoxyribonucleoside triphosphate biosynthetic process | 2 | 4 | 2.648e-02 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 112 | 3993 | 2.648e-02 |
| GO:BP | GO:0051754 | meiotic sister chromatid cohesion, centromeric | 2 | 4 | 2.648e-02 |
| GO:BP | GO:0006235 | dTTP biosynthetic process | 2 | 4 | 2.648e-02 |
| GO:BP | GO:0007079 | mitotic chromosome movement towards spindle pole | 2 | 4 | 2.648e-02 |
| GO:BP | GO:0046075 | dTTP metabolic process | 2 | 4 | 2.648e-02 |
| GO:BP | GO:1990086 | lens fiber cell apoptotic process | 2 | 4 | 2.648e-02 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 6 | 65 | 2.835e-02 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 4 | 28 | 2.913e-02 |
| GO:BP | GO:0033262 | regulation of nuclear cell cycle DNA replication | 3 | 14 | 2.996e-02 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 3 | 14 | 2.996e-02 |
| GO:BP | GO:0009888 | tissue development | 63 | 2032 | 3.049e-02 |
| GO:BP | GO:0006284 | base-excision repair | 5 | 46 | 3.049e-02 |
| GO:BP | GO:0016477 | cell migration | 50 | 1534 | 3.119e-02 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 7 | 89 | 3.198e-02 |
| GO:BP | GO:0035418 | protein localization to synapse | 7 | 89 | 3.198e-02 |
| GO:BP | GO:0000723 | telomere maintenance | 10 | 164 | 3.198e-02 |
| GO:BP | GO:0070977 | bone maturation | 4 | 29 | 3.253e-02 |
| GO:BP | GO:0010165 | response to X-ray | 4 | 29 | 3.253e-02 |
| GO:BP | GO:0006915 | apoptotic process | 60 | 1923 | 3.277e-02 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 5 | 47 | 3.298e-02 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 10 | 165 | 3.298e-02 |
| GO:BP | GO:0071772 | response to BMP | 10 | 165 | 3.298e-02 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 7 | 90 | 3.356e-02 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 8 | 114 | 3.365e-02 |
| GO:BP | GO:0071763 | nuclear membrane organization | 5 | 48 | 3.558e-02 |
| GO:BP | GO:0044027 | negative regulation of gene expression via chromosomal CpG island methylation | 3 | 15 | 3.558e-02 |
| GO:BP | GO:0043101 | purine-containing compound salvage | 3 | 15 | 3.558e-02 |
| GO:BP | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway | 3 | 15 | 3.558e-02 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 7 | 92 | 3.738e-02 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 7 | 92 | 3.738e-02 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 13 | 254 | 3.817e-02 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 21 | 506 | 3.817e-02 |
| GO:BP | GO:0097421 | liver regeneration | 4 | 31 | 4.040e-02 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 2 | 5 | 4.052e-02 |
| GO:BP | GO:0071962 | mitotic sister chromatid cohesion, centromeric | 2 | 5 | 4.052e-02 |
| GO:BP | GO:0031536 | positive regulation of exit from mitosis | 2 | 5 | 4.052e-02 |
| GO:BP | GO:0043137 | DNA replication, removal of RNA primer | 2 | 5 | 4.052e-02 |
| GO:BP | GO:0098535 | de novo centriole assembly involved in multi-ciliated epithelial cell differentiation | 2 | 5 | 4.052e-02 |
| GO:BP | GO:0140499 | negative regulation of mitotic spindle assembly checkpoint signaling | 2 | 5 | 4.052e-02 |
| GO:BP | GO:0090233 | negative regulation of spindle checkpoint | 2 | 5 | 4.052e-02 |
| GO:BP | GO:0051311 | meiotic metaphase chromosome alignment | 2 | 5 | 4.052e-02 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 10 | 171 | 4.078e-02 |
| GO:BP | GO:0050918 | positive chemotaxis | 6 | 71 | 4.097e-02 |
| GO:BP | GO:0012501 | programmed cell death | 61 | 1987 | 4.118e-02 |
| GO:BP | GO:0090399 | replicative senescence | 3 | 16 | 4.152e-02 |
| GO:BP | GO:0051782 | negative regulation of cell division | 3 | 16 | 4.152e-02 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 59 | 1912 | 4.241e-02 |
| GO:BP | GO:0008219 | cell death | 61 | 1991 | 4.268e-02 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 4 | 32 | 4.399e-02 |
| GO:BP | GO:0042311 | vasodilation | 5 | 51 | 4.406e-02 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 5 | 51 | 4.406e-02 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 5 | 51 | 4.406e-02 |
| GO:BP | GO:0090102 | cochlea development | 5 | 51 | 4.406e-02 |
| GO:BP | GO:0048468 | cell development | 83 | 2876 | 4.483e-02 |
| GO:BP | GO:0051261 | protein depolymerization | 8 | 121 | 4.529e-02 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 8 | 121 | 4.529e-02 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 93 | 3288 | 4.594e-02 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 8 | 122 | 4.738e-02 |
| GO:BP | GO:0002063 | chondrocyte development | 4 | 33 | 4.749e-02 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 21 | 519 | 4.749e-02 |
| GO:BP | GO:0046661 | male sex differentiation | 10 | 176 | 4.749e-02 |
| GO:BP | GO:0046653 | tetrahydrofolate metabolic process | 3 | 17 | 4.749e-02 |
| GO:BP | GO:0014841 | skeletal muscle satellite cell proliferation | 3 | 17 | 4.749e-02 |
| GO:BP | GO:0031573 | mitotic intra-S DNA damage checkpoint signaling | 3 | 17 | 4.749e-02 |
| GO:BP | GO:0048799 | animal organ maturation | 4 | 33 | 4.749e-02 |
| GO:BP | GO:0043094 | metabolic compound salvage | 4 | 33 | 4.749e-02 |
| GO:BP | GO:0043173 | nucleotide salvage | 3 | 17 | 4.749e-02 |
| GO:BP | GO:0007063 | regulation of sister chromatid cohesion | 3 | 17 | 4.749e-02 |
| GO:BP | GO:0085020 | protein K6-linked ubiquitination | 3 | 17 | 4.749e-02 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 8 | 123 | 4.863e-02 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 7 | 98 | 4.872e-02 |
| GO:BP | GO:0046785 | microtubule polymerization | 7 | 98 | 4.872e-02 |
| KEGG | KEGG:04110 | Cell cycle | 46 | 157 | 4.527e-33 |
| KEGG | KEGG:03030 | DNA replication | 16 | 36 | 3.615e-14 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 12 | 54 | 1.440e-06 |
| KEGG | KEGG:04114 | Oocyte meiosis | 17 | 131 | 4.452e-06 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 15 | 102 | 4.452e-06 |
| KEGG | KEGG:03440 | Homologous recombination | 10 | 41 | 4.452e-06 |
| KEGG | KEGG:04115 | p53 signaling pathway | 12 | 74 | 2.423e-05 |
| KEGG | KEGG:04218 | Cellular senescence | 16 | 155 | 1.585e-04 |
| KEGG | KEGG:04814 | Motor proteins | 17 | 193 | 5.981e-04 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 17 | 218 | 2.517e-03 |
| KEGG | KEGG:03410 | Base excision repair | 7 | 44 | 3.885e-03 |
| KEGG | KEGG:03430 | Mismatch repair | 5 | 23 | 6.852e-03 |
| KEGG | KEGG:05222 | Small cell lung cancer | 9 | 92 | 1.611e-02 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 7 | 57 | 1.611e-02 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 8 | 84 | 3.409e-02 |
| KEGG | KEGG:04512 | ECM-receptor interaction | 8 | 89 | 4.604e-02 |
# write.csv(table_motif2_GOKEGG_genes, "data/new/table_motif2_GOKEGG_genes.csv")
#GO:BP
table_motif2_genes_GOBP_RUV <- table_motif2_GOKEGG_genes_RUV %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
# saveRDS(table_motif2_genes_GOBP, "data/table_motif2_genes_GOBP.RDS")
table_motif2_genes_GOBP_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("M2 Enriched GO:BP Terms RUVs")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))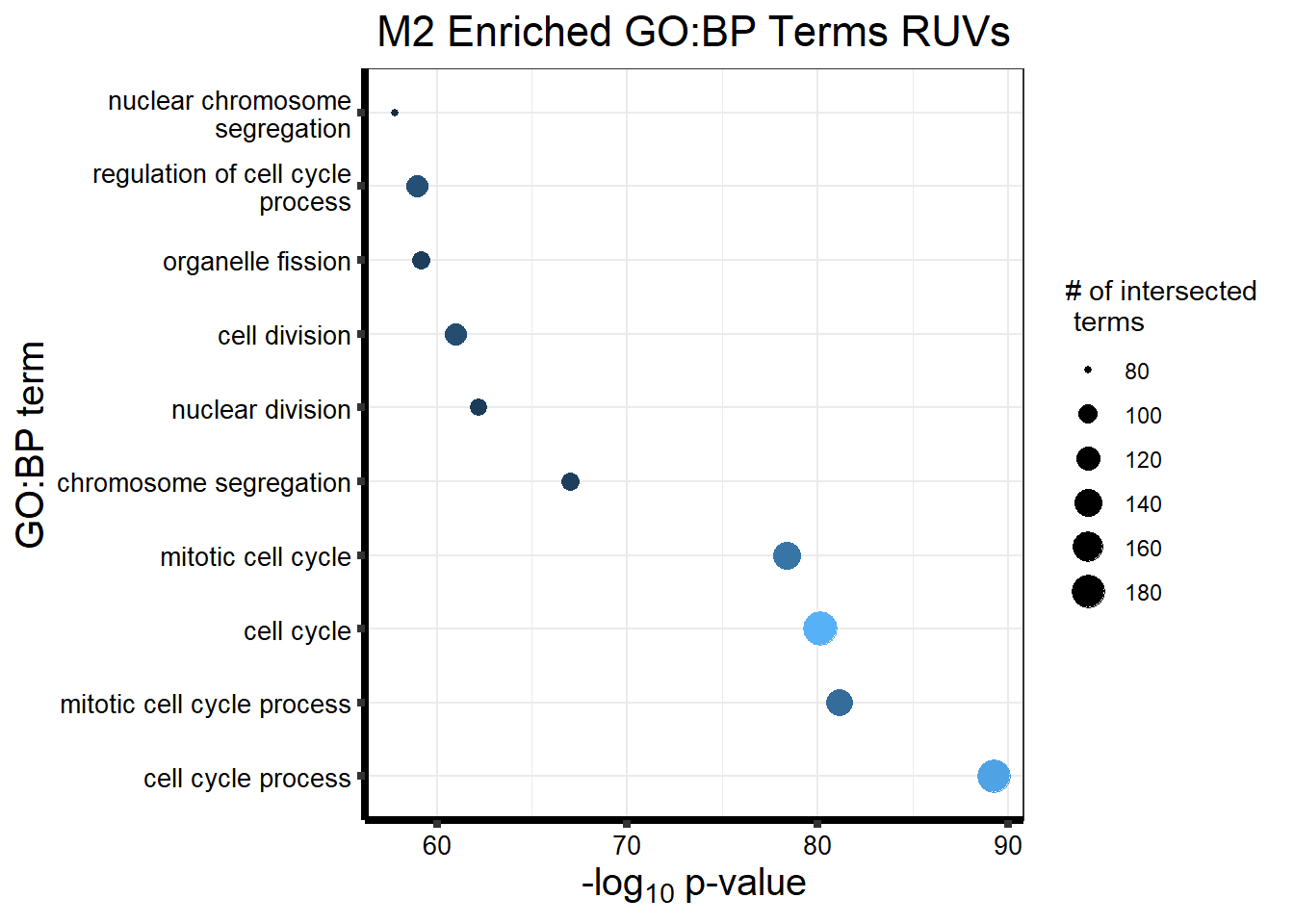
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#KEGG
table_motif2_genes_KEGG_RUV <- table_motif2_GOKEGG_genes_RUV %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_motif2_genes_KEGG_RUV %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("M2 DEGs Enriched KEGG Terms RUVs")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))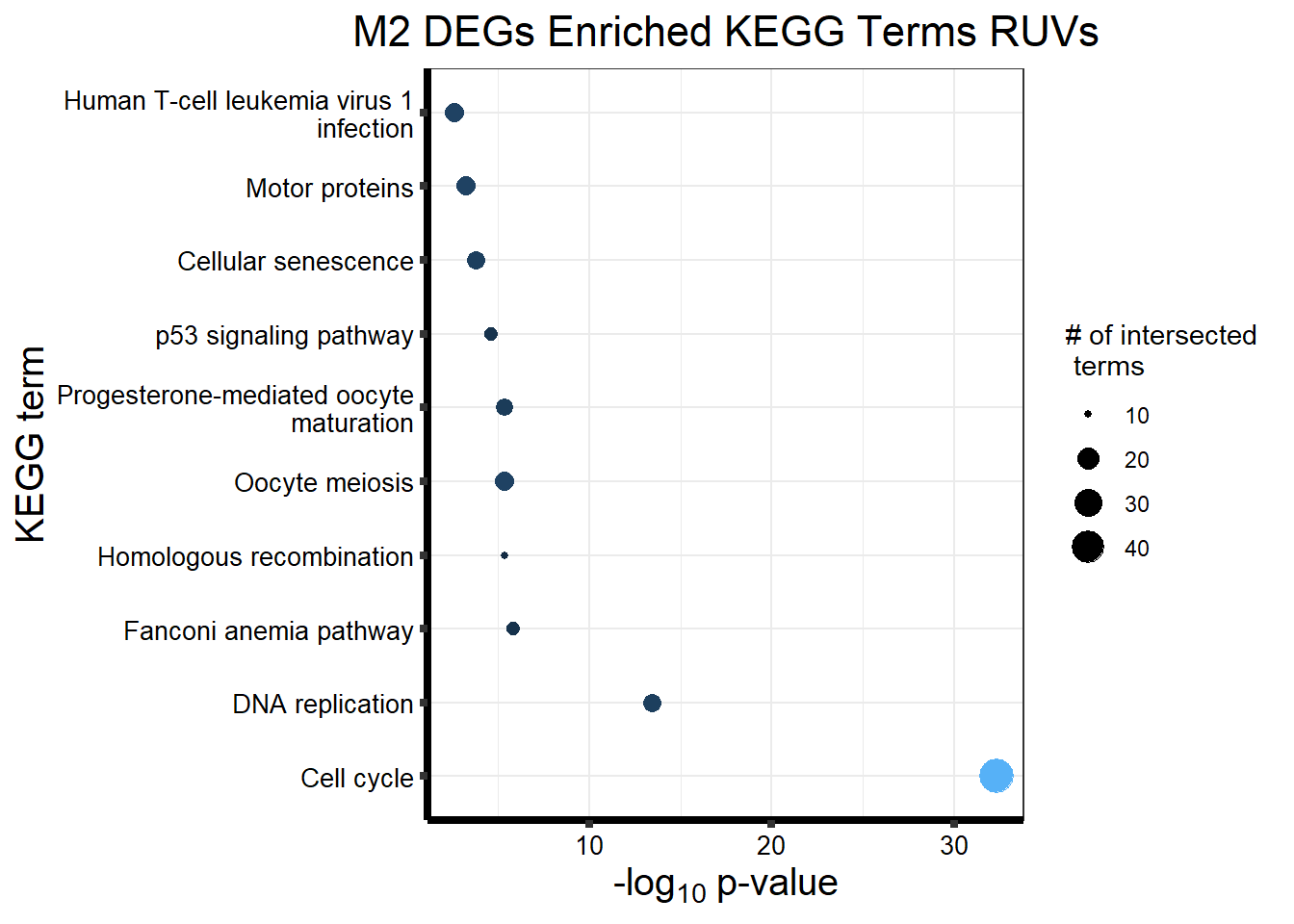
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#now take these final gene sets for each motif - clustlike + p.post consensus genes
#clust1_genes_RUV
#clust2_genes_RUV
# library(gprofiler2)
#####Motif 1 Gene Set#####
# motif1_genes_matrix_RUV_clust <- as.matrix(clust1_genes_RUV)
# colnames(motif1_genes_matrix_RUV_clust) <- c("entrezgene_ID")
# saveRDS(motif1_genes_matrix_RUV_clust, "data/new/RUV/CMF/motif1_genes_matrix_RUV_clustlike.RDS")
motif1_genes_matrix_RUV_clust <- readRDS("data/new/RUV/CMF/motif1_genes_matrix_RUV_clustlike.RDS")
length(motif1_genes_matrix_RUV_clust)[1] 13600#13600 genes in this set for motif 1
motif1_mat_GOKEGG_RUV_clust <- gost(query = motif1_genes_matrix_RUV_clust,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
motif1_GOKEGG_genes_RUV_clust <- gostplot(motif1_mat_GOKEGG_RUV_clust, capped = FALSE, interactive = TRUE)
motif1_GOKEGG_genes_RUV_clusttable_motif1_GOKEGG_genes_RUV_clust <- motif1_mat_GOKEGG_RUV_clust$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_motif1_GOKEGG_genes_RUV_clust %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0019538 | protein metabolic process | 3572 | 4721 | 6.575e-248 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 3961 | 5390 | 1.663e-235 |
| GO:BP | GO:0006996 | organelle organization | 2772 | 3594 | 5.707e-204 |
| GO:BP | GO:0043412 | macromolecule modification | 2363 | 3030 | 1.811e-180 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 4134 | 5920 | 2.252e-172 |
| GO:BP | GO:0048518 | positive regulation of biological process | 4329 | 6264 | 4.298e-169 |
| GO:BP | GO:0046907 | intracellular transport | 1214 | 1381 | 4.334e-164 |
| GO:BP | GO:0036211 | protein modification process | 2200 | 2846 | 7.300e-158 |
| GO:BP | GO:0051641 | cellular localization | 2713 | 3661 | 1.013e-154 |
| GO:BP | GO:0009056 | catabolic process | 2054 | 2639 | 4.245e-152 |
| GO:BP | GO:0019219 | regulation of nucleobase-containing compound metabolic process | 2911 | 3990 | 1.232e-151 |
| GO:BP | GO:0065007 | biological regulation | 7872 | 12743 | 8.913e-149 |
| GO:BP | GO:0051179 | localization | 3836 | 5555 | 2.391e-142 |
| GO:BP | GO:0050789 | regulation of biological process | 7630 | 12336 | 2.999e-140 |
| GO:BP | GO:0051252 | regulation of RNA metabolic process | 2693 | 3687 | 4.054e-139 |
| GO:BP | GO:0051649 | establishment of localization in cell | 1611 | 2010 | 2.010e-137 |
| GO:BP | GO:0050794 | regulation of cellular process | 7402 | 11946 | 4.870e-134 |
| GO:BP | GO:0006810 | transport | 3110 | 4407 | 4.569e-129 |
| GO:BP | GO:0033554 | cellular response to stress | 1474 | 1830 | 3.304e-128 |
| GO:BP | GO:0006351 | DNA-templated transcription | 2579 | 3549 | 1.662e-127 |
| GO:BP | GO:0009057 | macromolecule catabolic process | 1187 | 1417 | 1.738e-125 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 1864 | 2433 | 4.395e-124 |
| GO:BP | GO:0070727 | cellular macromolecule localization | 2086 | 2782 | 4.661e-123 |
| GO:BP | GO:0033036 | macromolecule localization | 2367 | 3228 | 9.817e-123 |
| GO:BP | GO:0008104 | protein localization | 2075 | 2770 | 1.402e-121 |
| GO:BP | GO:0006355 | regulation of DNA-templated transcription | 2470 | 3409 | 1.103e-118 |
| GO:BP | GO:2001141 | regulation of RNA biosynthetic process | 2481 | 3428 | 2.229e-118 |
| GO:BP | GO:0009893 | positive regulation of metabolic process | 2578 | 3597 | 2.059e-115 |
| GO:BP | GO:0019222 | regulation of metabolic process | 4610 | 7035 | 2.064e-113 |
| GO:BP | GO:0007275 | multicellular organism development | 3255 | 4727 | 3.294e-112 |
| GO:BP | GO:0023051 | regulation of signaling | 2494 | 3478 | 2.199e-111 |
| GO:BP | GO:0010646 | regulation of cell communication | 2498 | 3486 | 4.892e-111 |
| GO:BP | GO:0015031 | protein transport | 1180 | 1444 | 2.458e-109 |
| GO:BP | GO:0035556 | intracellular signal transduction | 2165 | 2965 | 1.692e-107 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 1450 | 1855 | 3.472e-106 |
| GO:BP | GO:0009966 | regulation of signal transduction | 2201 | 3034 | 1.188e-104 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 2355 | 3288 | 3.280e-103 |
| GO:BP | GO:0032502 | developmental process | 4297 | 6553 | 4.417e-103 |
| GO:BP | GO:0071705 | nitrogen compound transport | 1488 | 1923 | 7.698e-103 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 1799 | 2407 | 5.284e-102 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 1800 | 2410 | 1.296e-101 |
| GO:BP | GO:0048856 | anatomical structure development | 3968 | 5997 | 4.295e-101 |
| GO:BP | GO:0051234 | establishment of localization | 3337 | 4928 | 4.577e-100 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 2770 | 3993 | 1.190e-97 |
| GO:BP | GO:0030163 | protein catabolic process | 874 | 1030 | 2.765e-97 |
| GO:BP | GO:0048731 | system development | 2799 | 4053 | 2.940e-95 |
| GO:BP | GO:0006366 | transcription by RNA polymerase II | 1964 | 2711 | 6.088e-91 |
| GO:BP | GO:0060255 | regulation of macromolecule metabolic process | 4215 | 6492 | 4.354e-90 |
| GO:BP | GO:0009894 | regulation of catabolic process | 866 | 1040 | 2.886e-87 |
| GO:BP | GO:0045935 | positive regulation of nucleobase-containing compound metabolic process | 1525 | 2036 | 4.086e-86 |
| GO:BP | GO:0051603 | proteolysis involved in protein catabolic process | 683 | 786 | 5.449e-85 |
| GO:BP | GO:0006357 | regulation of transcription by RNA polymerase II | 1863 | 2578 | 4.008e-84 |
| GO:BP | GO:0016192 | vesicle-mediated transport | 1224 | 1592 | 5.152e-80 |
| GO:BP | GO:0007399 | nervous system development | 1864 | 2604 | 1.815e-78 |
| GO:BP | GO:0043687 | post-translational protein modification | 839 | 1027 | 3.781e-76 |
| GO:BP | GO:0006886 | intracellular protein transport | 596 | 686 | 9.810e-74 |
| GO:BP | GO:0051254 | positive regulation of RNA metabolic process | 1367 | 1835 | 1.107e-73 |
| GO:BP | GO:0070647 | protein modification by small protein conjugation or removal | 798 | 975 | 5.319e-73 |
| GO:BP | GO:0030030 | cell projection organization | 1234 | 1631 | 5.602e-73 |
| GO:BP | GO:0032446 | protein modification by small protein conjugation | 713 | 853 | 1.056e-72 |
| GO:BP | GO:0045184 | establishment of protein localization | 1427 | 1937 | 1.249e-71 |
| GO:BP | GO:0043632 | modification-dependent macromolecule catabolic process | 562 | 643 | 1.423e-71 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 3645 | 5629 | 2.042e-71 |
| GO:BP | GO:0120036 | plasma membrane bounded cell projection organization | 1201 | 1588 | 1.010e-70 |
| GO:BP | GO:0019941 | modification-dependent protein catabolic process | 557 | 638 | 1.581e-70 |
| GO:BP | GO:0048519 | negative regulation of biological process | 3757 | 5834 | 7.605e-70 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 1432 | 1953 | 1.287e-69 |
| GO:BP | GO:0061919 | process utilizing autophagic mechanism | 525 | 597 | 7.456e-69 |
| GO:BP | GO:0006914 | autophagy | 525 | 597 | 7.456e-69 |
| GO:BP | GO:0010498 | proteasomal protein catabolic process | 485 | 543 | 1.244e-68 |
| GO:BP | GO:0033043 | regulation of organelle organization | 907 | 1148 | 1.665e-68 |
| GO:BP | GO:0006511 | ubiquitin-dependent protein catabolic process | 547 | 628 | 1.897e-68 |
| GO:BP | GO:0006412 | translation | 614 | 727 | 8.566e-66 |
| GO:BP | GO:0042254 | ribosome biogenesis | 312 | 323 | 2.994e-65 |
| GO:BP | GO:0009891 | positive regulation of biosynthetic process | 1877 | 2696 | 1.271e-63 |
| GO:BP | GO:0070925 | organelle assembly | 829 | 1046 | 1.789e-63 |
| GO:BP | GO:1902680 | positive regulation of RNA biosynthetic process | 1255 | 1699 | 1.789e-63 |
| GO:BP | GO:0045893 | positive regulation of DNA-templated transcription | 1253 | 1697 | 3.430e-63 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 801 | 1011 | 4.123e-61 |
| GO:BP | GO:0010557 | positive regulation of macromolecule biosynthetic process | 1806 | 2593 | 4.190e-61 |
| GO:BP | GO:0008219 | cell death | 1433 | 1991 | 5.241e-61 |
| GO:BP | GO:0016567 | protein ubiquitination | 635 | 768 | 6.842e-61 |
| GO:BP | GO:0012501 | programmed cell death | 1429 | 1987 | 1.782e-60 |
| GO:BP | GO:0051716 | cellular response to stimulus | 4594 | 7376 | 1.806e-60 |
| GO:BP | GO:0009653 | anatomical structure morphogenesis | 1877 | 2713 | 2.144e-60 |
| GO:BP | GO:0006915 | apoptotic process | 1388 | 1923 | 3.553e-60 |
| GO:BP | GO:0032879 | regulation of localization | 1454 | 2029 | 4.878e-60 |
| GO:BP | GO:0006974 | DNA damage response | 726 | 908 | 3.782e-58 |
| GO:BP | GO:0044238 | primary metabolic process | 7310 | 12342 | 4.093e-58 |
| GO:BP | GO:0022008 | neurogenesis | 1286 | 1771 | 7.569e-58 |
| GO:BP | GO:0023057 | negative regulation of signaling | 1072 | 1435 | 7.610e-58 |
| GO:BP | GO:0009968 | negative regulation of signal transduction | 1001 | 1326 | 1.366e-57 |
| GO:BP | GO:0010648 | negative regulation of cell communication | 1072 | 1436 | 1.382e-57 |
| GO:BP | GO:0065008 | regulation of biological quality | 2009 | 2947 | 1.996e-57 |
| GO:BP | GO:0016236 | macroautophagy | 336 | 362 | 3.150e-57 |
| GO:BP | GO:0080135 | regulation of cellular response to stress | 453 | 519 | 4.695e-57 |
| GO:BP | GO:0050793 | regulation of developmental process | 1708 | 2457 | 1.601e-56 |
| GO:BP | GO:0009889 | regulation of biosynthetic process | 3681 | 5809 | 1.026e-55 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 1366 | 1912 | 8.064e-55 |
| GO:BP | GO:0048585 | negative regulation of response to stimulus | 1220 | 1680 | 1.088e-54 |
| GO:BP | GO:0006950 | response to stress | 2594 | 3948 | 2.765e-54 |
| GO:BP | GO:0007010 | cytoskeleton organization | 1122 | 1529 | 8.169e-54 |
| GO:BP | GO:0007005 | mitochondrion organization | 387 | 435 | 1.773e-53 |
| GO:BP | GO:0048869 | cellular developmental process | 2878 | 4438 | 1.811e-53 |
| GO:BP | GO:0030154 | cell differentiation | 2877 | 4437 | 2.248e-53 |
| GO:BP | GO:0060341 | regulation of cellular localization | 786 | 1013 | 5.061e-53 |
| GO:BP | GO:0016072 | rRNA metabolic process | 256 | 266 | 2.314e-52 |
| GO:BP | GO:0045934 | negative regulation of nucleobase-containing compound metabolic process | 1118 | 1529 | 2.680e-52 |
| GO:BP | GO:0006508 | proteolysis | 1090 | 1486 | 4.849e-52 |
| GO:BP | GO:0061024 | membrane organization | 655 | 821 | 1.178e-51 |
| GO:BP | GO:0019637 | organophosphate metabolic process | 825 | 1077 | 1.453e-51 |
| GO:BP | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 369 | 414 | 1.939e-51 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 843 | 1105 | 2.020e-51 |
| GO:BP | GO:0010556 | regulation of macromolecule biosynthetic process | 3564 | 5644 | 5.811e-51 |
| GO:BP | GO:0051248 | negative regulation of protein metabolic process | 555 | 677 | 1.139e-50 |
| GO:BP | GO:0065009 | regulation of molecular function | 1091 | 1496 | 5.031e-50 |
| GO:BP | GO:0051253 | negative regulation of RNA metabolic process | 1029 | 1400 | 1.196e-49 |
| GO:BP | GO:0048468 | cell development | 1935 | 2876 | 9.615e-49 |
| GO:BP | GO:0048699 | generation of neurons | 1110 | 1536 | 1.028e-47 |
| GO:BP | GO:0006364 | rRNA processing | 219 | 225 | 1.463e-47 |
| GO:BP | GO:0051247 | positive regulation of protein metabolic process | 788 | 1039 | 3.751e-46 |
| GO:BP | GO:0007049 | cell cycle | 1185 | 1663 | 4.383e-46 |
| GO:BP | GO:0010468 | regulation of gene expression | 3478 | 5536 | 5.152e-46 |
| GO:BP | GO:0032880 | regulation of protein localization | 701 | 907 | 5.711e-46 |
| GO:BP | GO:0031399 | regulation of protein modification process | 794 | 1049 | 6.074e-46 |
| GO:BP | GO:0030029 | actin filament-based process | 646 | 825 | 1.094e-45 |
| GO:BP | GO:0016310 | phosphorylation | 967 | 1320 | 2.004e-45 |
| GO:BP | GO:0044281 | small molecule metabolic process | 1253 | 1776 | 2.518e-45 |
| GO:BP | GO:0009896 | positive regulation of catabolic process | 455 | 546 | 3.831e-45 |
| GO:BP | GO:0072359 | circulatory system development | 854 | 1145 | 4.735e-45 |
| GO:BP | GO:0048193 | Golgi vesicle transport | 285 | 312 | 9.983e-45 |
| GO:BP | GO:0031175 | neuron projection development | 774 | 1023 | 1.251e-44 |
| GO:BP | GO:0016197 | endosomal transport | 268 | 290 | 1.542e-44 |
| GO:BP | GO:0030182 | neuron differentiation | 1047 | 1451 | 2.316e-44 |
| GO:BP | GO:0048666 | neuron development | 872 | 1177 | 4.989e-44 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 5077 | 8393 | 5.164e-44 |
| GO:BP | GO:1901135 | carbohydrate derivative metabolic process | 745 | 983 | 2.319e-43 |
| GO:BP | GO:0006259 | DNA metabolic process | 759 | 1005 | 3.161e-43 |
| GO:BP | GO:0007507 | heart development | 492 | 605 | 7.292e-43 |
| GO:BP | GO:0045892 | negative regulation of DNA-templated transcription | 934 | 1280 | 1.326e-42 |
| GO:BP | GO:1902679 | negative regulation of RNA biosynthetic process | 943 | 1296 | 3.530e-42 |
| GO:BP | GO:0050896 | response to stimulus | 5400 | 8999 | 7.766e-42 |
| GO:BP | GO:0031344 | regulation of cell projection organization | 526 | 658 | 1.054e-41 |
| GO:BP | GO:0043067 | regulation of programmed cell death | 1073 | 1506 | 2.004e-41 |
| GO:BP | GO:0030036 | actin cytoskeleton organization | 572 | 729 | 8.302e-41 |
| GO:BP | GO:1902532 | negative regulation of intracellular signal transduction | 527 | 662 | 8.333e-41 |
| GO:BP | GO:0042981 | regulation of apoptotic process | 1042 | 1462 | 2.986e-40 |
| GO:BP | GO:0000902 | cell morphogenesis | 745 | 996 | 8.102e-40 |
| GO:BP | GO:0048513 | animal organ development | 2023 | 3085 | 1.311e-39 |
| GO:BP | GO:0120035 | regulation of plasma membrane bounded cell projection organization | 511 | 642 | 1.681e-39 |
| GO:BP | GO:0010256 | endomembrane system organization | 491 | 613 | 2.647e-39 |
| GO:BP | GO:0140053 | mitochondrial gene expression | 166 | 168 | 2.916e-39 |
| GO:BP | GO:0034655 | nucleobase-containing compound catabolic process | 437 | 535 | 6.547e-39 |
| GO:BP | GO:0006468 | protein phosphorylation | 890 | 1227 | 8.285e-39 |
| GO:BP | GO:0010506 | regulation of autophagy | 318 | 367 | 2.157e-38 |
| GO:BP | GO:0090407 | organophosphate biosynthetic process | 492 | 619 | 1.083e-37 |
| GO:BP | GO:0051049 | regulation of transport | 1123 | 1607 | 2.520e-37 |
| GO:BP | GO:0071495 | cellular response to endogenous stimulus | 882 | 1223 | 7.427e-37 |
| GO:BP | GO:0048584 | positive regulation of response to stimulus | 1562 | 2331 | 7.655e-37 |
| GO:BP | GO:0018193 | peptidyl-amino acid modification | 535 | 686 | 9.403e-37 |
| GO:BP | GO:0097435 | supramolecular fiber organization | 650 | 861 | 1.019e-36 |
| GO:BP | GO:0061061 | muscle structure development | 539 | 693 | 1.921e-36 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 1047 | 1489 | 2.043e-36 |
| GO:BP | GO:0007167 | enzyme-linked receptor protein signaling pathway | 719 | 970 | 3.994e-36 |
| GO:BP | GO:0009967 | positive regulation of signal transduction | 1101 | 1578 | 4.391e-36 |
| GO:BP | GO:0006417 | regulation of translation | 324 | 380 | 4.861e-36 |
| GO:BP | GO:0051239 | regulation of multicellular organismal process | 1930 | 2956 | 8.509e-36 |
| GO:BP | GO:0023052 | signaling | 3977 | 6515 | 1.141e-35 |
| GO:BP | GO:0006281 | DNA repair | 489 | 621 | 1.700e-35 |
| GO:BP | GO:0006913 | nucleocytoplasmic transport | 290 | 334 | 2.396e-35 |
| GO:BP | GO:0051169 | nuclear transport | 290 | 334 | 2.396e-35 |
| GO:BP | GO:0023056 | positive regulation of signaling | 1232 | 1796 | 2.903e-35 |
| GO:BP | GO:0007154 | cell communication | 3987 | 6540 | 5.347e-35 |
| GO:BP | GO:0032501 | multicellular organismal process | 4424 | 7322 | 1.068e-34 |
| GO:BP | GO:0042176 | regulation of protein catabolic process | 314 | 369 | 1.445e-34 |
| GO:BP | GO:0010647 | positive regulation of cell communication | 1228 | 1795 | 2.577e-34 |
| GO:BP | GO:0006325 | chromatin organization | 659 | 884 | 2.621e-34 |
| GO:BP | GO:0031647 | regulation of protein stability | 289 | 335 | 4.279e-34 |
| GO:BP | GO:0051726 | regulation of cell cycle | 788 | 1087 | 4.876e-34 |
| GO:BP | GO:0030705 | cytoskeleton-dependent intracellular transport | 195 | 209 | 6.834e-34 |
| GO:BP | GO:0080134 | regulation of response to stress | 975 | 1387 | 1.052e-33 |
| GO:BP | GO:0009790 | embryo development | 817 | 1135 | 1.424e-33 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 537 | 701 | 4.180e-33 |
| GO:BP | GO:0006399 | tRNA metabolic process | 195 | 210 | 4.322e-33 |
| GO:BP | GO:0045944 | positive regulation of transcription by RNA polymerase II | 891 | 1256 | 6.269e-33 |
| GO:BP | GO:0055086 | nucleobase-containing small molecule metabolic process | 518 | 673 | 7.384e-33 |
| GO:BP | GO:0006753 | nucleoside phosphate metabolic process | 500 | 646 | 8.648e-33 |
| GO:BP | GO:0009987 | cellular process | 11205 | 20247 | 9.438e-33 |
| GO:BP | GO:0034330 | cell junction organization | 622 | 834 | 2.002e-32 |
| GO:BP | GO:0048858 | cell projection morphogenesis | 522 | 681 | 2.739e-32 |
| GO:BP | GO:0006091 | generation of precursor metabolites and energy | 386 | 478 | 3.189e-32 |
| GO:BP | GO:0120039 | plasma membrane bounded cell projection morphogenesis | 518 | 676 | 5.672e-32 |
| GO:BP | GO:0033365 | protein localization to organelle | 858 | 1209 | 9.395e-32 |
| GO:BP | GO:0022402 | cell cycle process | 902 | 1280 | 1.139e-31 |
| GO:BP | GO:0031400 | negative regulation of protein modification process | 295 | 349 | 2.336e-31 |
| GO:BP | GO:0030031 | cell projection assembly | 475 | 613 | 2.455e-31 |
| GO:BP | GO:0034976 | response to endoplasmic reticulum stress | 238 | 270 | 3.002e-31 |
| GO:BP | GO:0051640 | organelle localization | 479 | 620 | 4.825e-31 |
| GO:BP | GO:0000209 | protein polyubiquitination | 245 | 280 | 5.196e-31 |
| GO:BP | GO:0040007 | growth | 679 | 929 | 6.276e-31 |
| GO:BP | GO:0000278 | mitotic cell cycle | 655 | 892 | 8.723e-31 |
| GO:BP | GO:0016050 | vesicle organization | 313 | 376 | 9.791e-31 |
| GO:BP | GO:0120031 | plasma membrane bounded cell projection assembly | 465 | 600 | 1.060e-30 |
| GO:BP | GO:0016043 | cellular component organization | 4877 | 8184 | 1.306e-30 |
| GO:BP | GO:0051604 | protein maturation | 424 | 539 | 1.538e-30 |
| GO:BP | GO:0070848 | response to growth factor | 544 | 721 | 1.687e-30 |
| GO:BP | GO:0007166 | cell surface receptor signaling pathway | 1824 | 2819 | 2.402e-30 |
| GO:BP | GO:0097190 | apoptotic signaling pathway | 468 | 606 | 2.953e-30 |
| GO:BP | GO:0071363 | cellular response to growth factor stimulus | 522 | 689 | 5.091e-30 |
| GO:BP | GO:0048812 | neuron projection morphogenesis | 503 | 660 | 5.217e-30 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 391 | 492 | 7.629e-30 |
| GO:BP | GO:0010970 | transport along microtubule | 167 | 178 | 7.988e-30 |
| GO:BP | GO:1901137 | carbohydrate derivative biosynthetic process | 459 | 594 | 8.971e-30 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 675 | 928 | 1.010e-29 |
| GO:BP | GO:0141188 | nucleic acid catabolic process | 288 | 343 | 1.396e-29 |
| GO:BP | GO:0007033 | vacuole organization | 213 | 239 | 1.567e-29 |
| GO:BP | GO:0032543 | mitochondrial translation | 128 | 130 | 1.567e-29 |
| GO:BP | GO:0034504 | protein localization to nucleus | 270 | 318 | 2.264e-29 |
| GO:BP | GO:0000122 | negative regulation of transcription by RNA polymerase II | 680 | 938 | 3.022e-29 |
| GO:BP | GO:0031346 | positive regulation of cell projection organization | 297 | 357 | 4.523e-29 |
| GO:BP | GO:0002181 | cytoplasmic translation | 156 | 165 | 5.878e-29 |
| GO:BP | GO:1903311 | regulation of mRNA metabolic process | 265 | 312 | 7.048e-29 |
| GO:BP | GO:0006402 | mRNA catabolic process | 228 | 261 | 1.260e-28 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 405 | 517 | 1.852e-28 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 346 | 430 | 2.683e-28 |
| GO:BP | GO:1901873 | regulation of post-translational protein modification | 215 | 244 | 3.557e-28 |
| GO:BP | GO:0006401 | RNA catabolic process | 266 | 315 | 3.931e-28 |
| GO:BP | GO:0016241 | regulation of macroautophagy | 170 | 184 | 3.935e-28 |
| GO:BP | GO:0035295 | tube development | 779 | 1101 | 4.500e-28 |
| GO:BP | GO:0051650 | establishment of vesicle localization | 182 | 200 | 5.939e-28 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 642 | 884 | 6.319e-28 |
| GO:BP | GO:0016055 | Wnt signaling pathway | 364 | 458 | 8.786e-28 |
| GO:BP | GO:0051648 | vesicle localization | 196 | 219 | 9.654e-28 |
| GO:BP | GO:0007264 | small GTPase-mediated signal transduction | 395 | 505 | 1.658e-27 |
| GO:BP | GO:0022603 | regulation of anatomical structure morphogenesis | 607 | 831 | 1.827e-27 |
| GO:BP | GO:0048667 | cell morphogenesis involved in neuron differentiation | 457 | 599 | 2.035e-27 |
| GO:BP | GO:0010975 | regulation of neuron projection development | 355 | 446 | 2.509e-27 |
| GO:BP | GO:1902903 | regulation of supramolecular fiber organization | 312 | 383 | 2.680e-27 |
| GO:BP | GO:0009888 | tissue development | 1343 | 2032 | 3.001e-27 |
| GO:BP | GO:0062197 | cellular response to chemical stress | 266 | 317 | 3.117e-27 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 391 | 500 | 3.383e-27 |
| GO:BP | GO:0048646 | anatomical structure formation involved in morphogenesis | 837 | 1199 | 3.459e-27 |
| GO:BP | GO:1903320 | regulation of protein modification by small protein conjugation or removal | 210 | 239 | 3.777e-27 |
| GO:BP | GO:0007165 | signal transduction | 3633 | 6002 | 4.274e-27 |
| GO:BP | GO:0044089 | positive regulation of cellular component biogenesis | 394 | 505 | 4.839e-27 |
| GO:BP | GO:0072521 | purine-containing compound metabolic process | 432 | 563 | 7.649e-27 |
| GO:BP | GO:0016049 | cell growth | 380 | 485 | 9.941e-27 |
| GO:BP | GO:1901698 | response to nitrogen compound | 761 | 1080 | 1.522e-26 |
| GO:BP | GO:0001701 | in utero embryonic development | 331 | 413 | 1.752e-26 |
| GO:BP | GO:0008654 | phospholipid biosynthetic process | 222 | 257 | 2.339e-26 |
| GO:BP | GO:0008033 | tRNA processing | 133 | 139 | 3.096e-26 |
| GO:BP | GO:0045595 | regulation of cell differentiation | 1064 | 1576 | 3.798e-26 |
| GO:BP | GO:0051094 | positive regulation of developmental process | 922 | 1343 | 3.862e-26 |
| GO:BP | GO:0035239 | tube morphogenesis | 633 | 879 | 6.628e-26 |
| GO:BP | GO:0006979 | response to oxidative stress | 321 | 400 | 7.224e-26 |
| GO:BP | GO:1990778 | protein localization to cell periphery | 297 | 365 | 7.887e-26 |
| GO:BP | GO:0030162 | regulation of proteolysis | 325 | 406 | 7.887e-26 |
| GO:BP | GO:0006282 | regulation of DNA repair | 198 | 225 | 8.562e-26 |
| GO:BP | GO:0060537 | muscle tissue development | 341 | 430 | 1.044e-25 |
| GO:BP | GO:0099111 | microtubule-based transport | 194 | 220 | 1.567e-25 |
| GO:BP | GO:0043009 | chordate embryonic development | 499 | 671 | 1.945e-25 |
| GO:BP | GO:0006403 | RNA localization | 179 | 200 | 2.403e-25 |
| GO:BP | GO:0042592 | homeostatic process | 1157 | 1736 | 2.906e-25 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 512 | 692 | 3.094e-25 |
| GO:BP | GO:0014706 | striated muscle tissue development | 325 | 408 | 3.986e-25 |
| GO:BP | GO:0006354 | DNA-templated transcription elongation | 128 | 134 | 4.981e-25 |
| GO:BP | GO:0035966 | response to topologically incorrect protein | 146 | 157 | 5.941e-25 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 679 | 957 | 6.551e-25 |
| GO:BP | GO:0072659 | protein localization to plasma membrane | 244 | 291 | 6.864e-25 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 795 | 1144 | 7.637e-25 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 678 | 956 | 8.566e-25 |
| GO:BP | GO:0043254 | regulation of protein-containing complex assembly | 333 | 421 | 9.739e-25 |
| GO:BP | GO:0009117 | nucleotide metabolic process | 381 | 493 | 1.040e-24 |
| GO:BP | GO:0042274 | ribosomal small subunit biogenesis | 103 | 104 | 1.445e-24 |
| GO:BP | GO:0007017 | microtubule-based process | 704 | 999 | 1.575e-24 |
| GO:BP | GO:1903050 | regulation of proteolysis involved in protein catabolic process | 204 | 236 | 2.599e-24 |
| GO:BP | GO:0006163 | purine nucleotide metabolic process | 338 | 430 | 3.206e-24 |
| GO:BP | GO:0032386 | regulation of intracellular transport | 225 | 266 | 5.148e-24 |
| GO:BP | GO:0022411 | cellular component disassembly | 348 | 446 | 6.970e-24 |
| GO:BP | GO:0043069 | negative regulation of programmed cell death | 643 | 905 | 8.234e-24 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 542 | 745 | 8.356e-24 |
| GO:BP | GO:0009895 | negative regulation of catabolic process | 290 | 361 | 1.679e-23 |
| GO:BP | GO:0050808 | synapse organization | 424 | 563 | 1.978e-23 |
| GO:BP | GO:1903829 | positive regulation of protein localization | 382 | 499 | 2.221e-23 |
| GO:BP | GO:0006338 | chromatin remodeling | 528 | 725 | 2.378e-23 |
| GO:BP | GO:0050821 | protein stabilization | 194 | 224 | 2.412e-23 |
| GO:BP | GO:0016482 | cytosolic transport | 132 | 141 | 2.563e-23 |
| GO:BP | GO:0048738 | cardiac muscle tissue development | 201 | 234 | 3.319e-23 |
| GO:BP | GO:1902533 | positive regulation of intracellular signal transduction | 770 | 1113 | 3.711e-23 |
| GO:BP | GO:0005975 | carbohydrate metabolic process | 422 | 561 | 3.771e-23 |
| GO:BP | GO:0006839 | mitochondrial transport | 160 | 178 | 3.783e-23 |
| GO:BP | GO:0042692 | muscle cell differentiation | 319 | 405 | 3.931e-23 |
| GO:BP | GO:0006457 | protein folding | 190 | 219 | 4.542e-23 |
| GO:BP | GO:0007015 | actin filament organization | 356 | 461 | 5.597e-23 |
| GO:BP | GO:0044092 | negative regulation of molecular function | 416 | 553 | 7.939e-23 |
| GO:BP | GO:2000026 | regulation of multicellular organismal development | 955 | 1418 | 7.984e-23 |
| GO:BP | GO:0008610 | lipid biosynthetic process | 529 | 729 | 8.195e-23 |
| GO:BP | GO:0016477 | cell migration | 1024 | 1534 | 1.479e-22 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 592 | 830 | 1.524e-22 |
| GO:BP | GO:0043065 | positive regulation of apoptotic process | 399 | 528 | 1.555e-22 |
| GO:BP | GO:0048589 | developmental growth | 486 | 663 | 1.779e-22 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 240 | 291 | 1.878e-22 |
| GO:BP | GO:0015980 | energy derivation by oxidation of organic compounds | 277 | 345 | 2.146e-22 |
| GO:BP | GO:0032970 | regulation of actin filament-based process | 298 | 376 | 2.146e-22 |
| GO:BP | GO:0034599 | cellular response to oxidative stress | 209 | 247 | 2.326e-22 |
| GO:BP | GO:0001558 | regulation of cell growth | 318 | 406 | 2.510e-22 |
| GO:BP | GO:0048729 | tissue morphogenesis | 454 | 614 | 2.893e-22 |
| GO:BP | GO:0000956 | nuclear-transcribed mRNA catabolic process | 120 | 127 | 3.303e-22 |
| GO:BP | GO:0030334 | regulation of cell migration | 672 | 960 | 3.476e-22 |
| GO:BP | GO:0051168 | nuclear export | 152 | 169 | 4.595e-22 |
| GO:BP | GO:0043066 | negative regulation of apoptotic process | 618 | 874 | 4.724e-22 |
| GO:BP | GO:0008152 | metabolic process | 8044 | 14136 | 6.934e-22 |
| GO:BP | GO:0043484 | regulation of RNA splicing | 165 | 187 | 8.482e-22 |
| GO:BP | GO:0040008 | regulation of growth | 446 | 604 | 1.139e-21 |
| GO:BP | GO:0007169 | cell surface receptor protein tyrosine kinase signaling pathway | 462 | 629 | 1.172e-21 |
| GO:BP | GO:0031929 | TOR signaling | 150 | 167 | 1.241e-21 |
| GO:BP | GO:0060322 | head development | 574 | 806 | 1.326e-21 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 488 | 670 | 1.356e-21 |
| GO:BP | GO:0061136 | regulation of proteasomal protein catabolic process | 172 | 197 | 1.437e-21 |
| GO:BP | GO:0038202 | TORC1 signaling | 100 | 103 | 2.810e-21 |
| GO:BP | GO:0072655 | establishment of protein localization to mitochondrion | 116 | 123 | 2.909e-21 |
| GO:BP | GO:0070585 | protein localization to mitochondrion | 123 | 132 | 3.056e-21 |
| GO:BP | GO:0006368 | transcription elongation by RNA polymerase II | 112 | 118 | 3.349e-21 |
| GO:BP | GO:0043414 | macromolecule methylation | 119 | 127 | 4.012e-21 |
| GO:BP | GO:0006986 | response to unfolded protein | 129 | 140 | 4.391e-21 |
| GO:BP | GO:0043068 | positive regulation of programmed cell death | 407 | 546 | 4.696e-21 |
| GO:BP | GO:0045017 | glycerolipid biosynthetic process | 209 | 250 | 5.152e-21 |
| GO:BP | GO:0007417 | central nervous system development | 730 | 1061 | 6.165e-21 |
| GO:BP | GO:0098657 | import into cell | 644 | 922 | 7.065e-21 |
| GO:BP | GO:0071559 | response to transforming growth factor beta | 222 | 269 | 7.434e-21 |
| GO:BP | GO:0010563 | negative regulation of phosphorus metabolic process | 246 | 304 | 8.809e-21 |
| GO:BP | GO:0045936 | negative regulation of phosphate metabolic process | 246 | 304 | 8.809e-21 |
| GO:BP | GO:0098876 | vesicle-mediated transport to the plasma membrane | 140 | 155 | 9.299e-21 |
| GO:BP | GO:0009451 | RNA modification | 154 | 174 | 1.151e-20 |
| GO:BP | GO:0019693 | ribose phosphate metabolic process | 297 | 380 | 1.330e-20 |
| GO:BP | GO:0042326 | negative regulation of phosphorylation | 215 | 260 | 2.052e-20 |
| GO:BP | GO:2000145 | regulation of cell motility | 704 | 1022 | 2.218e-20 |
| GO:BP | GO:0070887 | cellular response to chemical stimulus | 1405 | 2192 | 2.218e-20 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 553 | 779 | 2.611e-20 |
| GO:BP | GO:0031032 | actomyosin structure organization | 181 | 212 | 2.732e-20 |
| GO:BP | GO:0070201 | regulation of establishment of protein localization | 399 | 537 | 3.297e-20 |
| GO:BP | GO:0001944 | vasculature development | 542 | 762 | 3.336e-20 |
| GO:BP | GO:0071560 | cellular response to transforming growth factor beta stimulus | 216 | 262 | 3.455e-20 |
| GO:BP | GO:0031396 | regulation of protein ubiquitination | 170 | 197 | 4.060e-20 |
| GO:BP | GO:0006897 | endocytosis | 508 | 709 | 5.501e-20 |
| GO:BP | GO:0007034 | vacuolar transport | 153 | 174 | 6.945e-20 |
| GO:BP | GO:0099003 | vesicle-mediated transport in synapse | 212 | 257 | 7.064e-20 |
| GO:BP | GO:0032984 | protein-containing complex disassembly | 212 | 257 | 7.064e-20 |
| GO:BP | GO:0009259 | ribonucleotide metabolic process | 290 | 372 | 8.055e-20 |
| GO:BP | GO:0060627 | regulation of vesicle-mediated transport | 394 | 531 | 8.907e-20 |
| GO:BP | GO:0006644 | phospholipid metabolic process | 303 | 392 | 1.101e-19 |
| GO:BP | GO:0032006 | regulation of TOR signaling | 132 | 146 | 1.132e-19 |
| GO:BP | GO:2001233 | regulation of apoptotic signaling pathway | 295 | 380 | 1.137e-19 |
| GO:BP | GO:0032259 | methylation | 197 | 236 | 1.141e-19 |
| GO:BP | GO:0007006 | mitochondrial membrane organization | 109 | 116 | 1.248e-19 |
| GO:BP | GO:0030111 | regulation of Wnt signaling pathway | 262 | 331 | 1.433e-19 |
| GO:BP | GO:1903432 | regulation of TORC1 signaling | 93 | 96 | 1.463e-19 |
| GO:BP | GO:0060828 | regulation of canonical Wnt signaling pathway | 210 | 255 | 1.596e-19 |
| GO:BP | GO:0097193 | intrinsic apoptotic signaling pathway | 249 | 312 | 1.721e-19 |
| GO:BP | GO:0009150 | purine ribonucleotide metabolic process | 275 | 351 | 2.315e-19 |
| GO:BP | GO:0051656 | establishment of organelle localization | 367 | 491 | 2.458e-19 |
| GO:BP | GO:0060070 | canonical Wnt signaling pathway | 248 | 311 | 2.520e-19 |
| GO:BP | GO:0006400 | tRNA modification | 92 | 95 | 2.566e-19 |
| GO:BP | GO:1902115 | regulation of organelle assembly | 195 | 234 | 2.638e-19 |
| GO:BP | GO:0018205 | peptidyl-lysine modification | 127 | 140 | 3.004e-19 |
| GO:BP | GO:0046474 | glycerophospholipid biosynthetic process | 178 | 210 | 3.009e-19 |
| GO:BP | GO:0015931 | nucleobase-containing compound transport | 197 | 237 | 3.010e-19 |
| GO:BP | GO:0040012 | regulation of locomotion | 727 | 1067 | 3.649e-19 |
| GO:BP | GO:0036503 | ERAD pathway | 103 | 109 | 4.415e-19 |
| GO:BP | GO:0045732 | positive regulation of protein catabolic process | 177 | 209 | 4.661e-19 |
| GO:BP | GO:0002009 | morphogenesis of an epithelium | 374 | 503 | 4.814e-19 |
| GO:BP | GO:0006626 | protein targeting to mitochondrion | 99 | 104 | 4.954e-19 |
| GO:BP | GO:0007409 | axonogenesis | 348 | 463 | 5.150e-19 |
| GO:BP | GO:0061564 | axon development | 390 | 528 | 5.266e-19 |
| GO:BP | GO:0001933 | negative regulation of protein phosphorylation | 202 | 245 | 6.559e-19 |
| GO:BP | GO:0009141 | nucleoside triphosphate metabolic process | 223 | 276 | 1.047e-18 |
| GO:BP | GO:0009144 | purine nucleoside triphosphate metabolic process | 212 | 260 | 1.071e-18 |
| GO:BP | GO:0009199 | ribonucleoside triphosphate metabolic process | 212 | 260 | 1.071e-18 |
| GO:BP | GO:0034329 | cell junction assembly | 373 | 503 | 1.162e-18 |
| GO:BP | GO:0007420 | brain development | 532 | 754 | 1.182e-18 |
| GO:BP | GO:0006629 | lipid metabolic process | 921 | 1391 | 1.186e-18 |
| GO:BP | GO:0016032 | viral process | 321 | 423 | 1.204e-18 |
| GO:BP | GO:0006352 | DNA-templated transcription initiation | 177 | 210 | 1.323e-18 |
| GO:BP | GO:0007178 | cell surface receptor protein serine/threonine kinase signaling pathway | 282 | 364 | 1.350e-18 |
| GO:BP | GO:0001568 | blood vessel development | 518 | 732 | 1.398e-18 |
| GO:BP | GO:0044782 | cilium organization | 326 | 431 | 1.494e-18 |
| GO:BP | GO:0099173 | postsynapse organization | 202 | 246 | 1.641e-18 |
| GO:BP | GO:0051236 | establishment of RNA localization | 143 | 163 | 2.098e-18 |
| GO:BP | GO:0032535 | regulation of cellular component size | 276 | 356 | 2.792e-18 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 457 | 637 | 3.704e-18 |
| GO:BP | GO:0000422 | autophagy of mitochondrion | 99 | 105 | 3.815e-18 |
| GO:BP | GO:0006892 | post-Golgi vesicle-mediated transport | 105 | 113 | 6.614e-18 |
| GO:BP | GO:0060271 | cilium assembly | 306 | 403 | 7.685e-18 |
| GO:BP | GO:0032784 | regulation of DNA-templated transcription elongation | 94 | 99 | 7.715e-18 |
| GO:BP | GO:0050658 | RNA transport | 140 | 160 | 8.700e-18 |
| GO:BP | GO:0050657 | nucleic acid transport | 140 | 160 | 8.700e-18 |
| GO:BP | GO:1905037 | autophagosome organization | 114 | 125 | 9.524e-18 |
| GO:BP | GO:0048870 | cell motility | 1156 | 1793 | 9.543e-18 |
| GO:BP | GO:0045739 | positive regulation of DNA repair | 123 | 137 | 9.694e-18 |
| GO:BP | GO:0032204 | regulation of telomere maintenance | 104 | 112 | 1.110e-17 |
| GO:BP | GO:0042147 | retrograde transport, endosome to Golgi | 97 | 103 | 1.122e-17 |
| GO:BP | GO:0031503 | protein-containing complex localization | 179 | 215 | 1.168e-17 |
| GO:BP | GO:0032956 | regulation of actin cytoskeleton organization | 260 | 334 | 1.189e-17 |
| GO:BP | GO:0061013 | regulation of mRNA catabolic process | 167 | 198 | 1.252e-17 |
| GO:BP | GO:0007032 | endosome organization | 93 | 98 | 1.313e-17 |
| GO:BP | GO:0099504 | synaptic vesicle cycle | 183 | 221 | 1.508e-17 |
| GO:BP | GO:0060562 | epithelial tube morphogenesis | 261 | 336 | 1.688e-17 |
| GO:BP | GO:0009205 | purine ribonucleoside triphosphate metabolic process | 205 | 253 | 1.723e-17 |
| GO:BP | GO:1901700 | response to oxygen-containing compound | 1081 | 1669 | 1.962e-17 |
| GO:BP | GO:0043487 | regulation of RNA stability | 163 | 193 | 2.500e-17 |
| GO:BP | GO:0045333 | cellular respiration | 197 | 242 | 3.061e-17 |
| GO:BP | GO:0051146 | striated muscle cell differentiation | 237 | 301 | 3.305e-17 |
| GO:BP | GO:0000045 | autophagosome assembly | 108 | 118 | 4.084e-17 |
| GO:BP | GO:0035967 | cellular response to topologically incorrect protein | 101 | 109 | 5.362e-17 |
| GO:BP | GO:0006414 | translational elongation | 78 | 80 | 5.684e-17 |
| GO:BP | GO:2000058 | regulation of ubiquitin-dependent protein catabolic process | 146 | 170 | 6.598e-17 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 505 | 720 | 6.671e-17 |
| GO:BP | GO:1901699 | cellular response to nitrogen compound | 458 | 645 | 7.378e-17 |
| GO:BP | GO:0035051 | cardiocyte differentiation | 132 | 151 | 1.054e-16 |
| GO:BP | GO:0016358 | dendrite development | 196 | 242 | 1.094e-16 |
| GO:BP | GO:0009100 | glycoprotein metabolic process | 282 | 371 | 1.449e-16 |
| GO:BP | GO:0141091 | transforming growth factor beta receptor superfamily signaling pathway | 264 | 344 | 1.855e-16 |
| GO:BP | GO:0043488 | regulation of mRNA stability | 153 | 181 | 2.414e-16 |
| GO:BP | GO:0090092 | regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 213 | 268 | 2.538e-16 |
| GO:BP | GO:0051028 | mRNA transport | 116 | 130 | 3.049e-16 |
| GO:BP | GO:0032869 | cellular response to insulin stimulus | 171 | 207 | 3.357e-16 |
| GO:BP | GO:0006360 | transcription by RNA polymerase I | 70 | 71 | 3.518e-16 |
| GO:BP | GO:0034243 | regulation of transcription elongation by RNA polymerase II | 83 | 87 | 3.944e-16 |
| GO:BP | GO:0031669 | cellular response to nutrient levels | 199 | 248 | 4.260e-16 |
| GO:BP | GO:0003007 | heart morphogenesis | 211 | 266 | 5.398e-16 |
| GO:BP | GO:0055007 | cardiac muscle cell differentiation | 106 | 117 | 5.476e-16 |
| GO:BP | GO:0090287 | regulation of cellular response to growth factor stimulus | 247 | 320 | 5.936e-16 |
| GO:BP | GO:0044770 | cell cycle phase transition | 384 | 532 | 5.949e-16 |
| GO:BP | GO:0010508 | positive regulation of autophagy | 138 | 161 | 7.860e-16 |
| GO:BP | GO:0006302 | double-strand break repair | 246 | 319 | 8.418e-16 |
| GO:BP | GO:0007179 | transforming growth factor beta receptor signaling pathway | 171 | 208 | 8.515e-16 |
| GO:BP | GO:0022604 | regulation of cell morphogenesis | 197 | 246 | 9.190e-16 |
| GO:BP | GO:0051170 | import into nucleus | 145 | 171 | 9.481e-16 |
| GO:BP | GO:0009725 | response to hormone | 616 | 907 | 9.563e-16 |
| GO:BP | GO:0072384 | organelle transport along microtubule | 85 | 90 | 1.035e-15 |
| GO:BP | GO:0070936 | protein K48-linked ubiquitination | 92 | 99 | 1.043e-15 |
| GO:BP | GO:0007030 | Golgi organization | 132 | 153 | 1.181e-15 |
| GO:BP | GO:0043603 | amide metabolic process | 334 | 455 | 1.262e-15 |
| GO:BP | GO:0006383 | transcription by RNA polymerase III | 62 | 62 | 1.344e-15 |
| GO:BP | GO:0110053 | regulation of actin filament organization | 208 | 263 | 1.640e-15 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 267 | 352 | 1.706e-15 |
| GO:BP | GO:0051258 | protein polymerization | 222 | 284 | 1.800e-15 |
| GO:BP | GO:0042594 | response to starvation | 178 | 219 | 1.809e-15 |
| GO:BP | GO:0009892 | negative regulation of metabolic process | 2058 | 3375 | 2.620e-15 |
| GO:BP | GO:0045862 | positive regulation of proteolysis | 177 | 218 | 2.705e-15 |
| GO:BP | GO:0006082 | organic acid metabolic process | 619 | 915 | 2.830e-15 |
| GO:BP | GO:0006661 | phosphatidylinositol biosynthetic process | 114 | 129 | 3.084e-15 |
| GO:BP | GO:0043436 | oxoacid metabolic process | 615 | 909 | 3.446e-15 |
| GO:BP | GO:0006650 | glycerophospholipid metabolic process | 238 | 309 | 3.446e-15 |
| GO:BP | GO:0032868 | response to insulin | 212 | 270 | 3.595e-15 |
| GO:BP | GO:0051050 | positive regulation of transport | 577 | 847 | 3.771e-15 |
| GO:BP | GO:0006888 | endoplasmic reticulum to Golgi vesicle-mediated transport | 119 | 136 | 3.771e-15 |
| GO:BP | GO:0018105 | peptidyl-serine phosphorylation | 146 | 174 | 4.917e-15 |
| GO:BP | GO:0046034 | ATP metabolic process | 184 | 229 | 5.130e-15 |
| GO:BP | GO:0051348 | negative regulation of transferase activity | 141 | 167 | 5.384e-15 |
| GO:BP | GO:0009267 | cellular response to starvation | 148 | 177 | 5.722e-15 |
| GO:BP | GO:2000142 | regulation of DNA-templated transcription initiation | 78 | 82 | 6.073e-15 |
| GO:BP | GO:0003012 | muscle system process | 321 | 438 | 7.327e-15 |
| GO:BP | GO:0006606 | protein import into nucleus | 140 | 166 | 8.318e-15 |
| GO:BP | GO:0018209 | peptidyl-serine modification | 154 | 186 | 8.571e-15 |
| GO:BP | GO:0019752 | carboxylic acid metabolic process | 600 | 887 | 8.571e-15 |
| GO:BP | GO:0006900 | vesicle budding from membrane | 81 | 86 | 8.885e-15 |
| GO:BP | GO:1901701 | cellular response to oxygen-containing compound | 778 | 1182 | 9.013e-15 |
| GO:BP | GO:0043434 | response to peptide hormone | 322 | 440 | 9.168e-15 |
| GO:BP | GO:0050684 | regulation of mRNA processing | 117 | 134 | 9.540e-15 |
| GO:BP | GO:0051301 | cell division | 459 | 658 | 1.023e-14 |
| GO:BP | GO:0150115 | cell-substrate junction organization | 94 | 103 | 1.037e-14 |
| GO:BP | GO:0001510 | RNA methylation | 77 | 81 | 1.039e-14 |
| GO:BP | GO:0090066 | regulation of anatomical structure size | 360 | 500 | 1.041e-14 |
| GO:BP | GO:0061014 | positive regulation of mRNA catabolic process | 97 | 107 | 1.068e-14 |
| GO:BP | GO:1901293 | nucleoside phosphate biosynthetic process | 240 | 314 | 1.252e-14 |
| GO:BP | GO:0007041 | lysosomal transport | 119 | 137 | 1.271e-14 |
| GO:BP | GO:0055001 | muscle cell development | 162 | 198 | 1.291e-14 |
| GO:BP | GO:1903313 | positive regulation of mRNA metabolic process | 124 | 144 | 1.366e-14 |
| GO:BP | GO:0033108 | mitochondrial respiratory chain complex assembly | 108 | 122 | 1.411e-14 |
| GO:BP | GO:0045597 | positive regulation of cell differentiation | 594 | 879 | 1.604e-14 |
| GO:BP | GO:0048514 | blood vessel morphogenesis | 444 | 635 | 1.630e-14 |
| GO:BP | GO:0042273 | ribosomal large subunit biogenesis | 68 | 70 | 1.662e-14 |
| GO:BP | GO:0007517 | muscle organ development | 273 | 365 | 1.662e-14 |
| GO:BP | GO:0008283 | cell population proliferation | 1268 | 2015 | 1.741e-14 |
| GO:BP | GO:0051338 | regulation of transferase activity | 358 | 498 | 1.806e-14 |
| GO:BP | GO:0051056 | regulation of small GTPase mediated signal transduction | 235 | 307 | 1.823e-14 |
| GO:BP | GO:0051240 | positive regulation of multicellular organismal process | 1064 | 1667 | 1.895e-14 |
| GO:BP | GO:0000723 | telomere maintenance | 138 | 164 | 1.911e-14 |
| GO:BP | GO:0005996 | monosaccharide metabolic process | 197 | 250 | 2.021e-14 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 354 | 492 | 2.098e-14 |
| GO:BP | GO:1902905 | positive regulation of supramolecular fiber organization | 147 | 177 | 2.256e-14 |
| GO:BP | GO:0071375 | cellular response to peptide hormone stimulus | 240 | 315 | 2.344e-14 |
| GO:BP | GO:0050807 | regulation of synapse organization | 236 | 309 | 2.478e-14 |
| GO:BP | GO:0051093 | negative regulation of developmental process | 621 | 925 | 2.679e-14 |
| GO:BP | GO:0032870 | cellular response to hormone stimulus | 445 | 638 | 2.859e-14 |
| GO:BP | GO:0009060 | aerobic respiration | 162 | 199 | 3.085e-14 |
| GO:BP | GO:0060260 | regulation of transcription initiation by RNA polymerase II | 71 | 74 | 3.223e-14 |
| GO:BP | GO:0070085 | glycosylation | 193 | 245 | 4.139e-14 |
| GO:BP | GO:0008088 | axo-dendritic transport | 78 | 83 | 4.320e-14 |
| GO:BP | GO:0046486 | glycerolipid metabolic process | 293 | 398 | 5.117e-14 |
| GO:BP | GO:0001667 | ameboidal-type cell migration | 186 | 235 | 5.637e-14 |
| GO:BP | GO:0051223 | regulation of protein transport | 321 | 442 | 5.661e-14 |
| GO:BP | GO:0055002 | striated muscle cell development | 142 | 171 | 6.948e-14 |
| GO:BP | GO:0006367 | transcription initiation at RNA polymerase II promoter | 142 | 171 | 6.948e-14 |
| GO:BP | GO:0030522 | intracellular receptor signaling pathway | 279 | 377 | 7.811e-14 |
| GO:BP | GO:0060560 | developmental growth involved in morphogenesis | 189 | 240 | 8.497e-14 |
| GO:BP | GO:0003013 | circulatory system process | 421 | 602 | 8.552e-14 |
| GO:BP | GO:0007249 | canonical NF-kappaB signal transduction | 234 | 308 | 9.213e-14 |
| GO:BP | GO:0019725 | cellular homeostasis | 564 | 835 | 9.793e-14 |
| GO:BP | GO:0051017 | actin filament bundle assembly | 134 | 160 | 1.056e-13 |
| GO:BP | GO:0043086 | negative regulation of catalytic activity | 224 | 293 | 1.063e-13 |
| GO:BP | GO:0050803 | regulation of synapse structure or activity | 239 | 316 | 1.151e-13 |
| GO:BP | GO:0034620 | cellular response to unfolded protein | 86 | 94 | 1.212e-13 |
| GO:BP | GO:1901888 | regulation of cell junction assembly | 204 | 263 | 1.252e-13 |
| GO:BP | GO:0061157 | mRNA destabilization | 92 | 102 | 1.267e-13 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 311 | 428 | 1.339e-13 |
| GO:BP | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 121 | 142 | 1.578e-13 |
| GO:BP | GO:0003205 | cardiac chamber development | 140 | 169 | 1.587e-13 |
| GO:BP | GO:0051336 | regulation of hydrolase activity | 262 | 352 | 1.650e-13 |
| GO:BP | GO:0003015 | heart process | 197 | 253 | 1.800e-13 |
| GO:BP | GO:0060491 | regulation of cell projection assembly | 164 | 204 | 1.801e-13 |
| GO:BP | GO:0019080 | viral gene expression | 97 | 109 | 1.854e-13 |
| GO:BP | GO:0009101 | glycoprotein biosynthetic process | 228 | 300 | 1.891e-13 |
| GO:BP | GO:0000165 | MAPK cascade | 507 | 744 | 2.260e-13 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 184 | 234 | 2.484e-13 |
| GO:BP | GO:0032206 | positive regulation of telomere maintenance | 71 | 75 | 2.651e-13 |
| GO:BP | GO:0030490 | maturation of SSU-rRNA | 53 | 53 | 2.710e-13 |
| GO:BP | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | 112 | 130 | 3.034e-13 |
| GO:BP | GO:2001242 | regulation of intrinsic apoptotic signaling pathway | 145 | 177 | 3.297e-13 |
| GO:BP | GO:0007044 | cell-substrate junction assembly | 87 | 96 | 3.436e-13 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 389 | 554 | 3.554e-13 |
| GO:BP | GO:0030335 | positive regulation of cell migration | 392 | 559 | 3.845e-13 |
| GO:BP | GO:0120032 | regulation of plasma membrane bounded cell projection assembly | 162 | 202 | 3.870e-13 |
| GO:BP | GO:0043087 | regulation of GTPase activity | 162 | 202 | 3.870e-13 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 121 | 143 | 4.585e-13 |
| GO:BP | GO:1903052 | positive regulation of proteolysis involved in protein catabolic process | 116 | 136 | 4.785e-13 |
| GO:BP | GO:0061572 | actin filament bundle organization | 135 | 163 | 4.833e-13 |
| GO:BP | GO:0040011 | locomotion | 813 | 1255 | 4.833e-13 |
| GO:BP | GO:0000959 | mitochondrial RNA metabolic process | 52 | 52 | 4.871e-13 |
| GO:BP | GO:0048871 | multicellular organismal-level homeostasis | 555 | 825 | 4.921e-13 |
| GO:BP | GO:0048588 | developmental cell growth | 178 | 226 | 4.930e-13 |
| GO:BP | GO:0006486 | protein glycosylation | 178 | 226 | 4.930e-13 |
| GO:BP | GO:0043413 | macromolecule glycosylation | 178 | 226 | 4.930e-13 |
| GO:BP | GO:2000144 | positive regulation of DNA-templated transcription initiation | 66 | 69 | 5.026e-13 |
| GO:BP | GO:0140238 | presynaptic endocytosis | 80 | 87 | 5.048e-13 |
| GO:BP | GO:0036465 | synaptic vesicle recycling | 86 | 95 | 5.571e-13 |
| GO:BP | GO:0043549 | regulation of kinase activity | 311 | 431 | 5.866e-13 |
| GO:BP | GO:1901875 | positive regulation of post-translational protein modification | 113 | 132 | 5.905e-13 |
| GO:BP | GO:0072524 | pyridine-containing compound metabolic process | 139 | 169 | 5.963e-13 |
| GO:BP | GO:0032786 | positive regulation of DNA-templated transcription, elongation | 57 | 58 | 6.140e-13 |
| GO:BP | GO:0046488 | phosphatidylinositol metabolic process | 134 | 162 | 7.209e-13 |
| GO:BP | GO:0050779 | RNA destabilization | 94 | 106 | 7.712e-13 |
| GO:BP | GO:0070482 | response to oxygen levels | 255 | 344 | 8.054e-13 |
| GO:BP | GO:1903844 | regulation of cellular response to transforming growth factor beta stimulus | 122 | 145 | 8.258e-13 |
| GO:BP | GO:0032456 | endocytic recycling | 79 | 86 | 8.397e-13 |
| GO:BP | GO:0032007 | negative regulation of TOR signaling | 65 | 68 | 8.656e-13 |
| GO:BP | GO:1903322 | positive regulation of protein modification by small protein conjugation or removal | 112 | 131 | 9.175e-13 |
| GO:BP | GO:0035148 | tube formation | 124 | 148 | 9.661e-13 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 188 | 242 | 1.037e-12 |
| GO:BP | GO:0006413 | translational initiation | 109 | 127 | 1.144e-12 |
| GO:BP | GO:0000302 | response to reactive oxygen species | 159 | 199 | 1.193e-12 |
| GO:BP | GO:0019827 | stem cell population maintenance | 146 | 180 | 1.257e-12 |
| GO:BP | GO:0048041 | focal adhesion assembly | 78 | 85 | 1.401e-12 |
| GO:BP | GO:0060047 | heart contraction | 189 | 244 | 1.465e-12 |
| GO:BP | GO:2001234 | negative regulation of apoptotic signaling pathway | 181 | 232 | 1.483e-12 |
| GO:BP | GO:0031589 | cell-substrate adhesion | 262 | 356 | 1.483e-12 |
| GO:BP | GO:2000781 | positive regulation of double-strand break repair | 84 | 93 | 1.491e-12 |
| GO:BP | GO:0021915 | neural tube development | 132 | 160 | 1.628e-12 |
| GO:BP | GO:0051960 | regulation of nervous system development | 326 | 457 | 1.696e-12 |
| GO:BP | GO:0010821 | regulation of mitochondrion organization | 100 | 115 | 2.053e-12 |
| GO:BP | GO:0055085 | transmembrane transport | 1000 | 1578 | 2.053e-12 |
| GO:BP | GO:0016239 | positive regulation of macroautophagy | 80 | 88 | 2.454e-12 |
| GO:BP | GO:0140352 | export from cell | 614 | 928 | 2.688e-12 |
| GO:BP | GO:0072522 | purine-containing compound biosynthetic process | 197 | 257 | 2.700e-12 |
| GO:BP | GO:0044283 | small molecule biosynthetic process | 410 | 593 | 2.834e-12 |
| GO:BP | GO:0033157 | regulation of intracellular protein transport | 126 | 152 | 2.945e-12 |
| GO:BP | GO:0016311 | dephosphorylation | 171 | 218 | 2.967e-12 |
| GO:BP | GO:0048284 | organelle fusion | 135 | 165 | 2.983e-12 |
| GO:BP | GO:0043542 | endothelial cell migration | 173 | 221 | 2.983e-12 |
| GO:BP | GO:0031345 | negative regulation of cell projection organization | 148 | 184 | 3.086e-12 |
| GO:BP | GO:0007029 | endoplasmic reticulum organization | 91 | 103 | 3.189e-12 |
| GO:BP | GO:0033673 | negative regulation of kinase activity | 121 | 145 | 3.411e-12 |
| GO:BP | GO:1905475 | regulation of protein localization to membrane | 154 | 193 | 3.542e-12 |
| GO:BP | GO:0045727 | positive regulation of translation | 116 | 138 | 3.847e-12 |
| GO:BP | GO:0048488 | synaptic vesicle endocytosis | 76 | 83 | 3.847e-12 |
| GO:BP | GO:0031667 | response to nutrient levels | 358 | 510 | 3.920e-12 |
| GO:BP | GO:0019318 | hexose metabolic process | 176 | 226 | 4.273e-12 |
| GO:BP | GO:0099175 | regulation of postsynapse organization | 125 | 151 | 4.413e-12 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 134 | 164 | 4.429e-12 |
| GO:BP | GO:0061077 | chaperone-mediated protein folding | 69 | 74 | 4.932e-12 |
| GO:BP | GO:0007040 | lysosome organization | 98 | 113 | 5.078e-12 |
| GO:BP | GO:1901800 | positive regulation of proteasomal protein catabolic process | 98 | 113 | 5.078e-12 |
| GO:BP | GO:0080171 | lytic vacuole organization | 98 | 113 | 5.078e-12 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 140 | 173 | 5.565e-12 |
| GO:BP | GO:0034614 | cellular response to reactive oxygen species | 122 | 147 | 5.884e-12 |
| GO:BP | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | 53 | 54 | 6.032e-12 |
| GO:BP | GO:2000147 | positive regulation of cell motility | 405 | 587 | 6.311e-12 |
| GO:BP | GO:0010594 | regulation of endothelial cell migration | 137 | 169 | 7.712e-12 |
| GO:BP | GO:1903828 | negative regulation of protein localization | 168 | 215 | 8.670e-12 |
| GO:BP | GO:0072175 | epithelial tube formation | 114 | 136 | 8.996e-12 |
| GO:BP | GO:0046496 | nicotinamide nucleotide metabolic process | 132 | 162 | 9.848e-12 |
| GO:BP | GO:0019362 | pyridine nucleotide metabolic process | 132 | 162 | 9.848e-12 |
| GO:BP | GO:0036293 | response to decreased oxygen levels | 234 | 316 | 1.012e-11 |
| GO:BP | GO:0098727 | maintenance of cell number | 147 | 184 | 1.012e-11 |
| GO:BP | GO:0060606 | tube closure | 80 | 89 | 1.065e-11 |
| GO:BP | GO:0045444 | fat cell differentiation | 185 | 241 | 1.130e-11 |
| GO:BP | GO:0051051 | negative regulation of transport | 295 | 412 | 1.176e-11 |
| GO:BP | GO:0044057 | regulation of system process | 383 | 553 | 1.187e-11 |
| GO:BP | GO:0043122 | regulation of canonical NF-kappaB signal transduction | 215 | 287 | 1.246e-11 |
| GO:BP | GO:0060261 | positive regulation of transcription initiation by RNA polymerase II | 60 | 63 | 1.319e-11 |
| GO:BP | GO:0098732 | macromolecule deacylation | 56 | 58 | 1.349e-11 |
| GO:BP | GO:0048762 | mesenchymal cell differentiation | 192 | 252 | 1.429e-11 |
| GO:BP | GO:0010605 | negative regulation of macromolecule metabolic process | 1904 | 3157 | 1.440e-11 |
| GO:BP | GO:0031123 | RNA 3’-end processing | 85 | 96 | 1.473e-11 |
| GO:BP | GO:0051495 | positive regulation of cytoskeleton organization | 148 | 186 | 1.522e-11 |
| GO:BP | GO:0060485 | mesenchyme development | 236 | 320 | 1.645e-11 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 201 | 266 | 1.672e-11 |
| GO:BP | GO:0001843 | neural tube closure | 79 | 88 | 1.726e-11 |
| GO:BP | GO:0008286 | insulin receptor signaling pathway | 105 | 124 | 1.949e-11 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 288 | 402 | 1.950e-11 |
| GO:BP | GO:0042177 | negative regulation of protein catabolic process | 100 | 117 | 2.021e-11 |
| GO:BP | GO:0000423 | mitophagy | 66 | 71 | 2.356e-11 |
| GO:BP | GO:1903008 | organelle disassembly | 66 | 71 | 2.356e-11 |
| GO:BP | GO:0007416 | synapse assembly | 196 | 259 | 2.498e-11 |
| GO:BP | GO:0032940 | secretion by cell | 565 | 854 | 2.501e-11 |
| GO:BP | GO:0051668 | localization within membrane | 607 | 924 | 2.530e-11 |
| GO:BP | GO:0048878 | chemical homeostasis | 678 | 1043 | 2.614e-11 |
| GO:BP | GO:0048638 | regulation of developmental growth | 229 | 310 | 2.650e-11 |
| GO:BP | GO:0030968 | endoplasmic reticulum unfolded protein response | 72 | 79 | 2.876e-11 |
| GO:BP | GO:0007007 | inner mitochondrial membrane organization | 45 | 45 | 2.946e-11 |
| GO:BP | GO:0090148 | membrane fission | 45 | 45 | 2.946e-11 |
| GO:BP | GO:0006903 | vesicle targeting | 62 | 66 | 3.175e-11 |
| GO:BP | GO:0050770 | regulation of axonogenesis | 120 | 146 | 3.367e-11 |
| GO:BP | GO:0040017 | positive regulation of locomotion | 412 | 603 | 3.530e-11 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 272 | 378 | 3.615e-11 |
| GO:BP | GO:0030336 | negative regulation of cell migration | 217 | 292 | 3.710e-11 |
| GO:BP | GO:1901874 | negative regulation of post-translational protein modification | 83 | 94 | 3.771e-11 |
| GO:BP | GO:0014020 | primary neural tube formation | 83 | 94 | 3.771e-11 |
| GO:BP | GO:0038127 | ERBB signaling pathway | 108 | 129 | 4.137e-11 |
| GO:BP | GO:0071806 | protein transmembrane transport | 68 | 74 | 4.474e-11 |
| GO:BP | GO:0001841 | neural tube formation | 88 | 101 | 4.481e-11 |
| GO:BP | GO:0055006 | cardiac cell development | 77 | 86 | 4.527e-11 |
| GO:BP | GO:0060048 | cardiac muscle contraction | 117 | 142 | 4.527e-11 |
| GO:BP | GO:1905897 | regulation of response to endoplasmic reticulum stress | 77 | 86 | 4.527e-11 |
| GO:BP | GO:0010976 | positive regulation of neuron projection development | 128 | 158 | 4.668e-11 |
| GO:BP | GO:0017148 | negative regulation of translation | 110 | 132 | 4.742e-11 |
| GO:BP | GO:0003279 | cardiac septum development | 93 | 108 | 4.803e-11 |
| GO:BP | GO:0001525 | angiogenesis | 377 | 547 | 4.915e-11 |
| GO:BP | GO:0051098 | regulation of binding | 165 | 213 | 4.919e-11 |
| GO:BP | GO:0031334 | positive regulation of protein-containing complex assembly | 157 | 201 | 4.967e-11 |
| GO:BP | GO:0048813 | dendrite morphogenesis | 119 | 145 | 4.967e-11 |
| GO:BP | GO:0008285 | negative regulation of cell population proliferation | 474 | 706 | 5.587e-11 |
| GO:BP | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | 49 | 50 | 5.827e-11 |
| GO:BP | GO:0006469 | negative regulation of protein kinase activity | 114 | 138 | 6.055e-11 |
| GO:BP | GO:0048872 | homeostasis of number of cells | 242 | 332 | 6.482e-11 |
| GO:BP | GO:0098930 | axonal transport | 64 | 69 | 6.549e-11 |
| GO:BP | GO:0043543 | protein acylation | 79 | 89 | 6.697e-11 |
| GO:BP | GO:0072665 | protein localization to vacuole | 79 | 89 | 6.697e-11 |
| GO:BP | GO:1901292 | nucleoside phosphate catabolic process | 146 | 185 | 6.718e-11 |
| GO:BP | GO:1904951 | positive regulation of establishment of protein localization | 240 | 329 | 6.902e-11 |
| GO:BP | GO:0010595 | positive regulation of endothelial cell migration | 92 | 107 | 7.465e-11 |
| GO:BP | GO:0006006 | glucose metabolic process | 145 | 184 | 9.749e-11 |
| GO:BP | GO:0046434 | organophosphate catabolic process | 186 | 246 | 1.009e-10 |
| GO:BP | GO:0030258 | lipid modification | 153 | 196 | 1.009e-10 |
| GO:BP | GO:0032388 | positive regulation of intracellular transport | 115 | 140 | 1.009e-10 |
| GO:BP | GO:0032271 | regulation of protein polymerization | 155 | 199 | 1.009e-10 |
| GO:BP | GO:0051090 | regulation of DNA-binding transcription factor activity | 235 | 322 | 1.061e-10 |
| GO:BP | GO:1901136 | carbohydrate derivative catabolic process | 193 | 257 | 1.181e-10 |
| GO:BP | GO:1904263 | positive regulation of TORC1 signaling | 52 | 54 | 1.222e-10 |
| GO:BP | GO:0001666 | response to hypoxia | 222 | 302 | 1.248e-10 |
| GO:BP | GO:0006635 | fatty acid beta-oxidation | 69 | 76 | 1.271e-10 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 301 | 427 | 1.326e-10 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 1051 | 1685 | 1.342e-10 |
| GO:BP | GO:0099177 | regulation of trans-synaptic signaling | 355 | 514 | 1.353e-10 |
| GO:BP | GO:0046822 | regulation of nucleocytoplasmic transport | 98 | 116 | 1.378e-10 |
| GO:BP | GO:0048598 | embryonic morphogenesis | 420 | 620 | 1.392e-10 |
| GO:BP | GO:0008016 | regulation of heart contraction | 158 | 204 | 1.410e-10 |
| GO:BP | GO:0006941 | striated muscle contraction | 148 | 189 | 1.417e-10 |
| GO:BP | GO:0009411 | response to UV | 125 | 155 | 1.475e-10 |
| GO:BP | GO:0072523 | purine-containing compound catabolic process | 125 | 155 | 1.475e-10 |
| GO:BP | GO:0006405 | RNA export from nucleus | 80 | 91 | 1.513e-10 |
| GO:BP | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 59 | 63 | 1.513e-10 |
| GO:BP | GO:0003229 | ventricular cardiac muscle tissue development | 59 | 63 | 1.513e-10 |
| GO:BP | GO:0042255 | ribosome assembly | 59 | 63 | 1.513e-10 |
| GO:BP | GO:0045927 | positive regulation of growth | 192 | 256 | 1.599e-10 |
| GO:BP | GO:0040013 | negative regulation of locomotion | 237 | 326 | 1.627e-10 |
| GO:BP | GO:0045022 | early endosome to late endosome transport | 42 | 42 | 1.679e-10 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 131 | 164 | 1.694e-10 |
| GO:BP | GO:0050804 | modulation of chemical synaptic transmission | 354 | 513 | 1.694e-10 |
| GO:BP | GO:0048024 | regulation of mRNA splicing, via spliceosome | 95 | 112 | 1.745e-10 |
| GO:BP | GO:0009887 | animal organ morphogenesis | 673 | 1042 | 1.778e-10 |
| GO:BP | GO:0098927 | vesicle-mediated transport between endosomal compartments | 47 | 48 | 1.779e-10 |
| GO:BP | GO:0006458 | ‘de novo’ protein folding | 47 | 48 | 1.779e-10 |
| GO:BP | GO:1904262 | negative regulation of TORC1 signaling | 47 | 48 | 1.779e-10 |
| GO:BP | GO:0006893 | Golgi to plasma membrane transport | 62 | 67 | 1.801e-10 |
| GO:BP | GO:0016073 | snRNA metabolic process | 55 | 58 | 1.896e-10 |
| GO:BP | GO:0046903 | secretion | 643 | 992 | 2.020e-10 |
| GO:BP | GO:0061337 | cardiac conduction | 82 | 94 | 2.039e-10 |
| GO:BP | GO:0030038 | contractile actin filament bundle assembly | 92 | 108 | 2.206e-10 |
| GO:BP | GO:0043149 | stress fiber assembly | 92 | 108 | 2.206e-10 |
| GO:BP | GO:1903321 | negative regulation of protein modification by small protein conjugation or removal | 79 | 90 | 2.388e-10 |
| GO:BP | GO:2000146 | negative regulation of cell motility | 225 | 308 | 2.431e-10 |
| GO:BP | GO:0030097 | hemopoiesis | 633 | 976 | 2.496e-10 |
| GO:BP | GO:0060284 | regulation of cell development | 556 | 847 | 2.496e-10 |
| GO:BP | GO:0010810 | regulation of cell-substrate adhesion | 166 | 217 | 2.514e-10 |
| GO:BP | GO:0006936 | muscle contraction | 251 | 349 | 2.563e-10 |
| GO:BP | GO:0030178 | negative regulation of Wnt signaling pathway | 136 | 172 | 2.669e-10 |
| GO:BP | GO:0009611 | response to wounding | 387 | 568 | 2.725e-10 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 156 | 202 | 2.754e-10 |
| GO:BP | GO:0002262 | myeloid cell homeostasis | 142 | 181 | 2.791e-10 |
| GO:BP | GO:0032790 | ribosome disassembly | 41 | 41 | 2.975e-10 |
| GO:BP | GO:0032509 | endosome transport via multivesicular body sorting pathway | 41 | 41 | 2.975e-10 |
| GO:BP | GO:0009314 | response to radiation | 306 | 437 | 2.975e-10 |
| GO:BP | GO:0031122 | cytoplasmic microtubule organization | 61 | 66 | 2.987e-10 |
| GO:BP | GO:0090257 | regulation of muscle system process | 175 | 231 | 2.988e-10 |
| GO:BP | GO:0006356 | regulation of transcription by RNA polymerase I | 46 | 47 | 3.101e-10 |
| GO:BP | GO:0006984 | ER-nucleus signaling pathway | 46 | 47 | 3.101e-10 |
| GO:BP | GO:0010628 | positive regulation of gene expression | 772 | 1212 | 3.194e-10 |
| GO:BP | GO:0055082 | intracellular chemical homeostasis | 485 | 730 | 3.428e-10 |
| GO:BP | GO:0030307 | positive regulation of cell growth | 129 | 162 | 3.523e-10 |
| GO:BP | GO:0060429 | epithelium development | 787 | 1238 | 3.588e-10 |
| GO:BP | GO:0003231 | cardiac ventricle development | 105 | 127 | 3.593e-10 |
| GO:BP | GO:0008361 | regulation of cell size | 143 | 183 | 3.990e-10 |
| GO:BP | GO:0046390 | ribose phosphate biosynthetic process | 143 | 183 | 3.990e-10 |
| GO:BP | GO:1900180 | regulation of protein localization to nucleus | 118 | 146 | 4.053e-10 |
| GO:BP | GO:0001503 | ossification | 301 | 430 | 4.546e-10 |
| GO:BP | GO:0001838 | embryonic epithelial tube formation | 102 | 123 | 4.830e-10 |
| GO:BP | GO:0031397 | negative regulation of protein ubiquitination | 69 | 77 | 5.243e-10 |
| GO:BP | GO:0030488 | tRNA methylation | 40 | 40 | 5.341e-10 |
| GO:BP | GO:0032200 | telomere organization | 148 | 191 | 5.651e-10 |
| GO:BP | GO:0046777 | protein autophosphorylation | 140 | 179 | 5.686e-10 |
| GO:BP | GO:0009201 | ribonucleoside triphosphate biosynthetic process | 99 | 119 | 6.459e-10 |
| GO:BP | GO:0006643 | membrane lipid metabolic process | 163 | 214 | 6.804e-10 |
| GO:BP | GO:0043547 | positive regulation of GTPase activity | 110 | 135 | 7.224e-10 |
| GO:BP | GO:0090090 | negative regulation of canonical Wnt signaling pathway | 112 | 138 | 7.788e-10 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 112 | 138 | 7.788e-10 |
| GO:BP | GO:0055013 | cardiac muscle cell development | 71 | 80 | 7.814e-10 |
| GO:BP | GO:2000060 | positive regulation of ubiquitin-dependent protein catabolic process | 89 | 105 | 8.078e-10 |
| GO:BP | GO:0048863 | stem cell differentiation | 183 | 245 | 8.313e-10 |
| GO:BP | GO:1990542 | mitochondrial transmembrane transport | 84 | 98 | 8.313e-10 |
| GO:BP | GO:0046785 | microtubule polymerization | 84 | 98 | 8.313e-10 |
| GO:BP | GO:0030239 | myofibril assembly | 68 | 76 | 8.493e-10 |
| GO:BP | GO:0150116 | regulation of cell-substrate junction organization | 62 | 68 | 8.888e-10 |
| GO:BP | GO:0043244 | regulation of protein-containing complex disassembly | 105 | 128 | 8.990e-10 |
| GO:BP | GO:0006997 | nucleus organization | 118 | 147 | 9.256e-10 |
| GO:BP | GO:0060996 | dendritic spine development | 86 | 101 | 1.027e-09 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 173 | 230 | 1.044e-09 |
| GO:BP | GO:0170036 | import into the mitochondrion | 48 | 50 | 1.068e-09 |
| GO:BP | GO:0035601 | protein deacylation | 48 | 50 | 1.068e-09 |
| GO:BP | GO:0001822 | kidney development | 232 | 322 | 1.068e-09 |
| GO:BP | GO:0071985 | multivesicular body sorting pathway | 48 | 50 | 1.068e-09 |
| GO:BP | GO:0009166 | nucleotide catabolic process | 128 | 162 | 1.094e-09 |
| GO:BP | GO:0003206 | cardiac chamber morphogenesis | 102 | 124 | 1.218e-09 |
| GO:BP | GO:0009142 | nucleoside triphosphate biosynthetic process | 104 | 127 | 1.343e-09 |
| GO:BP | GO:0072001 | renal system development | 238 | 332 | 1.369e-09 |
| GO:BP | GO:0051345 | positive regulation of hydrolase activity | 157 | 206 | 1.391e-09 |
| GO:BP | GO:1904375 | regulation of protein localization to cell periphery | 119 | 149 | 1.411e-09 |
| GO:BP | GO:0044091 | membrane biogenesis | 61 | 67 | 1.463e-09 |
| GO:BP | GO:0009062 | fatty acid catabolic process | 90 | 107 | 1.464e-09 |
| GO:BP | GO:0009260 | ribonucleotide biosynthetic process | 137 | 176 | 1.642e-09 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 266 | 377 | 1.675e-09 |
| GO:BP | GO:0019395 | fatty acid oxidation | 92 | 110 | 1.708e-09 |
| GO:BP | GO:0090174 | organelle membrane fusion | 101 | 123 | 1.825e-09 |
| GO:BP | GO:0042073 | intraciliary transport | 47 | 49 | 1.851e-09 |
| GO:BP | GO:0062012 | regulation of small molecule metabolic process | 230 | 320 | 1.860e-09 |
| GO:BP | GO:0031333 | negative regulation of protein-containing complex assembly | 114 | 142 | 1.884e-09 |
| GO:BP | GO:0045596 | negative regulation of cell differentiation | 446 | 671 | 1.921e-09 |
| GO:BP | GO:0032008 | positive regulation of TOR signaling | 66 | 74 | 2.227e-09 |
| GO:BP | GO:1901796 | regulation of signal transduction by p53 class mediator | 89 | 106 | 2.243e-09 |
| GO:BP | GO:0099536 | synaptic signaling | 527 | 807 | 2.662e-09 |
| GO:BP | GO:0030177 | positive regulation of Wnt signaling pathway | 115 | 144 | 2.898e-09 |
| GO:BP | GO:0051084 | ‘de novo’ post-translational protein folding | 42 | 43 | 3.010e-09 |
| GO:BP | GO:0030308 | negative regulation of cell growth | 135 | 174 | 3.333e-09 |
| GO:BP | GO:1990138 | neuron projection extension | 133 | 171 | 3.346e-09 |
| GO:BP | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway | 88 | 105 | 3.447e-09 |
| GO:BP | GO:0043112 | receptor metabolic process | 65 | 73 | 3.618e-09 |
| GO:BP | GO:1903051 | negative regulation of proteolysis involved in protein catabolic process | 65 | 73 | 3.618e-09 |
| GO:BP | GO:0071709 | membrane assembly | 56 | 61 | 3.863e-09 |
| GO:BP | GO:0061951 | establishment of protein localization to plasma membrane | 59 | 65 | 3.998e-09 |
| GO:BP | GO:0051241 | negative regulation of multicellular organismal process | 692 | 1087 | 4.046e-09 |
| GO:BP | GO:0006906 | vesicle fusion | 99 | 121 | 4.088e-09 |
| GO:BP | GO:0007160 | cell-matrix adhesion | 176 | 237 | 4.121e-09 |
| GO:BP | GO:0008154 | actin polymerization or depolymerization | 146 | 191 | 4.227e-09 |
| GO:BP | GO:0006119 | oxidative phosphorylation | 116 | 146 | 4.347e-09 |
| GO:BP | GO:0021537 | telencephalon development | 206 | 284 | 4.545e-09 |
| GO:BP | GO:0009152 | purine ribonucleotide biosynthetic process | 126 | 161 | 4.736e-09 |
| GO:BP | GO:0032787 | monocarboxylic acid metabolic process | 411 | 616 | 4.783e-09 |
| GO:BP | GO:0106027 | neuron projection organization | 80 | 94 | 4.832e-09 |
| GO:BP | GO:0009145 | purine nucleoside triphosphate biosynthetic process | 94 | 114 | 5.023e-09 |
| GO:BP | GO:0099537 | trans-synaptic signaling | 510 | 781 | 5.215e-09 |
| GO:BP | GO:0090100 | positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 87 | 104 | 5.223e-09 |
| GO:BP | GO:0006476 | protein deacetylation | 41 | 42 | 5.268e-09 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 233 | 327 | 5.284e-09 |
| GO:BP | GO:0006790 | sulfur compound metabolic process | 221 | 308 | 5.345e-09 |
| GO:BP | GO:0030100 | regulation of endocytosis | 214 | 297 | 5.500e-09 |
| GO:BP | GO:0043038 | amino acid activation | 45 | 47 | 5.526e-09 |
| GO:BP | GO:1990928 | response to amino acid starvation | 52 | 56 | 5.727e-09 |
| GO:BP | GO:0086065 | cell communication involved in cardiac conduction | 52 | 56 | 5.727e-09 |
| GO:BP | GO:0006289 | nucleotide-excision repair | 72 | 83 | 5.883e-09 |
| GO:BP | GO:0030099 | myeloid cell differentiation | 305 | 443 | 6.004e-09 |
| GO:BP | GO:0051276 | chromosome organization | 385 | 574 | 6.328e-09 |
| GO:BP | GO:0044088 | regulation of vacuole organization | 55 | 60 | 6.328e-09 |
| GO:BP | GO:0007004 | telomere maintenance via telomerase | 55 | 60 | 6.328e-09 |
| GO:BP | GO:0099518 | vesicle cytoskeletal trafficking | 58 | 64 | 6.476e-09 |
| GO:BP | GO:0007254 | JNK cascade | 135 | 175 | 6.479e-09 |
| GO:BP | GO:0007224 | smoothened signaling pathway | 119 | 151 | 6.485e-09 |
| GO:BP | GO:1902904 | negative regulation of supramolecular fiber organization | 123 | 157 | 6.635e-09 |
| GO:BP | GO:0022900 | electron transport chain | 102 | 126 | 6.957e-09 |
| GO:BP | GO:0120034 | positive regulation of plasma membrane bounded cell projection assembly | 93 | 113 | 7.437e-09 |
| GO:BP | GO:0048675 | axon extension | 93 | 113 | 7.437e-09 |
| GO:BP | GO:0009206 | purine ribonucleoside triphosphate biosynthetic process | 93 | 113 | 7.437e-09 |
| GO:BP | GO:0022406 | membrane docking | 74 | 86 | 7.499e-09 |
| GO:BP | GO:0006195 | purine nucleotide catabolic process | 110 | 138 | 8.481e-09 |
| GO:BP | GO:0010212 | response to ionizing radiation | 112 | 141 | 8.757e-09 |
| GO:BP | GO:0006470 | protein dephosphorylation | 114 | 144 | 8.988e-09 |
| GO:BP | GO:0034101 | erythrocyte homeostasis | 116 | 147 | 9.173e-09 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 118 | 150 | 9.302e-09 |
| GO:BP | GO:0090101 | negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 118 | 150 | 9.302e-09 |
| GO:BP | GO:0043039 | tRNA aminoacylation | 44 | 46 | 9.448e-09 |
| GO:BP | GO:0007020 | microtubule nucleation | 44 | 46 | 9.448e-09 |
| GO:BP | GO:1904356 | regulation of telomere maintenance via telomere lengthening | 51 | 55 | 9.516e-09 |
| GO:BP | GO:0001952 | regulation of cell-matrix adhesion | 99 | 122 | 9.553e-09 |
| GO:BP | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 35 | 35 | 9.885e-09 |
| GO:BP | GO:0000460 | maturation of 5.8S rRNA | 35 | 35 | 9.885e-09 |
| GO:BP | GO:0042060 | wound healing | 298 | 433 | 9.888e-09 |
| GO:BP | GO:0008015 | blood circulation | 350 | 518 | 1.043e-08 |
| GO:BP | GO:0090109 | regulation of cell-substrate junction assembly | 57 | 63 | 1.050e-08 |
| GO:BP | GO:0051893 | regulation of focal adhesion assembly | 57 | 63 | 1.050e-08 |
| GO:BP | GO:1903522 | regulation of blood circulation | 187 | 256 | 1.103e-08 |
| GO:BP | GO:0060541 | respiratory system development | 167 | 225 | 1.192e-08 |
| GO:BP | GO:0022904 | respiratory electron transport chain | 94 | 115 | 1.211e-08 |
| GO:BP | GO:0016052 | carbohydrate catabolic process | 133 | 173 | 1.271e-08 |
| GO:BP | GO:0042982 | amyloid precursor protein metabolic process | 65 | 74 | 1.292e-08 |
| GO:BP | GO:0051494 | negative regulation of cytoskeleton organization | 121 | 155 | 1.342e-08 |
| GO:BP | GO:0006164 | purine nucleotide biosynthetic process | 146 | 193 | 1.454e-08 |
| GO:BP | GO:0071383 | cellular response to steroid hormone stimulus | 159 | 213 | 1.469e-08 |
| GO:BP | GO:0032922 | circadian regulation of gene expression | 62 | 70 | 1.475e-08 |
| GO:BP | GO:0006406 | mRNA export from nucleus | 62 | 70 | 1.475e-08 |
| GO:BP | GO:0007173 | epidermal growth factor receptor signaling pathway | 89 | 108 | 1.494e-08 |
| GO:BP | GO:0031398 | positive regulation of protein ubiquitination | 89 | 108 | 1.494e-08 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 239 | 339 | 1.516e-08 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 239 | 339 | 1.516e-08 |
| GO:BP | GO:0098916 | anterograde trans-synaptic signaling | 504 | 775 | 1.523e-08 |
| GO:BP | GO:0007268 | chemical synaptic transmission | 504 | 775 | 1.523e-08 |
| GO:BP | GO:0031110 | regulation of microtubule polymerization or depolymerization | 82 | 98 | 1.554e-08 |
| GO:BP | GO:0010761 | fibroblast migration | 50 | 54 | 1.576e-08 |
| GO:BP | GO:0009303 | rRNA transcription | 39 | 40 | 1.585e-08 |
| GO:BP | GO:0035329 | hippo signaling | 53 | 58 | 1.705e-08 |
| GO:BP | GO:0006611 | protein export from nucleus | 56 | 62 | 1.707e-08 |
| GO:BP | GO:0006109 | regulation of carbohydrate metabolic process | 134 | 175 | 1.728e-08 |
| GO:BP | GO:0090151 | establishment of protein localization to mitochondrial membrane | 34 | 34 | 1.752e-08 |
| GO:BP | GO:0072344 | rescue of stalled ribosome | 34 | 34 | 1.752e-08 |
| GO:BP | GO:0034063 | stress granule assembly | 34 | 34 | 1.752e-08 |
| GO:BP | GO:0030048 | actin filament-based movement | 108 | 136 | 1.752e-08 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 132 | 172 | 1.758e-08 |
| GO:BP | GO:0110020 | regulation of actomyosin structure organization | 84 | 101 | 1.765e-08 |
| GO:BP | GO:0009165 | nucleotide biosynthetic process | 169 | 229 | 1.923e-08 |
| GO:BP | GO:0010833 | telomere maintenance via telomere lengthening | 64 | 73 | 2.031e-08 |
| GO:BP | GO:0090263 | positive regulation of canonical Wnt signaling pathway | 88 | 107 | 2.222e-08 |
| GO:BP | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 69 | 80 | 2.225e-08 |
| GO:BP | GO:0060411 | cardiac septum morphogenesis | 61 | 69 | 2.343e-08 |
| GO:BP | GO:0010977 | negative regulation of neuron projection development | 105 | 132 | 2.464e-08 |
| GO:BP | GO:0001837 | epithelial to mesenchymal transition | 131 | 171 | 2.469e-08 |
| GO:BP | GO:0071453 | cellular response to oxygen levels | 127 | 165 | 2.572e-08 |
| GO:BP | GO:0046854 | phosphatidylinositol phosphate biosynthetic process | 58 | 65 | 2.607e-08 |
| GO:BP | GO:0007155 | cell adhesion | 945 | 1530 | 2.616e-08 |
| GO:BP | GO:0051646 | mitochondrion localization | 49 | 53 | 2.619e-08 |
| GO:BP | GO:0034198 | cellular response to amino acid starvation | 49 | 53 | 2.619e-08 |
| GO:BP | GO:0030010 | establishment of cell polarity | 123 | 159 | 2.634e-08 |
| GO:BP | GO:0019058 | viral life cycle | 220 | 310 | 2.671e-08 |
| GO:BP | GO:0046467 | membrane lipid biosynthetic process | 117 | 150 | 2.673e-08 |
| GO:BP | GO:0060998 | regulation of dendritic spine development | 55 | 61 | 2.765e-08 |
| GO:BP | GO:0006446 | regulation of translational initiation | 71 | 83 | 2.768e-08 |
| GO:BP | GO:0097061 | dendritic spine organization | 71 | 83 | 2.768e-08 |
| GO:BP | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 52 | 57 | 2.781e-08 |
| GO:BP | GO:0051651 | maintenance of location in cell | 155 | 208 | 2.814e-08 |
| GO:BP | GO:0032231 | regulation of actin filament bundle assembly | 85 | 103 | 2.977e-08 |
| GO:BP | GO:0051402 | neuron apoptotic process | 211 | 296 | 3.066e-08 |
| GO:BP | GO:0010507 | negative regulation of autophagy | 78 | 93 | 3.071e-08 |
| GO:BP | GO:0036257 | multivesicular body organization | 33 | 33 | 3.123e-08 |
| GO:BP | GO:0008333 | endosome to lysosome transport | 63 | 72 | 3.197e-08 |
| GO:BP | GO:0097191 | extrinsic apoptotic signaling pathway | 169 | 230 | 3.258e-08 |
| GO:BP | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 80 | 96 | 3.537e-08 |
| GO:BP | GO:0006513 | protein monoubiquitination | 45 | 48 | 3.724e-08 |
| GO:BP | GO:0072583 | clathrin-dependent endocytosis | 45 | 48 | 3.724e-08 |
| GO:BP | GO:0016331 | morphogenesis of embryonic epithelium | 118 | 152 | 3.769e-08 |
| GO:BP | GO:1903076 | regulation of protein localization to plasma membrane | 91 | 112 | 3.876e-08 |
| GO:BP | GO:0099068 | postsynapse assembly | 75 | 89 | 4.012e-08 |
| GO:BP | GO:0051235 | maintenance of location | 161 | 218 | 4.156e-08 |
| GO:BP | GO:0061512 | protein localization to cilium | 65 | 75 | 4.224e-08 |
| GO:BP | GO:0009299 | mRNA transcription | 48 | 52 | 4.344e-08 |
| GO:BP | GO:0034612 | response to tumor necrosis factor | 168 | 229 | 4.377e-08 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 340 | 506 | 4.448e-08 |
| GO:BP | GO:0006754 | ATP biosynthetic process | 84 | 102 | 4.448e-08 |
| GO:BP | GO:0045995 | regulation of embryonic development | 77 | 92 | 4.667e-08 |
| GO:BP | GO:0006418 | tRNA aminoacylation for protein translation | 41 | 43 | 4.696e-08 |
| GO:BP | GO:0000966 | RNA 5’-end processing | 37 | 38 | 4.807e-08 |
| GO:BP | GO:0043500 | muscle adaptation | 86 | 105 | 4.883e-08 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 86 | 105 | 4.883e-08 |
| GO:BP | GO:0000380 | alternative mRNA splicing, via spliceosome | 62 | 71 | 5.048e-08 |
| GO:BP | GO:0030218 | erythrocyte differentiation | 107 | 136 | 5.277e-08 |
| GO:BP | GO:0051147 | regulation of muscle cell differentiation | 111 | 142 | 5.360e-08 |
| GO:BP | GO:0006898 | receptor-mediated endocytosis | 190 | 264 | 5.483e-08 |
| GO:BP | GO:0099022 | vesicle tethering | 32 | 32 | 5.577e-08 |
| GO:BP | GO:0036258 | multivesicular body assembly | 32 | 32 | 5.577e-08 |
| GO:BP | GO:0043491 | phosphatidylinositol 3-kinase/protein kinase B signal transduction | 212 | 299 | 5.719e-08 |
| GO:BP | GO:0010232 | vascular transport | 74 | 88 | 6.094e-08 |
| GO:BP | GO:0150104 | transport across blood-brain barrier | 74 | 88 | 6.094e-08 |
| GO:BP | GO:0014812 | muscle cell migration | 74 | 88 | 6.094e-08 |
| GO:BP | GO:0051962 | positive regulation of nervous system development | 210 | 296 | 6.203e-08 |
| GO:BP | GO:0042789 | mRNA transcription by RNA polymerase II | 44 | 47 | 6.242e-08 |
| GO:BP | GO:0032981 | mitochondrial respiratory chain complex I assembly | 56 | 63 | 6.658e-08 |
| GO:BP | GO:0006278 | RNA-templated DNA biosynthetic process | 56 | 63 | 6.658e-08 |
| GO:BP | GO:0010257 | NADH dehydrogenase complex assembly | 56 | 63 | 6.658e-08 |
| GO:BP | GO:0051492 | regulation of stress fiber assembly | 76 | 91 | 7.050e-08 |
| GO:BP | GO:0008366 | axon ensheathment | 122 | 159 | 7.101e-08 |
| GO:BP | GO:0007272 | ensheathment of neurons | 122 | 159 | 7.101e-08 |
| GO:BP | GO:0055008 | cardiac muscle tissue morphogenesis | 53 | 59 | 7.234e-08 |
| GO:BP | GO:0009154 | purine ribonucleotide catabolic process | 102 | 129 | 7.287e-08 |
| GO:BP | GO:0030323 | respiratory tube development | 150 | 202 | 7.380e-08 |
| GO:BP | GO:0043401 | steroid hormone receptor signaling pathway | 108 | 138 | 7.561e-08 |
| GO:BP | GO:0005977 | glycogen metabolic process | 71 | 84 | 7.891e-08 |
| GO:BP | GO:0006303 | double-strand break repair via nonhomologous end joining | 61 | 70 | 7.925e-08 |
| GO:BP | GO:0017004 | cytochrome complex assembly | 40 | 42 | 7.987e-08 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 133 | 176 | 8.209e-08 |
| GO:BP | GO:0045324 | late endosome to vacuole transport | 36 | 37 | 8.334e-08 |
| GO:BP | GO:0050769 | positive regulation of neurogenesis | 178 | 246 | 8.472e-08 |
| GO:BP | GO:0072666 | establishment of protein localization to vacuole | 58 | 66 | 9.352e-08 |
| GO:BP | GO:0030155 | regulation of cell adhesion | 511 | 794 | 9.530e-08 |
| GO:BP | GO:0050773 | regulation of dendrite development | 82 | 100 | 9.860e-08 |
| GO:BP | GO:0043409 | negative regulation of MAPK cascade | 121 | 158 | 9.917e-08 |
| GO:BP | GO:0006359 | regulation of transcription by RNA polymerase III | 31 | 31 | 9.945e-08 |
| GO:BP | GO:2000736 | regulation of stem cell differentiation | 68 | 80 | 1.012e-07 |
| GO:BP | GO:0036294 | cellular response to decreased oxygen levels | 115 | 149 | 1.047e-07 |
| GO:BP | GO:0042149 | cellular response to glucose starvation | 43 | 46 | 1.047e-07 |
| GO:BP | GO:0032210 | regulation of telomere maintenance via telomerase | 43 | 46 | 1.047e-07 |
| GO:BP | GO:0034644 | cellular response to UV | 75 | 90 | 1.061e-07 |
| GO:BP | GO:0046328 | regulation of JNK cascade | 109 | 140 | 1.073e-07 |
| GO:BP | GO:0006887 | exocytosis | 249 | 360 | 1.083e-07 |
| GO:BP | GO:0001649 | osteoblast differentiation | 170 | 234 | 1.124e-07 |
| GO:BP | GO:0010717 | regulation of epithelial to mesenchymal transition | 86 | 106 | 1.148e-07 |
| GO:BP | GO:0003300 | cardiac muscle hypertrophy | 70 | 83 | 1.204e-07 |
| GO:BP | GO:0016925 | protein sumoylation | 60 | 69 | 1.245e-07 |
| GO:BP | GO:0070534 | protein K63-linked ubiquitination | 60 | 69 | 1.245e-07 |
| GO:BP | GO:0051865 | protein autoubiquitination | 65 | 76 | 1.280e-07 |
| GO:BP | GO:0045926 | negative regulation of growth | 166 | 228 | 1.290e-07 |
| GO:BP | GO:0010906 | regulation of glucose metabolic process | 79 | 96 | 1.332e-07 |
| GO:BP | GO:0043123 | positive regulation of canonical NF-kappaB signal transduction | 157 | 214 | 1.342e-07 |
| GO:BP | GO:0042552 | myelination | 120 | 157 | 1.377e-07 |
| GO:BP | GO:0044042 | glucan metabolic process | 72 | 86 | 1.401e-07 |
| GO:BP | GO:0016242 | negative regulation of macroautophagy | 35 | 36 | 1.452e-07 |
| GO:BP | GO:0009261 | ribonucleotide catabolic process | 106 | 136 | 1.526e-07 |
| GO:BP | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway | 131 | 174 | 1.552e-07 |
| GO:BP | GO:0050772 | positive regulation of axonogenesis | 67 | 79 | 1.553e-07 |
| GO:BP | GO:0072526 | pyridine-containing compound catabolic process | 83 | 102 | 1.576e-07 |
| GO:BP | GO:0005976 | polysaccharide metabolic process | 83 | 102 | 1.576e-07 |
| GO:BP | GO:1903312 | negative regulation of mRNA metabolic process | 83 | 102 | 1.576e-07 |
| GO:BP | GO:0045937 | positive regulation of phosphate metabolic process | 370 | 560 | 1.707e-07 |
| GO:BP | GO:0010562 | positive regulation of phosphorus metabolic process | 370 | 560 | 1.707e-07 |
| GO:BP | GO:0051963 | regulation of synapse assembly | 125 | 165 | 1.739e-07 |
| GO:BP | GO:0018279 | protein N-linked glycosylation via asparagine | 42 | 45 | 1.752e-07 |
| GO:BP | GO:1903573 | negative regulation of response to endoplasmic reticulum stress | 42 | 45 | 1.752e-07 |
| GO:BP | GO:0043408 | regulation of MAPK cascade | 416 | 637 | 1.862e-07 |
| GO:BP | GO:0048511 | rhythmic process | 214 | 305 | 1.875e-07 |
| GO:BP | GO:0097479 | synaptic vesicle localization | 51 | 57 | 1.880e-07 |
| GO:BP | GO:0032507 | maintenance of protein location in cell | 51 | 57 | 1.880e-07 |
| GO:BP | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 145 | 196 | 1.920e-07 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 91 | 114 | 1.947e-07 |
| GO:BP | GO:0043393 | regulation of protein binding | 91 | 114 | 1.947e-07 |
| GO:BP | GO:2000785 | regulation of autophagosome assembly | 45 | 49 | 1.950e-07 |
| GO:BP | GO:0010720 | positive regulation of cell development | 308 | 458 | 1.984e-07 |
| GO:BP | GO:0042063 | gliogenesis | 243 | 352 | 2.070e-07 |
| GO:BP | GO:2001235 | positive regulation of apoptotic signaling pathway | 107 | 138 | 2.129e-07 |
| GO:BP | GO:0032989 | cellular anatomical entity morphogenesis | 107 | 138 | 2.129e-07 |
| GO:BP | GO:0009185 | ribonucleoside diphosphate metabolic process | 99 | 126 | 2.129e-07 |
| GO:BP | GO:0010927 | cellular component assembly involved in morphogenesis | 107 | 138 | 2.129e-07 |
| GO:BP | GO:0033059 | cellular pigmentation | 56 | 64 | 2.324e-07 |
| GO:BP | GO:0031113 | regulation of microtubule polymerization | 56 | 64 | 2.324e-07 |
| GO:BP | GO:0015986 | proton motive force-driven ATP synthesis | 66 | 78 | 2.368e-07 |
| GO:BP | GO:0030324 | lung development | 146 | 198 | 2.436e-07 |
| GO:BP | GO:0045943 | positive regulation of transcription by RNA polymerase I | 34 | 35 | 2.515e-07 |
| GO:BP | GO:0016051 | carbohydrate biosynthetic process | 142 | 192 | 2.745e-07 |
| GO:BP | GO:0000910 | cytokinesis | 142 | 192 | 2.745e-07 |
| GO:BP | GO:0060395 | SMAD protein signal transduction | 68 | 81 | 2.786e-07 |
| GO:BP | GO:0007611 | learning or memory | 199 | 282 | 2.868e-07 |
| GO:BP | GO:0030900 | forebrain development | 286 | 423 | 2.882e-07 |
| GO:BP | GO:0009179 | purine ribonucleoside diphosphate metabolic process | 92 | 116 | 2.898e-07 |
| GO:BP | GO:0009135 | purine nucleoside diphosphate metabolic process | 92 | 116 | 2.898e-07 |
| GO:BP | GO:0034440 | lipid oxidation | 92 | 116 | 2.898e-07 |
| GO:BP | GO:0009225 | nucleotide-sugar metabolic process | 41 | 44 | 2.923e-07 |
| GO:BP | GO:0090114 | COPII-coated vesicle budding | 41 | 44 | 2.923e-07 |
| GO:BP | GO:1902414 | protein localization to cell junction | 94 | 119 | 2.956e-07 |
| GO:BP | GO:1901799 | negative regulation of proteasomal protein catabolic process | 50 | 56 | 3.012e-07 |
| GO:BP | GO:0042987 | amyloid precursor protein catabolic process | 50 | 56 | 3.012e-07 |
| GO:BP | GO:0006487 | protein N-linked glycosylation | 63 | 74 | 3.020e-07 |
| GO:BP | GO:0042776 | proton motive force-driven mitochondrial ATP synthesis | 58 | 67 | 3.028e-07 |
| GO:BP | GO:0034762 | regulation of transmembrane transport | 298 | 443 | 3.163e-07 |
| GO:BP | GO:0033135 | regulation of peptidyl-serine phosphorylation | 79 | 97 | 3.167e-07 |
| GO:BP | GO:0016601 | Rac protein signal transduction | 44 | 48 | 3.189e-07 |
| GO:BP | GO:0006479 | protein methylation | 44 | 48 | 3.189e-07 |
| GO:BP | GO:0006623 | protein targeting to vacuole | 44 | 48 | 3.189e-07 |
| GO:BP | GO:0008213 | protein alkylation | 44 | 48 | 3.189e-07 |
| GO:BP | GO:1902459 | positive regulation of stem cell population maintenance | 44 | 48 | 3.189e-07 |
| GO:BP | GO:0060173 | limb development | 143 | 194 | 3.450e-07 |
| GO:BP | GO:0048736 | appendage development | 143 | 194 | 3.450e-07 |
| GO:BP | GO:0009191 | ribonucleoside diphosphate catabolic process | 83 | 103 | 3.585e-07 |
| GO:BP | GO:0008637 | apoptotic mitochondrial changes | 83 | 103 | 3.585e-07 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 322 | 483 | 3.615e-07 |
| GO:BP | GO:0006520 | amino acid metabolic process | 208 | 297 | 3.615e-07 |
| GO:BP | GO:0097352 | autophagosome maturation | 55 | 63 | 3.631e-07 |
| GO:BP | GO:0006812 | monoatomic cation transport | 669 | 1069 | 3.688e-07 |
| GO:BP | GO:0010586 | miRNA metabolic process | 85 | 106 | 3.753e-07 |
| GO:BP | GO:0006901 | vesicle coating | 37 | 39 | 3.900e-07 |
| GO:BP | GO:0044743 | protein transmembrane import into intracellular organelle | 37 | 39 | 3.900e-07 |
| GO:BP | GO:0097581 | lamellipodium organization | 74 | 90 | 3.944e-07 |
| GO:BP | GO:0050890 | cognition | 227 | 328 | 4.213e-07 |
| GO:BP | GO:0009132 | nucleoside diphosphate metabolic process | 103 | 133 | 4.240e-07 |
| GO:BP | GO:0061462 | protein localization to lysosome | 52 | 59 | 4.251e-07 |
| GO:BP | GO:0030518 | nuclear receptor-mediated steroid hormone signaling pathway | 95 | 121 | 4.251e-07 |
| GO:BP | GO:0006937 | regulation of muscle contraction | 124 | 165 | 4.274e-07 |
| GO:BP | GO:0062125 | regulation of mitochondrial gene expression | 33 | 34 | 4.316e-07 |
| GO:BP | GO:0048732 | gland development | 304 | 454 | 4.504e-07 |
| GO:BP | GO:0003281 | ventricular septum development | 62 | 73 | 4.614e-07 |
| GO:BP | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | 57 | 66 | 4.702e-07 |
| GO:BP | GO:2000059 | negative regulation of ubiquitin-dependent protein catabolic process | 49 | 55 | 4.807e-07 |
| GO:BP | GO:0048489 | synaptic vesicle transport | 40 | 43 | 4.833e-07 |
| GO:BP | GO:0060999 | positive regulation of dendritic spine development | 40 | 43 | 4.833e-07 |
| GO:BP | GO:0072595 | maintenance of protein localization in organelle | 40 | 43 | 4.833e-07 |
| GO:BP | GO:0019364 | pyridine nucleotide catabolic process | 80 | 99 | 4.937e-07 |
| GO:BP | GO:0008089 | anterograde axonal transport | 46 | 51 | 5.179e-07 |
| GO:BP | GO:0055010 | ventricular cardiac muscle tissue morphogenesis | 43 | 47 | 5.228e-07 |
| GO:BP | GO:0032527 | protein exit from endoplasmic reticulum | 43 | 47 | 5.228e-07 |
| GO:BP | GO:0047496 | vesicle transport along microtubule | 43 | 47 | 5.228e-07 |
| GO:BP | GO:0014897 | striated muscle hypertrophy | 71 | 86 | 5.309e-07 |
| GO:BP | GO:0140056 | organelle localization by membrane tethering | 64 | 76 | 5.447e-07 |
| GO:BP | GO:0007266 | Rho protein signal transduction | 110 | 144 | 5.472e-07 |
| GO:BP | GO:1903513 | endoplasmic reticulum to cytosol transport | 28 | 28 | 5.525e-07 |
| GO:BP | GO:0030970 | retrograde protein transport, ER to cytosol | 28 | 28 | 5.525e-07 |
| GO:BP | GO:0036010 | protein localization to endosome | 28 | 28 | 5.525e-07 |
| GO:BP | GO:0055080 | monoatomic cation homeostasis | 395 | 606 | 5.549e-07 |
| GO:BP | GO:0003158 | endothelium development | 106 | 138 | 5.722e-07 |
| GO:BP | GO:0070252 | actin-mediated cell contraction | 88 | 111 | 5.766e-07 |
| GO:BP | GO:0035265 | organ growth | 123 | 164 | 5.808e-07 |
| GO:BP | GO:0008589 | regulation of smoothened signaling pathway | 73 | 89 | 5.821e-07 |
| GO:BP | GO:0072329 | monocarboxylic acid catabolic process | 98 | 126 | 6.017e-07 |
| GO:BP | GO:0007265 | Ras protein signal transduction | 96 | 123 | 6.025e-07 |
| GO:BP | GO:0030041 | actin filament polymerization | 121 | 161 | 6.062e-07 |
| GO:BP | GO:0009791 | post-embryonic development | 75 | 92 | 6.323e-07 |
| GO:BP | GO:1900182 | positive regulation of protein localization to nucleus | 77 | 95 | 6.798e-07 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 202 | 289 | 6.993e-07 |
| GO:BP | GO:2000036 | regulation of stem cell population maintenance | 61 | 72 | 7.039e-07 |
| GO:BP | GO:0042542 | response to hydrogen peroxide | 79 | 98 | 7.208e-07 |
| GO:BP | GO:0009416 | response to light stimulus | 223 | 323 | 7.321e-07 |
| GO:BP | GO:0061387 | regulation of extent of cell growth | 81 | 101 | 7.568e-07 |
| GO:BP | GO:0060538 | skeletal muscle organ development | 142 | 194 | 7.739e-07 |
| GO:BP | GO:0009134 | nucleoside diphosphate catabolic process | 85 | 107 | 8.131e-07 |
| GO:BP | GO:0055117 | regulation of cardiac muscle contraction | 63 | 75 | 8.288e-07 |
| GO:BP | GO:0141193 | nuclear receptor-mediated signaling pathway | 131 | 177 | 8.314e-07 |
| GO:BP | GO:0035592 | establishment of protein localization to extracellular region | 259 | 382 | 8.425e-07 |
| GO:BP | GO:0050801 | monoatomic ion homeostasis | 400 | 616 | 8.526e-07 |
| GO:BP | GO:0043534 | blood vessel endothelial cell migration | 93 | 119 | 8.573e-07 |
| GO:BP | GO:0014896 | muscle hypertrophy | 72 | 88 | 8.666e-07 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 138 | 188 | 8.781e-07 |
| GO:BP | GO:0035567 | non-canonical Wnt signaling pathway | 58 | 68 | 9.000e-07 |
| GO:BP | GO:0009306 | protein secretion | 257 | 379 | 9.237e-07 |
| GO:BP | GO:0006665 | sphingolipid metabolic process | 125 | 168 | 9.800e-07 |
| GO:BP | GO:0044282 | small molecule catabolic process | 252 | 371 | 9.829e-07 |
| GO:BP | GO:1904358 | positive regulation of telomere maintenance via telomere lengthening | 27 | 27 | 9.852e-07 |
| GO:BP | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway | 27 | 27 | 9.852e-07 |
| GO:BP | GO:0033119 | negative regulation of RNA splicing | 27 | 27 | 9.852e-07 |
| GO:BP | GO:0001101 | response to acid chemical | 110 | 145 | 1.017e-06 |
| GO:BP | GO:0000725 | recombinational repair | 141 | 193 | 1.031e-06 |
| GO:BP | GO:0006942 | regulation of striated muscle contraction | 78 | 97 | 1.049e-06 |
| GO:BP | GO:0090316 | positive regulation of intracellular protein transport | 78 | 97 | 1.049e-06 |
| GO:BP | GO:0071456 | cellular response to hypoxia | 108 | 142 | 1.049e-06 |
| GO:BP | GO:0046032 | ADP catabolic process | 78 | 97 | 1.049e-06 |
| GO:BP | GO:0051222 | positive regulation of protein transport | 182 | 258 | 1.082e-06 |
| GO:BP | GO:0071229 | cellular response to acid chemical | 80 | 100 | 1.094e-06 |
| GO:BP | GO:0009181 | purine ribonucleoside diphosphate catabolic process | 80 | 100 | 1.094e-06 |
| GO:BP | GO:0009137 | purine nucleoside diphosphate catabolic process | 80 | 100 | 1.094e-06 |
| GO:BP | GO:0002011 | morphogenesis of an epithelial sheet | 55 | 64 | 1.128e-06 |
| GO:BP | GO:0034446 | substrate adhesion-dependent cell spreading | 82 | 103 | 1.133e-06 |
| GO:BP | GO:0006310 | DNA recombination | 237 | 347 | 1.156e-06 |
| GO:BP | GO:0051896 | regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 185 | 263 | 1.175e-06 |
| GO:BP | GO:0072657 | protein localization to membrane | 524 | 827 | 1.183e-06 |
| GO:BP | GO:0045445 | myoblast differentiation | 90 | 115 | 1.208e-06 |
| GO:BP | GO:0061448 | connective tissue development | 203 | 292 | 1.209e-06 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 115 | 153 | 1.237e-06 |
| GO:BP | GO:0014909 | smooth muscle cell migration | 62 | 74 | 1.247e-06 |
| GO:BP | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening | 31 | 32 | 1.286e-06 |
| GO:BP | GO:0071526 | semaphorin-plexin signaling pathway | 44 | 49 | 1.347e-06 |
| GO:BP | GO:0006497 | protein lipidation | 44 | 49 | 1.347e-06 |
| GO:BP | GO:0034248 | regulation of amide metabolic process | 57 | 67 | 1.379e-06 |
| GO:BP | GO:0048639 | positive regulation of developmental growth | 122 | 164 | 1.390e-06 |
| GO:BP | GO:0035196 | miRNA processing | 41 | 45 | 1.396e-06 |
| GO:BP | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process | 41 | 45 | 1.396e-06 |
| GO:BP | GO:0018107 | peptidyl-threonine phosphorylation | 64 | 77 | 1.420e-06 |
| GO:BP | GO:0043535 | regulation of blood vessel endothelial cell migration | 75 | 93 | 1.452e-06 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 207 | 299 | 1.491e-06 |
| GO:BP | GO:0071356 | cellular response to tumor necrosis factor | 150 | 208 | 1.567e-06 |
| GO:BP | GO:0007610 | behavior | 433 | 674 | 1.571e-06 |
| GO:BP | GO:0007369 | gastrulation | 143 | 197 | 1.580e-06 |
| GO:BP | GO:0043604 | amide biosynthetic process | 127 | 172 | 1.585e-06 |
| GO:BP | GO:0062207 | regulation of pattern recognition receptor signaling pathway | 127 | 172 | 1.585e-06 |
| GO:BP | GO:0006873 | intracellular monoatomic ion homeostasis | 349 | 533 | 1.590e-06 |
| GO:BP | GO:0060415 | muscle tissue morphogenesis | 59 | 70 | 1.631e-06 |
| GO:BP | GO:0141137 | positive regulation of gene expression, epigenetic | 49 | 56 | 1.666e-06 |
| GO:BP | GO:0006473 | protein acetylation | 49 | 56 | 1.666e-06 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 95 | 123 | 1.672e-06 |
| GO:BP | GO:0006515 | protein quality control for misfolded or incompletely synthesized proteins | 26 | 26 | 1.744e-06 |
| GO:BP | GO:0044272 | sulfur compound biosynthetic process | 112 | 149 | 1.764e-06 |
| GO:BP | GO:0098655 | monoatomic cation transmembrane transport | 552 | 877 | 1.921e-06 |
| GO:BP | GO:0046165 | alcohol biosynthetic process | 108 | 143 | 1.921e-06 |
| GO:BP | GO:0031648 | protein destabilization | 46 | 52 | 1.946e-06 |
| GO:BP | GO:0007098 | centrosome cycle | 106 | 140 | 1.995e-06 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 72 | 89 | 2.011e-06 |
| GO:BP | GO:0071692 | protein localization to extracellular region | 261 | 388 | 2.069e-06 |
| GO:BP | GO:0030003 | intracellular monoatomic cation homeostasis | 343 | 524 | 2.108e-06 |
| GO:BP | GO:0043523 | regulation of neuron apoptotic process | 171 | 242 | 2.108e-06 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 171 | 242 | 2.108e-06 |
| GO:BP | GO:0006998 | nuclear envelope organization | 51 | 59 | 2.167e-06 |
| GO:BP | GO:0032092 | positive regulation of protein binding | 43 | 48 | 2.167e-06 |
| GO:BP | GO:0001881 | receptor recycling | 43 | 48 | 2.167e-06 |
| GO:BP | GO:0043200 | response to amino acid | 100 | 131 | 2.192e-06 |
| GO:BP | GO:0006096 | glycolytic process | 76 | 95 | 2.209e-06 |
| GO:BP | GO:0051085 | chaperone cofactor-dependent protein refolding | 30 | 31 | 2.215e-06 |
| GO:BP | GO:0019082 | viral protein processing | 30 | 31 | 2.215e-06 |
| GO:BP | GO:0051259 | protein complex oligomerization | 189 | 271 | 2.224e-06 |
| GO:BP | GO:1903146 | regulation of autophagy of mitochondrion | 40 | 44 | 2.275e-06 |
| GO:BP | GO:0097485 | neuron projection guidance | 169 | 239 | 2.280e-06 |
| GO:BP | GO:0070646 | protein modification by small protein removal | 113 | 151 | 2.293e-06 |
| GO:BP | GO:0043502 | regulation of muscle adaptation | 65 | 79 | 2.354e-06 |
| GO:BP | GO:0039531 | regulation of cytoplasmic pattern recognition receptor signaling pathway | 92 | 119 | 2.374e-06 |
| GO:BP | GO:0046031 | ADP metabolic process | 82 | 104 | 2.377e-06 |
| GO:BP | GO:1901652 | response to peptide | 601 | 962 | 2.404e-06 |
| GO:BP | GO:0046902 | regulation of mitochondrial membrane permeability | 48 | 55 | 2.622e-06 |
| GO:BP | GO:0019216 | regulation of lipid metabolic process | 219 | 320 | 2.625e-06 |
| GO:BP | GO:0003179 | heart valve morphogenesis | 53 | 62 | 2.686e-06 |
| GO:BP | GO:0043467 | regulation of generation of precursor metabolites and energy | 103 | 136 | 2.840e-06 |
| GO:BP | GO:1900076 | regulation of cellular response to insulin stimulus | 60 | 72 | 2.840e-06 |
| GO:BP | GO:0051881 | regulation of mitochondrial membrane potential | 60 | 72 | 2.840e-06 |
| GO:BP | GO:0045598 | regulation of fat cell differentiation | 101 | 133 | 2.944e-06 |
| GO:BP | GO:0007411 | axon guidance | 168 | 238 | 2.949e-06 |
| GO:BP | GO:0048545 | response to steroid hormone | 231 | 340 | 3.057e-06 |
| GO:BP | GO:0046364 | monosaccharide biosynthetic process | 73 | 91 | 3.082e-06 |
| GO:BP | GO:0045214 | sarcomere organization | 45 | 51 | 3.085e-06 |
| GO:BP | GO:1903241 | U2-type prespliceosome assembly | 25 | 25 | 3.090e-06 |
| GO:BP | GO:0086064 | cell communication by electrical coupling involved in cardiac conduction | 25 | 25 | 3.090e-06 |
| GO:BP | GO:0000470 | maturation of LSU-rRNA | 25 | 25 | 3.090e-06 |
| GO:BP | GO:1904292 | regulation of ERAD pathway | 25 | 25 | 3.090e-06 |
| GO:BP | GO:0007269 | neurotransmitter secretion | 112 | 150 | 3.116e-06 |
| GO:BP | GO:0099643 | signal release from synapse | 112 | 150 | 3.116e-06 |
| GO:BP | GO:0016180 | snRNA processing | 33 | 35 | 3.146e-06 |
| GO:BP | GO:0000154 | rRNA modification | 33 | 35 | 3.146e-06 |
| GO:BP | GO:0019068 | virion assembly | 33 | 35 | 3.146e-06 |
| GO:BP | GO:0006891 | intra-Golgi vesicle-mediated transport | 33 | 35 | 3.146e-06 |
| GO:BP | GO:0001764 | neuron migration | 137 | 189 | 3.146e-06 |
| GO:BP | GO:0050775 | positive regulation of dendrite morphogenesis | 33 | 35 | 3.146e-06 |
| GO:BP | GO:0043954 | cellular component maintenance | 62 | 75 | 3.153e-06 |
| GO:BP | GO:0003170 | heart valve development | 62 | 75 | 3.153e-06 |
| GO:BP | GO:0006090 | pyruvate metabolic process | 95 | 124 | 3.173e-06 |
| GO:BP | GO:0045185 | maintenance of protein location | 55 | 65 | 3.188e-06 |
| GO:BP | GO:0099072 | regulation of postsynaptic membrane neurotransmitter receptor levels | 93 | 121 | 3.243e-06 |
| GO:BP | GO:0045861 | negative regulation of proteolysis | 110 | 147 | 3.243e-06 |
| GO:BP | GO:0008064 | regulation of actin polymerization or depolymerization | 110 | 147 | 3.243e-06 |
| GO:BP | GO:0048278 | vesicle docking | 50 | 58 | 3.323e-06 |
| GO:BP | GO:0006664 | glycolipid metabolic process | 83 | 106 | 3.390e-06 |
| GO:BP | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 42 | 47 | 3.440e-06 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 292 | 441 | 3.551e-06 |
| GO:BP | GO:0034097 | response to cytokine | 591 | 947 | 3.629e-06 |
| GO:BP | GO:1903214 | regulation of protein targeting to mitochondrion | 39 | 43 | 3.675e-06 |
| GO:BP | GO:0086001 | cardiac muscle cell action potential | 66 | 81 | 3.760e-06 |
| GO:BP | GO:0030516 | regulation of axon extension | 68 | 84 | 4.028e-06 |
| GO:BP | GO:0043462 | regulation of ATP-dependent activity | 52 | 61 | 4.107e-06 |
| GO:BP | GO:0001654 | eye development | 276 | 415 | 4.147e-06 |
| GO:BP | GO:0018210 | peptidyl-threonine modification | 70 | 87 | 4.251e-06 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 245 | 364 | 4.312e-06 |
| GO:BP | GO:0006836 | neurotransmitter transport | 151 | 212 | 4.728e-06 |
| GO:BP | GO:1902017 | regulation of cilium assembly | 61 | 74 | 4.736e-06 |
| GO:BP | GO:0042775 | mitochondrial ATP synthesis coupled electron transport | 78 | 99 | 4.781e-06 |
| GO:BP | GO:0016079 | synaptic vesicle exocytosis | 78 | 99 | 4.781e-06 |
| GO:BP | GO:0042773 | ATP synthesis coupled electron transport | 78 | 99 | 4.781e-06 |
| GO:BP | GO:0014037 | Schwann cell differentiation | 44 | 50 | 4.858e-06 |
| GO:BP | GO:0098815 | modulation of excitatory postsynaptic potential | 44 | 50 | 4.858e-06 |
| GO:BP | GO:0008360 | regulation of cell shape | 105 | 140 | 4.881e-06 |
| GO:BP | GO:1903533 | regulation of protein targeting | 63 | 77 | 5.169e-06 |
| GO:BP | GO:0003151 | outflow tract morphogenesis | 63 | 77 | 5.169e-06 |
| GO:BP | GO:0050435 | amyloid-beta metabolic process | 49 | 57 | 5.183e-06 |
| GO:BP | GO:0051058 | negative regulation of small GTPase mediated signal transduction | 49 | 57 | 5.183e-06 |
| GO:BP | GO:0051654 | establishment of mitochondrion localization | 32 | 34 | 5.304e-06 |
| GO:BP | GO:0006734 | NADH metabolic process | 32 | 34 | 5.304e-06 |
| GO:BP | GO:0042790 | nucleolar large rRNA transcription by RNA polymerase I | 24 | 24 | 5.465e-06 |
| GO:BP | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 24 | 24 | 5.465e-06 |
| GO:BP | GO:0032212 | positive regulation of telomere maintenance via telomerase | 24 | 24 | 5.465e-06 |
| GO:BP | GO:0060759 | regulation of response to cytokine stimulus | 128 | 176 | 5.503e-06 |
| GO:BP | GO:0031124 | mRNA 3’-end processing | 41 | 46 | 5.525e-06 |
| GO:BP | GO:0045667 | regulation of osteoblast differentiation | 99 | 131 | 5.525e-06 |
| GO:BP | GO:0002183 | cytoplasmic translational initiation | 41 | 46 | 5.525e-06 |
| GO:BP | GO:0034394 | protein localization to cell surface | 56 | 67 | 5.620e-06 |
| GO:BP | GO:0002221 | pattern recognition receptor signaling pathway | 185 | 267 | 5.748e-06 |
| GO:BP | GO:1902991 | regulation of amyloid precursor protein catabolic process | 38 | 42 | 5.968e-06 |
| GO:BP | GO:0010662 | regulation of striated muscle cell apoptotic process | 38 | 42 | 5.968e-06 |
| GO:BP | GO:0043001 | Golgi to plasma membrane protein transport | 38 | 42 | 5.968e-06 |
| GO:BP | GO:0010762 | regulation of fibroblast migration | 35 | 38 | 5.968e-06 |
| GO:BP | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 35 | 38 | 5.968e-06 |
| GO:BP | GO:0003018 | vascular process in circulatory system | 188 | 272 | 6.074e-06 |
| GO:BP | GO:0060997 | dendritic spine morphogenesis | 51 | 60 | 6.280e-06 |
| GO:BP | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress | 51 | 60 | 6.280e-06 |
| GO:BP | GO:0048259 | regulation of receptor-mediated endocytosis | 91 | 119 | 6.284e-06 |
| GO:BP | GO:0019083 | viral transcription | 46 | 53 | 6.358e-06 |
| GO:BP | GO:0045815 | transcription initiation-coupled chromatin remodeling | 46 | 53 | 6.358e-06 |
| GO:BP | GO:0002831 | regulation of response to biotic stimulus | 350 | 540 | 6.416e-06 |
| GO:BP | GO:2001236 | regulation of extrinsic apoptotic signaling pathway | 115 | 156 | 6.447e-06 |
| GO:BP | GO:0098662 | inorganic cation transmembrane transport | 535 | 854 | 6.473e-06 |
| GO:BP | GO:0090311 | regulation of protein deacetylation | 28 | 29 | 6.493e-06 |
| GO:BP | GO:0045116 | protein neddylation | 28 | 29 | 6.493e-06 |
| GO:BP | GO:0071230 | cellular response to amino acid stimulus | 73 | 92 | 6.532e-06 |
| GO:BP | GO:0032273 | positive regulation of protein polymerization | 73 | 92 | 6.532e-06 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 73 | 92 | 6.532e-06 |
| GO:BP | GO:1903509 | liposaccharide metabolic process | 83 | 107 | 6.728e-06 |
| GO:BP | GO:0009408 | response to heat | 79 | 101 | 6.771e-06 |
| GO:BP | GO:0002027 | regulation of heart rate | 79 | 101 | 6.771e-06 |
| GO:BP | GO:0007219 | Notch signaling pathway | 134 | 186 | 7.142e-06 |
| GO:BP | GO:0030832 | regulation of actin filament length | 111 | 150 | 7.229e-06 |
| GO:BP | GO:1903053 | regulation of extracellular matrix organization | 53 | 63 | 7.358e-06 |
| GO:BP | GO:1903747 | regulation of establishment of protein localization to mitochondrion | 43 | 49 | 7.615e-06 |
| GO:BP | GO:0043536 | positive regulation of blood vessel endothelial cell migration | 48 | 56 | 7.968e-06 |
| GO:BP | GO:0007519 | skeletal muscle tissue development | 130 | 180 | 8.308e-06 |
| GO:BP | GO:0051261 | protein depolymerization | 92 | 121 | 8.448e-06 |
| GO:BP | GO:0006767 | water-soluble vitamin metabolic process | 55 | 66 | 8.448e-06 |
| GO:BP | GO:0010632 | regulation of epithelial cell migration | 55 | 66 | 8.448e-06 |
| GO:BP | GO:0010657 | muscle cell apoptotic process | 66 | 82 | 8.554e-06 |
| GO:BP | GO:0035107 | appendage morphogenesis | 114 | 155 | 8.643e-06 |
| GO:BP | GO:0035108 | limb morphogenesis | 114 | 155 | 8.643e-06 |
| GO:BP | GO:0010658 | striated muscle cell apoptotic process | 40 | 45 | 8.793e-06 |
| GO:BP | GO:0061001 | regulation of dendritic spine morphogenesis | 40 | 45 | 8.793e-06 |
| GO:BP | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway | 40 | 45 | 8.793e-06 |
| GO:BP | GO:0010644 | cell communication by electrical coupling | 31 | 33 | 8.824e-06 |
| GO:BP | GO:0008053 | mitochondrial fusion | 31 | 33 | 8.824e-06 |
| GO:BP | GO:0048199 | vesicle targeting, to, from or within Golgi | 31 | 33 | 8.824e-06 |
| GO:BP | GO:0045446 | endothelial cell differentiation | 88 | 115 | 8.905e-06 |
| GO:BP | GO:0043473 | pigmentation | 88 | 115 | 8.905e-06 |
| GO:BP | GO:1902369 | negative regulation of RNA catabolic process | 72 | 91 | 9.379e-06 |
| GO:BP | GO:0019646 | aerobic electron transport chain | 72 | 91 | 9.379e-06 |
| GO:BP | GO:1901879 | regulation of protein depolymerization | 72 | 91 | 9.379e-06 |
| GO:BP | GO:0090559 | regulation of membrane permeability | 57 | 69 | 9.389e-06 |
| GO:BP | GO:0002028 | regulation of sodium ion transport | 57 | 69 | 9.389e-06 |
| GO:BP | GO:1905477 | positive regulation of protein localization to membrane | 82 | 106 | 9.432e-06 |
| GO:BP | GO:0044804 | nucleophagy | 23 | 23 | 9.562e-06 |
| GO:BP | GO:0016237 | microautophagy | 23 | 23 | 9.562e-06 |
| GO:BP | GO:0000963 | mitochondrial RNA processing | 23 | 23 | 9.562e-06 |
| GO:BP | GO:0050686 | negative regulation of mRNA processing | 23 | 23 | 9.562e-06 |
| GO:BP | GO:0090042 | tubulin deacetylation | 23 | 23 | 9.562e-06 |
| GO:BP | GO:0006739 | NADP metabolic process | 37 | 41 | 9.562e-06 |
| GO:BP | GO:0010770 | positive regulation of cell morphogenesis | 34 | 37 | 9.710e-06 |
| GO:BP | GO:1901524 | regulation of mitophagy | 34 | 37 | 9.710e-06 |
| GO:BP | GO:0007018 | microtubule-based movement | 312 | 478 | 9.844e-06 |
| GO:BP | GO:0032409 | regulation of transporter activity | 131 | 182 | 1.002e-05 |
| GO:BP | GO:0035264 | multicellular organism growth | 117 | 160 | 1.006e-05 |
| GO:BP | GO:1902600 | proton transmembrane transport | 136 | 190 | 1.045e-05 |
| GO:BP | GO:0046395 | carboxylic acid catabolic process | 171 | 246 | 1.051e-05 |
| GO:BP | GO:0016054 | organic acid catabolic process | 171 | 246 | 1.051e-05 |
| GO:BP | GO:0007051 | spindle organization | 146 | 206 | 1.094e-05 |
| GO:BP | GO:0001763 | morphogenesis of a branching structure | 146 | 206 | 1.094e-05 |
| GO:BP | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated | 27 | 28 | 1.105e-05 |
| GO:BP | GO:0047497 | mitochondrion transport along microtubule | 27 | 28 | 1.105e-05 |
| GO:BP | GO:0009081 | branched-chain amino acid metabolic process | 27 | 28 | 1.105e-05 |
| GO:BP | GO:2000819 | regulation of nucleotide-excision repair | 27 | 28 | 1.105e-05 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 52 | 62 | 1.105e-05 |
| GO:BP | GO:0010001 | glial cell differentiation | 182 | 264 | 1.139e-05 |
| GO:BP | GO:0034220 | monoatomic ion transmembrane transport | 646 | 1048 | 1.139e-05 |
| GO:BP | GO:0045911 | positive regulation of DNA recombination | 63 | 78 | 1.172e-05 |
| GO:BP | GO:0000271 | polysaccharide biosynthetic process | 47 | 55 | 1.215e-05 |
| GO:BP | GO:0086003 | cardiac muscle cell contraction | 65 | 81 | 1.230e-05 |
| GO:BP | GO:0048814 | regulation of dendrite morphogenesis | 54 | 65 | 1.255e-05 |
| GO:BP | GO:0009615 | response to virus | 273 | 414 | 1.313e-05 |
| GO:BP | GO:0019319 | hexose biosynthetic process | 69 | 87 | 1.313e-05 |
| GO:BP | GO:0150063 | visual system development | 276 | 419 | 1.316e-05 |
| GO:BP | GO:0051100 | negative regulation of binding | 77 | 99 | 1.348e-05 |
| GO:BP | GO:0043648 | dicarboxylic acid metabolic process | 77 | 99 | 1.348e-05 |
| GO:BP | GO:0034205 | amyloid-beta formation | 39 | 44 | 1.392e-05 |
| GO:BP | GO:0032101 | regulation of response to external stimulus | 665 | 1082 | 1.393e-05 |
| GO:BP | GO:1905515 | non-motile cilium assembly | 56 | 68 | 1.393e-05 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 49 | 58 | 1.449e-05 |
| GO:BP | GO:0048880 | sensory system development | 280 | 426 | 1.449e-05 |
| GO:BP | GO:1903955 | positive regulation of protein targeting to mitochondrion | 30 | 32 | 1.471e-05 |
| GO:BP | GO:0006811 | monoatomic ion transport | 780 | 1282 | 1.506e-05 |
| GO:BP | GO:0060412 | ventricular septum morphogenesis | 36 | 40 | 1.549e-05 |
| GO:BP | GO:0061008 | hepaticobiliary system development | 110 | 150 | 1.636e-05 |
| GO:BP | GO:0006695 | cholesterol biosynthetic process | 51 | 61 | 1.678e-05 |
| GO:BP | GO:1902653 | secondary alcohol biosynthetic process | 51 | 61 | 1.678e-05 |
| GO:BP | GO:0051898 | negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 51 | 61 | 1.678e-05 |
| GO:BP | GO:0048025 | negative regulation of mRNA splicing, via spliceosome | 22 | 22 | 1.697e-05 |
| GO:BP | GO:0006390 | mitochondrial transcription | 22 | 22 | 1.697e-05 |
| GO:BP | GO:0009083 | branched-chain amino acid catabolic process | 22 | 22 | 1.697e-05 |
| GO:BP | GO:0140861 | DNA repair-dependent chromatin remodeling | 22 | 22 | 1.697e-05 |
| GO:BP | GO:0006631 | fatty acid metabolic process | 264 | 400 | 1.729e-05 |
| GO:BP | GO:0043242 | negative regulation of protein-containing complex disassembly | 66 | 83 | 1.848e-05 |
| GO:BP | GO:0060419 | heart growth | 66 | 83 | 1.848e-05 |
| GO:BP | GO:0051099 | positive regulation of binding | 80 | 104 | 1.848e-05 |
| GO:BP | GO:0032365 | intracellular lipid transport | 41 | 47 | 1.872e-05 |
| GO:BP | GO:1902116 | negative regulation of organelle assembly | 41 | 47 | 1.872e-05 |
| GO:BP | GO:0050919 | negative chemotaxis | 41 | 47 | 1.872e-05 |
| GO:BP | GO:0090398 | cellular senescence | 78 | 101 | 1.877e-05 |
| GO:BP | GO:0048864 | stem cell development | 68 | 86 | 1.881e-05 |
| GO:BP | GO:0060390 | regulation of SMAD protein signal transduction | 53 | 64 | 1.881e-05 |
| GO:BP | GO:0006112 | energy reserve metabolic process | 76 | 98 | 1.899e-05 |
| GO:BP | GO:0009226 | nucleotide-sugar biosynthetic process | 26 | 27 | 1.899e-05 |
| GO:BP | GO:0042026 | protein refolding | 26 | 27 | 1.899e-05 |
| GO:BP | GO:0007059 | chromosome segregation | 280 | 427 | 1.907e-05 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 157 | 225 | 1.935e-05 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 93 | 124 | 1.993e-05 |
| GO:BP | GO:0045088 | regulation of innate immune response | 293 | 449 | 2.067e-05 |
| GO:BP | GO:0014910 | regulation of smooth muscle cell migration | 55 | 67 | 2.069e-05 |
| GO:BP | GO:0046890 | regulation of lipid biosynthetic process | 130 | 182 | 2.071e-05 |
| GO:BP | GO:0071478 | cellular response to radiation | 130 | 182 | 2.071e-05 |
| GO:BP | GO:0010038 | response to metal ion | 234 | 351 | 2.131e-05 |
| GO:BP | GO:0022898 | regulation of transmembrane transporter activity | 123 | 171 | 2.155e-05 |
| GO:BP | GO:0007612 | learning | 116 | 160 | 2.193e-05 |
| GO:BP | GO:0046839 | phospholipid dephosphorylation | 38 | 43 | 2.206e-05 |
| GO:BP | GO:0014044 | Schwann cell development | 38 | 43 | 2.206e-05 |
| GO:BP | GO:0003208 | cardiac ventricle morphogenesis | 57 | 70 | 2.231e-05 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 171 | 248 | 2.249e-05 |
| GO:BP | GO:0002064 | epithelial cell development | 153 | 219 | 2.302e-05 |
| GO:BP | GO:0043010 | camera-type eye development | 241 | 363 | 2.423e-05 |
| GO:BP | GO:1901185 | negative regulation of ERBB signaling pathway | 29 | 31 | 2.459e-05 |
| GO:BP | GO:0150117 | positive regulation of cell-substrate junction organization | 29 | 31 | 2.459e-05 |
| GO:BP | GO:0065002 | intracellular protein transmembrane transport | 29 | 31 | 2.459e-05 |
| GO:BP | GO:0032205 | negative regulation of telomere maintenance | 35 | 39 | 2.484e-05 |
| GO:BP | GO:0010665 | regulation of cardiac muscle cell apoptotic process | 35 | 39 | 2.484e-05 |
| GO:BP | GO:0070979 | protein K11-linked ubiquitination | 35 | 39 | 2.484e-05 |
| GO:BP | GO:0010660 | regulation of muscle cell apoptotic process | 61 | 76 | 2.484e-05 |
| GO:BP | GO:1905898 | positive regulation of response to endoplasmic reticulum stress | 35 | 39 | 2.484e-05 |
| GO:BP | GO:0001959 | regulation of cytokine-mediated signaling pathway | 119 | 165 | 2.493e-05 |
| GO:BP | GO:0010811 | positive regulation of cell-substrate adhesion | 94 | 126 | 2.579e-05 |
| GO:BP | GO:0070897 | transcription preinitiation complex assembly | 65 | 82 | 2.643e-05 |
| GO:BP | GO:0006066 | alcohol metabolic process | 240 | 362 | 2.953e-05 |
| GO:BP | GO:0090043 | regulation of tubulin deacetylation | 21 | 21 | 3.018e-05 |
| GO:BP | GO:0070301 | cellular response to hydrogen peroxide | 54 | 66 | 3.079e-05 |
| GO:BP | GO:0070129 | regulation of mitochondrial translation | 25 | 26 | 3.280e-05 |
| GO:BP | GO:0033617 | mitochondrial cytochrome c oxidase assembly | 25 | 26 | 3.280e-05 |
| GO:BP | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway | 25 | 26 | 3.280e-05 |
| GO:BP | GO:0016126 | sterol biosynthetic process | 56 | 69 | 3.301e-05 |
| GO:BP | GO:0043524 | negative regulation of neuron apoptotic process | 118 | 164 | 3.301e-05 |
| GO:BP | GO:1901184 | regulation of ERBB signaling pathway | 56 | 69 | 3.301e-05 |
| GO:BP | GO:0030833 | regulation of actin filament polymerization | 95 | 128 | 3.334e-05 |
| GO:BP | GO:0106106 | cold-induced thermogenesis | 111 | 153 | 3.353e-05 |
| GO:BP | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | 80 | 105 | 3.482e-05 |
| GO:BP | GO:0060840 | artery development | 80 | 105 | 3.482e-05 |
| GO:BP | GO:1990823 | response to leukemia inhibitory factor | 80 | 105 | 3.482e-05 |
| GO:BP | GO:0010659 | cardiac muscle cell apoptotic process | 37 | 42 | 3.517e-05 |
| GO:BP | GO:0002244 | hematopoietic progenitor cell differentiation | 102 | 139 | 3.524e-05 |
| GO:BP | GO:0030879 | mammary gland development | 102 | 139 | 3.524e-05 |
| GO:BP | GO:0016485 | protein processing | 156 | 225 | 3.655e-05 |
| GO:BP | GO:0043489 | RNA stabilization | 62 | 78 | 3.738e-05 |
| GO:BP | GO:0048644 | muscle organ morphogenesis | 62 | 78 | 3.738e-05 |
| GO:BP | GO:0002218 | activation of innate immune response | 205 | 305 | 3.769e-05 |
| GO:BP | GO:0060612 | adipose tissue development | 49 | 59 | 3.796e-05 |
| GO:BP | GO:0019915 | lipid storage | 72 | 93 | 3.803e-05 |
| GO:BP | GO:0170062 | nutrient storage | 72 | 93 | 3.803e-05 |
| GO:BP | GO:0014033 | neural crest cell differentiation | 72 | 93 | 3.803e-05 |
| GO:BP | GO:0001889 | liver development | 107 | 147 | 3.836e-05 |
| GO:BP | GO:0007422 | peripheral nervous system development | 70 | 90 | 3.836e-05 |
| GO:BP | GO:0006094 | gluconeogenesis | 66 | 84 | 3.836e-05 |
| GO:BP | GO:2000628 | regulation of miRNA metabolic process | 70 | 90 | 3.836e-05 |
| GO:BP | GO:0051046 | regulation of secretion | 391 | 617 | 3.891e-05 |
| GO:BP | GO:0032781 | positive regulation of ATP-dependent activity | 34 | 38 | 4.006e-05 |
| GO:BP | GO:0046856 | phosphatidylinositol dephosphorylation | 28 | 30 | 4.106e-05 |
| GO:BP | GO:0008535 | respiratory chain complex IV assembly | 28 | 30 | 4.106e-05 |
| GO:BP | GO:1903055 | positive regulation of extracellular matrix organization | 28 | 30 | 4.106e-05 |
| GO:BP | GO:0032233 | positive regulation of actin filament bundle assembly | 51 | 62 | 4.177e-05 |
| GO:BP | GO:0042733 | embryonic digit morphogenesis | 51 | 62 | 4.177e-05 |
| GO:BP | GO:1902882 | regulation of response to oxidative stress | 31 | 34 | 4.278e-05 |
| GO:BP | GO:0032412 | regulation of monoatomic ion transmembrane transporter activity | 117 | 163 | 4.304e-05 |
| GO:BP | GO:0010171 | body morphogenesis | 44 | 52 | 4.372e-05 |
| GO:BP | GO:0051496 | positive regulation of stress fiber assembly | 44 | 52 | 4.372e-05 |
| GO:BP | GO:0001890 | placenta development | 110 | 152 | 4.404e-05 |
| GO:BP | GO:0046824 | positive regulation of nucleocytoplasmic transport | 53 | 65 | 4.518e-05 |
| GO:BP | GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 53 | 65 | 4.518e-05 |
| GO:BP | GO:0034605 | cellular response to heat | 53 | 65 | 4.518e-05 |
| GO:BP | GO:0002520 | immune system development | 142 | 203 | 4.521e-05 |
| GO:BP | GO:0001501 | skeletal system development | 345 | 540 | 4.912e-05 |
| GO:BP | GO:0042158 | lipoprotein biosynthetic process | 46 | 55 | 5.057e-05 |
| GO:BP | GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 57 | 71 | 5.057e-05 |
| GO:BP | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | 46 | 55 | 5.057e-05 |
| GO:BP | GO:0006260 | DNA replication | 191 | 283 | 5.205e-05 |
| GO:BP | GO:0010631 | epithelial cell migration | 73 | 95 | 5.235e-05 |
| GO:BP | GO:1903578 | regulation of ATP metabolic process | 59 | 74 | 5.235e-05 |
| GO:BP | GO:0051788 | response to misfolded protein | 20 | 20 | 5.325e-05 |
| GO:BP | GO:0006517 | protein deglycosylation | 20 | 20 | 5.325e-05 |
| GO:BP | GO:2000434 | regulation of protein neddylation | 20 | 20 | 5.325e-05 |
| GO:BP | GO:0042407 | cristae formation | 20 | 20 | 5.325e-05 |
| GO:BP | GO:0030509 | BMP signaling pathway | 111 | 154 | 5.394e-05 |
| GO:BP | GO:0046620 | regulation of organ growth | 69 | 89 | 5.396e-05 |
| GO:BP | GO:0035418 | protein localization to synapse | 69 | 89 | 5.396e-05 |
| GO:BP | GO:0061138 | morphogenesis of a branching epithelium | 133 | 189 | 5.409e-05 |
| GO:BP | GO:0014032 | neural crest cell development | 63 | 80 | 5.414e-05 |
| GO:BP | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator | 65 | 83 | 5.439e-05 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 65 | 83 | 5.439e-05 |
| GO:BP | GO:0035330 | regulation of hippo signaling | 36 | 41 | 5.504e-05 |
| GO:BP | GO:0000266 | mitochondrial fission | 36 | 41 | 5.504e-05 |
| GO:BP | GO:0031111 | negative regulation of microtubule polymerization or depolymerization | 36 | 41 | 5.504e-05 |
| GO:BP | GO:0038179 | neurotrophin signaling pathway | 36 | 41 | 5.504e-05 |
| GO:BP | GO:0035994 | response to muscle stretch | 24 | 25 | 5.545e-05 |
| GO:BP | GO:0140467 | integrated stress response signaling | 41 | 48 | 5.594e-05 |
| GO:BP | GO:0070849 | response to epidermal growth factor | 41 | 48 | 5.594e-05 |
| GO:BP | GO:0071763 | nuclear membrane organization | 41 | 48 | 5.594e-05 |
| GO:BP | GO:0085029 | extracellular matrix assembly | 41 | 48 | 5.594e-05 |
| GO:BP | GO:0007623 | circadian rhythm | 146 | 210 | 5.611e-05 |
| GO:BP | GO:1904950 | negative regulation of establishment of protein localization | 95 | 129 | 5.613e-05 |
| GO:BP | GO:0007613 | memory | 95 | 129 | 5.613e-05 |
| GO:BP | GO:0048568 | embryonic organ development | 299 | 463 | 5.630e-05 |
| GO:BP | GO:0042391 | regulation of membrane potential | 296 | 458 | 5.694e-05 |
| GO:BP | GO:1990845 | adaptive thermogenesis | 121 | 170 | 5.723e-05 |
| GO:BP | GO:0120161 | regulation of cold-induced thermogenesis | 109 | 151 | 5.742e-05 |
| GO:BP | GO:0030148 | sphingolipid biosynthetic process | 84 | 112 | 5.747e-05 |
| GO:BP | GO:0007267 | cell-cell signaling | 809 | 1342 | 6.006e-05 |
| GO:BP | GO:1903530 | regulation of secretion by cell | 358 | 563 | 6.044e-05 |
| GO:BP | GO:0033993 | response to lipid | 572 | 930 | 6.173e-05 |
| GO:BP | GO:1905039 | carboxylic acid transmembrane transport | 107 | 148 | 6.183e-05 |
| GO:BP | GO:1990830 | cellular response to leukemia inhibitory factor | 78 | 103 | 6.556e-05 |
| GO:BP | GO:1990090 | cellular response to nerve growth factor stimulus | 43 | 51 | 6.608e-05 |
| GO:BP | GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 43 | 51 | 6.608e-05 |
| GO:BP | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 89 | 120 | 6.704e-05 |
| GO:BP | GO:1902473 | regulation of protein localization to synapse | 27 | 29 | 6.779e-05 |
| GO:BP | GO:0010664 | negative regulation of striated muscle cell apoptotic process | 27 | 29 | 6.779e-05 |
| GO:BP | GO:0043247 | telomere maintenance in response to DNA damage | 30 | 33 | 6.913e-05 |
| GO:BP | GO:0045048 | protein insertion into ER membrane | 30 | 33 | 6.913e-05 |
| GO:BP | GO:0045806 | negative regulation of endocytosis | 54 | 67 | 6.969e-05 |
| GO:BP | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process | 38 | 44 | 7.050e-05 |
| GO:BP | GO:0098609 | cell-cell adhesion | 598 | 976 | 7.323e-05 |
| GO:BP | GO:0034763 | negative regulation of transmembrane transport | 85 | 114 | 7.476e-05 |
| GO:BP | GO:1901880 | negative regulation of protein depolymerization | 58 | 73 | 7.477e-05 |
| GO:BP | GO:0060324 | face development | 45 | 54 | 7.543e-05 |
| GO:BP | GO:0010611 | regulation of cardiac muscle hypertrophy | 45 | 54 | 7.543e-05 |
| GO:BP | GO:0045661 | regulation of myoblast differentiation | 60 | 76 | 7.604e-05 |
| GO:BP | GO:0001570 | vasculogenesis | 66 | 85 | 7.618e-05 |
| GO:BP | GO:0002200 | somatic diversification of immune receptors | 62 | 79 | 7.663e-05 |
| GO:BP | GO:0021543 | pallium development | 138 | 198 | 7.971e-05 |
| GO:BP | GO:0014888 | striated muscle adaptation | 40 | 47 | 8.622e-05 |
| GO:BP | GO:0021762 | substantia nigra development | 40 | 47 | 8.622e-05 |
| GO:BP | GO:0048753 | pigment granule organization | 35 | 40 | 8.665e-05 |
| GO:BP | GO:1903115 | regulation of actin filament-based movement | 35 | 40 | 8.665e-05 |
| GO:BP | GO:1900077 | negative regulation of cellular response to insulin stimulus | 35 | 40 | 8.665e-05 |
| GO:BP | GO:0033002 | muscle cell proliferation | 144 | 208 | 9.051e-05 |
| GO:BP | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway | 49 | 60 | 9.130e-05 |
| GO:BP | GO:1903825 | organic acid transmembrane transport | 109 | 152 | 9.225e-05 |
| GO:BP | GO:0090672 | telomerase RNA localization | 19 | 19 | 9.346e-05 |
| GO:BP | GO:0071025 | RNA surveillance | 19 | 19 | 9.346e-05 |
| GO:BP | GO:0090671 | telomerase RNA localization to Cajal body | 19 | 19 | 9.346e-05 |
| GO:BP | GO:0010968 | regulation of microtubule nucleation | 19 | 19 | 9.346e-05 |
| GO:BP | GO:0055003 | cardiac myofibril assembly | 19 | 19 | 9.346e-05 |
| GO:BP | GO:0090685 | RNA localization to nucleus | 19 | 19 | 9.346e-05 |
| GO:BP | GO:1904505 | regulation of telomere maintenance in response to DNA damage | 19 | 19 | 9.346e-05 |
| GO:BP | GO:0045945 | positive regulation of transcription by RNA polymerase III | 19 | 19 | 9.346e-05 |
| GO:BP | GO:0090670 | RNA localization to Cajal body | 19 | 19 | 9.346e-05 |
| GO:BP | GO:0003299 | muscle hypertrophy in response to stress | 23 | 24 | 9.396e-05 |
| GO:BP | GO:0007042 | lysosomal lumen acidification | 23 | 24 | 9.396e-05 |
| GO:BP | GO:0014887 | cardiac muscle adaptation | 23 | 24 | 9.396e-05 |
| GO:BP | GO:0051961 | negative regulation of nervous system development | 102 | 141 | 9.396e-05 |
| GO:BP | GO:0051457 | maintenance of protein location in nucleus | 23 | 24 | 9.396e-05 |
| GO:BP | GO:0014898 | cardiac muscle hypertrophy in response to stress | 23 | 24 | 9.396e-05 |
| GO:BP | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 95 | 130 | 9.396e-05 |
| GO:BP | GO:0043255 | regulation of carbohydrate biosynthetic process | 73 | 96 | 9.662e-05 |
| GO:BP | GO:0043062 | extracellular structure organization | 217 | 328 | 9.795e-05 |
| GO:BP | GO:0046825 | regulation of protein export from nucleus | 32 | 36 | 1.011e-04 |
| GO:BP | GO:1902003 | regulation of amyloid-beta formation | 32 | 36 | 1.011e-04 |
| GO:BP | GO:0007035 | vacuolar acidification | 32 | 36 | 1.011e-04 |
| GO:BP | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion | 32 | 36 | 1.011e-04 |
| GO:BP | GO:0006099 | tricarboxylic acid cycle | 32 | 36 | 1.011e-04 |
| GO:BP | GO:0048754 | branching morphogenesis of an epithelial tube | 112 | 157 | 1.020e-04 |
| GO:BP | GO:1901264 | carbohydrate derivative transport | 69 | 90 | 1.024e-04 |
| GO:BP | GO:0048167 | regulation of synaptic plasticity | 153 | 223 | 1.044e-04 |
| GO:BP | GO:0071901 | negative regulation of protein serine/threonine kinase activity | 57 | 72 | 1.067e-04 |
| GO:BP | GO:0032272 | negative regulation of protein polymerization | 57 | 72 | 1.067e-04 |
| GO:BP | GO:0006970 | response to osmotic stress | 65 | 84 | 1.067e-04 |
| GO:BP | GO:0031099 | regeneration | 140 | 202 | 1.070e-04 |
| GO:BP | GO:0140029 | exocytic process | 63 | 81 | 1.077e-04 |
| GO:BP | GO:0055017 | cardiac muscle tissue growth | 59 | 75 | 1.078e-04 |
| GO:BP | GO:1905168 | positive regulation of double-strand break repair via homologous recombination | 37 | 43 | 1.084e-04 |
| GO:BP | GO:0071364 | cellular response to epidermal growth factor stimulus | 37 | 43 | 1.084e-04 |
| GO:BP | GO:0005978 | glycogen biosynthetic process | 37 | 43 | 1.084e-04 |
| GO:BP | GO:0009250 | glucan biosynthetic process | 37 | 43 | 1.084e-04 |
| GO:BP | GO:0035751 | regulation of lysosomal lumen pH | 29 | 32 | 1.113e-04 |
| GO:BP | GO:0180046 | GPI anchored protein biosynthesis | 29 | 32 | 1.113e-04 |
| GO:BP | GO:0006622 | protein targeting to lysosome | 29 | 32 | 1.113e-04 |
| GO:BP | GO:0006505 | GPI anchor metabolic process | 29 | 32 | 1.113e-04 |
| GO:BP | GO:0098901 | regulation of cardiac muscle cell action potential | 26 | 28 | 1.113e-04 |
| GO:BP | GO:1901798 | positive regulation of signal transduction by p53 class mediator | 26 | 28 | 1.113e-04 |
| GO:BP | GO:0170034 | L-amino acid biosynthetic process | 44 | 53 | 1.119e-04 |
| GO:BP | GO:0009266 | response to temperature stimulus | 120 | 170 | 1.130e-04 |
| GO:BP | GO:0002040 | sprouting angiogenesis | 96 | 132 | 1.154e-04 |
| GO:BP | GO:0030198 | extracellular matrix organization | 216 | 327 | 1.178e-04 |
| GO:BP | GO:0002758 | innate immune response-activating signaling pathway | 190 | 284 | 1.189e-04 |
| GO:BP | GO:0098771 | inorganic ion homeostasis | 339 | 534 | 1.230e-04 |
| GO:BP | GO:0001704 | formation of primary germ layer | 94 | 129 | 1.238e-04 |
| GO:BP | GO:0043405 | regulation of MAP kinase activity | 85 | 115 | 1.278e-04 |
| GO:BP | GO:0045089 | positive regulation of innate immune response | 244 | 374 | 1.293e-04 |
| GO:BP | GO:0045229 | external encapsulating structure organization | 217 | 329 | 1.301e-04 |
| GO:BP | GO:0006284 | base-excision repair | 39 | 46 | 1.302e-04 |
| GO:BP | GO:0007031 | peroxisome organization | 34 | 39 | 1.342e-04 |
| GO:BP | GO:0071711 | basement membrane organization | 34 | 39 | 1.342e-04 |
| GO:BP | GO:0046627 | negative regulation of insulin receptor signaling pathway | 34 | 39 | 1.342e-04 |
| GO:BP | GO:0034389 | lipid droplet organization | 34 | 39 | 1.342e-04 |
| GO:BP | GO:0032438 | melanosome organization | 34 | 39 | 1.342e-04 |
| GO:BP | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | 34 | 39 | 1.342e-04 |
| GO:BP | GO:0045010 | actin nucleation | 50 | 62 | 1.402e-04 |
| GO:BP | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway | 50 | 62 | 1.402e-04 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 129 | 185 | 1.424e-04 |
| GO:BP | GO:1901890 | positive regulation of cell junction assembly | 81 | 109 | 1.430e-04 |
| GO:BP | GO:0098660 | inorganic ion transmembrane transport | 577 | 944 | 1.482e-04 |
| GO:BP | GO:0071345 | cellular response to cytokine stimulus | 525 | 854 | 1.491e-04 |
| GO:BP | GO:0006111 | regulation of gluconeogenesis | 41 | 49 | 1.503e-04 |
| GO:BP | GO:0023061 | signal release | 319 | 501 | 1.514e-04 |
| GO:BP | GO:0030032 | lamellipodium assembly | 56 | 71 | 1.523e-04 |
| GO:BP | GO:0061614 | miRNA transcription | 60 | 77 | 1.526e-04 |
| GO:BP | GO:0009247 | glycolipid biosynthetic process | 60 | 77 | 1.526e-04 |
| GO:BP | GO:0006740 | NADPH regeneration | 22 | 23 | 1.593e-04 |
| GO:BP | GO:0033962 | P-body assembly | 22 | 23 | 1.593e-04 |
| GO:BP | GO:1900151 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 22 | 23 | 1.593e-04 |
| GO:BP | GO:0034249 | negative regulation of amide metabolic process | 22 | 23 | 1.593e-04 |
| GO:BP | GO:0045879 | negative regulation of smoothened signaling pathway | 31 | 35 | 1.600e-04 |
| GO:BP | GO:0046329 | negative regulation of JNK cascade | 31 | 35 | 1.600e-04 |
| GO:BP | GO:0048147 | negative regulation of fibroblast proliferation | 31 | 35 | 1.600e-04 |
| GO:BP | GO:1904294 | positive regulation of ERAD pathway | 18 | 18 | 1.628e-04 |
| GO:BP | GO:0006743 | ubiquinone metabolic process | 18 | 18 | 1.628e-04 |
| GO:BP | GO:2000641 | regulation of early endosome to late endosome transport | 18 | 18 | 1.628e-04 |
| GO:BP | GO:0097494 | regulation of vesicle size | 18 | 18 | 1.628e-04 |
| GO:BP | GO:0071218 | cellular response to misfolded protein | 18 | 18 | 1.628e-04 |
| GO:BP | GO:0001522 | pseudouridine synthesis | 18 | 18 | 1.628e-04 |
| GO:BP | GO:0032239 | regulation of nucleobase-containing compound transport | 18 | 18 | 1.628e-04 |
| GO:BP | GO:0000028 | ribosomal small subunit assembly | 18 | 18 | 1.628e-04 |
| GO:BP | GO:0034727 | piecemeal microautophagy of the nucleus | 18 | 18 | 1.628e-04 |
| GO:BP | GO:0035278 | miRNA-mediated gene silencing by inhibition of translation | 18 | 18 | 1.628e-04 |
| GO:BP | GO:0035794 | positive regulation of mitochondrial membrane permeability | 36 | 42 | 1.664e-04 |
| GO:BP | GO:0097300 | programmed necrotic cell death | 43 | 52 | 1.666e-04 |
| GO:BP | GO:0051224 | negative regulation of protein transport | 84 | 114 | 1.702e-04 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 84 | 114 | 1.702e-04 |
| GO:BP | GO:0031623 | receptor internalization | 98 | 136 | 1.730e-04 |
| GO:BP | GO:0032606 | type I interferon production | 91 | 125 | 1.747e-04 |
| GO:BP | GO:0032479 | regulation of type I interferon production | 91 | 125 | 1.747e-04 |
| GO:BP | GO:0006506 | GPI anchor biosynthetic process | 28 | 31 | 1.783e-04 |
| GO:BP | GO:0070536 | protein K63-linked deubiquitination | 28 | 31 | 1.783e-04 |
| GO:BP | GO:0046685 | response to arsenic-containing substance | 28 | 31 | 1.783e-04 |
| GO:BP | GO:1903846 | positive regulation of cellular response to transforming growth factor beta stimulus | 28 | 31 | 1.783e-04 |
| GO:BP | GO:0016226 | iron-sulfur cluster assembly | 28 | 31 | 1.783e-04 |
| GO:BP | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway | 28 | 31 | 1.783e-04 |
| GO:BP | GO:0031163 | metallo-sulfur cluster assembly | 28 | 31 | 1.783e-04 |
| GO:BP | GO:1900101 | regulation of endoplasmic reticulum unfolded protein response | 28 | 31 | 1.783e-04 |
| GO:BP | GO:0034067 | protein localization to Golgi apparatus | 28 | 31 | 1.783e-04 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 103 | 144 | 1.791e-04 |
| GO:BP | GO:0002042 | cell migration involved in sprouting angiogenesis | 45 | 55 | 1.805e-04 |
| GO:BP | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling | 25 | 27 | 1.824e-04 |
| GO:BP | GO:0031365 | N-terminal protein amino acid modification | 25 | 27 | 1.824e-04 |
| GO:BP | GO:0031468 | nuclear membrane reassembly | 25 | 27 | 1.824e-04 |
| GO:BP | GO:0072006 | nephron development | 113 | 160 | 1.850e-04 |
| GO:BP | GO:0061025 | membrane fusion | 131 | 189 | 1.903e-04 |
| GO:BP | GO:0031102 | neuron projection regeneration | 47 | 58 | 1.925e-04 |
| GO:BP | GO:1904062 | regulation of monoatomic cation transmembrane transport | 189 | 284 | 1.954e-04 |
| GO:BP | GO:0017157 | regulation of exocytosis | 126 | 181 | 1.954e-04 |
| GO:BP | GO:0010959 | regulation of metal ion transport | 243 | 374 | 1.981e-04 |
| GO:BP | GO:0001824 | blastocyst development | 87 | 119 | 1.992e-04 |
| GO:BP | GO:0022612 | gland morphogenesis | 94 | 130 | 1.993e-04 |
| GO:BP | GO:1901342 | regulation of vasculature development | 198 | 299 | 2.008e-04 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 116 | 165 | 2.008e-04 |
| GO:BP | GO:0071772 | response to BMP | 116 | 165 | 2.008e-04 |
| GO:BP | GO:2001057 | reactive nitrogen species metabolic process | 63 | 82 | 2.052e-04 |
| GO:BP | GO:1905508 | protein localization to microtubule organizing center | 33 | 38 | 2.073e-04 |
| GO:BP | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 33 | 38 | 2.073e-04 |
| GO:BP | GO:0043470 | regulation of carbohydrate catabolic process | 51 | 64 | 2.077e-04 |
| GO:BP | GO:1903078 | positive regulation of protein localization to plasma membrane | 51 | 64 | 2.077e-04 |
| GO:BP | GO:0008643 | carbohydrate transport | 99 | 138 | 2.084e-04 |
| GO:BP | GO:1904377 | positive regulation of protein localization to cell periphery | 59 | 76 | 2.122e-04 |
| GO:BP | GO:1902893 | regulation of miRNA transcription | 59 | 76 | 2.122e-04 |
| GO:BP | GO:0002562 | somatic diversification of immune receptors via germline recombination within a single locus | 57 | 73 | 2.137e-04 |
| GO:BP | GO:0016444 | somatic cell DNA recombination | 57 | 73 | 2.137e-04 |
| GO:BP | GO:0072073 | kidney epithelium development | 109 | 154 | 2.178e-04 |
| GO:BP | GO:0050679 | positive regulation of epithelial cell proliferation | 140 | 204 | 2.214e-04 |
| GO:BP | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 40 | 48 | 2.224e-04 |
| GO:BP | GO:0060443 | mammary gland morphogenesis | 40 | 48 | 2.224e-04 |
| GO:BP | GO:0061383 | trabecula morphogenesis | 40 | 48 | 2.224e-04 |
| GO:BP | GO:0021987 | cerebral cortex development | 95 | 132 | 2.436e-04 |
| GO:BP | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | 42 | 51 | 2.471e-04 |
| GO:BP | GO:0060914 | heart formation | 30 | 34 | 2.511e-04 |
| GO:BP | GO:0086004 | regulation of cardiac muscle cell contraction | 30 | 34 | 2.511e-04 |
| GO:BP | GO:0034405 | response to fluid shear stress | 30 | 34 | 2.511e-04 |
| GO:BP | GO:0140058 | neuron projection arborization | 30 | 34 | 2.511e-04 |
| GO:BP | GO:2000008 | regulation of protein localization to cell surface | 35 | 41 | 2.548e-04 |
| GO:BP | GO:0014074 | response to purine-containing compound | 105 | 148 | 2.575e-04 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 110 | 156 | 2.589e-04 |
| GO:BP | GO:1902652 | secondary alcohol metabolic process | 110 | 156 | 2.589e-04 |
| GO:BP | GO:0001659 | temperature homeostasis | 128 | 185 | 2.629e-04 |
| GO:BP | GO:0050771 | negative regulation of axonogenesis | 44 | 54 | 2.661e-04 |
| GO:BP | GO:0035493 | SNARE complex assembly | 21 | 22 | 2.666e-04 |
| GO:BP | GO:0034472 | snRNA 3’-end processing | 21 | 22 | 2.666e-04 |
| GO:BP | GO:0061709 | reticulophagy | 21 | 22 | 2.666e-04 |
| GO:BP | GO:0021756 | striatum development | 21 | 22 | 2.666e-04 |
| GO:BP | GO:0035050 | embryonic heart tube development | 66 | 87 | 2.666e-04 |
| GO:BP | GO:0097502 | mannosylation | 21 | 22 | 2.666e-04 |
| GO:BP | GO:0044794 | positive regulation by host of viral process | 21 | 22 | 2.666e-04 |
| GO:BP | GO:0090130 | tissue migration | 77 | 104 | 2.712e-04 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 64 | 84 | 2.763e-04 |
| GO:BP | GO:1900542 | regulation of purine nucleotide metabolic process | 64 | 84 | 2.763e-04 |
| GO:BP | GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 46 | 57 | 2.797e-04 |
| GO:BP | GO:0099084 | postsynaptic specialization organization | 46 | 57 | 2.797e-04 |
| GO:BP | GO:0001954 | positive regulation of cell-matrix adhesion | 46 | 57 | 2.797e-04 |
| GO:BP | GO:0014743 | regulation of muscle hypertrophy | 46 | 57 | 2.797e-04 |
| GO:BP | GO:0031098 | stress-activated protein kinase signaling cascade | 46 | 57 | 2.797e-04 |
| GO:BP | GO:0035113 | embryonic appendage morphogenesis | 91 | 126 | 2.805e-04 |
| GO:BP | GO:0030326 | embryonic limb morphogenesis | 91 | 126 | 2.805e-04 |
| GO:BP | GO:0036507 | protein demannosylation | 17 | 17 | 2.834e-04 |
| GO:BP | GO:0036508 | protein alpha-1,2-demannosylation | 17 | 17 | 2.834e-04 |
| GO:BP | GO:0046209 | nitric oxide metabolic process | 62 | 81 | 2.834e-04 |
| GO:BP | GO:0006415 | translational termination | 17 | 17 | 2.834e-04 |
| GO:BP | GO:0060382 | regulation of DNA strand elongation | 17 | 17 | 2.834e-04 |
| GO:BP | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway | 17 | 17 | 2.834e-04 |
| GO:BP | GO:0010649 | regulation of cell communication by electrical coupling | 17 | 17 | 2.834e-04 |
| GO:BP | GO:0006895 | Golgi to endosome transport | 17 | 17 | 2.834e-04 |
| GO:BP | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 27 | 30 | 2.837e-04 |
| GO:BP | GO:0035891 | exit from host cell | 27 | 30 | 2.837e-04 |
| GO:BP | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 27 | 30 | 2.837e-04 |
| GO:BP | GO:0010765 | positive regulation of sodium ion transport | 27 | 30 | 2.837e-04 |
| GO:BP | GO:0019076 | viral release from host cell | 27 | 30 | 2.837e-04 |
| GO:BP | GO:0060420 | regulation of heart growth | 48 | 60 | 2.886e-04 |
| GO:BP | GO:0050768 | negative regulation of neurogenesis | 96 | 134 | 2.909e-04 |
| GO:BP | GO:0021782 | glial cell development | 96 | 134 | 2.909e-04 |
| GO:BP | GO:0034219 | carbohydrate transmembrane transport | 96 | 134 | 2.909e-04 |
| GO:BP | GO:2000300 | regulation of synaptic vesicle exocytosis | 37 | 44 | 2.930e-04 |
| GO:BP | GO:0042398 | modified amino acid biosynthetic process | 37 | 44 | 2.930e-04 |
| GO:BP | GO:0010907 | positive regulation of glucose metabolic process | 37 | 44 | 2.930e-04 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 170 | 254 | 2.932e-04 |
| GO:BP | GO:0046755 | viral budding | 24 | 26 | 2.950e-04 |
| GO:BP | GO:0048207 | vesicle targeting, rough ER to cis-Golgi | 24 | 26 | 2.950e-04 |
| GO:BP | GO:0048208 | COPII vesicle coating | 24 | 26 | 2.950e-04 |
| GO:BP | GO:0042180 | ketone metabolic process | 145 | 213 | 2.950e-04 |
| GO:BP | GO:0051894 | positive regulation of focal adhesion assembly | 24 | 26 | 2.950e-04 |
| GO:BP | GO:0034453 | microtubule anchoring | 24 | 26 | 2.950e-04 |
| GO:BP | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process | 24 | 26 | 2.950e-04 |
| GO:BP | GO:0018345 | protein palmitoylation | 24 | 26 | 2.950e-04 |
| GO:BP | GO:0031167 | rRNA methylation | 24 | 26 | 2.950e-04 |
| GO:BP | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 52 | 66 | 2.975e-04 |
| GO:BP | GO:0090132 | epithelium migration | 73 | 98 | 2.978e-04 |
| GO:BP | GO:0016445 | somatic diversification of immunoglobulins | 54 | 69 | 2.981e-04 |
| GO:BP | GO:0042221 | response to chemical | 2243 | 3909 | 3.022e-04 |
| GO:BP | GO:0007423 | sensory organ development | 389 | 624 | 3.072e-04 |
| GO:BP | GO:0019079 | viral genome replication | 94 | 131 | 3.122e-04 |
| GO:BP | GO:0030001 | metal ion transport | 539 | 883 | 3.132e-04 |
| GO:BP | GO:2000765 | regulation of cytoplasmic translation | 32 | 37 | 3.162e-04 |
| GO:BP | GO:0071539 | protein localization to centrosome | 32 | 37 | 3.162e-04 |
| GO:BP | GO:0003161 | cardiac conduction system development | 32 | 37 | 3.162e-04 |
| GO:BP | GO:0043269 | regulation of monoatomic ion transport | 288 | 452 | 3.244e-04 |
| GO:BP | GO:0010656 | negative regulation of muscle cell apoptotic process | 39 | 47 | 3.280e-04 |
| GO:BP | GO:0045216 | cell-cell junction organization | 146 | 215 | 3.337e-04 |
| GO:BP | GO:0034765 | regulation of monoatomic ion transmembrane transport | 210 | 321 | 3.491e-04 |
| GO:BP | GO:2000377 | regulation of reactive oxygen species metabolic process | 102 | 144 | 3.549e-04 |
| GO:BP | GO:0044788 | modulation by host of viral process | 41 | 50 | 3.596e-04 |
| GO:BP | GO:0051489 | regulation of filopodium assembly | 41 | 50 | 3.596e-04 |
| GO:BP | GO:0008593 | regulation of Notch signaling pathway | 74 | 100 | 3.826e-04 |
| GO:BP | GO:0051588 | regulation of neurotransmitter transport | 74 | 100 | 3.826e-04 |
| GO:BP | GO:0045765 | regulation of angiogenesis | 193 | 293 | 3.839e-04 |
| GO:BP | GO:1902686 | mitochondrial outer membrane permeabilization involved in programmed cell death | 34 | 40 | 3.843e-04 |
| GO:BP | GO:0051403 | stress-activated MAPK cascade | 43 | 53 | 3.843e-04 |
| GO:BP | GO:1990089 | response to nerve growth factor | 43 | 53 | 3.843e-04 |
| GO:BP | GO:0032366 | intracellular sterol transport | 29 | 33 | 3.899e-04 |
| GO:BP | GO:2000463 | positive regulation of excitatory postsynaptic potential | 29 | 33 | 3.899e-04 |
| GO:BP | GO:0015749 | monosaccharide transmembrane transport | 88 | 122 | 3.930e-04 |
| GO:BP | GO:0071470 | cellular response to osmotic stress | 45 | 56 | 4.029e-04 |
| GO:BP | GO:0051298 | centrosome duplication | 57 | 74 | 4.122e-04 |
| GO:BP | GO:0008645 | hexose transmembrane transport | 86 | 119 | 4.246e-04 |
| GO:BP | GO:0002682 | regulation of immune system process | 936 | 1581 | 4.421e-04 |
| GO:BP | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway | 36 | 43 | 4.444e-04 |
| GO:BP | GO:1901661 | quinone metabolic process | 20 | 21 | 4.470e-04 |
| GO:BP | GO:0035268 | protein mannosylation | 20 | 21 | 4.470e-04 |
| GO:BP | GO:0071786 | endoplasmic reticulum tubular network organization | 20 | 21 | 4.470e-04 |
| GO:BP | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion | 20 | 21 | 4.470e-04 |
| GO:BP | GO:0018394 | peptidyl-lysine acetylation | 20 | 21 | 4.470e-04 |
| GO:BP | GO:0007026 | negative regulation of microtubule depolymerization | 26 | 29 | 4.546e-04 |
| GO:BP | GO:0009649 | entrainment of circadian clock | 26 | 29 | 4.546e-04 |
| GO:BP | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation | 26 | 29 | 4.546e-04 |
| GO:BP | GO:0090140 | regulation of mitochondrial fission | 26 | 29 | 4.546e-04 |
| GO:BP | GO:1903421 | regulation of synaptic vesicle recycling | 26 | 29 | 4.546e-04 |
| GO:BP | GO:1902683 | regulation of receptor localization to synapse | 26 | 29 | 4.546e-04 |
| GO:BP | GO:1904659 | D-glucose transmembrane transport | 84 | 116 | 4.552e-04 |
| GO:BP | GO:0001892 | embryonic placenta development | 66 | 88 | 4.743e-04 |
| GO:BP | GO:0035020 | regulation of Rac protein signal transduction | 23 | 25 | 4.879e-04 |
| GO:BP | GO:0060155 | platelet dense granule organization | 23 | 25 | 4.879e-04 |
| GO:BP | GO:0044090 | positive regulation of vacuole organization | 23 | 25 | 4.879e-04 |
| GO:BP | GO:0070316 | regulation of G0 to G1 transition | 31 | 36 | 4.922e-04 |
| GO:BP | GO:0003254 | regulation of membrane depolarization | 38 | 46 | 4.922e-04 |
| GO:BP | GO:1905710 | positive regulation of membrane permeability | 38 | 46 | 4.922e-04 |
| GO:BP | GO:1902745 | positive regulation of lamellipodium organization | 31 | 36 | 4.922e-04 |
| GO:BP | GO:0006779 | porphyrin-containing compound biosynthetic process | 31 | 36 | 4.922e-04 |
| GO:BP | GO:0033014 | tetrapyrrole biosynthetic process | 31 | 36 | 4.922e-04 |
| GO:BP | GO:0016579 | protein deubiquitination | 94 | 132 | 4.922e-04 |
| GO:BP | GO:1900102 | negative regulation of endoplasmic reticulum unfolded protein response | 16 | 16 | 4.924e-04 |
| GO:BP | GO:0051153 | regulation of striated muscle cell differentiation | 64 | 85 | 4.924e-04 |
| GO:BP | GO:0030242 | autophagy of peroxisome | 16 | 16 | 4.924e-04 |
| GO:BP | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 16 | 16 | 4.924e-04 |
| GO:BP | GO:1901663 | quinone biosynthetic process | 16 | 16 | 4.924e-04 |
| GO:BP | GO:0034497 | protein localization to phagophore assembly site | 16 | 16 | 4.924e-04 |
| GO:BP | GO:0043328 | protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway | 16 | 16 | 4.924e-04 |
| GO:BP | GO:2000232 | regulation of rRNA processing | 16 | 16 | 4.924e-04 |
| GO:BP | GO:0099116 | tRNA 5’-end processing | 16 | 16 | 4.924e-04 |
| GO:BP | GO:0060546 | negative regulation of necroptotic process | 16 | 16 | 4.924e-04 |
| GO:BP | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 16 | 16 | 4.924e-04 |
| GO:BP | GO:0006140 | regulation of nucleotide metabolic process | 64 | 85 | 4.924e-04 |
| GO:BP | GO:1900034 | regulation of cellular response to heat | 16 | 16 | 4.924e-04 |
| GO:BP | GO:1903358 | regulation of Golgi organization | 16 | 16 | 4.924e-04 |
| GO:BP | GO:0006744 | ubiquinone biosynthetic process | 16 | 16 | 4.924e-04 |
| GO:BP | GO:0045785 | positive regulation of cell adhesion | 308 | 488 | 5.002e-04 |
| GO:BP | GO:0071396 | cellular response to lipid | 381 | 613 | 5.104e-04 |
| GO:BP | GO:0032387 | negative regulation of intracellular transport | 40 | 49 | 5.275e-04 |
| GO:BP | GO:0051205 | protein insertion into membrane | 40 | 49 | 5.275e-04 |
| GO:BP | GO:0030521 | androgen receptor signaling pathway | 40 | 49 | 5.275e-04 |
| GO:BP | GO:0002833 | positive regulation of response to biotic stimulus | 256 | 400 | 5.292e-04 |
| GO:BP | GO:0097120 | receptor localization to synapse | 60 | 79 | 5.337e-04 |
| GO:BP | GO:1902373 | negative regulation of mRNA catabolic process | 60 | 79 | 5.337e-04 |
| GO:BP | GO:0008203 | cholesterol metabolic process | 102 | 145 | 5.365e-04 |
| GO:BP | GO:0062013 | positive regulation of small molecule metabolic process | 102 | 145 | 5.365e-04 |
| GO:BP | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity | 42 | 52 | 5.580e-04 |
| GO:BP | GO:0006672 | ceramide metabolic process | 78 | 107 | 5.584e-04 |
| GO:BP | GO:0038066 | p38MAPK cascade | 44 | 55 | 5.803e-04 |
| GO:BP | GO:0060323 | head morphogenesis | 33 | 39 | 5.811e-04 |
| GO:BP | GO:0007616 | long-term memory | 33 | 39 | 5.811e-04 |
| GO:BP | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 33 | 39 | 5.811e-04 |
| GO:BP | GO:0043506 | regulation of JUN kinase activity | 33 | 39 | 5.811e-04 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 54 | 70 | 5.823e-04 |
| GO:BP | GO:0045669 | positive regulation of osteoblast differentiation | 54 | 70 | 5.823e-04 |
| GO:BP | GO:0045600 | positive regulation of fat cell differentiation | 52 | 67 | 5.923e-04 |
| GO:BP | GO:0046626 | regulation of insulin receptor signaling pathway | 52 | 67 | 5.923e-04 |
| GO:BP | GO:0016447 | somatic recombination of immunoglobulin gene segments | 50 | 64 | 5.976e-04 |
| GO:BP | GO:0042306 | regulation of protein import into nucleus | 48 | 61 | 5.976e-04 |
| GO:BP | GO:0006783 | heme biosynthetic process | 28 | 32 | 6.069e-04 |
| GO:BP | GO:0070570 | regulation of neuron projection regeneration | 28 | 32 | 6.069e-04 |
| GO:BP | GO:0062098 | regulation of programmed necrotic cell death | 28 | 32 | 6.069e-04 |
| GO:BP | GO:0086019 | cell-cell signaling involved in cardiac conduction | 28 | 32 | 6.069e-04 |
| GO:BP | GO:0090504 | epiboly | 35 | 42 | 6.617e-04 |
| GO:BP | GO:1904861 | excitatory synapse assembly | 35 | 42 | 6.617e-04 |
| GO:BP | GO:0072594 | establishment of protein localization to organelle | 408 | 661 | 6.737e-04 |
| GO:BP | GO:0051225 | spindle assembly | 96 | 136 | 6.949e-04 |
| GO:BP | GO:0006575 | modified amino acid metabolic process | 122 | 178 | 7.064e-04 |
| GO:BP | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining | 25 | 28 | 7.263e-04 |
| GO:BP | GO:0150052 | regulation of postsynapse assembly | 37 | 45 | 7.295e-04 |
| GO:BP | GO:0048640 | negative regulation of developmental growth | 70 | 95 | 7.298e-04 |
| GO:BP | GO:0006098 | pentose-phosphate shunt | 19 | 20 | 7.487e-04 |
| GO:BP | GO:0006684 | sphingomyelin metabolic process | 19 | 20 | 7.487e-04 |
| GO:BP | GO:0071498 | cellular response to fluid shear stress | 19 | 20 | 7.487e-04 |
| GO:BP | GO:1901201 | regulation of extracellular matrix assembly | 19 | 20 | 7.487e-04 |
| GO:BP | GO:0046782 | regulation of viral transcription | 19 | 20 | 7.487e-04 |
| GO:BP | GO:0036499 | PERK-mediated unfolded protein response | 19 | 20 | 7.487e-04 |
| GO:BP | GO:0061003 | positive regulation of dendritic spine morphogenesis | 19 | 20 | 7.487e-04 |
| GO:BP | GO:0002097 | tRNA wobble base modification | 19 | 20 | 7.487e-04 |
| GO:BP | GO:0030150 | protein import into mitochondrial matrix | 19 | 20 | 7.487e-04 |
| GO:BP | GO:2000786 | positive regulation of autophagosome assembly | 19 | 20 | 7.487e-04 |
| GO:BP | GO:0043276 | anoikis | 30 | 35 | 7.552e-04 |
| GO:BP | GO:0031114 | regulation of microtubule depolymerization | 30 | 35 | 7.552e-04 |
| GO:BP | GO:0031952 | regulation of protein autophosphorylation | 30 | 35 | 7.552e-04 |
| GO:BP | GO:0000819 | sister chromatid segregation | 156 | 234 | 7.585e-04 |
| GO:BP | GO:1905314 | semi-lunar valve development | 39 | 48 | 7.759e-04 |
| GO:BP | GO:0097106 | postsynaptic density organization | 39 | 48 | 7.759e-04 |
| GO:BP | GO:0034331 | cell junction maintenance | 39 | 48 | 7.759e-04 |
| GO:BP | GO:0002090 | regulation of receptor internalization | 55 | 72 | 7.896e-04 |
| GO:BP | GO:0000027 | ribosomal large subunit assembly | 22 | 24 | 7.896e-04 |
| GO:BP | GO:0043153 | entrainment of circadian clock by photoperiod | 22 | 24 | 7.896e-04 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 55 | 72 | 7.896e-04 |
| GO:BP | GO:0051131 | chaperone-mediated protein complex assembly | 22 | 24 | 7.896e-04 |
| GO:BP | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process | 22 | 24 | 7.896e-04 |
| GO:BP | GO:0006809 | nitric oxide biosynthetic process | 55 | 72 | 7.896e-04 |
| GO:BP | GO:0050708 | regulation of protein secretion | 177 | 269 | 7.945e-04 |
| GO:BP | GO:1902743 | regulation of lamellipodium organization | 41 | 51 | 8.105e-04 |
| GO:BP | GO:0048009 | insulin-like growth factor receptor signaling pathway | 41 | 51 | 8.105e-04 |
| GO:BP | GO:0048255 | mRNA stabilization | 53 | 69 | 8.113e-04 |
| GO:BP | GO:0051123 | RNA polymerase II preinitiation complex assembly | 53 | 69 | 8.113e-04 |
| GO:BP | GO:0007528 | neuromuscular junction development | 43 | 54 | 8.326e-04 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 43 | 54 | 8.326e-04 |
| GO:BP | GO:0006110 | regulation of glycolytic process | 43 | 54 | 8.326e-04 |
| GO:BP | GO:0033121 | regulation of purine nucleotide catabolic process | 43 | 54 | 8.326e-04 |
| GO:BP | GO:0030811 | regulation of nucleotide catabolic process | 43 | 54 | 8.326e-04 |
| GO:BP | GO:0061245 | establishment or maintenance of bipolar cell polarity | 45 | 57 | 8.445e-04 |
| GO:BP | GO:0001738 | morphogenesis of a polarized epithelium | 45 | 57 | 8.445e-04 |
| GO:BP | GO:0035088 | establishment or maintenance of apical/basal cell polarity | 45 | 57 | 8.445e-04 |
| GO:BP | GO:0000041 | transition metal ion transport | 73 | 100 | 8.461e-04 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 193 | 296 | 8.599e-04 |
| GO:BP | GO:0046831 | regulation of RNA export from nucleus | 15 | 15 | 8.600e-04 |
| GO:BP | GO:0006999 | nuclear pore organization | 15 | 15 | 8.600e-04 |
| GO:BP | GO:0071027 | nuclear RNA surveillance | 15 | 15 | 8.600e-04 |
| GO:BP | GO:0033750 | ribosome localization | 15 | 15 | 8.600e-04 |
| GO:BP | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening | 15 | 15 | 8.600e-04 |
| GO:BP | GO:1901836 | regulation of transcription of nucleolar large rRNA by RNA polymerase I | 15 | 15 | 8.600e-04 |
| GO:BP | GO:0061307 | cardiac neural crest cell differentiation involved in heart development | 15 | 15 | 8.600e-04 |
| GO:BP | GO:1904507 | positive regulation of telomere maintenance in response to DNA damage | 15 | 15 | 8.600e-04 |
| GO:BP | GO:0061308 | cardiac neural crest cell development involved in heart development | 15 | 15 | 8.600e-04 |
| GO:BP | GO:1905666 | regulation of protein localization to endosome | 15 | 15 | 8.600e-04 |
| GO:BP | GO:0034616 | response to laminar fluid shear stress | 15 | 15 | 8.600e-04 |
| GO:BP | GO:0000054 | ribosomal subunit export from nucleus | 15 | 15 | 8.600e-04 |
| GO:BP | GO:0060413 | atrial septum morphogenesis | 15 | 15 | 8.600e-04 |
| GO:BP | GO:0048659 | smooth muscle cell proliferation | 100 | 143 | 8.707e-04 |
| GO:BP | GO:0045023 | G0 to G1 transition | 32 | 38 | 8.720e-04 |
| GO:BP | GO:0006516 | glycoprotein catabolic process | 32 | 38 | 8.720e-04 |
| GO:BP | GO:1902110 | positive regulation of mitochondrial membrane permeability involved in apoptotic process | 32 | 38 | 8.720e-04 |
| GO:BP | GO:0035383 | thioester metabolic process | 71 | 97 | 8.989e-04 |
| GO:BP | GO:0046330 | positive regulation of JNK cascade | 71 | 97 | 8.989e-04 |
| GO:BP | GO:0006637 | acyl-CoA metabolic process | 71 | 97 | 8.989e-04 |
| GO:BP | GO:0046928 | regulation of neurotransmitter secretion | 62 | 83 | 9.028e-04 |
| GO:BP | GO:0030947 | regulation of vascular endothelial growth factor receptor signaling pathway | 27 | 31 | 9.430e-04 |
| GO:BP | GO:0051491 | positive regulation of filopodium assembly | 27 | 31 | 9.430e-04 |
| GO:BP | GO:0033500 | carbohydrate homeostasis | 173 | 263 | 9.480e-04 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 88 | 124 | 9.543e-04 |
| GO:BP | GO:0021954 | central nervous system neuron development | 69 | 94 | 9.608e-04 |
| GO:BP | GO:0030641 | regulation of cellular pH | 69 | 94 | 9.608e-04 |
| GO:BP | GO:0071248 | cellular response to metal ion | 130 | 192 | 9.628e-04 |
| GO:BP | GO:0044319 | wound healing, spreading of cells | 34 | 41 | 9.771e-04 |
| GO:BP | GO:1902305 | regulation of sodium ion transmembrane transport | 34 | 41 | 9.771e-04 |
| GO:BP | GO:0045454 | cell redox homeostasis | 34 | 41 | 9.771e-04 |
| GO:BP | GO:0090505 | epiboly involved in wound healing | 34 | 41 | 9.771e-04 |
| GO:BP | GO:0032411 | positive regulation of transporter activity | 58 | 77 | 9.960e-04 |
| GO:BP | GO:0009743 | response to carbohydrate | 162 | 245 | 1.029e-03 |
| GO:BP | GO:0050680 | negative regulation of epithelial cell proliferation | 96 | 137 | 1.036e-03 |
| GO:BP | GO:0072164 | mesonephric tubule development | 74 | 102 | 1.039e-03 |
| GO:BP | GO:0072163 | mesonephric epithelium development | 74 | 102 | 1.039e-03 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 56 | 74 | 1.040e-03 |
| GO:BP | GO:0045834 | positive regulation of lipid metabolic process | 91 | 129 | 1.041e-03 |
| GO:BP | GO:0045646 | regulation of erythrocyte differentiation | 36 | 44 | 1.062e-03 |
| GO:BP | GO:0070373 | negative regulation of ERK1 and ERK2 cascade | 52 | 68 | 1.122e-03 |
| GO:BP | GO:1901607 | alpha-amino acid biosynthetic process | 52 | 68 | 1.122e-03 |
| GO:BP | GO:0010634 | positive regulation of epithelial cell migration | 38 | 47 | 1.125e-03 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 38 | 47 | 1.125e-03 |
| GO:BP | GO:0140014 | mitotic nuclear division | 184 | 282 | 1.132e-03 |
| GO:BP | GO:0016233 | telomere capping | 24 | 27 | 1.138e-03 |
| GO:BP | GO:0048011 | neurotrophin TRK receptor signaling pathway | 24 | 27 | 1.138e-03 |
| GO:BP | GO:0060544 | regulation of necroptotic process | 24 | 27 | 1.138e-03 |
| GO:BP | GO:0043507 | positive regulation of JUN kinase activity | 24 | 27 | 1.138e-03 |
| GO:BP | GO:0010721 | negative regulation of cell development | 175 | 267 | 1.139e-03 |
| GO:BP | GO:0042593 | glucose homeostasis | 172 | 262 | 1.139e-03 |
| GO:BP | GO:0006654 | phosphatidic acid biosynthetic process | 29 | 34 | 1.144e-03 |
| GO:BP | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator | 29 | 34 | 1.144e-03 |
| GO:BP | GO:0018198 | peptidyl-cysteine modification | 29 | 34 | 1.144e-03 |
| GO:BP | GO:0051647 | nucleus localization | 29 | 34 | 1.144e-03 |
| GO:BP | GO:1902235 | regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 29 | 34 | 1.144e-03 |
| GO:BP | GO:0033013 | tetrapyrrole metabolic process | 50 | 65 | 1.147e-03 |
| GO:BP | GO:0031103 | axon regeneration | 40 | 50 | 1.160e-03 |
| GO:BP | GO:0060976 | coronary vasculature development | 40 | 50 | 1.160e-03 |
| GO:BP | GO:1900024 | regulation of substrate adhesion-dependent cell spreading | 48 | 62 | 1.171e-03 |
| GO:BP | GO:0051148 | negative regulation of muscle cell differentiation | 48 | 62 | 1.171e-03 |
| GO:BP | GO:0032648 | regulation of interferon-beta production | 46 | 59 | 1.186e-03 |
| GO:BP | GO:0032608 | interferon-beta production | 46 | 59 | 1.186e-03 |
| GO:BP | GO:0086002 | cardiac muscle cell action potential involved in contraction | 46 | 59 | 1.186e-03 |
| GO:BP | GO:0031529 | ruffle organization | 44 | 56 | 1.190e-03 |
| GO:BP | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 44 | 56 | 1.190e-03 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 97 | 139 | 1.205e-03 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 158 | 239 | 1.233e-03 |
| GO:BP | GO:0099514 | synaptic vesicle cytoskeletal transport | 18 | 19 | 1.233e-03 |
| GO:BP | GO:1902992 | negative regulation of amyloid precursor protein catabolic process | 18 | 19 | 1.233e-03 |
| GO:BP | GO:0099517 | synaptic vesicle transport along microtubule | 18 | 19 | 1.233e-03 |
| GO:BP | GO:0018022 | peptidyl-lysine methylation | 18 | 19 | 1.233e-03 |
| GO:BP | GO:0046931 | pore complex assembly | 18 | 19 | 1.233e-03 |
| GO:BP | GO:0097284 | hepatocyte apoptotic process | 18 | 19 | 1.233e-03 |
| GO:BP | GO:0035269 | protein O-linked mannosylation | 18 | 19 | 1.233e-03 |
| GO:BP | GO:0035720 | intraciliary anterograde transport | 18 | 19 | 1.233e-03 |
| GO:BP | GO:0110154 | RNA decapping | 18 | 19 | 1.233e-03 |
| GO:BP | GO:0048490 | anterograde synaptic vesicle transport | 18 | 19 | 1.233e-03 |
| GO:BP | GO:1903232 | melanosome assembly | 18 | 19 | 1.233e-03 |
| GO:BP | GO:1902774 | late endosome to lysosome transport | 21 | 23 | 1.263e-03 |
| GO:BP | GO:0003283 | atrial septum development | 21 | 23 | 1.263e-03 |
| GO:BP | GO:0039702 | viral budding via host ESCRT complex | 21 | 23 | 1.263e-03 |
| GO:BP | GO:0035331 | negative regulation of hippo signaling | 21 | 23 | 1.263e-03 |
| GO:BP | GO:0006851 | mitochondrial calcium ion transmembrane transport | 21 | 23 | 1.263e-03 |
| GO:BP | GO:1901983 | regulation of protein acetylation | 21 | 23 | 1.263e-03 |
| GO:BP | GO:0030901 | midbrain development | 68 | 93 | 1.267e-03 |
| GO:BP | GO:0033209 | tumor necrosis factor-mediated signaling pathway | 80 | 112 | 1.305e-03 |
| GO:BP | GO:0003230 | cardiac atrium development | 31 | 37 | 1.307e-03 |
| GO:BP | GO:0051592 | response to calcium ion | 95 | 136 | 1.308e-03 |
| GO:BP | GO:0051453 | regulation of intracellular pH | 66 | 90 | 1.358e-03 |
| GO:BP | GO:0001657 | ureteric bud development | 73 | 101 | 1.358e-03 |
| GO:BP | GO:0048844 | artery morphogenesis | 55 | 73 | 1.417e-03 |
| GO:BP | GO:0003143 | embryonic heart tube morphogenesis | 55 | 73 | 1.417e-03 |
| GO:BP | GO:0086091 | regulation of heart rate by cardiac conduction | 33 | 40 | 1.450e-03 |
| GO:BP | GO:0060391 | positive regulation of SMAD protein signal transduction | 33 | 40 | 1.450e-03 |
| GO:BP | GO:1904738 | vascular associated smooth muscle cell migration | 26 | 30 | 1.456e-03 |
| GO:BP | GO:1904357 | negative regulation of telomere maintenance via telomere lengthening | 26 | 30 | 1.456e-03 |
| GO:BP | GO:0000002 | mitochondrial genome maintenance | 26 | 30 | 1.456e-03 |
| GO:BP | GO:0001778 | plasma membrane repair | 26 | 30 | 1.456e-03 |
| GO:BP | GO:0032367 | intracellular cholesterol transport | 26 | 30 | 1.456e-03 |
| GO:BP | GO:0032414 | positive regulation of ion transmembrane transporter activity | 53 | 70 | 1.478e-03 |
| GO:BP | GO:0046847 | filopodium assembly | 53 | 70 | 1.478e-03 |
| GO:BP | GO:0060972 | left/right pattern formation | 101 | 146 | 1.481e-03 |
| GO:BP | GO:1904353 | regulation of telomere capping | 14 | 14 | 1.491e-03 |
| GO:BP | GO:0098734 | macromolecule depalmitoylation | 14 | 14 | 1.491e-03 |
| GO:BP | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation | 14 | 14 | 1.491e-03 |
| GO:BP | GO:0007008 | outer mitochondrial membrane organization | 14 | 14 | 1.491e-03 |
| GO:BP | GO:1901985 | positive regulation of protein acetylation | 14 | 14 | 1.491e-03 |
| GO:BP | GO:1901203 | positive regulation of extracellular matrix assembly | 14 | 14 | 1.491e-03 |
| GO:BP | GO:0035970 | peptidyl-threonine dephosphorylation | 14 | 14 | 1.491e-03 |
| GO:BP | GO:0051645 | Golgi localization | 14 | 14 | 1.491e-03 |
| GO:BP | GO:0072393 | microtubule anchoring at microtubule organizing center | 14 | 14 | 1.491e-03 |
| GO:BP | GO:0045040 | protein insertion into mitochondrial outer membrane | 14 | 14 | 1.491e-03 |
| GO:BP | GO:0035721 | intraciliary retrograde transport | 14 | 14 | 1.491e-03 |
| GO:BP | GO:1905668 | positive regulation of protein localization to endosome | 14 | 14 | 1.491e-03 |
| GO:BP | GO:1904350 | regulation of protein catabolic process in the vacuole | 14 | 14 | 1.491e-03 |
| GO:BP | GO:0032933 | SREBP signaling pathway | 14 | 14 | 1.491e-03 |
| GO:BP | GO:0035791 | platelet-derived growth factor receptor-beta signaling pathway | 14 | 14 | 1.491e-03 |
| GO:BP | GO:0001885 | endothelial cell development | 51 | 67 | 1.527e-03 |
| GO:BP | GO:0016042 | lipid catabolic process | 226 | 354 | 1.537e-03 |
| GO:BP | GO:1990000 | amyloid fibril formation | 35 | 43 | 1.538e-03 |
| GO:BP | GO:0030149 | sphingolipid catabolic process | 35 | 43 | 1.538e-03 |
| GO:BP | GO:0009066 | aspartate family amino acid metabolic process | 35 | 43 | 1.538e-03 |
| GO:BP | GO:0099563 | modification of synaptic structure | 35 | 43 | 1.538e-03 |
| GO:BP | GO:0030902 | hindbrain development | 112 | 164 | 1.540e-03 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 69 | 95 | 1.559e-03 |
| GO:BP | GO:0048524 | positive regulation of viral process | 49 | 64 | 1.576e-03 |
| GO:BP | GO:0030042 | actin filament depolymerization | 45 | 58 | 1.656e-03 |
| GO:BP | GO:0030837 | negative regulation of actin filament polymerization | 45 | 58 | 1.656e-03 |
| GO:BP | GO:0042181 | ketone biosynthetic process | 39 | 49 | 1.656e-03 |
| GO:BP | GO:0140374 | antiviral innate immune response | 43 | 55 | 1.674e-03 |
| GO:BP | GO:0030838 | positive regulation of actin filament polymerization | 41 | 52 | 1.675e-03 |
| GO:BP | GO:0009890 | negative regulation of biosynthetic process | 1620 | 2813 | 1.681e-03 |
| GO:BP | GO:0010565 | regulation of ketone metabolic process | 89 | 127 | 1.690e-03 |
| GO:BP | GO:0051607 | defense response to virus | 205 | 319 | 1.721e-03 |
| GO:BP | GO:0021591 | ventricular system development | 28 | 33 | 1.726e-03 |
| GO:BP | GO:0060421 | positive regulation of heart growth | 28 | 33 | 1.726e-03 |
| GO:BP | GO:0031116 | positive regulation of microtubule polymerization | 28 | 33 | 1.726e-03 |
| GO:BP | GO:0060325 | face morphogenesis | 28 | 33 | 1.726e-03 |
| GO:BP | GO:0060993 | kidney morphogenesis | 72 | 100 | 1.774e-03 |
| GO:BP | GO:0006493 | protein O-linked glycosylation | 72 | 100 | 1.774e-03 |
| GO:BP | GO:0003181 | atrioventricular valve morphogenesis | 23 | 26 | 1.775e-03 |
| GO:BP | GO:0061744 | motor behavior | 23 | 26 | 1.775e-03 |
| GO:BP | GO:0005980 | glycogen catabolic process | 23 | 26 | 1.775e-03 |
| GO:BP | GO:1905207 | regulation of cardiocyte differentiation | 23 | 26 | 1.775e-03 |
| GO:BP | GO:0007039 | protein catabolic process in the vacuole | 23 | 26 | 1.775e-03 |
| GO:BP | GO:0046323 | D-glucose import | 56 | 75 | 1.821e-03 |
| GO:BP | GO:0007229 | integrin-mediated signaling pathway | 82 | 116 | 1.850e-03 |
| GO:BP | GO:0051260 | protein homooligomerization | 136 | 204 | 1.872e-03 |
| GO:BP | GO:0048661 | positive regulation of smooth muscle cell proliferation | 63 | 86 | 1.926e-03 |
| GO:BP | GO:0099010 | modification of postsynaptic structure | 30 | 36 | 1.954e-03 |
| GO:BP | GO:0006301 | postreplication repair | 30 | 36 | 1.954e-03 |
| GO:BP | GO:0035459 | vesicle cargo loading | 30 | 36 | 1.954e-03 |
| GO:BP | GO:0006904 | vesicle docking involved in exocytosis | 30 | 36 | 1.954e-03 |
| GO:BP | GO:0070306 | lens fiber cell differentiation | 30 | 36 | 1.954e-03 |
| GO:BP | GO:0090630 | activation of GTPase activity | 30 | 36 | 1.954e-03 |
| GO:BP | GO:0032885 | regulation of polysaccharide biosynthetic process | 30 | 36 | 1.954e-03 |
| GO:BP | GO:0051150 | regulation of smooth muscle cell differentiation | 30 | 36 | 1.954e-03 |
| GO:BP | GO:0030199 | collagen fibril organization | 52 | 69 | 2.014e-03 |
| GO:BP | GO:0006879 | intracellular iron ion homeostasis | 52 | 69 | 2.014e-03 |
| GO:BP | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis | 20 | 22 | 2.036e-03 |
| GO:BP | GO:0031954 | positive regulation of protein autophosphorylation | 20 | 22 | 2.036e-03 |
| GO:BP | GO:1900407 | regulation of cellular response to oxidative stress | 20 | 22 | 2.036e-03 |
| GO:BP | GO:0032042 | mitochondrial DNA metabolic process | 20 | 22 | 2.036e-03 |
| GO:BP | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | 20 | 22 | 2.036e-03 |
| GO:BP | GO:1900242 | regulation of synaptic vesicle endocytosis | 20 | 22 | 2.036e-03 |
| GO:BP | GO:0032785 | negative regulation of DNA-templated transcription, elongation | 20 | 22 | 2.036e-03 |
| GO:BP | GO:1905146 | lysosomal protein catabolic process | 20 | 22 | 2.036e-03 |
| GO:BP | GO:0008090 | retrograde axonal transport | 20 | 22 | 2.036e-03 |
| GO:BP | GO:0043555 | regulation of translation in response to stress | 20 | 22 | 2.036e-03 |
| GO:BP | GO:0086026 | atrial cardiac muscle cell to AV node cell signaling | 17 | 18 | 2.037e-03 |
| GO:BP | GO:0086014 | atrial cardiac muscle cell action potential | 17 | 18 | 2.037e-03 |
| GO:BP | GO:0090110 | COPII-coated vesicle cargo loading | 17 | 18 | 2.037e-03 |
| GO:BP | GO:0072673 | lamellipodium morphogenesis | 17 | 18 | 2.037e-03 |
| GO:BP | GO:0086066 | atrial cardiac muscle cell to AV node cell communication | 17 | 18 | 2.037e-03 |
| GO:BP | GO:0043923 | positive regulation by host of viral transcription | 17 | 18 | 2.037e-03 |
| GO:BP | GO:0061333 | renal tubule morphogenesis | 61 | 83 | 2.037e-03 |
| GO:BP | GO:0062099 | negative regulation of programmed necrotic cell death | 17 | 18 | 2.037e-03 |
| GO:BP | GO:0019896 | axonal transport of mitochondrion | 17 | 18 | 2.037e-03 |
| GO:BP | GO:1902430 | negative regulation of amyloid-beta formation | 17 | 18 | 2.037e-03 |
| GO:BP | GO:0061323 | cell proliferation involved in heart morphogenesis | 17 | 18 | 2.037e-03 |
| GO:BP | GO:2000136 | regulation of cell proliferation involved in heart morphogenesis | 17 | 18 | 2.037e-03 |
| GO:BP | GO:0045722 | positive regulation of gluconeogenesis | 17 | 18 | 2.037e-03 |
| GO:BP | GO:0015936 | coenzyme A metabolic process | 17 | 18 | 2.037e-03 |
| GO:BP | GO:0016125 | sterol metabolic process | 109 | 160 | 2.090e-03 |
| GO:BP | GO:0051125 | regulation of actin nucleation | 32 | 39 | 2.112e-03 |
| GO:BP | GO:0120033 | negative regulation of plasma membrane bounded cell projection assembly | 32 | 39 | 2.112e-03 |
| GO:BP | GO:0072593 | reactive oxygen species metabolic process | 152 | 231 | 2.119e-03 |
| GO:BP | GO:0031347 | regulation of defense response | 489 | 809 | 2.119e-03 |
| GO:BP | GO:0046394 | carboxylic acid biosynthetic process | 205 | 320 | 2.157e-03 |
| GO:BP | GO:1990874 | vascular associated smooth muscle cell proliferation | 48 | 63 | 2.164e-03 |
| GO:BP | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway | 48 | 63 | 2.164e-03 |
| GO:BP | GO:0006518 | peptide metabolic process | 48 | 63 | 2.164e-03 |
| GO:BP | GO:0048660 | regulation of smooth muscle cell proliferation | 96 | 139 | 2.201e-03 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 96 | 139 | 2.201e-03 |
| GO:BP | GO:0051156 | glucose 6-phosphate metabolic process | 25 | 29 | 2.218e-03 |
| GO:BP | GO:0003171 | atrioventricular valve development | 25 | 29 | 2.218e-03 |
| GO:BP | GO:0048679 | regulation of axon regeneration | 25 | 29 | 2.218e-03 |
| GO:BP | GO:1903034 | regulation of response to wounding | 115 | 170 | 2.219e-03 |
| GO:BP | GO:0019432 | triglyceride biosynthetic process | 34 | 42 | 2.230e-03 |
| GO:BP | GO:0043124 | negative regulation of canonical NF-kappaB signal transduction | 46 | 60 | 2.231e-03 |
| GO:BP | GO:0080164 | regulation of nitric oxide metabolic process | 46 | 60 | 2.231e-03 |
| GO:BP | GO:0099172 | presynapse organization | 46 | 60 | 2.231e-03 |
| GO:BP | GO:0009749 | response to glucose | 138 | 208 | 2.275e-03 |
| GO:BP | GO:0001755 | neural crest cell migration | 44 | 57 | 2.288e-03 |
| GO:BP | GO:0030865 | cortical cytoskeleton organization | 44 | 57 | 2.288e-03 |
| GO:BP | GO:0051452 | intracellular pH reduction | 44 | 57 | 2.288e-03 |
| GO:BP | GO:1902895 | positive regulation of miRNA transcription | 44 | 57 | 2.288e-03 |
| GO:BP | GO:0009746 | response to hexose | 141 | 213 | 2.288e-03 |
| GO:BP | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway | 71 | 99 | 2.289e-03 |
| GO:BP | GO:0008652 | amino acid biosynthetic process | 57 | 77 | 2.293e-03 |
| GO:BP | GO:0045663 | positive regulation of myoblast differentiation | 36 | 45 | 2.301e-03 |
| GO:BP | GO:0021695 | cerebellar cortex development | 42 | 54 | 2.327e-03 |
| GO:BP | GO:0055021 | regulation of cardiac muscle tissue growth | 42 | 54 | 2.327e-03 |
| GO:BP | GO:0046466 | membrane lipid catabolic process | 38 | 48 | 2.339e-03 |
| GO:BP | GO:0120163 | negative regulation of cold-induced thermogenesis | 38 | 48 | 2.339e-03 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 40 | 51 | 2.342e-03 |
| GO:BP | GO:0046463 | acylglycerol biosynthetic process | 40 | 51 | 2.342e-03 |
| GO:BP | GO:0046460 | neutral lipid biosynthetic process | 40 | 51 | 2.342e-03 |
| GO:BP | GO:0046578 | regulation of Ras protein signal transduction | 40 | 51 | 2.342e-03 |
| GO:BP | GO:0055088 | lipid homeostasis | 113 | 167 | 2.425e-03 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 116 | 172 | 2.492e-03 |
| GO:BP | GO:2001257 | regulation of cation channel activity | 62 | 85 | 2.523e-03 |
| GO:BP | GO:0034284 | response to monosaccharide | 145 | 220 | 2.533e-03 |
| GO:BP | GO:0060348 | bone development | 145 | 220 | 2.533e-03 |
| GO:BP | GO:1901655 | cellular response to ketone | 79 | 112 | 2.557e-03 |
| GO:BP | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 27 | 32 | 2.559e-03 |
| GO:BP | GO:0097707 | ferroptosis | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0035542 | regulation of SNARE complex assembly | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0001682 | tRNA 5’-leader removal | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0090646 | mitochondrial tRNA processing | 13 | 13 | 2.559e-03 |
| GO:BP | GO:1904380 | endoplasmic reticulum mannose trimming | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0006465 | signal peptide processing | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 27 | 32 | 2.559e-03 |
| GO:BP | GO:0090158 | endoplasmic reticulum membrane organization | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0003174 | mitral valve development | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0031958 | nuclear receptor-mediated corticosteroid signaling pathway | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0061795 | Golgi lumen acidification | 13 | 13 | 2.559e-03 |
| GO:BP | GO:1904152 | regulation of retrograde protein transport, ER to cytosol | 13 | 13 | 2.559e-03 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 53 | 71 | 2.559e-03 |
| GO:BP | GO:0033235 | positive regulation of protein sumoylation | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0043558 | regulation of translational initiation in response to stress | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0034333 | adherens junction assembly | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0009304 | tRNA transcription | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0090317 | negative regulation of intracellular protein transport | 27 | 32 | 2.559e-03 |
| GO:BP | GO:0034551 | mitochondrial respiratory chain complex III assembly | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0010867 | positive regulation of triglyceride biosynthetic process | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0045039 | protein insertion into mitochondrial inner membrane | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0017062 | respiratory chain complex III assembly | 13 | 13 | 2.559e-03 |
| GO:BP | GO:0060966 | regulation of gene silencing by regulatory ncRNA | 27 | 32 | 2.559e-03 |
| GO:BP | GO:0060674 | placenta blood vessel development | 27 | 32 | 2.559e-03 |
| GO:BP | GO:0006865 | amino acid transport | 111 | 164 | 2.636e-03 |
| GO:BP | GO:0032103 | positive regulation of response to external stimulus | 392 | 642 | 2.694e-03 |
| GO:BP | GO:1903779 | regulation of cardiac conduction | 22 | 25 | 2.739e-03 |
| GO:BP | GO:0007097 | nuclear migration | 22 | 25 | 2.739e-03 |
| GO:BP | GO:1901659 | glycosyl compound biosynthetic process | 22 | 25 | 2.739e-03 |
| GO:BP | GO:0070861 | regulation of protein exit from endoplasmic reticulum | 22 | 25 | 2.739e-03 |
| GO:BP | GO:0070841 | inclusion body assembly | 22 | 25 | 2.739e-03 |
| GO:BP | GO:0032400 | melanosome localization | 22 | 25 | 2.739e-03 |
| GO:BP | GO:0019751 | polyol metabolic process | 77 | 109 | 2.750e-03 |
| GO:BP | GO:0048144 | fibroblast proliferation | 77 | 109 | 2.750e-03 |
| GO:BP | GO:0009612 | response to mechanical stimulus | 146 | 222 | 2.751e-03 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 49 | 65 | 2.814e-03 |
| GO:BP | GO:2000630 | positive regulation of miRNA metabolic process | 49 | 65 | 2.814e-03 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 65 | 90 | 2.852e-03 |
| GO:BP | GO:0090162 | establishment of epithelial cell polarity | 29 | 35 | 2.853e-03 |
| GO:BP | GO:0014808 | release of sequestered calcium ion into cytosol by sarcoplasmic reticulum | 29 | 35 | 2.853e-03 |
| GO:BP | GO:0071482 | cellular response to light stimulus | 85 | 122 | 2.928e-03 |
| GO:BP | GO:0033619 | membrane protein proteolysis | 47 | 62 | 2.944e-03 |
| GO:BP | GO:0051851 | host-mediated perturbation of symbiont process | 75 | 106 | 2.995e-03 |
| GO:BP | GO:0001823 | mesonephros development | 75 | 106 | 2.995e-03 |
| GO:BP | GO:0061326 | renal tubule development | 75 | 106 | 2.995e-03 |
| GO:BP | GO:0090497 | mesenchymal cell migration | 45 | 59 | 3.063e-03 |
| GO:BP | GO:0098698 | postsynaptic specialization assembly | 31 | 38 | 3.069e-03 |
| GO:BP | GO:0045773 | positive regulation of axon extension | 31 | 38 | 3.069e-03 |
| GO:BP | GO:0031112 | positive regulation of microtubule polymerization or depolymerization | 31 | 38 | 3.069e-03 |
| GO:BP | GO:0039532 | negative regulation of cytoplasmic pattern recognition receptor signaling pathway | 43 | 56 | 3.164e-03 |
| GO:BP | GO:0170038 | proteinogenic amino acid biosynthetic process | 43 | 56 | 3.164e-03 |
| GO:BP | GO:0050792 | regulation of viral process | 107 | 158 | 3.176e-03 |
| GO:BP | GO:0001935 | endothelial cell proliferation | 107 | 158 | 3.176e-03 |
| GO:BP | GO:1901532 | regulation of hematopoietic progenitor cell differentiation | 33 | 41 | 3.213e-03 |
| GO:BP | GO:0072663 | establishment of protein localization to peroxisome | 19 | 21 | 3.223e-03 |
| GO:BP | GO:0010288 | response to lead ion | 19 | 21 | 3.223e-03 |
| GO:BP | GO:1902018 | negative regulation of cilium assembly | 19 | 21 | 3.223e-03 |
| GO:BP | GO:0072662 | protein localization to peroxisome | 19 | 21 | 3.223e-03 |
| GO:BP | GO:0045912 | negative regulation of carbohydrate metabolic process | 41 | 53 | 3.238e-03 |
| GO:BP | GO:0030201 | heparan sulfate proteoglycan metabolic process | 35 | 44 | 3.282e-03 |
| GO:BP | GO:0060338 | regulation of type I interferon-mediated signaling pathway | 35 | 44 | 3.282e-03 |
| GO:BP | GO:0097178 | ruffle assembly | 35 | 44 | 3.282e-03 |
| GO:BP | GO:0003176 | aortic valve development | 35 | 44 | 3.282e-03 |
| GO:BP | GO:0032506 | cytokinetic process | 39 | 50 | 3.286e-03 |
| GO:BP | GO:0060038 | cardiac muscle cell proliferation | 39 | 50 | 3.286e-03 |
| GO:BP | GO:0034766 | negative regulation of monoatomic ion transmembrane transport | 61 | 84 | 3.299e-03 |
| GO:BP | GO:0010463 | mesenchymal cell proliferation | 37 | 47 | 3.301e-03 |
| GO:BP | GO:0030073 | insulin secretion | 139 | 211 | 3.301e-03 |
| GO:BP | GO:0006103 | 2-oxoglutarate metabolic process | 16 | 17 | 3.335e-03 |
| GO:BP | GO:2000369 | regulation of clathrin-dependent endocytosis | 16 | 17 | 3.335e-03 |
| GO:BP | GO:0141198 | protein branched polyubiquitination | 16 | 17 | 3.335e-03 |
| GO:BP | GO:0002098 | tRNA wobble uridine modification | 16 | 17 | 3.335e-03 |
| GO:BP | GO:0070131 | positive regulation of mitochondrial translation | 16 | 17 | 3.335e-03 |
| GO:BP | GO:0007000 | nucleolus organization | 16 | 17 | 3.335e-03 |
| GO:BP | GO:0006991 | response to sterol depletion | 16 | 17 | 3.335e-03 |
| GO:BP | GO:0034349 | glial cell apoptotic process | 16 | 17 | 3.335e-03 |
| GO:BP | GO:0044790 | suppression of viral release by host | 16 | 17 | 3.335e-03 |
| GO:BP | GO:0071243 | cellular response to arsenic-containing substance | 16 | 17 | 3.335e-03 |
| GO:BP | GO:0003209 | cardiac atrium morphogenesis | 24 | 28 | 3.342e-03 |
| GO:BP | GO:0032801 | receptor catabolic process | 24 | 28 | 3.342e-03 |
| GO:BP | GO:0032469 | endoplasmic reticulum calcium ion homeostasis | 24 | 28 | 3.342e-03 |
| GO:BP | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity | 24 | 28 | 3.342e-03 |
| GO:BP | GO:1904752 | regulation of vascular associated smooth muscle cell migration | 24 | 28 | 3.342e-03 |
| GO:BP | GO:0099558 | maintenance of synapse structure | 24 | 28 | 3.342e-03 |
| GO:BP | GO:0010592 | positive regulation of lamellipodium assembly | 24 | 28 | 3.342e-03 |
| GO:BP | GO:0046513 | ceramide biosynthetic process | 52 | 70 | 3.414e-03 |
| GO:BP | GO:0048645 | animal organ formation | 52 | 70 | 3.414e-03 |
| GO:BP | GO:0032088 | negative regulation of NF-kappaB transcription factor activity | 52 | 70 | 3.414e-03 |
| GO:BP | GO:0046889 | positive regulation of lipid biosynthetic process | 66 | 92 | 3.431e-03 |
| GO:BP | GO:0035023 | regulation of Rho protein signal transduction | 64 | 89 | 3.731e-03 |
| GO:BP | GO:0050905 | neuromuscular process | 117 | 175 | 3.782e-03 |
| GO:BP | GO:0043271 | negative regulation of monoatomic ion transport | 79 | 113 | 3.798e-03 |
| GO:BP | GO:0021766 | hippocampus development | 69 | 97 | 3.829e-03 |
| GO:BP | GO:1903706 | regulation of hemopoiesis | 265 | 425 | 3.841e-03 |
| GO:BP | GO:0006885 | regulation of pH | 74 | 105 | 3.841e-03 |
| GO:BP | GO:0090288 | negative regulation of cellular response to growth factor stimulus | 74 | 105 | 3.841e-03 |
| GO:BP | GO:0060147 | regulation of post-transcriptional gene silencing | 26 | 31 | 3.859e-03 |
| GO:BP | GO:0045724 | positive regulation of cilium assembly | 26 | 31 | 3.859e-03 |
| GO:BP | GO:0051701 | biological process involved in interaction with host | 123 | 185 | 3.871e-03 |
| GO:BP | GO:0007368 | determination of left/right symmetry | 95 | 139 | 3.899e-03 |
| GO:BP | GO:0034502 | protein localization to chromosome | 87 | 126 | 3.906e-03 |
| GO:BP | GO:0043410 | positive regulation of MAPK cascade | 283 | 456 | 3.906e-03 |
| GO:BP | GO:0009755 | hormone-mediated signaling pathway | 147 | 225 | 3.946e-03 |
| GO:BP | GO:1904705 | regulation of vascular associated smooth muscle cell proliferation | 46 | 61 | 4.022e-03 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 569 | 954 | 4.036e-03 |
| GO:BP | GO:0032481 | positive regulation of type I interferon production | 55 | 75 | 4.059e-03 |
| GO:BP | GO:0010827 | regulation of D-glucose transmembrane transport | 55 | 75 | 4.059e-03 |
| GO:BP | GO:0015748 | organophosphate ester transport | 115 | 172 | 4.149e-03 |
| GO:BP | GO:0060021 | roof of mouth development | 67 | 94 | 4.150e-03 |
| GO:BP | GO:0016053 | organic acid biosynthetic process | 205 | 323 | 4.166e-03 |
| GO:BP | GO:0048534 | hematopoietic or lymphoid organ development | 72 | 102 | 4.181e-03 |
| GO:BP | GO:0045428 | regulation of nitric oxide biosynthetic process | 44 | 58 | 4.203e-03 |
| GO:BP | GO:0120009 | intermembrane lipid transfer | 28 | 34 | 4.222e-03 |
| GO:BP | GO:0086005 | ventricular cardiac muscle cell action potential | 28 | 34 | 4.222e-03 |
| GO:BP | GO:0030517 | negative regulation of axon extension | 28 | 34 | 4.222e-03 |
| GO:BP | GO:0097345 | mitochondrial outer membrane permeabilization | 28 | 34 | 4.222e-03 |
| GO:BP | GO:0060603 | mammary gland duct morphogenesis | 28 | 34 | 4.222e-03 |
| GO:BP | GO:1901673 | regulation of mitotic spindle assembly | 21 | 24 | 4.261e-03 |
| GO:BP | GO:0036119 | response to platelet-derived growth factor | 21 | 24 | 4.261e-03 |
| GO:BP | GO:0051560 | mitochondrial calcium ion homeostasis | 21 | 24 | 4.261e-03 |
| GO:BP | GO:1902993 | positive regulation of amyloid precursor protein catabolic process | 21 | 24 | 4.261e-03 |
| GO:BP | GO:0046835 | carbohydrate phosphorylation | 21 | 24 | 4.261e-03 |
| GO:BP | GO:0014912 | negative regulation of smooth muscle cell migration | 21 | 24 | 4.261e-03 |
| GO:BP | GO:0033233 | regulation of protein sumoylation | 21 | 24 | 4.261e-03 |
| GO:BP | GO:0042659 | regulation of cell fate specification | 21 | 24 | 4.261e-03 |
| GO:BP | GO:0003148 | outflow tract septum morphogenesis | 21 | 24 | 4.261e-03 |
| GO:BP | GO:0007626 | locomotory behavior | 145 | 222 | 4.309e-03 |
| GO:BP | GO:0051101 | regulation of DNA binding | 53 | 72 | 4.316e-03 |
| GO:BP | GO:0072028 | nephron morphogenesis | 60 | 83 | 4.320e-03 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 42 | 55 | 4.342e-03 |
| GO:BP | GO:0048286 | lung alveolus development | 42 | 55 | 4.342e-03 |
| GO:BP | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0140627 | ubiquitin-dependent protein catabolic process via the C-end degron rule pathway | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0031126 | sno(s)RNA 3’-end processing | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0061723 | glycophagy | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0110075 | regulation of ferroptosis | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0006448 | regulation of translational elongation | 12 | 12 | 4.400e-03 |
| GO:BP | GO:1904872 | regulation of telomerase RNA localization to Cajal body | 12 | 12 | 4.400e-03 |
| GO:BP | GO:1900153 | positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 12 | 12 | 4.400e-03 |
| GO:BP | GO:1905165 | regulation of lysosomal protein catabolic process | 12 | 12 | 4.400e-03 |
| GO:BP | GO:1902946 | protein localization to early endosome | 12 | 12 | 4.400e-03 |
| GO:BP | GO:1900370 | positive regulation of post-transcriptional gene silencing by RNA | 12 | 12 | 4.400e-03 |
| GO:BP | GO:1990253 | cellular response to leucine starvation | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0042797 | tRNA transcription by RNA polymerase III | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0050847 | progesterone receptor signaling pathway | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0097576 | vacuole fusion | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0042921 | nuclear receptor-mediated glucocorticoid signaling pathway | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0061309 | cardiac neural crest cell development involved in outflow tract morphogenesis | 12 | 12 | 4.400e-03 |
| GO:BP | GO:2000637 | positive regulation of miRNA-mediated gene silencing | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0000491 | small nucleolar ribonucleoprotein complex assembly | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0034454 | microtubule anchoring at centrosome | 12 | 12 | 4.400e-03 |
| GO:BP | GO:1904779 | regulation of protein localization to centrosome | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0038026 | reelin-mediated signaling pathway | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0098911 | regulation of ventricular cardiac muscle cell action potential | 12 | 12 | 4.400e-03 |
| GO:BP | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0060148 | positive regulation of post-transcriptional gene silencing | 12 | 12 | 4.400e-03 |
| GO:BP | GO:0003183 | mitral valve morphogenesis | 12 | 12 | 4.400e-03 |
| GO:BP | GO:1903514 | release of sequestered calcium ion into cytosol by endoplasmic reticulum | 30 | 37 | 4.407e-03 |
| GO:BP | GO:0042157 | lipoprotein metabolic process | 65 | 91 | 4.424e-03 |
| GO:BP | GO:0003197 | endocardial cushion development | 40 | 52 | 4.430e-03 |
| GO:BP | GO:0010332 | response to gamma radiation | 40 | 52 | 4.430e-03 |
| GO:BP | GO:0030510 | regulation of BMP signaling pathway | 75 | 107 | 4.438e-03 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 113 | 169 | 4.471e-03 |
| GO:BP | GO:0071616 | acyl-CoA biosynthetic process | 38 | 49 | 4.533e-03 |
| GO:BP | GO:0035384 | thioester biosynthetic process | 38 | 49 | 4.533e-03 |
| GO:BP | GO:0061082 | myeloid leukocyte cytokine production | 38 | 49 | 4.533e-03 |
| GO:BP | GO:0033120 | positive regulation of RNA splicing | 38 | 49 | 4.533e-03 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 51 | 69 | 4.536e-03 |
| GO:BP | GO:0070266 | necroptotic process | 32 | 40 | 4.540e-03 |
| GO:BP | GO:0010719 | negative regulation of epithelial to mesenchymal transition | 32 | 40 | 4.540e-03 |
| GO:BP | GO:0042886 | amide transport | 210 | 332 | 4.550e-03 |
| GO:BP | GO:0045913 | positive regulation of carbohydrate metabolic process | 58 | 80 | 4.583e-03 |
| GO:BP | GO:0006275 | regulation of DNA replication | 83 | 120 | 4.583e-03 |
| GO:BP | GO:0032964 | collagen biosynthetic process | 36 | 46 | 4.583e-03 |
| GO:BP | GO:0001825 | blastocyst formation | 36 | 46 | 4.583e-03 |
| GO:BP | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 36 | 46 | 4.583e-03 |
| GO:BP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 34 | 43 | 4.588e-03 |
| GO:BP | GO:0038084 | vascular endothelial growth factor signaling pathway | 34 | 43 | 4.588e-03 |
| GO:BP | GO:0007492 | endoderm development | 63 | 88 | 4.780e-03 |
| GO:BP | GO:0030301 | cholesterol transport | 73 | 104 | 4.835e-03 |
| GO:BP | GO:0001678 | intracellular glucose homeostasis | 114 | 171 | 4.977e-03 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 114 | 171 | 4.977e-03 |
| GO:BP | GO:0010866 | regulation of triglyceride biosynthetic process | 18 | 20 | 5.049e-03 |
| GO:BP | GO:0033137 | negative regulation of peptidyl-serine phosphorylation | 23 | 27 | 5.049e-03 |
| GO:BP | GO:0071480 | cellular response to gamma radiation | 23 | 27 | 5.049e-03 |
| GO:BP | GO:0009301 | snRNA transcription | 18 | 20 | 5.049e-03 |
| GO:BP | GO:0090141 | positive regulation of mitochondrial fission | 18 | 20 | 5.049e-03 |
| GO:BP | GO:0010614 | negative regulation of cardiac muscle hypertrophy | 23 | 27 | 5.049e-03 |
| GO:BP | GO:0009648 | photoperiodism | 23 | 27 | 5.049e-03 |
| GO:BP | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 18 | 20 | 5.049e-03 |
| GO:BP | GO:1990182 | exosomal secretion | 18 | 20 | 5.049e-03 |
| GO:BP | GO:0016188 | synaptic vesicle maturation | 23 | 27 | 5.049e-03 |
| GO:BP | GO:0009251 | glucan catabolic process | 23 | 27 | 5.049e-03 |
| GO:BP | GO:0055070 | copper ion homeostasis | 18 | 20 | 5.049e-03 |
| GO:BP | GO:0061620 | glycolytic process through glucose-6-phosphate | 18 | 20 | 5.049e-03 |
| GO:BP | GO:0032091 | negative regulation of protein binding | 47 | 63 | 5.091e-03 |
| GO:BP | GO:0015918 | sterol transport | 76 | 109 | 5.170e-03 |
| GO:BP | GO:0038061 | non-canonical NF-kappaB signal transduction | 84 | 122 | 5.285e-03 |
| GO:BP | GO:0001706 | endoderm formation | 45 | 60 | 5.390e-03 |
| GO:BP | GO:0003188 | heart valve formation | 15 | 16 | 5.439e-03 |
| GO:BP | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining | 15 | 16 | 5.439e-03 |
| GO:BP | GO:0030167 | proteoglycan catabolic process | 15 | 16 | 5.439e-03 |
| GO:BP | GO:0036444 | calcium import into the mitochondrion | 15 | 16 | 5.439e-03 |
| GO:BP | GO:0032897 | negative regulation of viral transcription | 15 | 16 | 5.439e-03 |
| GO:BP | GO:0036260 | RNA capping | 15 | 16 | 5.439e-03 |
| GO:BP | GO:2000725 | regulation of cardiac muscle cell differentiation | 15 | 16 | 5.439e-03 |
| GO:BP | GO:1990166 | protein localization to site of double-strand break | 15 | 16 | 5.439e-03 |
| GO:BP | GO:0098693 | regulation of synaptic vesicle cycle | 15 | 16 | 5.439e-03 |
| GO:BP | GO:0016074 | sno(s)RNA metabolic process | 15 | 16 | 5.439e-03 |
| GO:BP | GO:0110156 | mRNA methylguanosine-cap decapping | 15 | 16 | 5.439e-03 |
| GO:BP | GO:0045637 | regulation of myeloid cell differentiation | 139 | 213 | 5.583e-03 |
| GO:BP | GO:2000649 | regulation of sodium ion transmembrane transporter activity | 25 | 30 | 5.670e-03 |
| GO:BP | GO:0032835 | glomerulus development | 52 | 71 | 5.670e-03 |
| GO:BP | GO:0022011 | myelination in peripheral nervous system | 25 | 30 | 5.670e-03 |
| GO:BP | GO:1900368 | regulation of post-transcriptional gene silencing by regulatory ncRNA | 25 | 30 | 5.670e-03 |
| GO:BP | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 25 | 30 | 5.670e-03 |
| GO:BP | GO:2000629 | negative regulation of miRNA metabolic process | 25 | 30 | 5.670e-03 |
| GO:BP | GO:0048668 | collateral sprouting | 25 | 30 | 5.670e-03 |
| GO:BP | GO:0032292 | peripheral nervous system axon ensheathment | 25 | 30 | 5.670e-03 |
| GO:BP | GO:0046683 | response to organophosphorus | 90 | 132 | 5.720e-03 |
| GO:BP | GO:0072078 | nephron tubule morphogenesis | 57 | 79 | 6.047e-03 |
| GO:BP | GO:0051149 | positive regulation of muscle cell differentiation | 50 | 68 | 6.081e-03 |
| GO:BP | GO:0170033 | L-amino acid metabolic process | 116 | 175 | 6.107e-03 |
| GO:BP | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 39 | 51 | 6.149e-03 |
| GO:BP | GO:0010883 | regulation of lipid storage | 39 | 51 | 6.149e-03 |
| GO:BP | GO:0060249 | anatomical structure homeostasis | 172 | 269 | 6.303e-03 |
| GO:BP | GO:0042168 | heme metabolic process | 37 | 48 | 6.354e-03 |
| GO:BP | GO:0006687 | glycosphingolipid metabolic process | 48 | 65 | 6.498e-03 |
| GO:BP | GO:0046622 | positive regulation of organ growth | 35 | 45 | 6.498e-03 |
| GO:BP | GO:0002931 | response to ischemia | 48 | 65 | 6.498e-03 |
| GO:BP | GO:2000772 | regulation of cellular senescence | 35 | 45 | 6.498e-03 |
| GO:BP | GO:1904063 | negative regulation of cation transmembrane transport | 55 | 76 | 6.533e-03 |
| GO:BP | GO:0001941 | postsynaptic membrane organization | 33 | 42 | 6.542e-03 |
| GO:BP | GO:0090181 | regulation of cholesterol metabolic process | 31 | 39 | 6.542e-03 |
| GO:BP | GO:0015919 | peroxisomal membrane transport | 20 | 23 | 6.542e-03 |
| GO:BP | GO:0009167 | purine ribonucleoside monophosphate metabolic process | 33 | 42 | 6.542e-03 |
| GO:BP | GO:0099150 | regulation of postsynaptic specialization assembly | 20 | 23 | 6.542e-03 |
| GO:BP | GO:0003180 | aortic valve morphogenesis | 31 | 39 | 6.542e-03 |
| GO:BP | GO:0140112 | extracellular vesicle biogenesis | 20 | 23 | 6.542e-03 |
| GO:BP | GO:0042307 | positive regulation of protein import into nucleus | 33 | 42 | 6.542e-03 |
| GO:BP | GO:0043574 | peroxisomal transport | 20 | 23 | 6.542e-03 |
| GO:BP | GO:0032401 | establishment of melanosome localization | 20 | 23 | 6.542e-03 |
| GO:BP | GO:0046677 | response to antibiotic | 31 | 39 | 6.542e-03 |
| GO:BP | GO:0002062 | chondrocyte differentiation | 83 | 121 | 6.626e-03 |
| GO:BP | GO:0007009 | plasma membrane organization | 114 | 172 | 6.690e-03 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 70 | 100 | 6.736e-03 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 65 | 92 | 6.807e-03 |
| GO:BP | GO:0009161 | ribonucleoside monophosphate metabolic process | 46 | 62 | 6.894e-03 |
| GO:BP | GO:0008347 | glial cell migration | 46 | 62 | 6.894e-03 |
| GO:BP | GO:0002757 | immune response-activating signaling pathway | 303 | 494 | 7.048e-03 |
| GO:BP | GO:1901654 | response to ketone | 148 | 229 | 7.058e-03 |
| GO:BP | GO:1901605 | alpha-amino acid metabolic process | 139 | 214 | 7.279e-03 |
| GO:BP | GO:0071887 | leukocyte apoptotic process | 81 | 118 | 7.287e-03 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 81 | 118 | 7.287e-03 |
| GO:BP | GO:0072132 | mesenchyme morphogenesis | 44 | 59 | 7.323e-03 |
| GO:BP | GO:0048678 | response to axon injury | 58 | 81 | 7.323e-03 |
| GO:BP | GO:0072088 | nephron epithelium morphogenesis | 58 | 81 | 7.323e-03 |
| GO:BP | GO:1904018 | positive regulation of vasculature development | 112 | 169 | 7.360e-03 |
| GO:BP | GO:0001894 | tissue homeostasis | 171 | 268 | 7.376e-03 |
| GO:BP | GO:0048857 | neural nucleus development | 51 | 70 | 7.570e-03 |
| GO:BP | GO:0075522 | IRES-dependent viral translational initiation | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0090161 | Golgi ribbon formation | 11 | 11 | 7.623e-03 |
| GO:BP | GO:2000178 | negative regulation of neural precursor cell proliferation | 22 | 26 | 7.623e-03 |
| GO:BP | GO:2000050 | regulation of non-canonical Wnt signaling pathway | 22 | 26 | 7.623e-03 |
| GO:BP | GO:0070213 | protein auto-ADP-ribosylation | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0036490 | regulation of translation in response to endoplasmic reticulum stress | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0044829 | positive regulation by host of viral genome replication | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0019081 | viral translation | 22 | 26 | 7.623e-03 |
| GO:BP | GO:1905208 | negative regulation of cardiocyte differentiation | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0051469 | vesicle fusion with vacuole | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0007549 | sex-chromosome dosage compensation | 22 | 26 | 7.623e-03 |
| GO:BP | GO:0072110 | glomerular mesangial cell proliferation | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0070198 | protein localization to chromosome, telomeric region | 22 | 26 | 7.623e-03 |
| GO:BP | GO:1903564 | regulation of protein localization to cilium | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0071028 | nuclear mRNA surveillance | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0010613 | positive regulation of cardiac muscle hypertrophy | 22 | 26 | 7.623e-03 |
| GO:BP | GO:0000338 | protein deneddylation | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0051036 | regulation of endosome size | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0006983 | ER overload response | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0062028 | regulation of stress granule assembly | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0006307 | DNA alkylation repair | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0061763 | multivesicular body-lysosome fusion | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0051875 | pigment granule localization | 22 | 26 | 7.623e-03 |
| GO:BP | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane | 11 | 11 | 7.623e-03 |
| GO:BP | GO:1902547 | regulation of cellular response to vascular endothelial growth factor stimulus | 22 | 26 | 7.623e-03 |
| GO:BP | GO:0071044 | histone mRNA catabolic process | 11 | 11 | 7.623e-03 |
| GO:BP | GO:1903059 | regulation of protein lipidation | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0016479 | negative regulation of transcription by RNA polymerase I | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0022038 | corpus callosum development | 22 | 26 | 7.623e-03 |
| GO:BP | GO:0006686 | sphingomyelin biosynthetic process | 11 | 11 | 7.623e-03 |
| GO:BP | GO:0046324 | regulation of D-glucose import | 42 | 56 | 7.640e-03 |
| GO:BP | GO:0002208 | somatic diversification of immunoglobulins involved in immune response | 42 | 56 | 7.640e-03 |
| GO:BP | GO:0062237 | protein localization to postsynapse | 42 | 56 | 7.640e-03 |
| GO:BP | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 42 | 56 | 7.640e-03 |
| GO:BP | GO:0002204 | somatic recombination of immunoglobulin genes involved in immune response | 42 | 56 | 7.640e-03 |
| GO:BP | GO:0045190 | isotype switching | 42 | 56 | 7.640e-03 |
| GO:BP | GO:0021761 | limbic system development | 87 | 128 | 7.752e-03 |
| GO:BP | GO:0072080 | nephron tubule development | 71 | 102 | 7.781e-03 |
| GO:BP | GO:0097194 | execution phase of apoptosis | 56 | 78 | 7.840e-03 |
| GO:BP | GO:0006874 | intracellular calcium ion homeostasis | 192 | 304 | 7.854e-03 |
| GO:BP | GO:0098696 | regulation of neurotransmitter receptor localization to postsynaptic specialization membrane | 17 | 19 | 7.972e-03 |
| GO:BP | GO:0006491 | N-glycan processing | 17 | 19 | 7.972e-03 |
| GO:BP | GO:0010558 | negative regulation of macromolecule biosynthetic process | 1570 | 2745 | 7.972e-03 |
| GO:BP | GO:0097320 | plasma membrane tubulation | 17 | 19 | 7.972e-03 |
| GO:BP | GO:1903830 | magnesium ion transmembrane transport | 17 | 19 | 7.972e-03 |
| GO:BP | GO:0031053 | primary miRNA processing | 17 | 19 | 7.972e-03 |
| GO:BP | GO:0032253 | dense core granule localization | 17 | 19 | 7.972e-03 |
| GO:BP | GO:0090201 | negative regulation of release of cytochrome c from mitochondria | 17 | 19 | 7.972e-03 |
| GO:BP | GO:0061158 | 3’-UTR-mediated mRNA destabilization | 17 | 19 | 7.972e-03 |
| GO:BP | GO:0003215 | cardiac right ventricle morphogenesis | 17 | 19 | 7.972e-03 |
| GO:BP | GO:0034244 | negative regulation of transcription elongation by RNA polymerase II | 17 | 19 | 7.972e-03 |
| GO:BP | GO:0035372 | protein localization to microtubule | 17 | 19 | 7.972e-03 |
| GO:BP | GO:0060039 | pericardium development | 17 | 19 | 7.972e-03 |
| GO:BP | GO:0001947 | heart looping | 49 | 67 | 7.982e-03 |
| GO:BP | GO:0070050 | neuron cellular homeostasis | 49 | 67 | 7.982e-03 |
| GO:BP | GO:0007584 | response to nutrient | 113 | 171 | 7.991e-03 |
| GO:BP | GO:0030834 | regulation of actin filament depolymerization | 40 | 53 | 7.993e-03 |
| GO:BP | GO:0086009 | membrane repolarization | 40 | 53 | 7.993e-03 |
| GO:BP | GO:0034250 | positive regulation of amide metabolic process | 24 | 29 | 8.363e-03 |
| GO:BP | GO:0099054 | presynapse assembly | 38 | 50 | 8.363e-03 |
| GO:BP | GO:0060765 | regulation of androgen receptor signaling pathway | 24 | 29 | 8.363e-03 |
| GO:BP | GO:0045668 | negative regulation of osteoblast differentiation | 38 | 50 | 8.363e-03 |
| GO:BP | GO:0014741 | negative regulation of muscle hypertrophy | 24 | 29 | 8.363e-03 |
| GO:BP | GO:0055023 | positive regulation of cardiac muscle tissue growth | 24 | 29 | 8.363e-03 |
| GO:BP | GO:0043392 | negative regulation of DNA binding | 24 | 29 | 8.363e-03 |
| GO:BP | GO:0060964 | regulation of miRNA-mediated gene silencing | 24 | 29 | 8.363e-03 |
| GO:BP | GO:1903036 | positive regulation of response to wounding | 54 | 75 | 8.436e-03 |
| GO:BP | GO:0009799 | specification of symmetry | 99 | 148 | 8.440e-03 |
| GO:BP | GO:0009855 | determination of bilateral symmetry | 99 | 148 | 8.440e-03 |
| GO:BP | GO:0051216 | cartilage development | 138 | 213 | 8.493e-03 |
| GO:BP | GO:0032526 | response to retinoic acid | 77 | 112 | 8.601e-03 |
| GO:BP | GO:0008306 | associative learning | 64 | 91 | 8.621e-03 |
| GO:BP | GO:0033627 | cell adhesion mediated by integrin | 59 | 83 | 8.626e-03 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 167 | 262 | 8.638e-03 |
| GO:BP | GO:0006308 | DNA catabolic process | 26 | 32 | 8.848e-03 |
| GO:BP | GO:0016558 | protein import into peroxisome matrix | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0043248 | proteasome assembly | 14 | 15 | 8.848e-03 |
| GO:BP | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0006620 | post-translational protein targeting to endoplasmic reticulum membrane | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0018231 | peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0106057 | negative regulation of calcineurin-mediated signaling | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0018230 | peptidyl-L-cysteine S-palmitoylation | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0070986 | left/right axis specification | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0086069 | bundle of His cell to Purkinje myocyte communication | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0061684 | chaperone-mediated autophagy | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0140042 | lipid droplet formation | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0071501 | cellular response to sterol depletion | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0061635 | regulation of protein complex stability | 14 | 15 | 8.848e-03 |
| GO:BP | GO:0001936 | regulation of endothelial cell proliferation | 91 | 135 | 8.891e-03 |
| GO:BP | GO:0009126 | purine nucleoside monophosphate metabolic process | 34 | 44 | 8.915e-03 |
| GO:BP | GO:0061035 | regulation of cartilage development | 52 | 72 | 9.072e-03 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 32 | 41 | 9.072e-03 |
| GO:BP | GO:1904385 | cellular response to angiotensin | 28 | 35 | 9.072e-03 |
| GO:BP | GO:1900744 | regulation of p38MAPK cascade | 32 | 41 | 9.072e-03 |
| GO:BP | GO:0045746 | negative regulation of Notch signaling pathway | 32 | 41 | 9.072e-03 |
| GO:BP | GO:0070302 | regulation of stress-activated protein kinase signaling cascade | 32 | 41 | 9.072e-03 |
| GO:BP | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 28 | 35 | 9.072e-03 |
| GO:BP | GO:0061180 | mammary gland epithelium development | 52 | 72 | 9.072e-03 |
| GO:BP | GO:0046320 | regulation of fatty acid oxidation | 28 | 35 | 9.072e-03 |
| GO:BP | GO:0035904 | aorta development | 45 | 61 | 9.072e-03 |
| GO:BP | GO:0035337 | fatty-acyl-CoA metabolic process | 32 | 41 | 9.072e-03 |
| GO:BP | GO:0060043 | regulation of cardiac muscle cell proliferation | 30 | 38 | 9.137e-03 |
| GO:BP | GO:0046473 | phosphatidic acid metabolic process | 30 | 38 | 9.137e-03 |
| GO:BP | GO:0021955 | central nervous system neuron axonogenesis | 30 | 38 | 9.137e-03 |
| GO:BP | GO:1903532 | positive regulation of secretion by cell | 188 | 298 | 9.187e-03 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 219 | 351 | 9.230e-03 |
| GO:BP | GO:0021549 | cerebellum development | 75 | 109 | 9.354e-03 |
| GO:BP | GO:0015833 | peptide transport | 182 | 288 | 9.642e-03 |
| GO:BP | GO:0007632 | visual behavior | 43 | 58 | 9.675e-03 |
| GO:BP | GO:0070918 | regulatory ncRNA processing | 50 | 69 | 9.778e-03 |
| GO:BP | GO:0052547 | regulation of peptidase activity | 50 | 69 | 9.778e-03 |
| GO:BP | GO:0045947 | negative regulation of translational initiation | 19 | 22 | 9.924e-03 |
| GO:BP | GO:0036120 | cellular response to platelet-derived growth factor stimulus | 19 | 22 | 9.924e-03 |
| GO:BP | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity | 19 | 22 | 9.924e-03 |
| GO:BP | GO:0032482 | Rab protein signal transduction | 19 | 22 | 9.924e-03 |
| GO:BP | GO:0032252 | secretory granule localization | 19 | 22 | 9.924e-03 |
| GO:BP | GO:0035024 | negative regulation of Rho protein signal transduction | 19 | 22 | 9.924e-03 |
| GO:BP | GO:0032402 | melanosome transport | 19 | 22 | 9.924e-03 |
| GO:BP | GO:0032211 | negative regulation of telomere maintenance via telomerase | 19 | 22 | 9.924e-03 |
| GO:BP | GO:0048593 | camera-type eye morphogenesis | 92 | 137 | 9.937e-03 |
| GO:BP | GO:0022037 | metencephalon development | 81 | 119 | 1.012e-02 |
| GO:BP | GO:1904019 | epithelial cell apoptotic process | 60 | 85 | 1.018e-02 |
| GO:BP | GO:1903539 | protein localization to postsynaptic membrane | 41 | 55 | 1.026e-02 |
| GO:BP | GO:0006778 | porphyrin-containing compound metabolic process | 41 | 55 | 1.026e-02 |
| GO:BP | GO:0072009 | nephron epithelium development | 84 | 124 | 1.044e-02 |
| GO:BP | GO:0006694 | steroid biosynthetic process | 110 | 167 | 1.054e-02 |
| GO:BP | GO:1903169 | regulation of calcium ion transmembrane transport | 110 | 167 | 1.054e-02 |
| GO:BP | GO:0048592 | eye morphogenesis | 113 | 172 | 1.054e-02 |
| GO:BP | GO:0007052 | mitotic spindle organization | 90 | 134 | 1.096e-02 |
| GO:BP | GO:0050878 | regulation of body fluid levels | 233 | 376 | 1.109e-02 |
| GO:BP | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 21 | 25 | 1.142e-02 |
| GO:BP | GO:1902455 | negative regulation of stem cell population maintenance | 21 | 25 | 1.142e-02 |
| GO:BP | GO:2000209 | regulation of anoikis | 21 | 25 | 1.142e-02 |
| GO:BP | GO:0097150 | neuronal stem cell population maintenance | 21 | 25 | 1.142e-02 |
| GO:BP | GO:0060045 | positive regulation of cardiac muscle cell proliferation | 21 | 25 | 1.142e-02 |
| GO:BP | GO:0046580 | negative regulation of Ras protein signal transduction | 21 | 25 | 1.142e-02 |
| GO:BP | GO:0001919 | regulation of receptor recycling | 21 | 25 | 1.142e-02 |
| GO:BP | GO:0048169 | regulation of long-term neuronal synaptic plasticity | 21 | 25 | 1.142e-02 |
| GO:BP | GO:0062208 | positive regulation of pattern recognition receptor signaling pathway | 51 | 71 | 1.196e-02 |
| GO:BP | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | 35 | 46 | 1.203e-02 |
| GO:BP | GO:0071548 | response to dexamethasone | 35 | 46 | 1.203e-02 |
| GO:BP | GO:0060761 | negative regulation of response to cytokine stimulus | 61 | 87 | 1.205e-02 |
| GO:BP | GO:1901019 | regulation of calcium ion transmembrane transporter activity | 44 | 60 | 1.221e-02 |
| GO:BP | GO:0002381 | immunoglobulin production involved in immunoglobulin-mediated immune response | 44 | 60 | 1.221e-02 |
| GO:BP | GO:0043113 | receptor clustering | 44 | 60 | 1.221e-02 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 91 | 136 | 1.228e-02 |
| GO:BP | GO:0071902 | positive regulation of protein serine/threonine kinase activity | 91 | 136 | 1.228e-02 |
| GO:BP | GO:0046037 | GMP metabolic process | 23 | 28 | 1.237e-02 |
| GO:BP | GO:0001573 | ganglioside metabolic process | 23 | 28 | 1.237e-02 |
| GO:BP | GO:0010962 | regulation of glucan biosynthetic process | 23 | 28 | 1.237e-02 |
| GO:BP | GO:0005979 | regulation of glycogen biosynthetic process | 23 | 28 | 1.237e-02 |
| GO:BP | GO:0071549 | cellular response to dexamethasone stimulus | 23 | 28 | 1.237e-02 |
| GO:BP | GO:0039529 | RIG-I signaling pathway | 23 | 28 | 1.237e-02 |
| GO:BP | GO:1902894 | negative regulation of miRNA transcription | 23 | 28 | 1.237e-02 |
| GO:BP | GO:1900745 | positive regulation of p38MAPK cascade | 23 | 28 | 1.237e-02 |
| GO:BP | GO:2001259 | positive regulation of cation channel activity | 33 | 43 | 1.246e-02 |
| GO:BP | GO:0007595 | lactation | 33 | 43 | 1.246e-02 |
| GO:BP | GO:0051897 | positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction | 121 | 186 | 1.251e-02 |
| GO:BP | GO:0044539 | long-chain fatty acid import into cell | 16 | 18 | 1.257e-02 |
| GO:BP | GO:0061718 | glucose catabolic process to pyruvate | 16 | 18 | 1.257e-02 |
| GO:BP | GO:0035973 | aggrephagy | 16 | 18 | 1.257e-02 |
| GO:BP | GO:0071816 | tail-anchored membrane protein insertion into ER membrane | 16 | 18 | 1.257e-02 |
| GO:BP | GO:0010042 | response to manganese ion | 16 | 18 | 1.257e-02 |
| GO:BP | GO:0009208 | pyrimidine ribonucleoside triphosphate metabolic process | 16 | 18 | 1.257e-02 |
| GO:BP | GO:0061621 | canonical glycolysis | 16 | 18 | 1.257e-02 |
| GO:BP | GO:1900037 | regulation of cellular response to hypoxia | 16 | 18 | 1.257e-02 |
| GO:BP | GO:0060977 | coronary vasculature morphogenesis | 16 | 18 | 1.257e-02 |
| GO:BP | GO:0002573 | myeloid leukocyte differentiation | 153 | 240 | 1.266e-02 |
| GO:BP | GO:0045766 | positive regulation of angiogenesis | 109 | 166 | 1.269e-02 |
| GO:BP | GO:0090199 | regulation of release of cytochrome c from mitochondria | 31 | 40 | 1.278e-02 |
| GO:BP | GO:0014911 | positive regulation of smooth muscle cell migration | 31 | 40 | 1.278e-02 |
| GO:BP | GO:0043407 | negative regulation of MAP kinase activity | 31 | 40 | 1.278e-02 |
| GO:BP | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation | 31 | 40 | 1.278e-02 |
| GO:BP | GO:0097107 | postsynaptic density assembly | 25 | 31 | 1.284e-02 |
| GO:BP | GO:0061440 | kidney vasculature development | 25 | 31 | 1.284e-02 |
| GO:BP | GO:0061437 | renal system vasculature development | 25 | 31 | 1.284e-02 |
| GO:BP | GO:1904814 | regulation of protein localization to chromosome, telomeric region | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0034635 | glutathione transport | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0006625 | protein targeting to peroxisome | 10 | 10 | 1.289e-02 |
| GO:BP | GO:1902966 | positive regulation of protein localization to early endosome | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0098884 | postsynaptic neurotransmitter receptor internalization | 27 | 34 | 1.289e-02 |
| GO:BP | GO:0071166 | ribonucleoprotein complex localization | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0061626 | pharyngeal arch artery morphogenesis | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0070922 | RISC complex assembly | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0071425 | hematopoietic stem cell proliferation | 27 | 34 | 1.289e-02 |
| GO:BP | GO:0101024 | mitotic nuclear membrane organization | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0070831 | basement membrane assembly | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0015820 | L-leucine transport | 10 | 10 | 1.289e-02 |
| GO:BP | GO:1902965 | regulation of protein localization to early endosome | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0180012 | co-transcriptional RNA 3’-end processing, cleavage and polyadenylation pathway | 10 | 10 | 1.289e-02 |
| GO:BP | GO:2000234 | positive regulation of rRNA processing | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0034332 | adherens junction organization | 42 | 57 | 1.289e-02 |
| GO:BP | GO:0019673 | GDP-mannose metabolic process | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0032988 | protein-RNA complex disassembly | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0072124 | regulation of glomerular mesangial cell proliferation | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0010464 | regulation of mesenchymal cell proliferation | 27 | 34 | 1.289e-02 |
| GO:BP | GO:0007084 | mitotic nuclear membrane reassembly | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0000492 | box C/D snoRNP assembly | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0051409 | response to nitrosative stress | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0140239 | postsynaptic endocytosis | 27 | 34 | 1.289e-02 |
| GO:BP | GO:1904896 | ESCRT complex disassembly | 10 | 10 | 1.289e-02 |
| GO:BP | GO:1902513 | regulation of organelle transport along microtubule | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0072089 | stem cell proliferation | 83 | 123 | 1.289e-02 |
| GO:BP | GO:0031125 | rRNA 3’-end processing | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis | 10 | 10 | 1.289e-02 |
| GO:BP | GO:1904903 | ESCRT III complex disassembly | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0006084 | acetyl-CoA metabolic process | 27 | 34 | 1.289e-02 |
| GO:BP | GO:0010935 | regulation of macrophage cytokine production | 29 | 37 | 1.289e-02 |
| GO:BP | GO:0048194 | Golgi vesicle budding | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0051292 | nuclear pore complex assembly | 10 | 10 | 1.289e-02 |
| GO:BP | GO:1903242 | regulation of cardiac muscle hypertrophy in response to stress | 10 | 10 | 1.289e-02 |
| GO:BP | GO:2000766 | negative regulation of cytoplasmic translation | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0010612 | regulation of cardiac muscle adaptation | 10 | 10 | 1.289e-02 |
| GO:BP | GO:2000756 | regulation of peptidyl-lysine acetylation | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0015937 | coenzyme A biosynthetic process | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0006283 | transcription-coupled nucleotide-excision repair | 10 | 10 | 1.289e-02 |
| GO:BP | GO:1901838 | positive regulation of transcription of nucleolar large rRNA by RNA polymerase I | 10 | 10 | 1.289e-02 |
| GO:BP | GO:0034142 | toll-like receptor 4 signaling pathway | 42 | 57 | 1.289e-02 |
| GO:BP | GO:0010934 | macrophage cytokine production | 29 | 37 | 1.289e-02 |
| GO:BP | GO:0051047 | positive regulation of secretion | 201 | 322 | 1.292e-02 |
| GO:BP | GO:0050796 | regulation of insulin secretion | 116 | 178 | 1.357e-02 |
| GO:BP | GO:0048260 | positive regulation of receptor-mediated endocytosis | 40 | 54 | 1.366e-02 |
| GO:BP | GO:0045599 | negative regulation of fat cell differentiation | 40 | 54 | 1.366e-02 |
| GO:BP | GO:0072348 | sulfur compound transport | 47 | 65 | 1.369e-02 |
| GO:BP | GO:1990173 | protein localization to nucleoplasm | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0051127 | positive regulation of actin nucleation | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0019852 | L-ascorbic acid metabolic process | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0001921 | positive regulation of receptor recycling | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0051924 | regulation of calcium ion transport | 155 | 244 | 1.428e-02 |
| GO:BP | GO:0051775 | response to redox state | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0061430 | bone trabecula morphogenesis | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0015780 | nucleotide-sugar transmembrane transport | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0010763 | positive regulation of fibroblast migration | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0042983 | amyloid precursor protein biosynthetic process | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0015803 | branched-chain amino acid transport | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0042984 | regulation of amyloid precursor protein biosynthetic process | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0097468 | programmed cell death in response to reactive oxygen species | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0048388 | endosomal lumen acidification | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0030497 | fatty acid elongation | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0060816 | random inactivation of X chromosome | 13 | 14 | 1.428e-02 |
| GO:BP | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0043144 | sno(s)RNA processing | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0010635 | regulation of mitochondrial fusion | 13 | 14 | 1.428e-02 |
| GO:BP | GO:0008542 | visual learning | 38 | 51 | 1.448e-02 |
| GO:BP | GO:0048013 | ephrin receptor signaling pathway | 38 | 51 | 1.448e-02 |
| GO:BP | GO:0030168 | platelet activation | 93 | 140 | 1.483e-02 |
| GO:BP | GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 102 | 155 | 1.496e-02 |
| GO:BP | GO:0060216 | definitive hemopoiesis | 18 | 21 | 1.496e-02 |
| GO:BP | GO:1902284 | neuron projection extension involved in neuron projection guidance | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0010566 | regulation of ketone biosynthetic process | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0071322 | cellular response to carbohydrate stimulus | 108 | 165 | 1.496e-02 |
| GO:BP | GO:0036498 | IRE1-mediated unfolded protein response | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0140354 | lipid import into cell | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0072111 | cell proliferation involved in kidney development | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0086012 | membrane depolarization during cardiac muscle cell action potential | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0098760 | response to interleukin-7 | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0016556 | mRNA modification | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0097734 | extracellular exosome biogenesis | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0097062 | dendritic spine maintenance | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0009067 | aspartate family amino acid biosynthetic process | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0140896 | cGAS/STING signaling pathway | 18 | 21 | 1.496e-02 |
| GO:BP | GO:2001135 | regulation of endocytic recycling | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0048846 | axon extension involved in axon guidance | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0046426 | negative regulation of receptor signaling pathway via JAK-STAT | 18 | 21 | 1.496e-02 |
| GO:BP | GO:0034764 | positive regulation of transmembrane transport | 130 | 202 | 1.520e-02 |
| GO:BP | GO:0071277 | cellular response to calcium ion | 55 | 78 | 1.524e-02 |
| GO:BP | GO:0021575 | hindbrain morphogenesis | 36 | 48 | 1.528e-02 |
| GO:BP | GO:0018212 | peptidyl-tyrosine modification | 153 | 241 | 1.549e-02 |
| GO:BP | GO:0002683 | negative regulation of immune system process | 312 | 515 | 1.553e-02 |
| GO:BP | GO:0015807 | L-amino acid transport | 63 | 91 | 1.590e-02 |
| GO:BP | GO:0061458 | reproductive system development | 198 | 318 | 1.604e-02 |
| GO:BP | GO:0097237 | cellular response to toxic substance | 88 | 132 | 1.609e-02 |
| GO:BP | GO:0021587 | cerebellum morphogenesis | 34 | 45 | 1.613e-02 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 34 | 45 | 1.613e-02 |
| GO:BP | GO:0007596 | blood coagulation | 147 | 231 | 1.614e-02 |
| GO:BP | GO:0001508 | action potential | 112 | 172 | 1.623e-02 |
| GO:BP | GO:0009048 | dosage compensation by inactivation of X chromosome | 20 | 24 | 1.677e-02 |
| GO:BP | GO:1904889 | regulation of excitatory synapse assembly | 20 | 24 | 1.677e-02 |
| GO:BP | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway | 20 | 24 | 1.677e-02 |
| GO:BP | GO:0008334 | histone mRNA metabolic process | 20 | 24 | 1.677e-02 |
| GO:BP | GO:0061615 | glycolytic process through fructose-6-phosphate | 20 | 24 | 1.677e-02 |
| GO:BP | GO:0061484 | hematopoietic stem cell homeostasis | 20 | 24 | 1.677e-02 |
| GO:BP | GO:0051905 | establishment of pigment granule localization | 20 | 24 | 1.677e-02 |
| GO:BP | GO:0070296 | sarcoplasmic reticulum calcium ion transport | 32 | 42 | 1.688e-02 |
| GO:BP | GO:0032728 | positive regulation of interferon-beta production | 32 | 42 | 1.688e-02 |
| GO:BP | GO:1990776 | response to angiotensin | 32 | 42 | 1.688e-02 |
| GO:BP | GO:0007599 | hemostasis | 151 | 238 | 1.690e-02 |
| GO:BP | GO:0045191 | regulation of isotype switching | 30 | 39 | 1.754e-02 |
| GO:BP | GO:0010591 | regulation of lamellipodium assembly | 30 | 39 | 1.754e-02 |
| GO:BP | GO:0070884 | regulation of calcineurin-NFAT signaling cascade | 30 | 39 | 1.754e-02 |
| GO:BP | GO:1905476 | negative regulation of protein localization to membrane | 30 | 39 | 1.754e-02 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 30 | 39 | 1.754e-02 |
| GO:BP | GO:0099622 | cardiac muscle cell membrane repolarization | 30 | 39 | 1.754e-02 |
| GO:BP | GO:0032872 | regulation of stress-activated MAPK cascade | 30 | 39 | 1.754e-02 |
| GO:BP | GO:0014742 | positive regulation of muscle hypertrophy | 22 | 27 | 1.781e-02 |
| GO:BP | GO:0060343 | trabecula formation | 22 | 27 | 1.781e-02 |
| GO:BP | GO:0044827 | modulation by host of viral genome replication | 22 | 27 | 1.781e-02 |
| GO:BP | GO:0032878 | regulation of establishment or maintenance of cell polarity | 22 | 27 | 1.781e-02 |
| GO:BP | GO:0006638 | neutral lipid metabolic process | 89 | 134 | 1.783e-02 |
| GO:BP | GO:0051208 | sequestering of calcium ion | 89 | 134 | 1.783e-02 |
| GO:BP | GO:0097484 | dendrite extension | 28 | 36 | 1.803e-02 |
| GO:BP | GO:0060218 | hematopoietic stem cell differentiation | 28 | 36 | 1.803e-02 |
| GO:BP | GO:0017145 | stem cell division | 28 | 36 | 1.803e-02 |
| GO:BP | GO:0055074 | calcium ion homeostasis | 204 | 329 | 1.821e-02 |
| GO:BP | GO:2001222 | regulation of neuron migration | 39 | 53 | 1.825e-02 |
| GO:BP | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | 26 | 33 | 1.826e-02 |
| GO:BP | GO:0051642 | centrosome localization | 26 | 33 | 1.826e-02 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 26 | 33 | 1.826e-02 |
| GO:BP | GO:0061384 | heart trabecula morphogenesis | 26 | 33 | 1.826e-02 |
| GO:BP | GO:0015012 | heparan sulfate proteoglycan biosynthetic process | 26 | 33 | 1.826e-02 |
| GO:BP | GO:0001736 | establishment of planar polarity | 26 | 33 | 1.826e-02 |
| GO:BP | GO:0045648 | positive regulation of erythrocyte differentiation | 24 | 30 | 1.826e-02 |
| GO:BP | GO:0007164 | establishment of tissue polarity | 26 | 33 | 1.826e-02 |
| GO:BP | GO:1902807 | negative regulation of cell cycle G1/S phase transition | 44 | 61 | 1.939e-02 |
| GO:BP | GO:0032102 | negative regulation of response to external stimulus | 238 | 388 | 1.939e-02 |
| GO:BP | GO:0090083 | regulation of inclusion body assembly | 15 | 17 | 1.939e-02 |
| GO:BP | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity | 15 | 17 | 1.939e-02 |
| GO:BP | GO:0019184 | nonribosomal peptide biosynthetic process | 15 | 17 | 1.939e-02 |
| GO:BP | GO:1901096 | regulation of autophagosome maturation | 15 | 17 | 1.939e-02 |
| GO:BP | GO:0006878 | intracellular copper ion homeostasis | 15 | 17 | 1.939e-02 |
| GO:BP | GO:1903541 | regulation of exosomal secretion | 15 | 17 | 1.939e-02 |
| GO:BP | GO:0033151 | V(D)J recombination | 15 | 17 | 1.939e-02 |
| GO:BP | GO:0043217 | myelin maintenance | 15 | 17 | 1.939e-02 |
| GO:BP | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 15 | 17 | 1.939e-02 |
| GO:BP | GO:0071895 | odontoblast differentiation | 15 | 17 | 1.939e-02 |
| GO:BP | GO:0018126 | protein hydroxylation | 15 | 17 | 1.939e-02 |
| GO:BP | GO:0099638 | endosome to plasma membrane protein transport | 15 | 17 | 1.939e-02 |
| GO:BP | GO:0046365 | monosaccharide catabolic process | 37 | 50 | 1.946e-02 |
| GO:BP | GO:0014902 | myotube differentiation | 87 | 131 | 1.954e-02 |
| GO:BP | GO:1901657 | glycosyl compound metabolic process | 54 | 77 | 1.954e-02 |
| GO:BP | GO:0033363 | secretory granule organization | 49 | 69 | 1.970e-02 |
| GO:BP | GO:0048608 | reproductive structure development | 195 | 314 | 1.979e-02 |
| GO:BP | GO:0045055 | regulated exocytosis | 147 | 232 | 2.007e-02 |
| GO:BP | GO:0170039 | proteinogenic amino acid metabolic process | 115 | 178 | 2.077e-02 |
| GO:BP | GO:0006509 | membrane protein ectodomain proteolysis | 35 | 47 | 2.079e-02 |
| GO:BP | GO:0060711 | labyrinthine layer development | 35 | 47 | 2.079e-02 |
| GO:BP | GO:0002521 | leukocyte differentiation | 380 | 636 | 2.085e-02 |
| GO:BP | GO:0048168 | regulation of neuronal synaptic plasticity | 42 | 58 | 2.094e-02 |
| GO:BP | GO:0046148 | pigment biosynthetic process | 42 | 58 | 2.094e-02 |
| GO:BP | GO:0007389 | pattern specification process | 289 | 477 | 2.094e-02 |
| GO:BP | GO:0018108 | peptidyl-tyrosine phosphorylation | 151 | 239 | 2.094e-02 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 128 | 200 | 2.109e-02 |
| GO:BP | GO:0046660 | female sex differentiation | 82 | 123 | 2.132e-02 |
| GO:BP | GO:0014855 | striated muscle cell proliferation | 52 | 74 | 2.141e-02 |
| GO:BP | GO:0021885 | forebrain cell migration | 47 | 66 | 2.150e-02 |
| GO:BP | GO:0030282 | bone mineralization | 85 | 128 | 2.152e-02 |
| GO:BP | GO:0071331 | cellular response to hexose stimulus | 100 | 153 | 2.164e-02 |
| GO:BP | GO:0006639 | acylglycerol metabolic process | 88 | 133 | 2.165e-02 |
| GO:BP | GO:0032881 | regulation of polysaccharide metabolic process | 33 | 44 | 2.204e-02 |
| GO:BP | GO:1903170 | negative regulation of calcium ion transmembrane transport | 33 | 44 | 2.204e-02 |
| GO:BP | GO:0048285 | organelle fission | 302 | 500 | 2.204e-02 |
| GO:BP | GO:0070900 | mitochondrial tRNA modification | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0035520 | monoubiquitinated protein deubiquitination | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0140673 | transcription elongation-coupled chromatin remodeling | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0007021 | tubulin complex assembly | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0003162 | atrioventricular node development | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0062176 | R-loop processing | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0036500 | ATF6-mediated unfolded protein response | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein | 9 | 9 | 2.213e-02 |
| GO:BP | GO:1904153 | negative regulation of retrograde protein transport, ER to cytosol | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0042796 | snRNA transcription by RNA polymerase III | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0000467 | exonucleolytic trimming to generate mature 3’-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0031440 | regulation of mRNA 3’-end processing | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0034214 | protein hexamerization | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0090522 | vesicle tethering involved in exocytosis | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0006449 | regulation of translational termination | 9 | 9 | 2.213e-02 |
| GO:BP | GO:2000197 | regulation of ribonucleoprotein complex localization | 9 | 9 | 2.213e-02 |
| GO:BP | GO:2000169 | regulation of peptidyl-cysteine S-nitrosylation | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0097680 | double-strand break repair via classical nonhomologous end joining | 9 | 9 | 2.213e-02 |
| GO:BP | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0030200 | heparan sulfate proteoglycan catabolic process | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0000301 | retrograde transport, vesicle recycling within Golgi | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0180010 | co-transcriptional mRNA 3’-end processing, cleavage and polyadenylation pathway | 9 | 9 | 2.213e-02 |
| GO:BP | GO:1990481 | mRNA pseudouridine synthesis | 9 | 9 | 2.213e-02 |
| GO:BP | GO:1903332 | regulation of protein folding | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0160063 | multi-pass transmembrane protein insertion into ER membrane | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0016926 | protein desumoylation | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0031946 | regulation of glucocorticoid biosynthetic process | 9 | 9 | 2.213e-02 |
| GO:BP | GO:1900864 | mitochondrial RNA modification | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0048034 | heme O biosynthetic process | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0090119 | vesicle-mediated cholesterol transport | 9 | 9 | 2.213e-02 |
| GO:BP | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0048033 | heme O metabolic process | 9 | 9 | 2.213e-02 |
| GO:BP | GO:1904874 | positive regulation of telomerase RNA localization to Cajal body | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0034085 | establishment of sister chromatid cohesion | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0110076 | negative regulation of ferroptosis | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0010998 | regulation of translational initiation by eIF2 alpha phosphorylation | 9 | 9 | 2.213e-02 |
| GO:BP | GO:0048771 | tissue remodeling | 116 | 180 | 2.215e-02 |
| GO:BP | GO:0032410 | negative regulation of transporter activity | 40 | 55 | 2.232e-02 |
| GO:BP | GO:0006220 | pyrimidine nucleotide metabolic process | 40 | 55 | 2.232e-02 |
| GO:BP | GO:0070555 | response to interleukin-1 | 74 | 110 | 2.245e-02 |
| GO:BP | GO:2001044 | regulation of integrin-mediated signaling pathway | 17 | 20 | 2.259e-02 |
| GO:BP | GO:0046827 | positive regulation of protein export from nucleus | 17 | 20 | 2.259e-02 |
| GO:BP | GO:0098761 | cellular response to interleukin-7 | 17 | 20 | 2.259e-02 |
| GO:BP | GO:0015693 | magnesium ion transport | 17 | 20 | 2.259e-02 |
| GO:BP | GO:2001224 | positive regulation of neuron migration | 17 | 20 | 2.259e-02 |
| GO:BP | GO:0060973 | cell migration involved in heart development | 17 | 20 | 2.259e-02 |
| GO:BP | GO:0010421 | hydrogen peroxide-mediated programmed cell death | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0046349 | amino sugar biosynthetic process | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0045793 | positive regulation of cell size | 12 | 13 | 2.285e-02 |
| GO:BP | GO:1902902 | negative regulation of autophagosome assembly | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0140588 | chromatin looping | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0042761 | very long-chain fatty acid biosynthetic process | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0006054 | N-acetylneuraminate metabolic process | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0034498 | early endosome to Golgi transport | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0071233 | cellular response to L-leucine | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0006167 | AMP biosynthetic process | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0034982 | mitochondrial protein processing | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0032486 | Rap protein signal transduction | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0031987 | locomotion involved in locomotory behavior | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0031571 | mitotic G1 DNA damage checkpoint signaling | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0071679 | commissural neuron axon guidance | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0006361 | transcription initiation at RNA polymerase I promoter | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0070142 | synaptic vesicle budding | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0098935 | dendritic transport | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0061097 | regulation of protein tyrosine kinase activity | 31 | 41 | 2.285e-02 |
| GO:BP | GO:0042559 | pteridine-containing compound biosynthetic process | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0038203 | TORC2 signaling | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0061668 | mitochondrial ribosome assembly | 12 | 13 | 2.285e-02 |
| GO:BP | GO:0022029 | telencephalon cell migration | 45 | 63 | 2.288e-02 |
| GO:BP | GO:0043406 | positive regulation of MAP kinase activity | 50 | 71 | 2.288e-02 |
| GO:BP | GO:0071326 | cellular response to monosaccharide stimulus | 101 | 155 | 2.311e-02 |
| GO:BP | GO:0050714 | positive regulation of protein secretion | 98 | 150 | 2.324e-02 |
| GO:BP | GO:0071333 | cellular response to glucose stimulus | 98 | 150 | 2.324e-02 |
| GO:BP | GO:0050817 | coagulation | 150 | 238 | 2.381e-02 |
| GO:BP | GO:0045880 | positive regulation of smoothened signaling pathway | 29 | 38 | 2.382e-02 |
| GO:BP | GO:0043243 | positive regulation of protein-containing complex disassembly | 29 | 38 | 2.382e-02 |
| GO:BP | GO:1904707 | positive regulation of vascular associated smooth muscle cell proliferation | 29 | 38 | 2.382e-02 |
| GO:BP | GO:0010799 | regulation of peptidyl-threonine phosphorylation | 29 | 38 | 2.382e-02 |
| GO:BP | GO:0021696 | cerebellar cortex morphogenesis | 29 | 38 | 2.382e-02 |
| GO:BP | GO:1900181 | negative regulation of protein localization to nucleus | 29 | 38 | 2.382e-02 |
| GO:BP | GO:0048536 | spleen development | 29 | 38 | 2.382e-02 |
| GO:BP | GO:0001836 | release of cytochrome c from mitochondria | 38 | 52 | 2.382e-02 |
| GO:BP | GO:0034109 | homotypic cell-cell adhesion | 69 | 102 | 2.390e-02 |
| GO:BP | GO:0031349 | positive regulation of defense response | 305 | 506 | 2.398e-02 |
| GO:BP | GO:0106118 | regulation of sterol biosynthetic process | 19 | 23 | 2.438e-02 |
| GO:BP | GO:0046823 | negative regulation of nucleocytoplasmic transport | 19 | 23 | 2.438e-02 |
| GO:BP | GO:1905874 | regulation of postsynaptic density organization | 19 | 23 | 2.438e-02 |
| GO:BP | GO:0051904 | pigment granule transport | 19 | 23 | 2.438e-02 |
| GO:BP | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway | 19 | 23 | 2.438e-02 |
| GO:BP | GO:0003177 | pulmonary valve development | 19 | 23 | 2.438e-02 |
| GO:BP | GO:0045540 | regulation of cholesterol biosynthetic process | 19 | 23 | 2.438e-02 |
| GO:BP | GO:0060392 | negative regulation of SMAD protein signal transduction | 19 | 23 | 2.438e-02 |
| GO:BP | GO:0038128 | ERBB2 signaling pathway | 19 | 23 | 2.438e-02 |
| GO:BP | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway | 19 | 23 | 2.438e-02 |
| GO:BP | GO:0060444 | branching involved in mammary gland duct morphogenesis | 19 | 23 | 2.438e-02 |
| GO:BP | GO:0010878 | cholesterol storage | 19 | 23 | 2.438e-02 |
| GO:BP | GO:0006081 | aldehyde metabolic process | 53 | 76 | 2.440e-02 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 53 | 76 | 2.440e-02 |
| GO:BP | GO:0000038 | very long-chain fatty acid metabolic process | 27 | 35 | 2.466e-02 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 27 | 35 | 2.466e-02 |
| GO:BP | GO:0002684 | positive regulation of immune system process | 641 | 1099 | 2.521e-02 |
| GO:BP | GO:0002026 | regulation of the force of heart contraction | 21 | 26 | 2.531e-02 |
| GO:BP | GO:1990748 | cellular detoxification | 81 | 122 | 2.531e-02 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 21 | 26 | 2.531e-02 |
| GO:BP | GO:0019985 | translesion synthesis | 21 | 26 | 2.531e-02 |
| GO:BP | GO:0046339 | diacylglycerol metabolic process | 21 | 26 | 2.531e-02 |
| GO:BP | GO:0071684 | organism emergence from protective structure | 21 | 26 | 2.531e-02 |
| GO:BP | GO:0035313 | wound healing, spreading of epidermal cells | 21 | 26 | 2.531e-02 |
| GO:BP | GO:0035188 | hatching | 21 | 26 | 2.531e-02 |
| GO:BP | GO:2000737 | negative regulation of stem cell differentiation | 21 | 26 | 2.531e-02 |
| GO:BP | GO:0071108 | protein K48-linked deubiquitination | 25 | 32 | 2.531e-02 |
| GO:BP | GO:0006007 | glucose catabolic process | 21 | 26 | 2.531e-02 |
| GO:BP | GO:0001835 | blastocyst hatching | 21 | 26 | 2.531e-02 |
| GO:BP | GO:0046716 | muscle cell cellular homeostasis | 21 | 26 | 2.531e-02 |
| GO:BP | GO:0061028 | establishment of endothelial barrier | 36 | 49 | 2.546e-02 |
| GO:BP | GO:0072012 | glomerulus vasculature development | 23 | 29 | 2.553e-02 |
| GO:BP | GO:0000272 | polysaccharide catabolic process | 23 | 29 | 2.553e-02 |
| GO:BP | GO:0021680 | cerebellar Purkinje cell layer development | 23 | 29 | 2.553e-02 |
| GO:BP | GO:0010165 | response to X-ray | 23 | 29 | 2.553e-02 |
| GO:BP | GO:0035767 | endothelial cell chemotaxis | 23 | 29 | 2.553e-02 |
| GO:BP | GO:0051279 | regulation of release of sequestered calcium ion into cytosol | 56 | 81 | 2.571e-02 |
| GO:BP | GO:0030166 | proteoglycan biosynthetic process | 51 | 73 | 2.671e-02 |
| GO:BP | GO:0032963 | collagen metabolic process | 70 | 104 | 2.677e-02 |
| GO:BP | GO:0006826 | iron ion transport | 41 | 57 | 2.684e-02 |
| GO:BP | GO:0051055 | negative regulation of lipid biosynthetic process | 41 | 57 | 2.684e-02 |
| GO:BP | GO:0098900 | regulation of action potential | 41 | 57 | 2.684e-02 |
| GO:BP | GO:0033173 | calcineurin-NFAT signaling cascade | 34 | 46 | 2.731e-02 |
| GO:BP | GO:1900271 | regulation of long-term synaptic potentiation | 34 | 46 | 2.731e-02 |
| GO:BP | GO:0019320 | hexose catabolic process | 34 | 46 | 2.731e-02 |
| GO:BP | GO:0003016 | respiratory system process | 34 | 46 | 2.731e-02 |
| GO:BP | GO:0009410 | response to xenobiotic stimulus | 266 | 439 | 2.749e-02 |
| GO:BP | GO:0001775 | cell activation | 659 | 1132 | 2.761e-02 |
| GO:BP | GO:0002790 | peptide secretion | 163 | 261 | 2.767e-02 |
| GO:BP | GO:0021953 | central nervous system neuron differentiation | 136 | 215 | 2.833e-02 |
| GO:BP | GO:0006816 | calcium ion transport | 263 | 434 | 2.844e-02 |
| GO:BP | GO:0070371 | ERK1 and ERK2 cascade | 185 | 299 | 2.887e-02 |
| GO:BP | GO:0043647 | inositol phosphate metabolic process | 32 | 43 | 2.925e-02 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 32 | 43 | 2.925e-02 |
| GO:BP | GO:0072171 | mesonephric tubule morphogenesis | 49 | 70 | 2.930e-02 |
| GO:BP | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus | 49 | 70 | 2.930e-02 |
| GO:BP | GO:0051302 | regulation of cell division | 120 | 188 | 2.942e-02 |
| GO:BP | GO:0120162 | positive regulation of cold-induced thermogenesis | 68 | 101 | 2.951e-02 |
| GO:BP | GO:0001732 | formation of cytoplasmic translation initiation complex | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0031054 | pre-miRNA processing | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0031269 | pseudopodium assembly | 14 | 16 | 2.954e-02 |
| GO:BP | GO:1902307 | positive regulation of sodium ion transmembrane transport | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0097009 | energy homeostasis | 57 | 83 | 2.954e-02 |
| GO:BP | GO:0046036 | CTP metabolic process | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0072109 | glomerular mesangium development | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0006825 | copper ion transport | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0010633 | negative regulation of epithelial cell migration | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0072350 | tricarboxylic acid metabolic process | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0016078 | tRNA decay | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0042795 | snRNA transcription by RNA polymerase II | 14 | 16 | 2.954e-02 |
| GO:BP | GO:1905906 | regulation of amyloid fibril formation | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0098877 | neurotransmitter receptor transport to plasma membrane | 14 | 16 | 2.954e-02 |
| GO:BP | GO:1904754 | positive regulation of vascular associated smooth muscle cell migration | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0043201 | response to L-leucine | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0001960 | negative regulation of cytokine-mediated signaling pathway | 57 | 83 | 2.954e-02 |
| GO:BP | GO:0030033 | microvillus assembly | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0044849 | estrous cycle | 14 | 16 | 2.954e-02 |
| GO:BP | GO:1901526 | positive regulation of mitophagy | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0046753 | non-lytic viral release | 14 | 16 | 2.954e-02 |
| GO:BP | GO:0045824 | negative regulation of innate immune response | 60 | 88 | 3.069e-02 |
| GO:BP | GO:0002832 | negative regulation of response to biotic stimulus | 80 | 121 | 3.070e-02 |
| GO:BP | GO:0032355 | response to estradiol | 80 | 121 | 3.070e-02 |
| GO:BP | GO:0098739 | import across plasma membrane | 124 | 195 | 3.077e-02 |
| GO:BP | GO:0106056 | regulation of calcineurin-mediated signaling | 30 | 40 | 3.092e-02 |
| GO:BP | GO:0072337 | modified amino acid transport | 30 | 40 | 3.092e-02 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 52 | 75 | 3.093e-02 |
| GO:BP | GO:0051048 | negative regulation of secretion | 108 | 168 | 3.134e-02 |
| GO:BP | GO:0031297 | replication fork processing | 37 | 51 | 3.142e-02 |
| GO:BP | GO:0035987 | endodermal cell differentiation | 37 | 51 | 3.142e-02 |
| GO:BP | GO:0001656 | metanephros development | 63 | 93 | 3.158e-02 |
| GO:BP | GO:0035019 | somatic stem cell population maintenance | 47 | 67 | 3.194e-02 |
| GO:BP | GO:0030514 | negative regulation of BMP signaling pathway | 42 | 59 | 3.222e-02 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 42 | 59 | 3.222e-02 |
| GO:BP | GO:0045921 | positive regulation of exocytosis | 55 | 80 | 3.249e-02 |
| GO:BP | GO:0030072 | peptide hormone secretion | 159 | 255 | 3.261e-02 |
| GO:BP | GO:1900225 | regulation of NLRP3 inflammasome complex assembly | 28 | 37 | 3.267e-02 |
| GO:BP | GO:0031076 | embryonic camera-type eye development | 28 | 37 | 3.267e-02 |
| GO:BP | GO:0010874 | regulation of cholesterol efflux | 28 | 37 | 3.267e-02 |
| GO:BP | GO:0051154 | negative regulation of striated muscle cell differentiation | 28 | 37 | 3.267e-02 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 28 | 37 | 3.267e-02 |
| GO:BP | GO:1903531 | negative regulation of secretion by cell | 96 | 148 | 3.296e-02 |
| GO:BP | GO:0046879 | hormone secretion | 199 | 324 | 3.333e-02 |
| GO:BP | GO:0003184 | pulmonary valve morphogenesis | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0061099 | negative regulation of protein tyrosine kinase activity | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process | 16 | 19 | 3.363e-02 |
| GO:BP | GO:1904030 | negative regulation of cyclin-dependent protein kinase activity | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0006766 | vitamin metabolic process | 78 | 118 | 3.363e-02 |
| GO:BP | GO:0006857 | oligopeptide transport | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0043171 | peptide catabolic process | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0007162 | negative regulation of cell adhesion | 181 | 293 | 3.363e-02 |
| GO:BP | GO:0097401 | synaptic vesicle lumen acidification | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0009163 | nucleoside biosynthetic process | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0034404 | nucleobase-containing small molecule biosynthetic process | 16 | 19 | 3.363e-02 |
| GO:BP | GO:1903599 | positive regulation of autophagy of mitochondrion | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0034260 | negative regulation of GTPase activity | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0099151 | regulation of postsynaptic density assembly | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0048268 | clathrin coat assembly | 16 | 19 | 3.363e-02 |
| GO:BP | GO:1902004 | positive regulation of amyloid-beta formation | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0031644 | regulation of nervous system process | 78 | 118 | 3.363e-02 |
| GO:BP | GO:2000811 | negative regulation of anoikis | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0048311 | mitochondrion distribution | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0006607 | NLS-bearing protein import into nucleus | 16 | 19 | 3.363e-02 |
| GO:BP | GO:0030278 | regulation of ossification | 81 | 123 | 3.364e-02 |
| GO:BP | GO:0007435 | salivary gland morphogenesis | 26 | 34 | 3.407e-02 |
| GO:BP | GO:0032373 | positive regulation of sterol transport | 26 | 34 | 3.407e-02 |
| GO:BP | GO:0061081 | positive regulation of myeloid leukocyte cytokine production involved in immune response | 26 | 34 | 3.407e-02 |
| GO:BP | GO:0006536 | glutamate metabolic process | 26 | 34 | 3.407e-02 |
| GO:BP | GO:0019674 | NAD metabolic process | 26 | 34 | 3.407e-02 |
| GO:BP | GO:0032376 | positive regulation of cholesterol transport | 26 | 34 | 3.407e-02 |
| GO:BP | GO:0051965 | positive regulation of synapse assembly | 45 | 64 | 3.476e-02 |
| GO:BP | GO:0045807 | positive regulation of endocytosis | 100 | 155 | 3.517e-02 |
| GO:BP | GO:0009636 | response to toxic substance | 157 | 252 | 3.522e-02 |
| GO:BP | GO:0034311 | diol metabolic process | 24 | 31 | 3.541e-02 |
| GO:BP | GO:0032232 | negative regulation of actin filament bundle assembly | 24 | 31 | 3.541e-02 |
| GO:BP | GO:0045070 | positive regulation of viral genome replication | 24 | 31 | 3.541e-02 |
| GO:BP | GO:1900027 | regulation of ruffle assembly | 24 | 31 | 3.541e-02 |
| GO:BP | GO:0010458 | exit from mitosis | 24 | 31 | 3.541e-02 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 24 | 31 | 3.541e-02 |
| GO:BP | GO:0070588 | calcium ion transmembrane transport | 208 | 340 | 3.555e-02 |
| GO:BP | GO:1900273 | positive regulation of long-term synaptic potentiation | 18 | 22 | 3.567e-02 |
| GO:BP | GO:0016082 | synaptic vesicle priming | 18 | 22 | 3.567e-02 |
| GO:BP | GO:0036480 | neuron intrinsic apoptotic signaling pathway in response to oxidative stress | 18 | 22 | 3.567e-02 |
| GO:BP | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | 18 | 22 | 3.567e-02 |
| GO:BP | GO:0002089 | lens morphogenesis in camera-type eye | 18 | 22 | 3.567e-02 |
| GO:BP | GO:0046485 | ether lipid metabolic process | 18 | 22 | 3.567e-02 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 18 | 22 | 3.567e-02 |
| GO:BP | GO:0032012 | regulation of ARF protein signal transduction | 18 | 22 | 3.567e-02 |
| GO:BP | GO:0006662 | glycerol ether metabolic process | 18 | 22 | 3.567e-02 |
| GO:BP | GO:0032011 | ARF protein signal transduction | 18 | 22 | 3.567e-02 |
| GO:BP | GO:0034350 | regulation of glial cell apoptotic process | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0032793 | positive regulation of CREB transcription factor activity | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0006285 | base-excision repair, AP site formation | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0034139 | regulation of toll-like receptor 3 signaling pathway | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0000413 | protein peptidyl-prolyl isomerization | 22 | 28 | 3.614e-02 |
| GO:BP | GO:0034138 | toll-like receptor 3 signaling pathway | 20 | 25 | 3.614e-02 |
| GO:BP | GO:1904867 | protein localization to Cajal body | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0000305 | response to oxygen radical | 22 | 28 | 3.614e-02 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 70 | 105 | 3.614e-02 |
| GO:BP | GO:0010661 | positive regulation of muscle cell apoptotic process | 22 | 28 | 3.614e-02 |
| GO:BP | GO:0001783 | B cell apoptotic process | 20 | 25 | 3.614e-02 |
| GO:BP | GO:0032925 | regulation of activin receptor signaling pathway | 20 | 25 | 3.614e-02 |
| GO:BP | GO:0046039 | GTP metabolic process | 20 | 25 | 3.614e-02 |
| GO:BP | GO:0099515 | actin filament-based transport | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0051882 | mitochondrial depolarization | 20 | 25 | 3.614e-02 |
| GO:BP | GO:1903441 | protein localization to ciliary membrane | 11 | 12 | 3.614e-02 |
| GO:BP | GO:1903405 | protein localization to nuclear body | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0044346 | fibroblast apoptotic process | 20 | 25 | 3.614e-02 |
| GO:BP | GO:0060379 | cardiac muscle cell myoblast differentiation | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0060586 | multicellular organismal-level iron ion homeostasis | 22 | 28 | 3.614e-02 |
| GO:BP | GO:0001967 | suckling behavior | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0043403 | skeletal muscle tissue regeneration | 33 | 45 | 3.614e-02 |
| GO:BP | GO:0008611 | ether lipid biosynthetic process | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0071394 | cellular response to testosterone stimulus | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0042939 | tripeptide transport | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0061339 | establishment or maintenance of monopolar cell polarity | 20 | 25 | 3.614e-02 |
| GO:BP | GO:0042364 | water-soluble vitamin biosynthetic process | 11 | 12 | 3.614e-02 |
| GO:BP | GO:1903897 | regulation of PERK-mediated unfolded protein response | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0048755 | branching morphogenesis of a nerve | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0048499 | synaptic vesicle membrane organization | 22 | 28 | 3.614e-02 |
| GO:BP | GO:0035335 | peptidyl-tyrosine dephosphorylation | 20 | 25 | 3.614e-02 |
| GO:BP | GO:0003207 | cardiac chamber formation | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0002253 | activation of immune response | 334 | 560 | 3.614e-02 |
| GO:BP | GO:0034975 | protein folding in endoplasmic reticulum | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0033688 | regulation of osteoblast proliferation | 22 | 28 | 3.614e-02 |
| GO:BP | GO:1900152 | negative regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0021544 | subpallium development | 22 | 28 | 3.614e-02 |
| GO:BP | GO:1905671 | regulation of lysosome organization | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0046504 | glycerol ether biosynthetic process | 11 | 12 | 3.614e-02 |
| GO:BP | GO:2000009 | negative regulation of protein localization to cell surface | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0031340 | positive regulation of vesicle fusion | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0031293 | membrane protein intracellular domain proteolysis | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0006312 | mitotic recombination | 20 | 25 | 3.614e-02 |
| GO:BP | GO:0021670 | lateral ventricle development | 11 | 12 | 3.614e-02 |
| GO:BP | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway | 11 | 12 | 3.614e-02 |
| GO:BP | GO:0051282 | regulation of sequestering of calcium ion | 85 | 130 | 3.618e-02 |
| GO:BP | GO:0009064 | glutamine family amino acid metabolic process | 56 | 82 | 3.623e-02 |
| GO:BP | GO:0042246 | tissue regeneration | 56 | 82 | 3.623e-02 |
| GO:BP | GO:0061724 | lipophagy | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0062196 | regulation of lysosome size | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0000056 | ribosomal small subunit export from nucleus | 8 | 8 | 3.639e-02 |
| GO:BP | GO:1903943 | regulation of hepatocyte apoptotic process | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0090063 | positive regulation of microtubule nucleation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0036289 | peptidyl-serine autophosphorylation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0046984 | regulation of hemoglobin biosynthetic process | 8 | 8 | 3.639e-02 |
| GO:BP | GO:2000726 | negative regulation of cardiac muscle cell differentiation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:1902913 | positive regulation of neuroepithelial cell differentiation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0046492 | heme B metabolic process | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0003149 | membranous septum morphogenesis | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0036261 | 7-methylguanosine cap hypermethylation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:1903108 | regulation of mitochondrial transcription | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0006551 | L-leucine metabolic process | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0000965 | mitochondrial RNA 3’-end processing | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0106354 | tRNA surveillance | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0060509 | type I pneumocyte differentiation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:1903608 | protein localization to cytoplasmic stress granule | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0060600 | dichotomous subdivision of an epithelial terminal unit | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0043504 | mitochondrial DNA repair | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0061343 | cell adhesion involved in heart morphogenesis | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0051086 | chaperone mediated protein folding independent of cofactor | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0120180 | cell-substrate junction disassembly | 8 | 8 | 3.639e-02 |
| GO:BP | GO:1903147 | negative regulation of autophagy of mitochondrion | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0120181 | focal adhesion disassembly | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0006785 | heme B biosynthetic process | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0071051 | poly(A)-dependent snoRNA 3’-end processing | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0060675 | ureteric bud morphogenesis | 48 | 69 | 3.639e-02 |
| GO:BP | GO:0018202 | peptidyl-histidine modification | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0035434 | copper ion transmembrane transport | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0072033 | renal vesicle formation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:1905064 | negative regulation of vascular associated smooth muscle cell differentiation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0015959 | diadenosine polyphosphate metabolic process | 8 | 8 | 3.639e-02 |
| GO:BP | GO:1901401 | regulation of tetrapyrrole metabolic process | 8 | 8 | 3.639e-02 |
| GO:BP | GO:2000643 | positive regulation of early endosome to late endosome transport | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0006384 | transcription initiation at RNA polymerase III promoter | 8 | 8 | 3.639e-02 |
| GO:BP | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0031118 | rRNA pseudouridine synthesis | 8 | 8 | 3.639e-02 |
| GO:BP | GO:1901525 | negative regulation of mitophagy | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0033152 | immunoglobulin V(D)J recombination | 8 | 8 | 3.639e-02 |
| GO:BP | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0032906 | transforming growth factor beta2 production | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0010793 | regulation of mRNA export from nucleus | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0048227 | plasma membrane to endosome transport | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0032909 | regulation of transforming growth factor beta2 production | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0034775 | glutathione transmembrane transport | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0032347 | regulation of aldosterone biosynthetic process | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0010835 | regulation of protein ADP-ribosylation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0071499 | cellular response to laminar fluid shear stress | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0034475 | U4 snRNA 3’-end processing | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0150012 | positive regulation of neuron projection arborization | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0003223 | ventricular compact myocardium morphogenesis | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0031077 | post-embryonic camera-type eye development | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0032344 | regulation of aldosterone metabolic process | 8 | 8 | 3.639e-02 |
| GO:BP | GO:1904354 | negative regulation of telomere capping | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0051683 | establishment of Golgi localization | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0017185 | peptidyl-lysine hydroxylation | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0045008 | depyrimidination | 8 | 8 | 3.639e-02 |
| GO:BP | GO:0072330 | monocarboxylic acid biosynthetic process | 134 | 213 | 3.644e-02 |
| GO:BP | GO:0010596 | negative regulation of endothelial cell migration | 38 | 53 | 3.662e-02 |
| GO:BP | GO:0071260 | cellular response to mechanical stimulus | 51 | 74 | 3.768e-02 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 31 | 42 | 3.779e-02 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 65 | 97 | 3.833e-02 |
| GO:BP | GO:0061041 | regulation of wound healing | 86 | 132 | 3.892e-02 |
| GO:BP | GO:0060760 | positive regulation of response to cytokine stimulus | 46 | 66 | 3.939e-02 |
| GO:BP | GO:0021795 | cerebral cortex cell migration | 36 | 50 | 4.002e-02 |
| GO:BP | GO:0007019 | microtubule depolymerization | 36 | 50 | 4.002e-02 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 122 | 193 | 4.035e-02 |
| GO:BP | GO:1902107 | positive regulation of leukocyte differentiation | 122 | 193 | 4.035e-02 |
| GO:BP | GO:1903708 | positive regulation of hemopoiesis | 122 | 193 | 4.035e-02 |
| GO:BP | GO:0050685 | positive regulation of mRNA processing | 29 | 39 | 4.059e-02 |
| GO:BP | GO:0044409 | symbiont entry into host | 93 | 144 | 4.157e-02 |
| GO:BP | GO:0046718 | symbiont entry into host cell | 90 | 139 | 4.211e-02 |
| GO:BP | GO:0019217 | regulation of fatty acid metabolic process | 63 | 94 | 4.243e-02 |
| GO:BP | GO:1903725 | regulation of phospholipid metabolic process | 27 | 36 | 4.341e-02 |
| GO:BP | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway | 27 | 36 | 4.341e-02 |
| GO:BP | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | 27 | 36 | 4.341e-02 |
| GO:BP | GO:0009156 | ribonucleoside monophosphate biosynthetic process | 27 | 36 | 4.341e-02 |
| GO:BP | GO:0048332 | mesoderm morphogenesis | 52 | 76 | 4.344e-02 |
| GO:BP | GO:0048709 | oligodendrocyte differentiation | 72 | 109 | 4.349e-02 |
| GO:BP | GO:0008038 | neuron recognition | 34 | 47 | 4.356e-02 |
| GO:BP | GO:0031641 | regulation of myelination | 34 | 47 | 4.356e-02 |
| GO:BP | GO:0045778 | positive regulation of ossification | 39 | 55 | 4.422e-02 |
| GO:BP | GO:0055012 | ventricular cardiac muscle cell differentiation | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0051444 | negative regulation of ubiquitin-protein transferase activity | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0060026 | convergent extension | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0046761 | viral budding from plasma membrane | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0045176 | apical protein localization | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0051284 | positive regulation of sequestering of calcium ion | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0098969 | neurotransmitter receptor transport to postsynaptic membrane | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0006750 | glutathione biosynthetic process | 13 | 15 | 4.432e-02 |
| GO:BP | GO:1903894 | regulation of IRE1-mediated unfolded protein response | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0007176 | regulation of epidermal growth factor-activated receptor activity | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0009209 | pyrimidine ribonucleoside triphosphate biosynthetic process | 13 | 15 | 4.432e-02 |
| GO:BP | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0010715 | regulation of extracellular matrix disassembly | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0014029 | neural crest formation | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0150011 | regulation of neuron projection arborization | 13 | 15 | 4.432e-02 |
| GO:BP | GO:1903543 | positive regulation of exosomal secretion | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0099118 | microtubule-based protein transport | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0098840 | protein transport along microtubule | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0097212 | lysosomal membrane organization | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0006474 | N-terminal protein amino acid acetylation | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0090030 | regulation of steroid hormone biosynthetic process | 13 | 15 | 4.432e-02 |
| GO:BP | GO:2001028 | positive regulation of endothelial cell chemotaxis | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0008298 | intracellular mRNA localization | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0006047 | UDP-N-acetylglucosamine metabolic process | 13 | 15 | 4.432e-02 |
| GO:BP | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway | 25 | 33 | 4.568e-02 |
| GO:BP | GO:0033198 | response to ATP | 25 | 33 | 4.568e-02 |
| GO:BP | GO:0033687 | osteoblast proliferation | 25 | 33 | 4.568e-02 |
| GO:BP | GO:0051346 | negative regulation of hydrolase activity | 58 | 86 | 4.568e-02 |
| GO:BP | GO:0018208 | peptidyl-proline modification | 25 | 33 | 4.568e-02 |
| GO:BP | GO:0050806 | positive regulation of synaptic transmission | 114 | 180 | 4.602e-02 |
| GO:BP | GO:0009914 | hormone transport | 204 | 335 | 4.693e-02 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 32 | 44 | 4.697e-02 |
| GO:BP | GO:0048821 | erythrocyte development | 32 | 44 | 4.697e-02 |
| GO:BP | GO:0030501 | positive regulation of bone mineralization | 32 | 44 | 4.697e-02 |
| GO:BP | GO:0030835 | negative regulation of actin filament depolymerization | 32 | 44 | 4.697e-02 |
| GO:BP | GO:0010812 | negative regulation of cell-substrate adhesion | 42 | 60 | 4.735e-02 |
| GO:BP | GO:1904888 | cranial skeletal system development | 50 | 73 | 4.756e-02 |
| GO:BP | GO:0061371 | determination of heart left/right asymmetry | 50 | 73 | 4.756e-02 |
| GO:BP | GO:0003333 | amino acid transmembrane transport | 70 | 106 | 4.762e-02 |
| GO:BP | GO:0044344 | cellular response to fibroblast growth factor stimulus | 73 | 111 | 4.763e-02 |
| GO:BP | GO:0060055 | angiogenesis involved in wound healing | 23 | 30 | 4.786e-02 |
| GO:BP | GO:1903077 | negative regulation of protein localization to plasma membrane | 23 | 30 | 4.786e-02 |
| GO:BP | GO:0048261 | negative regulation of receptor-mediated endocytosis | 23 | 30 | 4.786e-02 |
| GO:BP | GO:0008299 | isoprenoid biosynthetic process | 23 | 30 | 4.786e-02 |
| GO:BP | GO:0051291 | protein heterooligomerization | 23 | 30 | 4.786e-02 |
| GO:BP | GO:0021952 | central nervous system projection neuron axonogenesis | 23 | 30 | 4.786e-02 |
| GO:BP | GO:0071577 | zinc ion transmembrane transport | 23 | 30 | 4.786e-02 |
| GO:BP | GO:0046621 | negative regulation of organ growth | 23 | 30 | 4.786e-02 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 197 | 323 | 4.786e-02 |
| GO:BP | GO:0045830 | positive regulation of isotype switching | 23 | 30 | 4.786e-02 |
| GO:BP | GO:0006541 | glutamine metabolic process | 23 | 30 | 4.786e-02 |
| GO:BP | GO:0030520 | estrogen receptor signaling pathway | 37 | 52 | 4.786e-02 |
| GO:BP | GO:0006044 | N-acetylglucosamine metabolic process | 15 | 18 | 4.885e-02 |
| GO:BP | GO:0046184 | aldehyde biosynthetic process | 15 | 18 | 4.885e-02 |
| GO:BP | GO:0010002 | cardioblast differentiation | 15 | 18 | 4.885e-02 |
| GO:BP | GO:1905038 | regulation of membrane lipid metabolic process | 15 | 18 | 4.885e-02 |
| GO:BP | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel | 15 | 18 | 4.885e-02 |
| GO:BP | GO:0055119 | relaxation of cardiac muscle | 15 | 18 | 4.885e-02 |
| GO:BP | GO:0003128 | heart field specification | 15 | 18 | 4.885e-02 |
| GO:BP | GO:1902570 | protein localization to nucleolus | 15 | 18 | 4.885e-02 |
| GO:BP | GO:0000303 | response to superoxide | 21 | 27 | 4.963e-02 |
| GO:BP | GO:1902914 | regulation of protein polyubiquitination | 21 | 27 | 4.963e-02 |
| GO:BP | GO:0021697 | cerebellar cortex formation | 21 | 27 | 4.963e-02 |
| GO:BP | GO:0018904 | ether metabolic process | 21 | 27 | 4.963e-02 |
| GO:BP | GO:0007498 | mesoderm development | 89 | 138 | 4.963e-02 |
| GO:BP | GO:0036344 | platelet morphogenesis | 21 | 27 | 4.963e-02 |
| GO:BP | GO:0006929 | substrate-dependent cell migration | 21 | 27 | 4.963e-02 |
| GO:BP | GO:0043525 | positive regulation of neuron apoptotic process | 45 | 65 | 4.985e-02 |
| GO:BP | GO:0140895 | cell surface toll-like receptor signaling pathway | 45 | 65 | 4.985e-02 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 102 | 160 | 4.986e-02 |
| KEGG | KEGG:04141 | Protein processing in endoplasmic reticulum | 161 | 168 | 5.294e-20 |
| KEGG | KEGG:04144 | Endocytosis | 224 | 248 | 9.101e-19 |
| KEGG | KEGG:04140 | Autophagy - animal | 157 | 165 | 9.101e-19 |
| KEGG | KEGG:05168 | Herpes simplex virus 1 infection | 419 | 506 | 3.468e-18 |
| KEGG | KEGG:04120 | Ubiquitin mediated proteolysis | 130 | 140 | 3.751e-13 |
| KEGG | KEGG:05014 | Amyotrophic lateral sclerosis | 300 | 363 | 1.152e-12 |
| KEGG | KEGG:05022 | Pathways of neurodegeneration - multiple diseases | 381 | 474 | 1.357e-12 |
| KEGG | KEGG:04137 | Mitophagy - animal | 98 | 103 | 7.432e-12 |
| KEGG | KEGG:05012 | Parkinson disease | 224 | 265 | 1.607e-11 |
| KEGG | KEGG:05010 | Alzheimer disease | 309 | 382 | 6.802e-11 |
| KEGG | KEGG:05016 | Huntington disease | 252 | 305 | 8.998e-11 |
| KEGG | KEGG:05132 | Salmonella infection | 208 | 249 | 9.046e-10 |
| KEGG | KEGG:04360 | Axon guidance | 156 | 181 | 1.913e-09 |
| KEGG | KEGG:04919 | Thyroid hormone signaling pathway | 109 | 121 | 3.660e-09 |
| KEGG | KEGG:05131 | Shigellosis | 204 | 246 | 4.362e-09 |
| KEGG | KEGG:04150 | mTOR signaling pathway | 135 | 155 | 6.227e-09 |
| KEGG | KEGG:04714 | Thermogenesis | 193 | 232 | 6.974e-09 |
| KEGG | KEGG:04722 | Neurotrophin signaling pathway | 106 | 118 | 7.758e-09 |
| KEGG | KEGG:03013 | Nucleocytoplasmic transport | 98 | 108 | 8.565e-09 |
| KEGG | KEGG:04142 | Lysosome | 116 | 131 | 9.088e-09 |
| KEGG | KEGG:05210 | Colorectal cancer | 80 | 86 | 1.467e-08 |
| KEGG | KEGG:04910 | Insulin signaling pathway | 120 | 137 | 2.048e-08 |
| KEGG | KEGG:05017 | Spinocerebellar ataxia | 123 | 141 | 2.189e-08 |
| KEGG | KEGG:03018 | RNA degradation | 74 | 79 | 2.347e-08 |
| KEGG | KEGG:03083 | Polycomb repressive complex | 78 | 84 | 2.558e-08 |
| KEGG | KEGG:05212 | Pancreatic cancer | 71 | 76 | 6.356e-08 |
| KEGG | KEGG:05220 | Chronic myeloid leukemia | 71 | 76 | 6.356e-08 |
| KEGG | KEGG:00970 | Aminoacyl-tRNA biosynthesis | 44 | 44 | 6.730e-08 |
| KEGG | KEGG:01100 | Metabolic pathways | 1094 | 1537 | 7.100e-08 |
| KEGG | KEGG:04012 | ErbB signaling pathway | 77 | 84 | 1.336e-07 |
| KEGG | KEGG:04510 | Focal adhesion | 167 | 202 | 1.538e-07 |
| KEGG | KEGG:04070 | Phosphatidylinositol signaling system | 87 | 97 | 2.067e-07 |
| KEGG | KEGG:05208 | Chemical carcinogenesis - reactive oxygen species | 180 | 221 | 3.718e-07 |
| KEGG | KEGG:04931 | Insulin resistance | 95 | 108 | 4.514e-07 |
| KEGG | KEGG:00510 | N-Glycan biosynthesis | 51 | 53 | 4.721e-07 |
| KEGG | KEGG:01521 | EGFR tyrosine kinase inhibitor resistance | 72 | 79 | 6.376e-07 |
| KEGG | KEGG:05213 | Endometrial cancer | 55 | 58 | 6.377e-07 |
| KEGG | KEGG:05020 | Prion disease | 215 | 271 | 1.009e-06 |
| KEGG | KEGG:05100 | Bacterial invasion of epithelial cells | 70 | 77 | 1.173e-06 |
| KEGG | KEGG:03010 | Ribosome | 128 | 153 | 1.540e-06 |
| KEGG | KEGG:05225 | Hepatocellular carcinoma | 137 | 166 | 2.931e-06 |
| KEGG | KEGG:04932 | Non-alcoholic fatty liver disease | 128 | 154 | 3.142e-06 |
| KEGG | KEGG:03020 | RNA polymerase | 34 | 34 | 3.370e-06 |
| KEGG | KEGG:03015 | mRNA surveillance pathway | 84 | 96 | 3.672e-06 |
| KEGG | KEGG:05415 | Diabetic cardiomyopathy | 163 | 202 | 4.013e-06 |
| KEGG | KEGG:05211 | Renal cell carcinoma | 62 | 68 | 4.140e-06 |
| KEGG | KEGG:05223 | Non-small cell lung cancer | 65 | 72 | 5.340e-06 |
| KEGG | KEGG:04520 | Adherens junction | 81 | 93 | 8.394e-06 |
| KEGG | KEGG:04933 | AGE-RAGE signaling pathway in diabetic complications | 86 | 100 | 1.277e-05 |
| KEGG | KEGG:04071 | Sphingolipid signaling pathway | 101 | 120 | 1.379e-05 |
| KEGG | KEGG:04152 | AMPK signaling pathway | 101 | 120 | 1.379e-05 |
| KEGG | KEGG:00310 | Lysine degradation | 57 | 63 | 2.034e-05 |
| KEGG | KEGG:05165 | Human papillomavirus infection | 252 | 330 | 2.213e-05 |
| KEGG | KEGG:04010 | MAPK signaling pathway | 231 | 301 | 2.948e-05 |
| KEGG | KEGG:03082 | ATP-dependent chromatin remodeling | 97 | 116 | 3.707e-05 |
| KEGG | KEGG:04390 | Hippo signaling pathway | 127 | 157 | 4.644e-05 |
| KEGG | KEGG:01200 | Carbon metabolism | 96 | 115 | 4.649e-05 |
| KEGG | KEGG:04211 | Longevity regulating pathway | 76 | 89 | 7.703e-05 |
| KEGG | KEGG:05222 | Small cell lung cancer | 78 | 92 | 1.024e-04 |
| KEGG | KEGG:04136 | Autophagy - other | 31 | 32 | 1.057e-04 |
| KEGG | KEGG:03022 | Basal transcription factors | 40 | 43 | 1.086e-04 |
| KEGG | KEGG:01212 | Fatty acid metabolism | 51 | 57 | 1.267e-04 |
| KEGG | KEGG:04218 | Cellular senescence | 124 | 155 | 1.484e-04 |
| KEGG | KEGG:03060 | Protein export | 30 | 31 | 1.484e-04 |
| KEGG | KEGG:05231 | Choline metabolism in cancer | 82 | 98 | 1.592e-04 |
| KEGG | KEGG:00562 | Inositol phosphate metabolism | 63 | 73 | 1.882e-04 |
| KEGG | KEGG:04210 | Apoptosis | 109 | 135 | 1.985e-04 |
| KEGG | KEGG:03050 | Proteasome | 42 | 46 | 1.985e-04 |
| KEGG | KEGG:04310 | Wnt signaling pathway | 137 | 174 | 2.346e-04 |
| KEGG | KEGG:05205 | Proteoglycans in cancer | 159 | 205 | 2.614e-04 |
| KEGG | KEGG:04146 | Peroxisome | 70 | 83 | 3.332e-04 |
| KEGG | KEGG:00520 | Amino sugar and nucleotide sugar metabolism | 44 | 49 | 3.363e-04 |
| KEGG | KEGG:05135 | Yersinia infection | 109 | 136 | 3.382e-04 |
| KEGG | KEGG:03250 | Viral life cycle - HIV-1 | 54 | 62 | 3.571e-04 |
| KEGG | KEGG:04261 | Adrenergic signaling in cardiomyocytes | 121 | 153 | 4.151e-04 |
| KEGG | KEGG:04810 | Regulation of actin cytoskeleton | 173 | 227 | 6.456e-04 |
| KEGG | KEGG:04350 | TGF-beta signaling pathway | 87 | 107 | 6.569e-04 |
| KEGG | KEGG:00513 | Various types of N-glycan biosynthesis | 38 | 42 | 6.857e-04 |
| KEGG | KEGG:01250 | Biosynthesis of nucleotide sugars | 34 | 37 | 7.303e-04 |
| KEGG | KEGG:01522 | Endocrine resistance | 78 | 95 | 7.466e-04 |
| KEGG | KEGG:05214 | Glioma | 63 | 75 | 8.654e-04 |
| KEGG | KEGG:00190 | Oxidative phosphorylation | 106 | 134 | 1.011e-03 |
| KEGG | KEGG:00020 | Citrate cycle (TCA cycle) | 28 | 30 | 1.434e-03 |
| KEGG | KEGG:04213 | Longevity regulating pathway - multiple species | 52 | 61 | 1.434e-03 |
| KEGG | KEGG:00280 | Valine, leucine and isoleucine degradation | 42 | 48 | 1.635e-03 |
| KEGG | KEGG:04340 | Hedgehog signaling pathway | 48 | 56 | 1.818e-03 |
| KEGG | KEGG:04710 | Circadian rhythm | 31 | 34 | 1.904e-03 |
| KEGG | KEGG:05161 | Hepatitis B | 125 | 162 | 1.909e-03 |
| KEGG | KEGG:05200 | Pathways in cancer | 377 | 527 | 2.096e-03 |
| KEGG | KEGG:05215 | Prostate cancer | 78 | 97 | 2.415e-03 |
| KEGG | KEGG:04148 | Efferocytosis | 120 | 156 | 2.935e-03 |
| KEGG | KEGG:05110 | Vibrio cholerae infection | 43 | 50 | 2.935e-03 |
| KEGG | KEGG:05216 | Thyroid cancer | 33 | 37 | 3.057e-03 |
| KEGG | KEGG:05418 | Fluid shear stress and atherosclerosis | 107 | 138 | 3.284e-03 |
| KEGG | KEGG:04130 | SNARE interactions in vesicular transport | 29 | 32 | 3.480e-03 |
| KEGG | KEGG:00900 | Terpenoid backbone biosynthesis | 21 | 22 | 3.547e-03 |
| KEGG | KEGG:05203 | Viral carcinogenesis | 151 | 201 | 4.068e-03 |
| KEGG | KEGG:04530 | Tight junction | 129 | 170 | 4.652e-03 |
| KEGG | KEGG:04330 | Notch signaling pathway | 50 | 60 | 4.734e-03 |
| KEGG | KEGG:00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 24 | 26 | 5.153e-03 |
| KEGG | KEGG:04728 | Dopaminergic synapse | 101 | 131 | 5.952e-03 |
| KEGG | KEGG:04370 | VEGF signaling pathway | 49 | 59 | 5.965e-03 |
| KEGG | KEGG:05120 | Epithelial cell signaling in Helicobacter pylori infection | 57 | 70 | 6.420e-03 |
| KEGG | KEGG:05230 | Central carbon metabolism in cancer | 57 | 70 | 6.420e-03 |
| KEGG | KEGG:04022 | cGMP-PKG signaling pathway | 124 | 164 | 6.758e-03 |
| KEGG | KEGG:04015 | Rap1 signaling pathway | 156 | 210 | 7.104e-03 |
| KEGG | KEGG:03420 | Nucleotide excision repair | 50 | 61 | 8.999e-03 |
| KEGG | KEGG:04068 | FoxO signaling pathway | 100 | 131 | 1.024e-02 |
| KEGG | KEGG:04922 | Glucagon signaling pathway | 83 | 107 | 1.024e-02 |
| KEGG | KEGG:05163 | Human cytomegalovirus infection | 164 | 223 | 1.097e-02 |
| KEGG | KEGG:04666 | Fc gamma R-mediated phagocytosis | 75 | 96 | 1.118e-02 |
| KEGG | KEGG:04720 | Long-term potentiation | 54 | 67 | 1.210e-02 |
| KEGG | KEGG:00062 | Fatty acid elongation | 24 | 27 | 1.550e-02 |
| KEGG | KEGG:01040 | Biosynthesis of unsaturated fatty acids | 24 | 27 | 1.550e-02 |
| KEGG | KEGG:05170 | Human immunodeficiency virus 1 infection | 154 | 210 | 1.652e-02 |
| KEGG | KEGG:05410 | Hypertrophic cardiomyopathy | 75 | 97 | 1.735e-02 |
| KEGG | KEGG:01524 | Platinum drug resistance | 57 | 72 | 1.894e-02 |
| KEGG | KEGG:04912 | GnRH signaling pathway | 72 | 93 | 1.894e-02 |
| KEGG | KEGG:04935 | Growth hormone synthesis, secretion and action | 91 | 120 | 1.961e-02 |
| KEGG | KEGG:05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 69 | 89 | 2.068e-02 |
| KEGG | KEGG:04066 | HIF-1 signaling pathway | 83 | 109 | 2.250e-02 |
| KEGG | KEGG:04926 | Relaxin signaling pathway | 97 | 129 | 2.271e-02 |
| KEGG | KEGG:05221 | Acute myeloid leukemia | 53 | 67 | 2.410e-02 |
| KEGG | KEGG:04928 | Parathyroid hormone synthesis, secretion and action | 80 | 105 | 2.410e-02 |
| KEGG | KEGG:00532 | Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate | 19 | 21 | 2.410e-02 |
| KEGG | KEGG:04540 | Gap junction | 68 | 88 | 2.410e-02 |
| KEGG | KEGG:03040 | Spliceosome | 128 | 174 | 2.492e-02 |
| KEGG | KEGG:04917 | Prolactin signaling pathway | 55 | 70 | 2.631e-02 |
| KEGG | KEGG:04920 | Adipocytokine signaling pathway | 55 | 70 | 2.631e-02 |
| KEGG | KEGG:04392 | Hippo signaling pathway - multiple species | 25 | 29 | 2.691e-02 |
| KEGG | KEGG:01210 | 2-Oxocarboxylic acid metabolism | 28 | 33 | 2.691e-02 |
| KEGG | KEGG:04962 | Vasopressin-regulated water reabsorption | 36 | 44 | 3.040e-02 |
| KEGG | KEGG:04668 | TNF signaling pathway | 85 | 113 | 3.240e-02 |
| KEGG | KEGG:05130 | Pathogenic Escherichia coli infection | 143 | 197 | 3.381e-02 |
| KEGG | KEGG:04215 | Apoptosis - multiple species | 27 | 32 | 3.462e-02 |
| KEGG | KEGG:05226 | Gastric cancer | 109 | 148 | 3.720e-02 |
| KEGG | KEGG:05414 | Dilated cardiomyopathy | 77 | 102 | 3.720e-02 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 157 | 218 | 3.789e-02 |
| KEGG | KEGG:04371 | Apelin signaling pathway | 102 | 138 | 3.789e-02 |
| KEGG | KEGG:00785 | Lipoic acid metabolism | 17 | 19 | 4.240e-02 |
| KEGG | KEGG:01230 | Biosynthesis of amino acids | 57 | 74 | 4.318e-02 |
# write.csv(table_motif1_GOKEGG_genes, "data/new/table_motif1_GOKEGG_genes.csv")
#GO:BP
table_motif1_genes_GOBP_RUV_clust <- table_motif1_GOKEGG_genes_RUV_clust %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
# saveRDS(table_motif1_genes_GOBP, "data/table_motif1_genes_GOBP.RDS")
table_motif1_genes_GOBP_RUV_clust %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("M1 Enriched GO:BP Terms RUVs clustlike")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))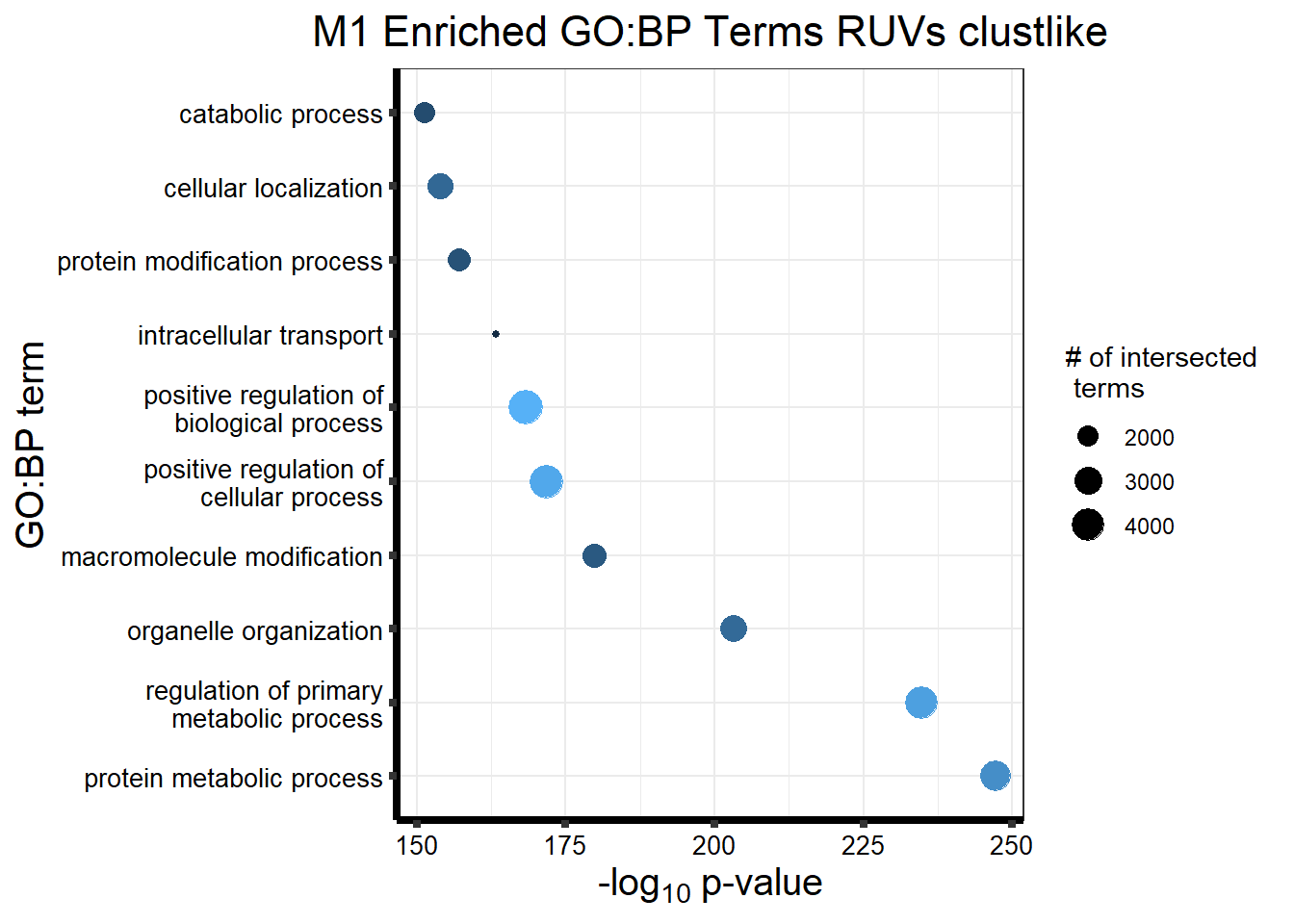
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#KEGG
table_motif1_genes_KEGG_RUV_clust <- table_motif1_GOKEGG_genes_RUV_clust %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_motif1_genes_KEGG_RUV_clust %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("M1 DEGs Enriched KEGG Terms RUVs clustlike")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))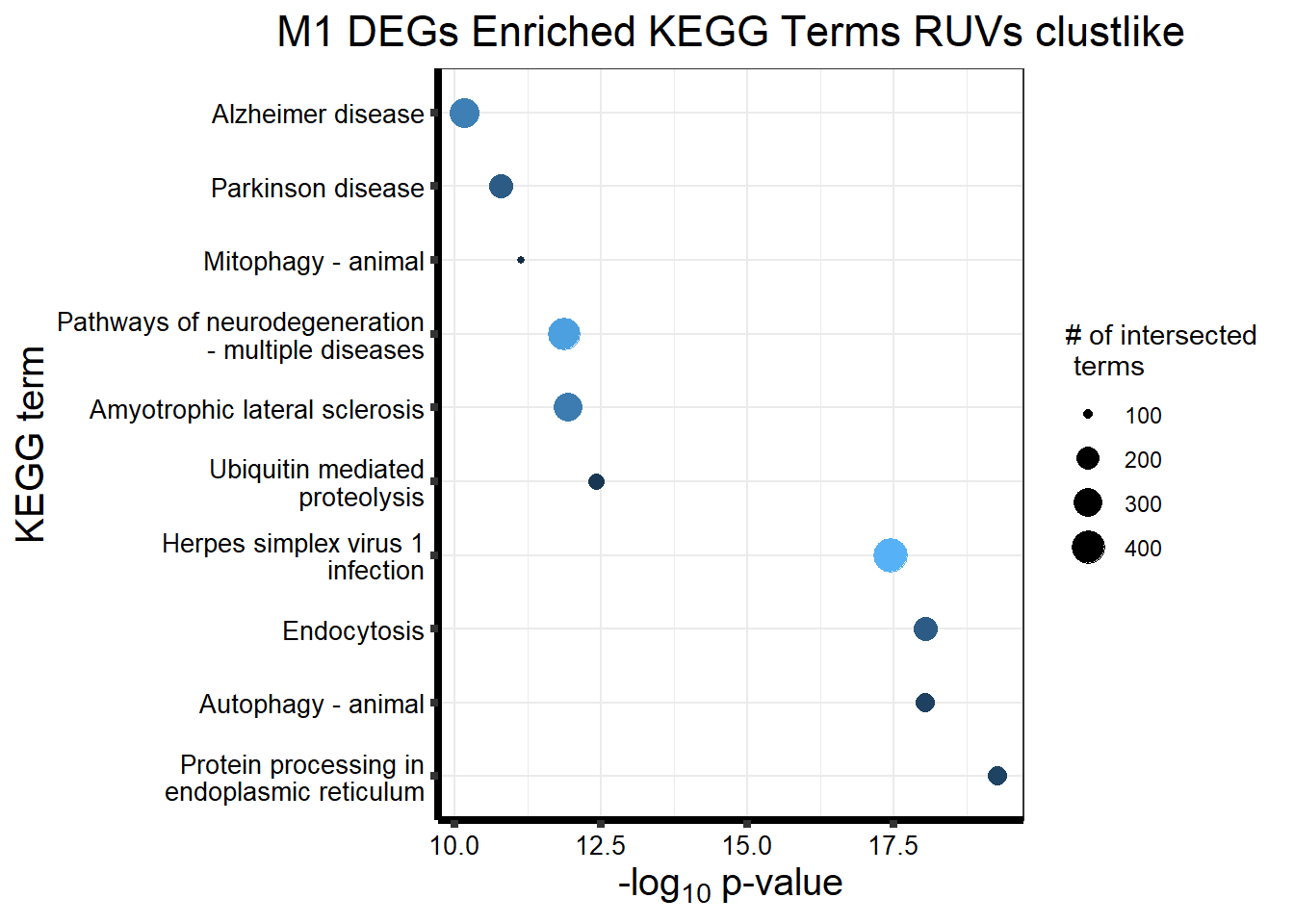
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#####Motif 2 Genes#####
# motif2_genes_matrix_RUV_clust <- as.matrix(clust2_genes_RUV)
# colnames(motif2_genes_matrix_RUV_clust) <- c("entrezgene_ID")
# saveRDS(motif2_genes_matrix_RUV_clust, "data/new/RUV/CMF/motif2_genes_matrix_RUV_clustlike.RDS")
motif2_genes_matrix_RUV_clust <- readRDS("data/new/RUV/CMF/motif2_genes_matrix_RUV_clustlike.RDS")
length(motif2_genes_matrix_RUV_clust)[1] 517#517 genes in this set for motif 2
motif2_mat_GOKEGG_RUV_clust <- gost(query = motif2_genes_matrix_RUV_clust,
organism = "hsapiens",
ordered_query = FALSE,
measure_underrepresentation = FALSE,
evcodes = FALSE,
user_threshold = 0.05,
correction_method = c("fdr"),
sources = c("GO:BP", "KEGG"))
motif2_GOKEGG_genes_RUV_clust <- gostplot(motif2_mat_GOKEGG_RUV_clust, capped = FALSE, interactive = TRUE)
motif2_GOKEGG_genes_RUV_clusttable_motif2_GOKEGG_genes_RUV_clust <- motif2_mat_GOKEGG_RUV_clust$result %>%
dplyr::select(c(source, term_id, term_name, intersection_size, term_size, p_value))
table_motif2_GOKEGG_genes_RUV_clust %>%
mutate_at(.vars = 6, .funs = scales::label_scientific(digits=4)) %>%
kableExtra::kable(.,) %>%
kableExtra::kable_paper("striped", full_width = FALSE) %>%
kableExtra::kable_styling(full_width = FALSE, position = "left", bootstrap_options = c("striped", "hover")) %>%
kableExtra::scroll_box(width = "100%", height = "400px")| source | term_id | term_name | intersection_size | term_size | p_value |
|---|---|---|---|---|---|
| GO:BP | GO:0022402 | cell cycle process | 176 | 1280 | 7.756e-89 |
| GO:BP | GO:1903047 | mitotic cell cycle process | 135 | 745 | 2.136e-81 |
| GO:BP | GO:0007049 | cell cycle | 186 | 1663 | 1.521e-79 |
| GO:BP | GO:0000278 | mitotic cell cycle | 142 | 892 | 1.736e-78 |
| GO:BP | GO:0007059 | chromosome segregation | 97 | 427 | 1.412e-66 |
| GO:BP | GO:0000280 | nuclear division | 95 | 452 | 7.896e-62 |
| GO:BP | GO:0051301 | cell division | 109 | 658 | 1.822e-60 |
| GO:BP | GO:0010564 | regulation of cell cycle process | 112 | 720 | 2.287e-59 |
| GO:BP | GO:0048285 | organelle fission | 96 | 500 | 7.533e-59 |
| GO:BP | GO:0098813 | nuclear chromosome segregation | 80 | 323 | 1.398e-57 |
| GO:BP | GO:0140014 | mitotic nuclear division | 73 | 282 | 8.759e-54 |
| GO:BP | GO:0051726 | regulation of cell cycle | 127 | 1087 | 1.616e-53 |
| GO:BP | GO:0051276 | chromosome organization | 95 | 574 | 3.075e-52 |
| GO:BP | GO:0000070 | mitotic sister chromatid segregation | 62 | 193 | 8.516e-52 |
| GO:BP | GO:0000819 | sister chromatid segregation | 66 | 234 | 3.728e-51 |
| GO:BP | GO:0044770 | cell cycle phase transition | 90 | 532 | 3.412e-50 |
| GO:BP | GO:0044772 | mitotic cell cycle phase transition | 79 | 428 | 1.318e-46 |
| GO:BP | GO:1901987 | regulation of cell cycle phase transition | 76 | 427 | 1.517e-43 |
| GO:BP | GO:0007346 | regulation of mitotic cell cycle | 79 | 492 | 6.046e-42 |
| GO:BP | GO:0006260 | DNA replication | 63 | 283 | 6.634e-42 |
| GO:BP | GO:0010948 | negative regulation of cell cycle process | 60 | 289 | 6.002e-38 |
| GO:BP | GO:1901990 | regulation of mitotic cell cycle phase transition | 63 | 327 | 6.715e-38 |
| GO:BP | GO:0000075 | cell cycle checkpoint signaling | 50 | 185 | 2.206e-37 |
| GO:BP | GO:1901988 | negative regulation of cell cycle phase transition | 55 | 248 | 2.667e-36 |
| GO:BP | GO:0051983 | regulation of chromosome segregation | 43 | 132 | 7.569e-36 |
| GO:BP | GO:0006261 | DNA-templated DNA replication | 46 | 160 | 1.146e-35 |
| GO:BP | GO:0045786 | negative regulation of cell cycle | 64 | 377 | 3.904e-35 |
| GO:BP | GO:1905818 | regulation of chromosome separation | 35 | 77 | 4.509e-35 |
| GO:BP | GO:0006259 | DNA metabolic process | 98 | 1005 | 1.276e-33 |
| GO:BP | GO:0007088 | regulation of mitotic nuclear division | 40 | 121 | 1.279e-33 |
| GO:BP | GO:0051304 | chromosome separation | 35 | 84 | 1.859e-33 |
| GO:BP | GO:0007093 | mitotic cell cycle checkpoint signaling | 41 | 136 | 1.156e-32 |
| GO:BP | GO:0051310 | metaphase chromosome alignment | 37 | 104 | 2.056e-32 |
| GO:BP | GO:0051783 | regulation of nuclear division | 42 | 149 | 3.681e-32 |
| GO:BP | GO:0090068 | positive regulation of cell cycle process | 52 | 262 | 8.072e-32 |
| GO:BP | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 43 | 169 | 6.576e-31 |
| GO:BP | GO:1901991 | negative regulation of mitotic cell cycle phase transition | 43 | 172 | 1.435e-30 |
| GO:BP | GO:0050000 | chromosome localization | 38 | 126 | 2.887e-30 |
| GO:BP | GO:0051303 | establishment of chromosome localization | 37 | 118 | 3.735e-30 |
| GO:BP | GO:0045930 | negative regulation of mitotic cell cycle | 47 | 225 | 9.682e-30 |
| GO:BP | GO:0010965 | regulation of mitotic sister chromatid separation | 29 | 62 | 1.724e-29 |
| GO:BP | GO:0007051 | spindle organization | 45 | 206 | 2.435e-29 |
| GO:BP | GO:0051321 | meiotic cell cycle | 52 | 297 | 4.361e-29 |
| GO:BP | GO:0051306 | mitotic sister chromatid separation | 29 | 65 | 9.234e-29 |
| GO:BP | GO:0045787 | positive regulation of cell cycle | 55 | 351 | 2.687e-28 |
| GO:BP | GO:0033045 | regulation of sister chromatid segregation | 34 | 107 | 6.417e-28 |
| GO:BP | GO:0033048 | negative regulation of mitotic sister chromatid segregation | 26 | 51 | 1.024e-27 |
| GO:BP | GO:0045841 | negative regulation of mitotic metaphase/anaphase transition | 26 | 51 | 1.024e-27 |
| GO:BP | GO:1903046 | meiotic cell cycle process | 45 | 224 | 1.024e-27 |
| GO:BP | GO:0033046 | negative regulation of sister chromatid segregation | 26 | 51 | 1.024e-27 |
| GO:BP | GO:2000816 | negative regulation of mitotic sister chromatid separation | 26 | 51 | 1.024e-27 |
| GO:BP | GO:0033047 | regulation of mitotic sister chromatid segregation | 27 | 57 | 1.094e-27 |
| GO:BP | GO:0051784 | negative regulation of nuclear division | 28 | 65 | 2.769e-27 |
| GO:BP | GO:0051985 | negative regulation of chromosome segregation | 26 | 53 | 3.451e-27 |
| GO:BP | GO:1905819 | negative regulation of chromosome separation | 26 | 53 | 3.451e-27 |
| GO:BP | GO:0045839 | negative regulation of mitotic nuclear division | 27 | 59 | 3.451e-27 |
| GO:BP | GO:1902100 | negative regulation of metaphase/anaphase transition of cell cycle | 26 | 53 | 3.451e-27 |
| GO:BP | GO:0006974 | DNA damage response | 84 | 908 | 4.955e-27 |
| GO:BP | GO:0007091 | metaphase/anaphase transition of mitotic cell cycle | 32 | 97 | 6.340e-27 |
| GO:BP | GO:0007052 | mitotic spindle organization | 36 | 134 | 7.920e-27 |
| GO:BP | GO:0007094 | mitotic spindle assembly checkpoint signaling | 25 | 49 | 1.009e-26 |
| GO:BP | GO:0071174 | mitotic spindle checkpoint signaling | 25 | 49 | 1.009e-26 |
| GO:BP | GO:0071173 | spindle assembly checkpoint signaling | 25 | 49 | 1.009e-26 |
| GO:BP | GO:0006281 | DNA repair | 69 | 621 | 1.336e-26 |
| GO:BP | GO:0044784 | metaphase/anaphase transition of cell cycle | 32 | 100 | 1.745e-26 |
| GO:BP | GO:0031577 | spindle checkpoint signaling | 25 | 50 | 1.887e-26 |
| GO:BP | GO:0030071 | regulation of mitotic metaphase/anaphase transition | 31 | 93 | 2.835e-26 |
| GO:BP | GO:0006996 | organelle organization | 180 | 3594 | 3.186e-26 |
| GO:BP | GO:1902099 | regulation of metaphase/anaphase transition of cell cycle | 31 | 96 | 8.462e-26 |
| GO:BP | GO:0140013 | meiotic nuclear division | 41 | 202 | 1.633e-25 |
| GO:BP | GO:0008608 | attachment of spindle microtubules to kinetochore | 25 | 54 | 2.149e-25 |
| GO:BP | GO:0033044 | regulation of chromosome organization | 43 | 242 | 2.480e-24 |
| GO:BP | GO:0000226 | microtubule cytoskeleton organization | 68 | 670 | 6.406e-24 |
| GO:BP | GO:0044839 | cell cycle G2/M phase transition | 34 | 156 | 4.003e-22 |
| GO:BP | GO:0000086 | G2/M transition of mitotic cell cycle | 32 | 139 | 1.407e-21 |
| GO:BP | GO:0007017 | microtubule-based process | 79 | 999 | 5.216e-21 |
| GO:BP | GO:2001251 | negative regulation of chromosome organization | 27 | 97 | 1.692e-20 |
| GO:BP | GO:0006270 | DNA replication initiation | 19 | 37 | 2.581e-20 |
| GO:BP | GO:0051225 | spindle assembly | 29 | 136 | 1.588e-18 |
| GO:BP | GO:0010389 | regulation of G2/M transition of mitotic cell cycle | 26 | 105 | 2.571e-18 |
| GO:BP | GO:0051656 | establishment of organelle localization | 51 | 491 | 4.099e-18 |
| GO:BP | GO:0061982 | meiosis I cell cycle process | 29 | 141 | 4.444e-18 |
| GO:BP | GO:0033554 | cellular response to stress | 105 | 1830 | 9.131e-18 |
| GO:BP | GO:0007080 | mitotic metaphase chromosome alignment | 21 | 63 | 1.064e-17 |
| GO:BP | GO:0033043 | regulation of organelle organization | 79 | 1148 | 2.239e-17 |
| GO:BP | GO:0051640 | organelle localization | 56 | 620 | 3.748e-17 |
| GO:BP | GO:0045132 | meiotic chromosome segregation | 24 | 96 | 5.189e-17 |
| GO:BP | GO:1902749 | regulation of cell cycle G2/M phase transition | 26 | 118 | 5.557e-17 |
| GO:BP | GO:0006302 | double-strand break repair | 40 | 319 | 5.879e-17 |
| GO:BP | GO:1905820 | positive regulation of chromosome separation | 16 | 32 | 7.715e-17 |
| GO:BP | GO:0006310 | DNA recombination | 41 | 347 | 1.850e-16 |
| GO:BP | GO:0000725 | recombinational repair | 31 | 193 | 3.945e-16 |
| GO:BP | GO:0051984 | positive regulation of chromosome segregation | 15 | 30 | 9.009e-16 |
| GO:BP | GO:0000724 | double-strand break repair via homologous recombination | 30 | 188 | 1.600e-15 |
| GO:BP | GO:0044786 | cell cycle DNA replication | 17 | 45 | 2.278e-15 |
| GO:BP | GO:0007010 | cytoskeleton organization | 89 | 1529 | 3.536e-15 |
| GO:BP | GO:0033260 | nuclear DNA replication | 16 | 41 | 1.019e-14 |
| GO:BP | GO:0140694 | membraneless organelle assembly | 43 | 430 | 1.271e-14 |
| GO:BP | GO:0007127 | meiosis I | 25 | 133 | 1.282e-14 |
| GO:BP | GO:0051716 | cellular response to stimulus | 252 | 7376 | 1.396e-14 |
| GO:BP | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore | 13 | 23 | 1.414e-14 |
| GO:BP | GO:0034508 | centromere complex assembly | 14 | 29 | 1.878e-14 |
| GO:BP | GO:0031570 | DNA integrity checkpoint signaling | 24 | 123 | 2.009e-14 |
| GO:BP | GO:1901989 | positive regulation of cell cycle phase transition | 23 | 114 | 3.794e-14 |
| GO:BP | GO:1901992 | positive regulation of mitotic cell cycle phase transition | 21 | 93 | 6.107e-14 |
| GO:BP | GO:0022414 | reproductive process | 89 | 1608 | 6.854e-14 |
| GO:BP | GO:0000727 | double-strand break repair via break-induced replication | 10 | 12 | 8.495e-14 |
| GO:BP | GO:0019953 | sexual reproduction | 71 | 1122 | 1.113e-13 |
| GO:BP | GO:0006275 | regulation of DNA replication | 23 | 120 | 1.181e-13 |
| GO:BP | GO:0051383 | kinetochore organization | 12 | 21 | 1.550e-13 |
| GO:BP | GO:0090329 | regulation of DNA-templated DNA replication | 15 | 40 | 1.592e-13 |
| GO:BP | GO:0035556 | intracellular signal transduction | 131 | 2965 | 2.346e-13 |
| GO:BP | GO:0045931 | positive regulation of mitotic cell cycle | 23 | 124 | 2.408e-13 |
| GO:BP | GO:0010639 | negative regulation of organelle organization | 36 | 352 | 1.628e-12 |
| GO:BP | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore | 12 | 25 | 2.417e-12 |
| GO:BP | GO:0090307 | mitotic spindle assembly | 18 | 76 | 2.471e-12 |
| GO:BP | GO:0030174 | regulation of DNA-templated DNA replication initiation | 10 | 15 | 3.298e-12 |
| GO:BP | GO:0044771 | meiotic cell cycle phase transition | 10 | 15 | 3.298e-12 |
| GO:BP | GO:1901976 | regulation of cell cycle checkpoint | 15 | 48 | 3.420e-12 |
| GO:BP | GO:0050896 | response to stimulus | 281 | 8999 | 8.591e-12 |
| GO:BP | GO:0000910 | cytokinesis | 26 | 192 | 9.081e-12 |
| GO:BP | GO:0000281 | mitotic cytokinesis | 19 | 95 | 1.260e-11 |
| GO:BP | GO:0051128 | regulation of cellular component organization | 110 | 2433 | 1.573e-11 |
| GO:BP | GO:1902969 | mitotic DNA replication | 10 | 17 | 1.952e-11 |
| GO:BP | GO:0051052 | regulation of DNA metabolic process | 41 | 500 | 3.991e-11 |
| GO:BP | GO:0090231 | regulation of spindle checkpoint | 11 | 25 | 8.420e-11 |
| GO:BP | GO:0051231 | spindle elongation | 9 | 14 | 9.241e-11 |
| GO:BP | GO:0032465 | regulation of cytokinesis | 18 | 94 | 1.108e-10 |
| GO:BP | GO:0007143 | female meiotic nuclear division | 13 | 42 | 1.671e-10 |
| GO:BP | GO:0007098 | centrosome cycle | 21 | 140 | 2.373e-10 |
| GO:BP | GO:1901970 | positive regulation of mitotic sister chromatid separation | 10 | 21 | 3.090e-10 |
| GO:BP | GO:0051256 | mitotic spindle midzone assembly | 8 | 11 | 3.443e-10 |
| GO:BP | GO:0000077 | DNA damage checkpoint signaling | 19 | 114 | 3.501e-10 |
| GO:BP | GO:0030261 | chromosome condensation | 13 | 45 | 4.328e-10 |
| GO:BP | GO:0016043 | cellular component organization | 256 | 8184 | 4.428e-10 |
| GO:BP | GO:0000022 | mitotic spindle elongation | 8 | 12 | 9.828e-10 |
| GO:BP | GO:0008283 | cell population proliferation | 92 | 2015 | 1.151e-09 |
| GO:BP | GO:0000082 | G1/S transition of mitotic cell cycle | 24 | 202 | 1.151e-09 |
| GO:BP | GO:0031023 | microtubule organizing center organization | 21 | 153 | 1.258e-09 |
| GO:BP | GO:1903504 | regulation of mitotic spindle checkpoint | 10 | 24 | 1.502e-09 |
| GO:BP | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint | 10 | 24 | 1.502e-09 |
| GO:BP | GO:0061640 | cytoskeleton-dependent cytokinesis | 19 | 124 | 1.507e-09 |
| GO:BP | GO:0051302 | regulation of cell division | 23 | 188 | 1.563e-09 |
| GO:BP | GO:0034502 | protein localization to chromosome | 19 | 126 | 1.980e-09 |
| GO:BP | GO:0071459 | protein localization to chromosome, centromeric region | 12 | 41 | 2.098e-09 |
| GO:BP | GO:0050789 | regulation of biological process | 345 | 12336 | 2.235e-09 |
| GO:BP | GO:0044774 | mitotic DNA integrity checkpoint signaling | 15 | 74 | 2.700e-09 |
| GO:BP | GO:0044843 | cell cycle G1/S phase transition | 25 | 230 | 2.898e-09 |
| GO:BP | GO:0051255 | spindle midzone assembly | 8 | 14 | 5.230e-09 |
| GO:BP | GO:0065004 | protein-DNA complex assembly | 25 | 241 | 7.797e-09 |
| GO:BP | GO:0071840 | cellular component organization or biogenesis | 256 | 8393 | 7.936e-09 |
| GO:BP | GO:0071824 | protein-DNA complex organization | 26 | 262 | 8.902e-09 |
| GO:BP | GO:0034080 | CENP-A containing chromatin assembly | 7 | 10 | 9.668e-09 |
| GO:BP | GO:0031055 | chromatin remodeling at centromere | 7 | 10 | 9.668e-09 |
| GO:BP | GO:0007062 | sister chromatid cohesion | 13 | 57 | 9.808e-09 |
| GO:BP | GO:0006271 | DNA strand elongation involved in DNA replication | 8 | 15 | 1.036e-08 |
| GO:BP | GO:1903083 | protein localization to condensed chromosome | 8 | 15 | 1.036e-08 |
| GO:BP | GO:0034501 | protein localization to kinetochore | 8 | 15 | 1.036e-08 |
| GO:BP | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle | 14 | 69 | 1.053e-08 |
| GO:BP | GO:0042770 | signal transduction in response to DNA damage | 21 | 173 | 1.132e-08 |
| GO:BP | GO:1902750 | negative regulation of cell cycle G2/M phase transition | 14 | 71 | 1.554e-08 |
| GO:BP | GO:0050794 | regulation of cellular process | 333 | 11946 | 2.253e-08 |
| GO:BP | GO:0006468 | protein phosphorylation | 63 | 1227 | 2.323e-08 |
| GO:BP | GO:0032467 | positive regulation of cytokinesis | 12 | 50 | 2.372e-08 |
| GO:BP | GO:0008315 | G2/MI transition of meiotic cell cycle | 6 | 7 | 2.460e-08 |
| GO:BP | GO:0051338 | regulation of transferase activity | 36 | 498 | 2.516e-08 |
| GO:BP | GO:0016310 | phosphorylation | 66 | 1320 | 2.528e-08 |
| GO:BP | GO:0048522 | positive regulation of cellular process | 194 | 5920 | 2.881e-08 |
| GO:BP | GO:0051382 | kinetochore assembly | 8 | 17 | 3.516e-08 |
| GO:BP | GO:0070925 | organelle assembly | 56 | 1046 | 4.870e-08 |
| GO:BP | GO:1901993 | regulation of meiotic cell cycle phase transition | 7 | 12 | 5.525e-08 |
| GO:BP | GO:0065007 | biological regulation | 348 | 12743 | 5.906e-08 |
| GO:BP | GO:0044818 | mitotic G2/M transition checkpoint | 12 | 55 | 7.338e-08 |
| GO:BP | GO:0051653 | spindle localization | 13 | 68 | 8.832e-08 |
| GO:BP | GO:0051445 | regulation of meiotic cell cycle | 13 | 68 | 8.832e-08 |
| GO:BP | GO:0035825 | homologous recombination | 13 | 68 | 8.832e-08 |
| GO:BP | GO:1905821 | positive regulation of chromosome condensation | 6 | 8 | 8.997e-08 |
| GO:BP | GO:0007076 | mitotic chromosome condensation | 8 | 19 | 9.976e-08 |
| GO:BP | GO:0090232 | positive regulation of spindle checkpoint | 7 | 13 | 1.121e-07 |
| GO:BP | GO:0044773 | mitotic DNA damage checkpoint signaling | 13 | 70 | 1.254e-07 |
| GO:BP | GO:0045005 | DNA-templated DNA replication maintenance of fidelity | 12 | 59 | 1.649e-07 |
| GO:BP | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle | 9 | 28 | 1.680e-07 |
| GO:BP | GO:0010638 | positive regulation of organelle organization | 34 | 492 | 2.119e-07 |
| GO:BP | GO:0051293 | establishment of spindle localization | 12 | 63 | 3.564e-07 |
| GO:BP | GO:0000212 | meiotic spindle organization | 8 | 22 | 3.826e-07 |
| GO:BP | GO:0090306 | meiotic spindle assembly | 7 | 15 | 3.892e-07 |
| GO:BP | GO:1902751 | positive regulation of cell cycle G2/M phase transition | 9 | 31 | 4.493e-07 |
| GO:BP | GO:0006950 | response to stress | 138 | 3948 | 5.713e-07 |
| GO:BP | GO:0048518 | positive regulation of biological process | 197 | 6264 | 5.937e-07 |
| GO:BP | GO:0050790 | regulation of catalytic activity | 49 | 928 | 7.588e-07 |
| GO:BP | GO:2000241 | regulation of reproductive process | 20 | 203 | 9.242e-07 |
| GO:BP | GO:0051781 | positive regulation of cell division | 14 | 98 | 1.014e-06 |
| GO:BP | GO:0071897 | DNA biosynthetic process | 18 | 172 | 1.740e-06 |
| GO:BP | GO:0051447 | negative regulation of meiotic cell cycle | 7 | 18 | 1.740e-06 |
| GO:BP | GO:0009314 | response to radiation | 30 | 437 | 1.772e-06 |
| GO:BP | GO:0051294 | establishment of spindle orientation | 10 | 47 | 1.793e-06 |
| GO:BP | GO:0051129 | negative regulation of cellular component organization | 40 | 701 | 1.937e-06 |
| GO:BP | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | 9 | 37 | 2.322e-06 |
| GO:BP | GO:2001252 | positive regulation of chromosome organization | 14 | 105 | 2.382e-06 |
| GO:BP | GO:0060623 | regulation of chromosome condensation | 6 | 12 | 2.409e-06 |
| GO:BP | GO:0007056 | spindle assembly involved in female meiosis | 6 | 12 | 2.409e-06 |
| GO:BP | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint | 6 | 12 | 2.409e-06 |
| GO:BP | GO:1901978 | positive regulation of cell cycle checkpoint | 7 | 19 | 2.582e-06 |
| GO:BP | GO:0048856 | anatomical structure development | 187 | 5997 | 3.371e-06 |
| GO:BP | GO:0000076 | DNA replication checkpoint signaling | 7 | 20 | 3.857e-06 |
| GO:BP | GO:0031297 | replication fork processing | 10 | 51 | 3.883e-06 |
| GO:BP | GO:1903490 | positive regulation of mitotic cytokinesis | 6 | 13 | 4.262e-06 |
| GO:BP | GO:0007144 | female meiosis I | 6 | 13 | 4.262e-06 |
| GO:BP | GO:0032502 | developmental process | 200 | 6553 | 4.478e-06 |
| GO:BP | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity | 12 | 80 | 4.985e-06 |
| GO:BP | GO:0070192 | chromosome organization involved in meiotic cell cycle | 11 | 66 | 5.266e-06 |
| GO:BP | GO:0007057 | spindle assembly involved in female meiosis I | 4 | 4 | 5.589e-06 |
| GO:BP | GO:0007275 | multicellular organism development | 154 | 4727 | 5.812e-06 |
| GO:BP | GO:0045143 | homologous chromosome segregation | 11 | 67 | 6.087e-06 |
| GO:BP | GO:0046385 | deoxyribose phosphate biosynthetic process | 6 | 14 | 6.980e-06 |
| GO:BP | GO:0090235 | regulation of metaphase plate congression | 6 | 14 | 6.980e-06 |
| GO:BP | GO:0009263 | deoxyribonucleotide biosynthetic process | 6 | 14 | 6.980e-06 |
| GO:BP | GO:0009265 | 2’-deoxyribonucleotide biosynthetic process | 6 | 14 | 6.980e-06 |
| GO:BP | GO:0040001 | establishment of mitotic spindle localization | 9 | 43 | 8.440e-06 |
| GO:BP | GO:0007292 | female gamete generation | 17 | 174 | 9.462e-06 |
| GO:BP | GO:0042127 | regulation of cell population proliferation | 70 | 1685 | 9.462e-06 |
| GO:BP | GO:0000731 | DNA synthesis involved in DNA repair | 9 | 44 | 1.026e-05 |
| GO:BP | GO:0051053 | negative regulation of DNA metabolic process | 15 | 139 | 1.249e-05 |
| GO:BP | GO:1902423 | regulation of attachment of mitotic spindle microtubules to kinetochore | 5 | 9 | 1.376e-05 |
| GO:BP | GO:0048519 | negative regulation of biological process | 180 | 5834 | 1.376e-05 |
| GO:BP | GO:0007154 | cell communication | 197 | 6540 | 1.550e-05 |
| GO:BP | GO:2000242 | negative regulation of reproductive process | 11 | 74 | 1.620e-05 |
| GO:BP | GO:0007131 | reciprocal meiotic recombination | 10 | 60 | 1.712e-05 |
| GO:BP | GO:0140527 | reciprocal homologous recombination | 10 | 60 | 1.712e-05 |
| GO:BP | GO:1902412 | regulation of mitotic cytokinesis | 6 | 16 | 1.712e-05 |
| GO:BP | GO:0045840 | positive regulation of mitotic nuclear division | 9 | 47 | 1.785e-05 |
| GO:BP | GO:0090224 | regulation of spindle organization | 9 | 48 | 2.144e-05 |
| GO:BP | GO:0007165 | signal transduction | 183 | 6002 | 2.303e-05 |
| GO:BP | GO:0065009 | regulation of molecular function | 63 | 1496 | 2.309e-05 |
| GO:BP | GO:1902315 | nuclear cell cycle DNA replication initiation | 4 | 5 | 2.445e-05 |
| GO:BP | GO:1902975 | mitotic DNA replication initiation | 4 | 5 | 2.445e-05 |
| GO:BP | GO:1902292 | cell cycle DNA replication initiation | 4 | 5 | 2.445e-05 |
| GO:BP | GO:0110030 | regulation of G2/MI transition of meiotic cell cycle | 4 | 5 | 2.445e-05 |
| GO:BP | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition | 6 | 17 | 2.477e-05 |
| GO:BP | GO:0051307 | meiotic chromosome separation | 5 | 10 | 2.522e-05 |
| GO:BP | GO:0045835 | negative regulation of meiotic nuclear division | 5 | 10 | 2.522e-05 |
| GO:BP | GO:0051649 | establishment of localization in cell | 78 | 2010 | 2.554e-05 |
| GO:BP | GO:0022616 | DNA strand elongation | 8 | 37 | 2.613e-05 |
| GO:BP | GO:0023052 | signaling | 195 | 6515 | 2.782e-05 |
| GO:BP | GO:0051785 | positive regulation of nuclear division | 10 | 64 | 2.979e-05 |
| GO:BP | GO:2000045 | regulation of G1/S transition of mitotic cell cycle | 15 | 150 | 3.003e-05 |
| GO:BP | GO:0045859 | regulation of protein kinase activity | 26 | 402 | 3.204e-05 |
| GO:BP | GO:1902101 | positive regulation of metaphase/anaphase transition of cell cycle | 6 | 18 | 3.513e-05 |
| GO:BP | GO:0043549 | regulation of kinase activity | 27 | 431 | 3.685e-05 |
| GO:BP | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore | 5 | 11 | 4.375e-05 |
| GO:BP | GO:1902806 | regulation of cell cycle G1/S phase transition | 16 | 176 | 4.712e-05 |
| GO:BP | GO:0008284 | positive regulation of cell population proliferation | 45 | 954 | 4.781e-05 |
| GO:BP | GO:0071900 | regulation of protein serine/threonine kinase activity | 19 | 242 | 4.995e-05 |
| GO:BP | GO:0048523 | negative regulation of cellular process | 172 | 5629 | 5.020e-05 |
| GO:BP | GO:0032886 | regulation of microtubule-based process | 20 | 266 | 5.281e-05 |
| GO:BP | GO:0006796 | phosphate-containing compound metabolic process | 88 | 2407 | 5.348e-05 |
| GO:BP | GO:0006793 | phosphorus metabolic process | 88 | 2410 | 5.603e-05 |
| GO:BP | GO:0006338 | chromatin remodeling | 37 | 725 | 6.529e-05 |
| GO:BP | GO:0010032 | meiotic chromosome condensation | 4 | 6 | 6.619e-05 |
| GO:BP | GO:1904666 | regulation of ubiquitin protein ligase activity | 6 | 20 | 6.732e-05 |
| GO:BP | GO:0010212 | response to ionizing radiation | 14 | 141 | 6.839e-05 |
| GO:BP | GO:0071478 | cellular response to radiation | 16 | 182 | 6.987e-05 |
| GO:BP | GO:0051347 | positive regulation of transferase activity | 21 | 296 | 7.260e-05 |
| GO:BP | GO:0007064 | mitotic sister chromatid cohesion | 7 | 31 | 8.195e-05 |
| GO:BP | GO:0070507 | regulation of microtubule cytoskeleton organization | 15 | 164 | 8.498e-05 |
| GO:BP | GO:0051130 | positive regulation of cellular component organization | 49 | 1105 | 8.854e-05 |
| GO:BP | GO:0060236 | regulation of mitotic spindle organization | 8 | 44 | 9.506e-05 |
| GO:BP | GO:0051298 | centrosome duplication | 10 | 74 | 1.056e-04 |
| GO:BP | GO:0007276 | gamete generation | 40 | 841 | 1.361e-04 |
| GO:BP | GO:0009792 | embryo development ending in birth or egg hatching | 35 | 692 | 1.396e-04 |
| GO:BP | GO:0048731 | system development | 130 | 4053 | 1.420e-04 |
| GO:BP | GO:1901994 | negative regulation of meiotic cell cycle phase transition | 4 | 7 | 1.439e-04 |
| GO:BP | GO:0044785 | metaphase/anaphase transition of meiotic cell cycle | 4 | 7 | 1.439e-04 |
| GO:BP | GO:0048609 | multicellular organismal reproductive process | 45 | 1000 | 1.478e-04 |
| GO:BP | GO:0001833 | inner cell mass cell proliferation | 5 | 14 | 1.629e-04 |
| GO:BP | GO:0040020 | regulation of meiotic nuclear division | 7 | 35 | 1.848e-04 |
| GO:BP | GO:0006325 | chromatin organization | 41 | 884 | 1.848e-04 |
| GO:BP | GO:0033316 | meiotic spindle assembly checkpoint signaling | 3 | 3 | 1.889e-04 |
| GO:BP | GO:0110032 | positive regulation of G2/MI transition of meiotic cell cycle | 3 | 3 | 1.889e-04 |
| GO:BP | GO:0071163 | DNA replication preinitiation complex assembly | 3 | 3 | 1.889e-04 |
| GO:BP | GO:0009411 | response to UV | 14 | 155 | 1.910e-04 |
| GO:BP | GO:0048513 | animal organ development | 104 | 3085 | 1.932e-04 |
| GO:BP | GO:0009628 | response to abiotic stimulus | 49 | 1144 | 2.103e-04 |
| GO:BP | GO:0032506 | cytokinetic process | 8 | 50 | 2.405e-04 |
| GO:BP | GO:0031109 | microtubule polymerization or depolymerization | 13 | 138 | 2.453e-04 |
| GO:BP | GO:0030010 | establishment of cell polarity | 14 | 159 | 2.510e-04 |
| GO:BP | GO:0000132 | establishment of mitotic spindle orientation | 7 | 37 | 2.626e-04 |
| GO:BP | GO:0009157 | deoxyribonucleoside monophosphate biosynthetic process | 4 | 8 | 2.661e-04 |
| GO:BP | GO:1905832 | positive regulation of spindle assembly | 4 | 8 | 2.661e-04 |
| GO:BP | GO:1902425 | positive regulation of attachment of mitotic spindle microtubules to kinetochore | 4 | 8 | 2.661e-04 |
| GO:BP | GO:0008156 | negative regulation of DNA replication | 6 | 26 | 3.185e-04 |
| GO:BP | GO:0051299 | centrosome separation | 5 | 16 | 3.225e-04 |
| GO:BP | GO:0043009 | chordate embryonic development | 33 | 671 | 4.106e-04 |
| GO:BP | GO:0043085 | positive regulation of catalytic activity | 29 | 554 | 4.146e-04 |
| GO:BP | GO:0031399 | regulation of protein modification process | 45 | 1049 | 4.470e-04 |
| GO:BP | GO:0000915 | actomyosin contractile ring assembly | 4 | 9 | 4.548e-04 |
| GO:BP | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis | 4 | 9 | 4.548e-04 |
| GO:BP | GO:1900264 | positive regulation of DNA-directed DNA polymerase activity | 4 | 9 | 4.548e-04 |
| GO:BP | GO:1904776 | regulation of protein localization to cell cortex | 4 | 9 | 4.548e-04 |
| GO:BP | GO:0062033 | positive regulation of mitotic sister chromatid segregation | 4 | 9 | 4.548e-04 |
| GO:BP | GO:0042325 | regulation of phosphorylation | 38 | 830 | 4.764e-04 |
| GO:BP | GO:0104004 | cellular response to environmental stimulus | 21 | 339 | 4.904e-04 |
| GO:BP | GO:0071214 | cellular response to abiotic stimulus | 21 | 339 | 4.904e-04 |
| GO:BP | GO:0009394 | 2’-deoxyribonucleotide metabolic process | 7 | 41 | 5.008e-04 |
| GO:BP | GO:0071479 | cellular response to ionizing radiation | 9 | 72 | 5.131e-04 |
| GO:BP | GO:0048144 | fibroblast proliferation | 11 | 109 | 5.438e-04 |
| GO:BP | GO:0051054 | positive regulation of DNA metabolic process | 19 | 291 | 5.629e-04 |
| GO:BP | GO:1902531 | regulation of intracellular signal transduction | 71 | 1953 | 5.731e-04 |
| GO:BP | GO:0019692 | deoxyribose phosphate metabolic process | 7 | 42 | 5.762e-04 |
| GO:BP | GO:0036297 | interstrand cross-link repair | 7 | 42 | 5.762e-04 |
| GO:BP | GO:0009262 | deoxyribonucleotide metabolic process | 7 | 42 | 5.762e-04 |
| GO:BP | GO:0001932 | regulation of protein phosphorylation | 36 | 779 | 6.151e-04 |
| GO:BP | GO:1901995 | positive regulation of meiotic cell cycle phase transition | 3 | 4 | 6.624e-04 |
| GO:BP | GO:0044779 | meiotic spindle checkpoint signaling | 3 | 4 | 6.624e-04 |
| GO:BP | GO:0045740 | positive regulation of DNA replication | 7 | 43 | 6.643e-04 |
| GO:BP | GO:0007099 | centriole replication | 7 | 43 | 6.643e-04 |
| GO:BP | GO:0044837 | actomyosin contractile ring organization | 4 | 10 | 6.979e-04 |
| GO:BP | GO:0033313 | meiotic cell cycle checkpoint signaling | 4 | 10 | 6.979e-04 |
| GO:BP | GO:1904668 | positive regulation of ubiquitin protein ligase activity | 4 | 10 | 6.979e-04 |
| GO:BP | GO:2000104 | negative regulation of DNA-templated DNA replication | 4 | 10 | 6.979e-04 |
| GO:BP | GO:0044093 | positive regulation of molecular function | 39 | 884 | 7.866e-04 |
| GO:BP | GO:1902808 | positive regulation of cell cycle G1/S phase transition | 8 | 61 | 9.369e-04 |
| GO:BP | GO:0033314 | mitotic DNA replication checkpoint signaling | 4 | 11 | 1.061e-03 |
| GO:BP | GO:0009966 | regulation of signal transduction | 99 | 3034 | 1.061e-03 |
| GO:BP | GO:0072697 | protein localization to cell cortex | 4 | 11 | 1.061e-03 |
| GO:BP | GO:0007163 | establishment or maintenance of cell polarity | 16 | 234 | 1.255e-03 |
| GO:BP | GO:0072331 | signal transduction by p53 class mediator | 13 | 164 | 1.307e-03 |
| GO:BP | GO:0098534 | centriole assembly | 7 | 48 | 1.327e-03 |
| GO:BP | GO:1902103 | negative regulation of metaphase/anaphase transition of meiotic cell cycle | 3 | 5 | 1.540e-03 |
| GO:BP | GO:1905133 | negative regulation of meiotic chromosome separation | 3 | 5 | 1.540e-03 |
| GO:BP | GO:1905784 | regulation of anaphase-promoting complex-dependent catabolic process | 3 | 5 | 1.540e-03 |
| GO:BP | GO:0001832 | blastocyst growth | 5 | 22 | 1.540e-03 |
| GO:BP | GO:0007019 | microtubule depolymerization | 7 | 50 | 1.706e-03 |
| GO:BP | GO:0019220 | regulation of phosphate metabolic process | 40 | 956 | 1.888e-03 |
| GO:BP | GO:0051174 | regulation of phosphorus metabolic process | 40 | 957 | 1.924e-03 |
| GO:BP | GO:0006301 | postreplication repair | 6 | 36 | 1.928e-03 |
| GO:BP | GO:0023051 | regulation of signaling | 109 | 3478 | 2.077e-03 |
| GO:BP | GO:0010646 | regulation of cell communication | 109 | 3486 | 2.269e-03 |
| GO:BP | GO:0009162 | deoxyribonucleoside monophosphate metabolic process | 5 | 24 | 2.329e-03 |
| GO:BP | GO:0051438 | regulation of ubiquitin-protein transferase activity | 6 | 38 | 2.600e-03 |
| GO:BP | GO:0071695 | anatomical structure maturation | 17 | 277 | 2.707e-03 |
| GO:BP | GO:0046605 | regulation of centrosome cycle | 7 | 54 | 2.717e-03 |
| GO:BP | GO:0048146 | positive regulation of fibroblast proliferation | 7 | 54 | 2.717e-03 |
| GO:BP | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 5 | 25 | 2.816e-03 |
| GO:BP | GO:1902102 | regulation of metaphase/anaphase transition of meiotic cell cycle | 3 | 6 | 2.832e-03 |
| GO:BP | GO:0090166 | Golgi disassembly | 3 | 6 | 2.832e-03 |
| GO:BP | GO:0006335 | DNA replication-dependent chromatin assembly | 3 | 6 | 2.832e-03 |
| GO:BP | GO:0090402 | oncogene-induced cell senescence | 3 | 6 | 2.832e-03 |
| GO:BP | GO:2000105 | positive regulation of DNA-templated DNA replication | 4 | 14 | 2.832e-03 |
| GO:BP | GO:1905132 | regulation of meiotic chromosome separation | 3 | 6 | 2.832e-03 |
| GO:BP | GO:0016321 | female meiosis chromosome segregation | 3 | 6 | 2.832e-03 |
| GO:BP | GO:0046073 | dTMP metabolic process | 3 | 6 | 2.832e-03 |
| GO:BP | GO:0007100 | mitotic centrosome separation | 4 | 14 | 2.832e-03 |
| GO:BP | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process | 3 | 6 | 2.832e-03 |
| GO:BP | GO:0042148 | DNA strand invasion | 3 | 6 | 2.832e-03 |
| GO:BP | GO:2000780 | negative regulation of double-strand break repair | 6 | 39 | 2.879e-03 |
| GO:BP | GO:0019985 | translesion synthesis | 5 | 26 | 3.298e-03 |
| GO:BP | GO:0009130 | pyrimidine nucleoside monophosphate biosynthetic process | 4 | 15 | 3.707e-03 |
| GO:BP | GO:0045738 | negative regulation of DNA repair | 6 | 41 | 3.788e-03 |
| GO:BP | GO:0048608 | reproductive structure development | 18 | 314 | 3.883e-03 |
| GO:BP | GO:0080090 | regulation of primary metabolic process | 155 | 5390 | 3.895e-03 |
| GO:BP | GO:0072698 | protein localization to microtubule cytoskeleton | 7 | 58 | 4.046e-03 |
| GO:BP | GO:0061458 | reproductive system development | 18 | 318 | 4.483e-03 |
| GO:BP | GO:0001824 | blastocyst development | 10 | 119 | 4.681e-03 |
| GO:BP | GO:0003006 | developmental process involved in reproduction | 41 | 1038 | 4.723e-03 |
| GO:BP | GO:0048308 | organelle inheritance | 4 | 16 | 4.736e-03 |
| GO:BP | GO:0048313 | Golgi inheritance | 4 | 16 | 4.736e-03 |
| GO:BP | GO:0030154 | cell differentiation | 131 | 4437 | 4.753e-03 |
| GO:BP | GO:0009124 | nucleoside monophosphate biosynthetic process | 6 | 43 | 4.785e-03 |
| GO:BP | GO:0072528 | pyrimidine-containing compound biosynthetic process | 6 | 43 | 4.785e-03 |
| GO:BP | GO:0048869 | cellular developmental process | 131 | 4438 | 4.785e-03 |
| GO:BP | GO:2000779 | regulation of double-strand break repair | 11 | 143 | 5.072e-03 |
| GO:BP | GO:0051246 | regulation of protein metabolic process | 64 | 1855 | 5.072e-03 |
| GO:BP | GO:0045137 | development of primary sexual characteristics | 15 | 243 | 5.425e-03 |
| GO:BP | GO:0090398 | cellular senescence | 9 | 101 | 5.763e-03 |
| GO:BP | GO:0051641 | cellular localization | 111 | 3661 | 5.787e-03 |
| GO:BP | GO:0032147 | activation of protein kinase activity | 7 | 62 | 5.866e-03 |
| GO:BP | GO:0044380 | protein localization to cytoskeleton | 7 | 62 | 5.866e-03 |
| GO:BP | GO:0036211 | protein modification process | 90 | 2846 | 5.993e-03 |
| GO:BP | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle | 6 | 45 | 5.993e-03 |
| GO:BP | GO:0001701 | in utero embryonic development | 21 | 413 | 5.993e-03 |
| GO:BP | GO:0099607 | lateral attachment of mitotic spindle microtubules to kinetochore | 2 | 2 | 5.994e-03 |
| GO:BP | GO:0110029 | negative regulation of meiosis I | 2 | 2 | 5.994e-03 |
| GO:BP | GO:1990505 | mitotic DNA replication maintenance of fidelity | 2 | 2 | 5.994e-03 |
| GO:BP | GO:1902298 | cell cycle DNA replication maintenance of fidelity | 2 | 2 | 5.994e-03 |
| GO:BP | GO:0140274 | repair of kinetochore microtubule attachment defect | 2 | 2 | 5.994e-03 |
| GO:BP | GO:1990426 | mitotic recombination-dependent replication fork processing | 2 | 2 | 5.994e-03 |
| GO:BP | GO:0140273 | repair of mitotic kinetochore microtubule attachment defect | 2 | 2 | 5.994e-03 |
| GO:BP | GO:0031619 | homologous chromosome orientation in meiotic metaphase I | 2 | 2 | 5.994e-03 |
| GO:BP | GO:1905318 | meiosis I spindle assembly checkpoint signaling | 2 | 2 | 5.994e-03 |
| GO:BP | GO:1905325 | regulation of meiosis I spindle assembly checkpoint | 2 | 2 | 5.994e-03 |
| GO:BP | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway | 2 | 2 | 5.994e-03 |
| GO:BP | GO:0014846 | esophagus smooth muscle contraction | 2 | 2 | 5.994e-03 |
| GO:BP | GO:0006997 | nucleus organization | 11 | 147 | 6.065e-03 |
| GO:BP | GO:0010569 | regulation of double-strand break repair via homologous recombination | 8 | 83 | 6.646e-03 |
| GO:BP | GO:0007077 | mitotic nuclear membrane disassembly | 3 | 8 | 6.873e-03 |
| GO:BP | GO:0010458 | exit from mitosis | 5 | 31 | 6.982e-03 |
| GO:BP | GO:0007096 | regulation of exit from mitosis | 4 | 18 | 7.085e-03 |
| GO:BP | GO:0006939 | smooth muscle contraction | 9 | 105 | 7.202e-03 |
| GO:BP | GO:0009790 | embryo development | 43 | 1135 | 7.343e-03 |
| GO:BP | GO:0032501 | multicellular organismal process | 199 | 7322 | 7.376e-03 |
| GO:BP | GO:0045814 | negative regulation of gene expression, epigenetic | 13 | 200 | 7.397e-03 |
| GO:BP | GO:0031507 | heterochromatin formation | 11 | 151 | 7.430e-03 |
| GO:BP | GO:0051493 | regulation of cytoskeleton organization | 24 | 517 | 8.468e-03 |
| GO:BP | GO:0006221 | pyrimidine nucleotide biosynthetic process | 5 | 33 | 9.168e-03 |
| GO:BP | GO:0051642 | centrosome localization | 5 | 33 | 9.168e-03 |
| GO:BP | GO:0061842 | microtubule organizing center localization | 5 | 33 | 9.168e-03 |
| GO:BP | GO:0031100 | animal organ regeneration | 7 | 68 | 9.543e-03 |
| GO:BP | GO:0046601 | positive regulation of centriole replication | 3 | 9 | 9.791e-03 |
| GO:BP | GO:0046602 | regulation of mitotic centrosome separation | 3 | 9 | 9.791e-03 |
| GO:BP | GO:0090169 | regulation of spindle assembly | 5 | 34 | 1.045e-02 |
| GO:BP | GO:2000243 | positive regulation of reproductive process | 8 | 90 | 1.088e-02 |
| GO:BP | GO:0034644 | cellular response to UV | 8 | 90 | 1.088e-02 |
| GO:BP | GO:0042311 | vasodilation | 6 | 51 | 1.088e-02 |
| GO:BP | GO:1902410 | mitotic cytokinetic process | 5 | 35 | 1.185e-02 |
| GO:BP | GO:0008406 | gonad development | 14 | 238 | 1.196e-02 |
| GO:BP | GO:0010332 | response to gamma radiation | 6 | 52 | 1.199e-02 |
| GO:BP | GO:0007548 | sex differentiation | 16 | 294 | 1.234e-02 |
| GO:BP | GO:0051443 | positive regulation of ubiquitin-protein transferase activity | 4 | 21 | 1.248e-02 |
| GO:BP | GO:0021700 | developmental maturation | 18 | 352 | 1.260e-02 |
| GO:BP | GO:0001556 | oocyte maturation | 5 | 36 | 1.332e-02 |
| GO:BP | GO:0110025 | DNA strand resection involved in replication fork processing | 3 | 10 | 1.336e-02 |
| GO:BP | GO:0016477 | cell migration | 53 | 1534 | 1.399e-02 |
| GO:BP | GO:0007129 | homologous chromosome pairing at meiosis | 6 | 54 | 1.440e-02 |
| GO:BP | GO:0009129 | pyrimidine nucleoside monophosphate metabolic process | 4 | 22 | 1.470e-02 |
| GO:BP | GO:0007018 | microtubule-based movement | 22 | 478 | 1.470e-02 |
| GO:BP | GO:2000001 | regulation of DNA damage checkpoint | 4 | 22 | 1.470e-02 |
| GO:BP | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process | 2 | 3 | 1.616e-02 |
| GO:BP | GO:0072527 | pyrimidine-containing compound metabolic process | 7 | 75 | 1.616e-02 |
| GO:BP | GO:0006231 | dTMP biosynthetic process | 2 | 3 | 1.616e-02 |
| GO:BP | GO:0034421 | post-translational protein acetylation | 2 | 3 | 1.616e-02 |
| GO:BP | GO:0071312 | cellular response to alkaloid | 5 | 38 | 1.662e-02 |
| GO:BP | GO:0071392 | cellular response to estradiol stimulus | 5 | 38 | 1.662e-02 |
| GO:BP | GO:0007399 | nervous system development | 81 | 2604 | 1.690e-02 |
| GO:BP | GO:0007140 | male meiotic nuclear division | 6 | 56 | 1.691e-02 |
| GO:BP | GO:0009888 | tissue development | 66 | 2032 | 1.691e-02 |
| GO:BP | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle | 6 | 56 | 1.691e-02 |
| GO:BP | GO:0000018 | regulation of DNA recombination | 10 | 144 | 1.700e-02 |
| GO:BP | GO:0046599 | regulation of centriole replication | 4 | 23 | 1.700e-02 |
| GO:BP | GO:0009219 | pyrimidine deoxyribonucleotide metabolic process | 4 | 23 | 1.700e-02 |
| GO:BP | GO:0048511 | rhythmic process | 16 | 305 | 1.709e-02 |
| GO:BP | GO:0009176 | pyrimidine deoxyribonucleoside monophosphate metabolic process | 3 | 11 | 1.709e-02 |
| GO:BP | GO:0070601 | centromeric sister chromatid cohesion | 3 | 11 | 1.709e-02 |
| GO:BP | GO:1990918 | double-strand break repair involved in meiotic recombination | 3 | 11 | 1.709e-02 |
| GO:BP | GO:0009650 | UV protection | 3 | 11 | 1.709e-02 |
| GO:BP | GO:0072711 | cellular response to hydroxyurea | 3 | 11 | 1.709e-02 |
| GO:BP | GO:0048583 | regulation of response to stimulus | 116 | 3993 | 1.732e-02 |
| GO:BP | GO:0006282 | regulation of DNA repair | 13 | 225 | 1.938e-02 |
| GO:BP | GO:0006029 | proteoglycan metabolic process | 8 | 100 | 1.980e-02 |
| GO:BP | GO:0009123 | nucleoside monophosphate metabolic process | 7 | 79 | 2.094e-02 |
| GO:BP | GO:0006998 | nuclear envelope organization | 6 | 59 | 2.149e-02 |
| GO:BP | GO:0035295 | tube development | 40 | 1101 | 2.154e-02 |
| GO:BP | GO:0072710 | response to hydroxyurea | 3 | 12 | 2.187e-02 |
| GO:BP | GO:0042276 | error-prone translesion synthesis | 3 | 12 | 2.187e-02 |
| GO:BP | GO:0014831 | gastro-intestinal system smooth muscle contraction | 3 | 12 | 2.187e-02 |
| GO:BP | GO:0051081 | nuclear membrane disassembly | 3 | 12 | 2.187e-02 |
| GO:BP | GO:0071493 | cellular response to UV-B | 3 | 12 | 2.187e-02 |
| GO:BP | GO:0071168 | protein localization to chromatin | 6 | 60 | 2.308e-02 |
| GO:BP | GO:0008584 | male gonad development | 10 | 152 | 2.430e-02 |
| GO:BP | GO:1902807 | negative regulation of cell cycle G1/S phase transition | 6 | 61 | 2.501e-02 |
| GO:BP | GO:0046546 | development of primary male sexual characteristics | 10 | 153 | 2.538e-02 |
| GO:BP | GO:0006760 | folic acid-containing compound metabolic process | 4 | 26 | 2.580e-02 |
| GO:BP | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | 4 | 26 | 2.580e-02 |
| GO:BP | GO:0006915 | apoptotic process | 62 | 1923 | 2.625e-02 |
| GO:BP | GO:0030866 | cortical actin cytoskeleton organization | 5 | 43 | 2.707e-02 |
| GO:BP | GO:0030397 | membrane disassembly | 3 | 13 | 2.736e-02 |
| GO:BP | GO:1903251 | multi-ciliated epithelial cell differentiation | 3 | 13 | 2.736e-02 |
| GO:BP | GO:0031401 | positive regulation of protein modification process | 26 | 637 | 2.765e-02 |
| GO:BP | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | 7 | 84 | 2.846e-02 |
| GO:BP | GO:0009416 | response to light stimulus | 16 | 323 | 2.846e-02 |
| GO:BP | GO:0046075 | dTTP metabolic process | 2 | 4 | 2.851e-02 |
| GO:BP | GO:0051296 | establishment of meiotic spindle orientation | 2 | 4 | 2.851e-02 |
| GO:BP | GO:1901563 | response to camptothecin | 2 | 4 | 2.851e-02 |
| GO:BP | GO:0051754 | meiotic sister chromatid cohesion, centromeric | 2 | 4 | 2.851e-02 |
| GO:BP | GO:1905448 | positive regulation of mitochondrial ATP synthesis coupled electron transport | 2 | 4 | 2.851e-02 |
| GO:BP | GO:0009212 | pyrimidine deoxyribonucleoside triphosphate biosynthetic process | 2 | 4 | 2.851e-02 |
| GO:BP | GO:0072757 | cellular response to camptothecin | 2 | 4 | 2.851e-02 |
| GO:BP | GO:0007079 | mitotic chromosome movement towards spindle pole | 2 | 4 | 2.851e-02 |
| GO:BP | GO:0006235 | dTTP biosynthetic process | 2 | 4 | 2.851e-02 |
| GO:BP | GO:0043060 | meiotic metaphase I homologous chromosome alignment | 2 | 4 | 2.851e-02 |
| GO:BP | GO:1990086 | lens fiber cell apoptotic process | 2 | 4 | 2.851e-02 |
| GO:BP | GO:0009987 | cellular process | 467 | 20247 | 2.998e-02 |
| GO:BP | GO:0048469 | cell maturation | 12 | 211 | 3.014e-02 |
| GO:BP | GO:0045860 | positive regulation of protein kinase activity | 13 | 239 | 3.030e-02 |
| GO:BP | GO:0043412 | macromolecule modification | 90 | 3030 | 3.185e-02 |
| GO:BP | GO:0048870 | cell motility | 58 | 1793 | 3.207e-02 |
| GO:BP | GO:0001934 | positive regulation of protein phosphorylation | 21 | 483 | 3.207e-02 |
| GO:BP | GO:0051446 | positive regulation of meiotic cell cycle | 4 | 28 | 3.236e-02 |
| GO:BP | GO:0051145 | smooth muscle cell differentiation | 6 | 65 | 3.239e-02 |
| GO:BP | GO:0040029 | epigenetic regulation of gene expression | 15 | 299 | 3.239e-02 |
| GO:BP | GO:0051177 | meiotic sister chromatid cohesion | 3 | 14 | 3.251e-02 |
| GO:BP | GO:0033262 | regulation of nuclear cell cycle DNA replication | 3 | 14 | 3.251e-02 |
| GO:BP | GO:0012501 | programmed cell death | 63 | 1987 | 3.347e-02 |
| GO:BP | GO:0141124 | intracellular signaling cassette | 61 | 1912 | 3.378e-02 |
| GO:BP | GO:0006284 | base-excision repair | 5 | 46 | 3.427e-02 |
| GO:BP | GO:0008219 | cell death | 63 | 1991 | 3.481e-02 |
| GO:BP | GO:0010165 | response to X-ray | 4 | 29 | 3.614e-02 |
| GO:BP | GO:0070977 | bone maturation | 4 | 29 | 3.614e-02 |
| GO:BP | GO:0050678 | regulation of epithelial cell proliferation | 17 | 364 | 3.665e-02 |
| GO:BP | GO:0010604 | positive regulation of macromolecule metabolic process | 96 | 3288 | 3.684e-02 |
| GO:BP | GO:1902117 | positive regulation of organelle assembly | 7 | 89 | 3.684e-02 |
| GO:BP | GO:0035418 | protein localization to synapse | 7 | 89 | 3.684e-02 |
| GO:BP | GO:0010824 | regulation of centrosome duplication | 5 | 47 | 3.705e-02 |
| GO:BP | GO:0000723 | telomere maintenance | 10 | 164 | 3.818e-02 |
| GO:BP | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway | 3 | 15 | 3.873e-02 |
| GO:BP | GO:0044027 | negative regulation of gene expression via chromosomal CpG island methylation | 3 | 15 | 3.873e-02 |
| GO:BP | GO:0043101 | purine-containing compound salvage | 3 | 15 | 3.873e-02 |
| GO:BP | GO:0048145 | regulation of fibroblast proliferation | 7 | 90 | 3.873e-02 |
| GO:BP | GO:0071772 | response to BMP | 10 | 165 | 3.930e-02 |
| GO:BP | GO:0042752 | regulation of circadian rhythm | 8 | 114 | 3.930e-02 |
| GO:BP | GO:0071773 | cellular response to BMP stimulus | 10 | 165 | 3.930e-02 |
| GO:BP | GO:0071763 | nuclear membrane organization | 5 | 48 | 3.985e-02 |
| GO:BP | GO:0051179 | localization | 151 | 5555 | 3.985e-02 |
| GO:BP | GO:0044087 | regulation of cellular component biogenesis | 36 | 1011 | 4.290e-02 |
| GO:BP | GO:0071962 | mitotic sister chromatid cohesion, centromeric | 2 | 5 | 4.297e-02 |
| GO:BP | GO:0140499 | negative regulation of mitotic spindle assembly checkpoint signaling | 2 | 5 | 4.297e-02 |
| GO:BP | GO:0001938 | positive regulation of endothelial cell proliferation | 7 | 92 | 4.297e-02 |
| GO:BP | GO:0098535 | de novo centriole assembly involved in multi-ciliated epithelial cell differentiation | 2 | 5 | 4.297e-02 |
| GO:BP | GO:0043137 | DNA replication, removal of RNA primer | 2 | 5 | 4.297e-02 |
| GO:BP | GO:2000278 | regulation of DNA biosynthetic process | 7 | 92 | 4.297e-02 |
| GO:BP | GO:0006297 | nucleotide-excision repair, DNA gap filling | 2 | 5 | 4.297e-02 |
| GO:BP | GO:0051311 | meiotic metaphase chromosome alignment | 2 | 5 | 4.297e-02 |
| GO:BP | GO:0031536 | positive regulation of exit from mitosis | 2 | 5 | 4.297e-02 |
| GO:BP | GO:0090233 | negative regulation of spindle checkpoint | 2 | 5 | 4.297e-02 |
| GO:BP | GO:0097421 | liver regeneration | 4 | 31 | 4.383e-02 |
| GO:BP | GO:0009719 | response to endogenous stimulus | 49 | 1489 | 4.436e-02 |
| GO:BP | GO:0090399 | replicative senescence | 3 | 16 | 4.507e-02 |
| GO:BP | GO:0051782 | negative regulation of cell division | 3 | 16 | 4.507e-02 |
| GO:BP | GO:0033674 | positive regulation of kinase activity | 13 | 254 | 4.618e-02 |
| GO:BP | GO:0050918 | positive chemotaxis | 6 | 71 | 4.639e-02 |
| GO:BP | GO:0061351 | neural precursor cell proliferation | 10 | 171 | 4.851e-02 |
| GO:BP | GO:0042558 | pteridine-containing compound metabolic process | 4 | 32 | 4.857e-02 |
| GO:BP | GO:2000145 | regulation of cell motility | 36 | 1022 | 4.892e-02 |
| GO:BP | GO:0050673 | epithelial cell proliferation | 19 | 441 | 4.920e-02 |
| GO:BP | GO:2000573 | positive regulation of DNA biosynthetic process | 5 | 51 | 4.921e-02 |
| GO:BP | GO:0042327 | positive regulation of phosphorylation | 21 | 506 | 4.921e-02 |
| GO:BP | GO:0045910 | negative regulation of DNA recombination | 5 | 51 | 4.921e-02 |
| GO:BP | GO:0090102 | cochlea development | 5 | 51 | 4.921e-02 |
| GO:BP | GO:0050767 | regulation of neurogenesis | 17 | 378 | 4.921e-02 |
| KEGG | KEGG:04110 | Cell cycle | 47 | 157 | 1.969e-33 |
| KEGG | KEGG:03030 | DNA replication | 16 | 36 | 7.231e-14 |
| KEGG | KEGG:03460 | Fanconi anemia pathway | 12 | 54 | 2.356e-06 |
| KEGG | KEGG:04114 | Oocyte meiosis | 17 | 131 | 7.617e-06 |
| KEGG | KEGG:03440 | Homologous recombination | 10 | 41 | 7.617e-06 |
| KEGG | KEGG:04914 | Progesterone-mediated oocyte maturation | 15 | 102 | 7.617e-06 |
| KEGG | KEGG:04115 | p53 signaling pathway | 12 | 74 | 3.877e-05 |
| KEGG | KEGG:04218 | Cellular senescence | 17 | 155 | 6.260e-05 |
| KEGG | KEGG:04814 | Motor proteins | 18 | 193 | 2.823e-04 |
| KEGG | KEGG:05166 | Human T-cell leukemia virus 1 infection | 17 | 218 | 4.291e-03 |
| KEGG | KEGG:03410 | Base excision repair | 7 | 44 | 5.154e-03 |
| KEGG | KEGG:03430 | Mismatch repair | 5 | 23 | 8.520e-03 |
| KEGG | KEGG:00240 | Pyrimidine metabolism | 7 | 57 | 2.191e-02 |
| KEGG | KEGG:05222 | Small cell lung cancer | 9 | 92 | 2.217e-02 |
| KEGG | KEGG:01232 | Nucleotide metabolism | 8 | 84 | 4.533e-02 |
| KEGG | KEGG:04151 | PI3K-Akt signaling pathway | 20 | 356 | 4.856e-02 |
# write.csv(table_motif2_GOKEGG_genes, "data/new/table_motif2_GOKEGG_genes.csv")
#GO:BP
table_motif2_genes_GOBP_RUV_clust <- table_motif2_GOKEGG_genes_RUV_clust %>%
dplyr::filter(source=="GO:BP") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
# saveRDS(table_motif2_genes_GOBP, "data/table_motif2_genes_GOBP.RDS")
table_motif2_genes_GOBP_RUV_clust %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("M2 Enriched GO:BP Terms RUVs clustlike")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("GO:BP term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))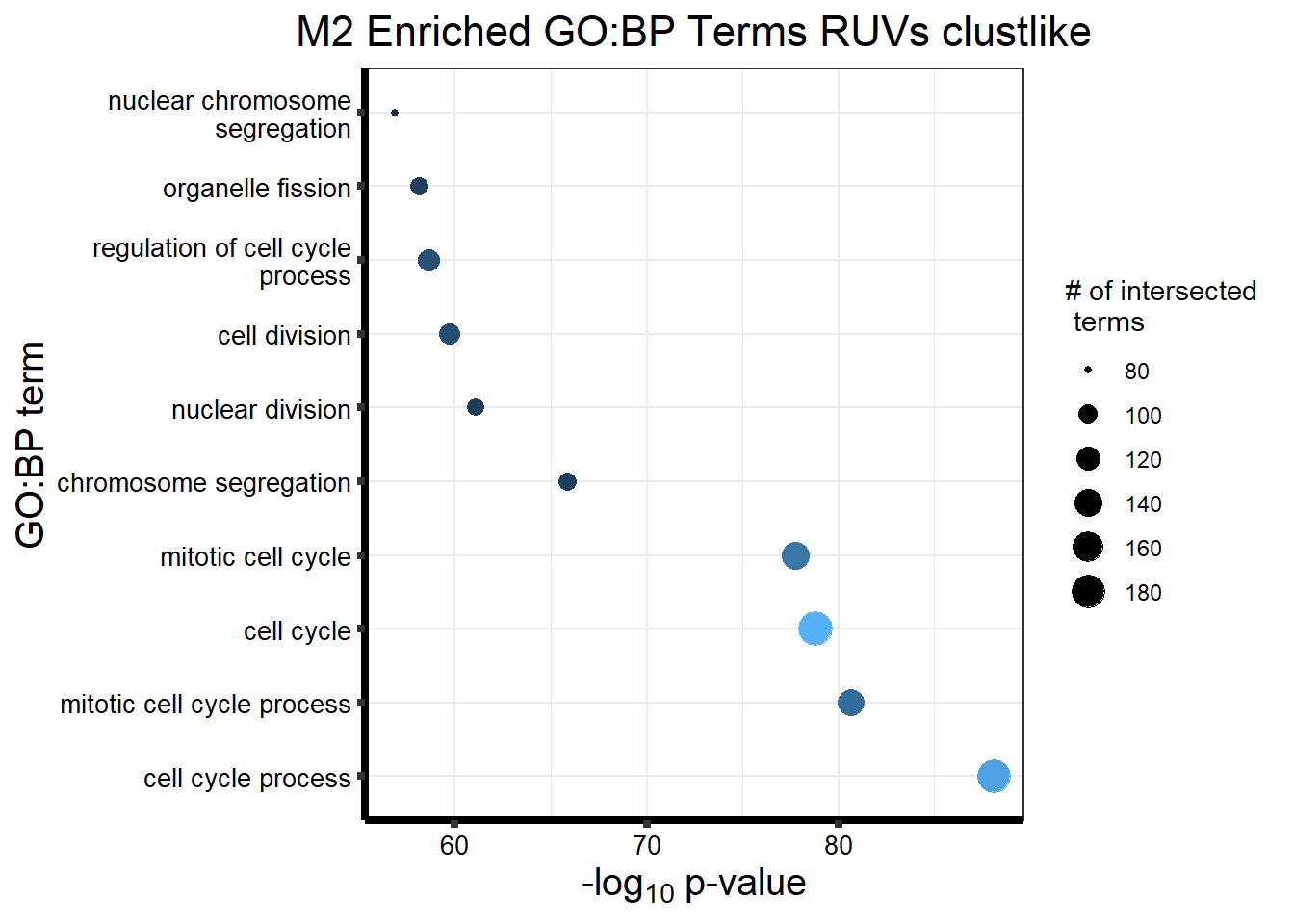
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#KEGG
table_motif2_genes_KEGG_RUV_clust <- table_motif2_GOKEGG_genes_RUV_clust %>%
dplyr::filter(source=="KEGG") %>%
dplyr::select(p_value, term_name, intersection_size) %>%
dplyr::slice_min(., n=10, order_by=p_value) %>%
mutate(log_val = -log10(p_value))
table_motif2_genes_KEGG_RUV_clust %>% ggplot(., aes(x = log_val, y = reorder(term_name, p_value), col= intersection_size)) +
geom_point(aes(size = intersection_size)) +
ggtitle("M2 DEGs Enriched KEGG Terms RUVs clustlike")+
xlab(expression("-log"[10]~"p-value"))+
guides(col="none", size= guide_legend(title = "# of intersected \n terms"))+
ylab("KEGG term")+
scale_y_discrete(labels = scales::label_wrap(30))+
theme_bw()+
theme(plot.title = element_text(size = rel(1.5), hjust = 0.5),
axis.title = element_text(size = 15, colour = "black"),
axis.ticks = element_line(linewidth = 1.5),
axis.line = element_line(linewidth = 1.5),
axis.text = element_text(size = 10, colour = "black", angle = 0),
strip.text = element_text(size = 15, colour = "black", face = "bold"))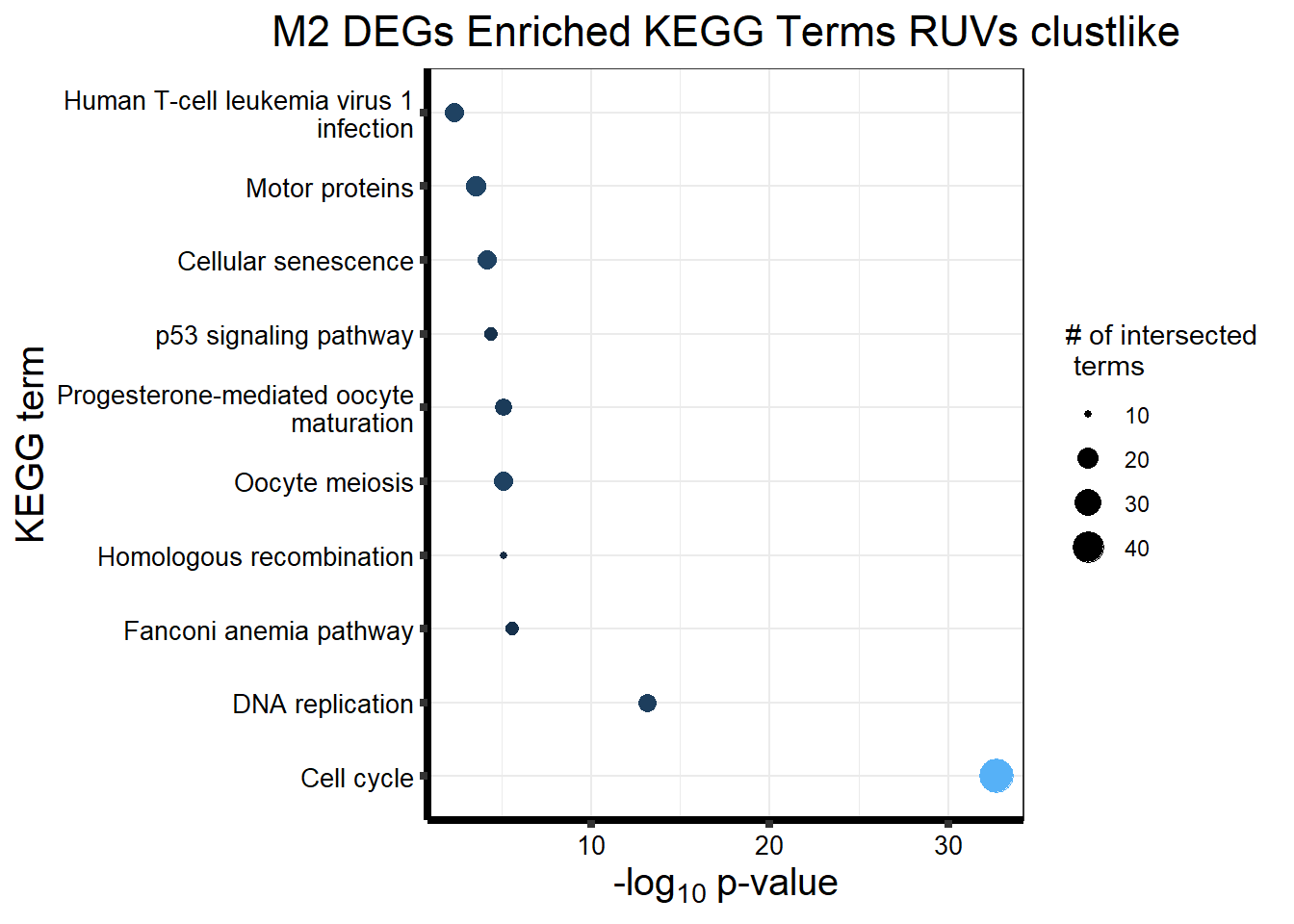
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
##Attempt Cormotif with Norm Counts Only
# #now since I'm having some strange outputs with the transformed data
# #I want to try and just feed in the normalized counts only
#
normcounts_CMF_rep <- as.data.frame(set1$normalizedCounts)
#
#first want to cut out the replicate samples as not needed for Cormotif
normcounts_CMF <- as.data.frame(normcounts_CMF_rep) %>%
dplyr::select(-(contains("6R")))
# #14319 genes in 36 columns, set up for plugging into Cormotif
# #now I need to set up my groupid and compid accordingly
# #put together my group id and comparison id to make the correct comparisons between experimental conditions
# #groupid tells which experimental conditions are grouped together
# #compid tells which experimental conditions should be compared against one another
# ##ie DOX24T vs DMSO24T matched control
#groupid that doesn't require changing order of matrix:
# groupid_other <- c(
# DOX_24T_1 = 1,
# DMSO_24T_1 = 4,
# DOX_24R_1 = 2,
# DMSO_24R_1 = 5,
# DOX_144R_1 = 3,
# DMSO_144R_1 = 6,
#
# DOX_24T_2 = 1,
# DMSO_24T_2 = 4,
# DOX_24R_2 = 2,
# DMSO_24R_2 = 5,
# DOX_144R_2 = 3,
# DMSO_144R_2 = 6,
#
# DOX_24T_3 = 1,
# DMSO_24T_3 = 4,
# DOX_24R_3 = 2,
# DMSO_24R_3 = 5,
# DOX_144R_3 = 3,
# DMSO_144R_3 = 6,
#
# DOX_24T_4 = 1,
# DMSO_24T_4 = 4,
# DOX_24R_4 = 2,
# DMSO_24R_4 = 5,
# DOX_144R_4 = 3,
# DMSO_144R_4 = 6,
#
# DOX_24T_5 = 1,
# DMSO_24T_5 = 4,
# DOX_24R_5 = 2,
# DMSO_24R_5 = 5,
# DOX_144R_5 = 3,
# DMSO_144R_5 = 6,
#
# DOX_24T_6 = 1,
# DMSO_24T_6 = 4,
# DOX_24R_6 = 2,
# DMSO_24R_6 = 5,
# DOX_144R_6 = 3,
# DMSO_144R_6 = 6
# )
groupid_other <- readRDS("data/new/RUV/groupid_other_unrearranged.RDS")
compid <- data.frame(Cond1 = c(1, 3, 5), Cond2 = c(2, 4, 6))
compid1 <- data.frame(Cond1 = c(1, 2, 3), Cond2 = c(4, 5, 6))###Run Cormotif Norm Counts Only
# #set the seed the same as the previous one to ensure consistency
#
# #fit Cormotif model
# # set.seed(19191)
# # #only set the seed ONCE
# #
# # motif.fit_RUV_NC_test <- cormotiffit(
# # exprs = exprs.NC_CMF_RUV,
# # groupid = groupid_other,
# # compid = compid,
# # K = 1:10,
# # max.iter = 1000,
# # BIC = TRUE,
# # runtype = "counts"
# # )
#
#saveRDS(motif.fit_RUV_NC_test, "data/new/RUV/motif.fit_RUVnormcounts_compid_250630.RDS")
motif.fit_RUV_NC_test <- readRDS("data/new/RUV/motif.fit_RUVnormcounts_250630.RDS")###Plot Cormotif Norm Counts Only
# #plot the AIC and BIC (we're interested in the Bayesian most of all)
plotIC(motif.fit_RUV_NC_test)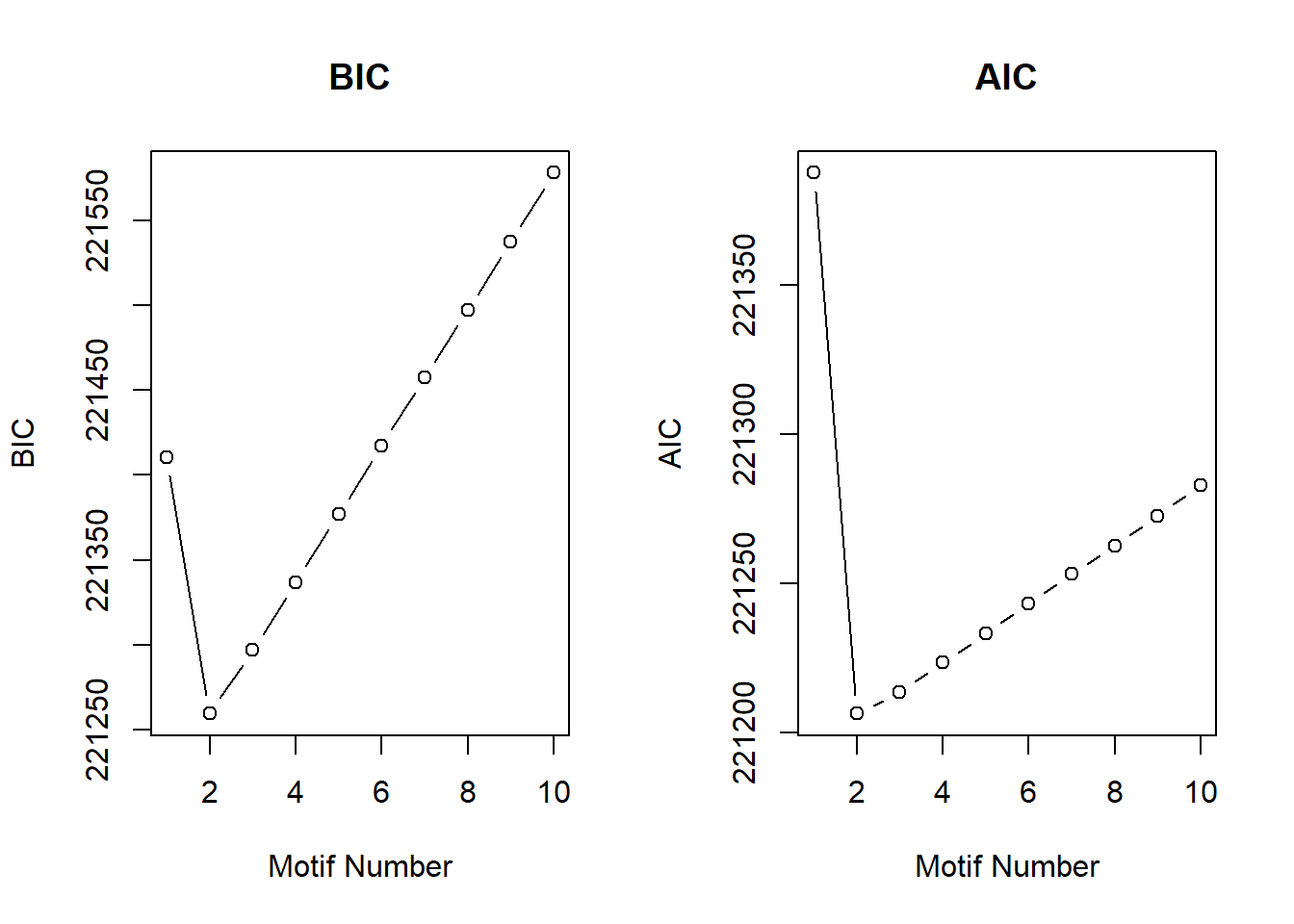
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#
# #visual check of the lowest BIC
# motif.fit_RUV_NC_test$bic
#
# #plot the motifs
plotMotif(motif.fit_RUV_NC_test)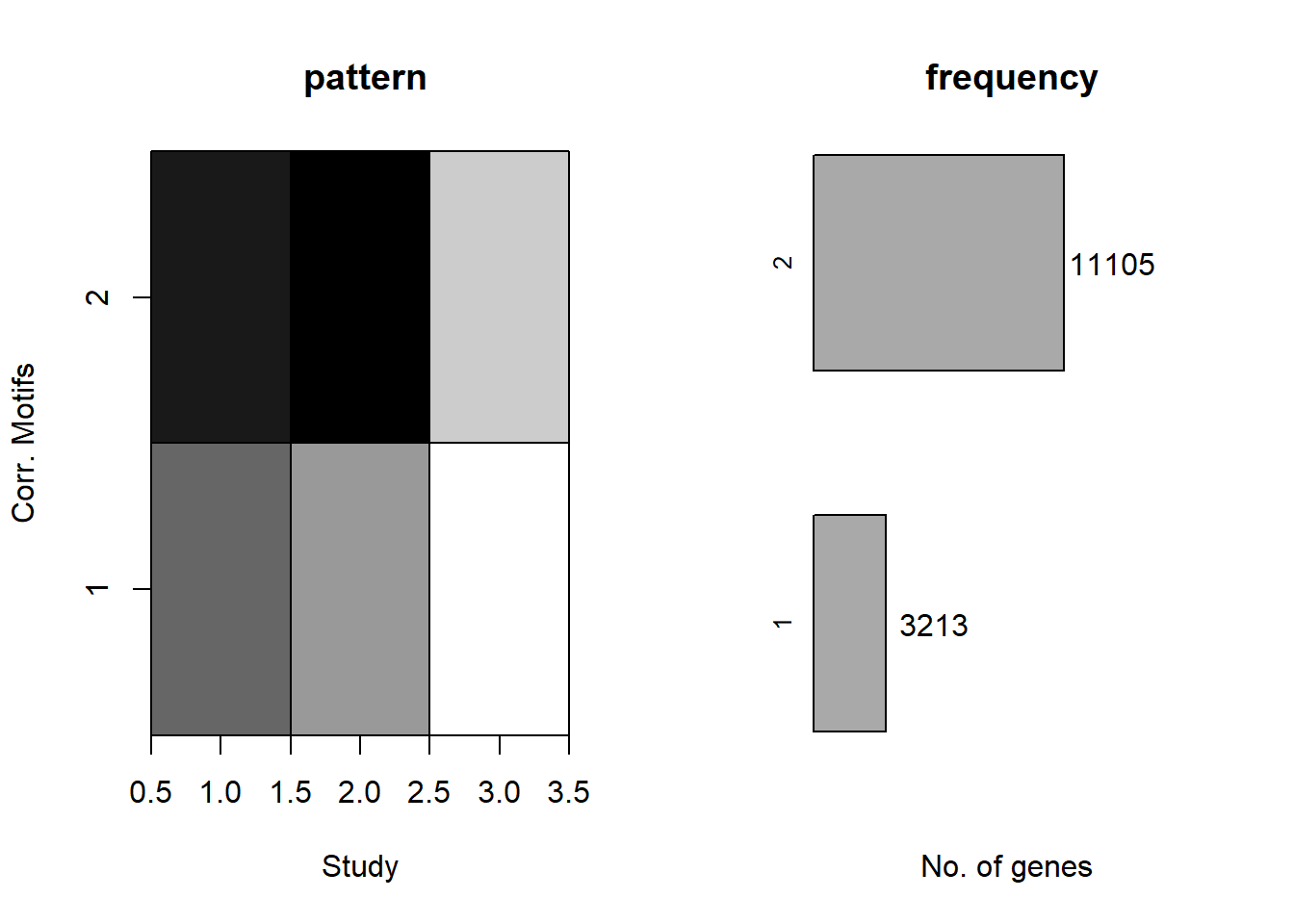
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
#
#plot the probability legend
myColors <- rev(c("#FFFFFF", "#E6E6E6" ,"#CCCCCC", "#B3B3B3", "#999999", "#808080", "#666666","#4C4C4C", "#333333", "#191919","#000000"))
plot.new()
legend('bottomleft',fill=myColors, legend =rev(c("0", "0.1", "0.2", "0.3", "0.4", "0.5", "0.6", "0.7", "0.8","0.9", "1")), box.col="white",title = "Probability\nlegend", horiz=FALSE,title.cex=.8)
| Version | Author | Date |
|---|---|---|
| 0b1ffad | emmapfort | 2025-07-02 |
Now that I’ve finished up with Cormotif, I want to make comparisons across DEGs and Cormotif analysis to identify DEGs within my motifs I also want to find out if any of my DEGs within a certain category (DDR, DIC, p53 target genes) are enriched in a motif.
#testing for each motif whether DNA damage response genes are enriched in that motif compared to all other tested genes
DNA_damage_genes <- readRDS("data/new/RUV/DDR_genes_entrezid.RDS")
total_DNA_damage_genes <- length(DNA_damage_genes) #Total number of DNA damage genes
#should be 65 genes
#define my Cormotif gene lists
CMF_genes <- list(
"Motif1" = motif1_genes_matrix, "Motif2" = motif2_genes_matrix, "Motif3" = motif3_genes_matrix
)
all_tested_genes <- unique(unlist(CMF_genes))
#this has the 12087 used for clustlike+ p.post together
#usual gene set is 14319
#compute my sets
ddr_set <- unique(na.omit(DNA_damage_genes))
motif_enrichment <- purrr::map_dfr(names(CMF_genes), function(motif_name) {
motif_genes <- unique(CMF_genes[[motif_name]])
# #print results
# print(motif_enrichment)
# Count in motif
in_motif_ddr <- sum(motif_genes %in% ddr_set)
#number of genes inside the motif that are DDR
in_motif_non_ddr <- length(motif_genes) - in_motif_ddr
#number of genes inside motif that are NOT DDR
# Count outside motif
other_genes <- setdiff(all_tested_genes, motif_genes)
#checking the difference between all genes and motif genes
out_motif_ddr <- sum(other_genes %in% ddr_set)
#all genes outside motif that are DDR
out_motif_non_ddr <- length(other_genes) - out_motif_ddr
#all genes outside the motif that are NOT DDR
# Build contingency table
contingency_ddr_cmf <- matrix(c(in_motif_ddr, in_motif_non_ddr,
out_motif_ddr, out_motif_non_ddr),
nrow = 2, byrow = TRUE)
#print results to check size of contingency table
print(contingency_ddr_cmf)
# Perform fisher's exact test - sample size too small for chi-square
fisher_test_ddr_cmf <- fisher.test(contingency_ddr_cmf)
p_value <- fisher_test_ddr_cmf$p.value
# Output summary
tibble(
Motif = motif_name,
In_Motif_DDR = in_motif_ddr,
In_Motif_NonDDR = in_motif_non_ddr,
Out_Motif_DDR = out_motif_ddr,
Out_Motif_NonDDR = out_motif_non_ddr,
Total_Genes = length(motif_genes),
DNA_Damage_Proportion = in_motif_ddr / length(motif_genes) * 100,
P_Value = fisher_test_ddr_cmf$p.value
)
}) %>%
mutate(Significance = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)) [,1] [,2]
[1,] 14 5624
[2,] 32 6417
[,1] [,2]
[1,] 22 6196
[2,] 24 5845
[,1] [,2]
[1,] 10 221
[2,] 36 11820# Recreate motif_long from motif_enrichment (with fixed column names)
motif_long <- motif_enrichment %>%
select(Motif, In_Motif_DDR, In_Motif_NonDDR, P_Value) %>% # include P_Value
pivot_longer(cols = c(In_Motif_DDR, In_Motif_NonDDR),
names_to = "Category", values_to = "Count") %>%
mutate(
Category = ifelse(Category == "In_Motif_DDR", "Yes", "No"),
Fraction = Count / ave(Count, Motif, FUN = sum),
# Add significance stars inline
Significant = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)
)
#now I want to make sure that the yes is at the top so I have to reorder
motif_long$Category <- factor(motif_long$Category, levels = c("Yes", "No"))
#ensure that the stars are also on top for significance
label_positions <- motif_long %>%
filter(Category == "Yes") %>%
mutate(y_pos = Fraction + 1.01) # 101% above the bar for visual clarity
# Plot with corrected star positioning
ggplot(motif_long, aes(x = Motif, y = Fraction, fill = Category)) +
geom_bar(stat = "identity", position = "stack") +
geom_text(
data = label_positions %>% filter(Significant != ""),
aes(x = Motif, y = y_pos, label = Significant),
size = 6,
color = "black",
fontface = "bold"
) +
scale_y_continuous(labels = scales::percent_format(scale = 1), limits = c(0, 1.08)) +
scale_fill_manual(values = c("Yes" = "#e41a1c", "No" = "#377eb8")) +
labs(
title = "Proportion of DNA Damage Genes per Cormotif Motif",
x = "Cormotif Motif",
y = "Percentage",
fill = "DDR Gene"
) +
theme_minimal() +
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
plot.title = element_text(size = 16, hjust = 0.5)
)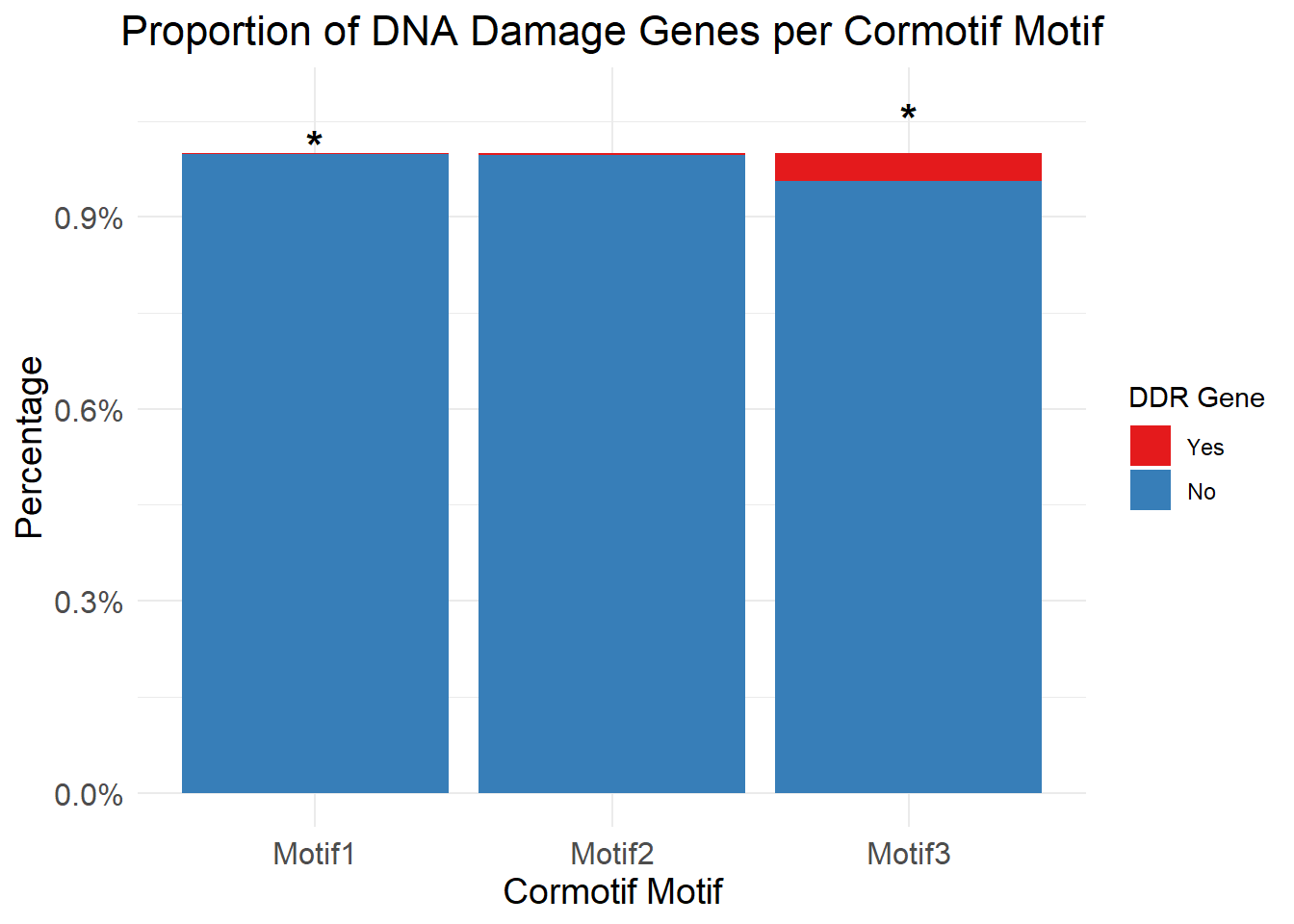
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#print the proportions so I know the exact percentages and p values
print(motif_long)# A tibble: 6 × 6
Motif P_Value Category Count Fraction Significant
<chr> <dbl> <fct> <int> <dbl> <chr>
1 Motif1 0.0370 Yes 14 0.00248 "*"
2 Motif1 0.0370 No 5624 0.998 "*"
3 Motif2 0.659 Yes 22 0.00354 ""
4 Motif2 0.659 No 6196 0.996 ""
5 Motif3 0.0000000119 Yes 10 0.0433 "*"
6 Motif3 0.0000000119 No 221 0.957 "*" DOX_cardiotox_genes <- readRDS("data/new/RUV/DIC_genes_entrezid.RDS")
total_DIC_genes <- length(DOX_cardiotox_genes) #Total number of DNA damage genes
#should be 29
#define my Cormotif gene lists
CMF_genes <- list(
"Motif1" = motif1_genes_matrix, "Motif2" = motif2_genes_matrix, "Motif3" = motif3_genes_matrix
)
all_tested_genes <- unique(unlist(CMF_genes))
#this has the 12087 used for clustlike+ p.post together
#usual gene set is 14319
#compute my sets
dic_set <- unique(na.omit(DOX_cardiotox_genes))
motif_enrichment_dic <- purrr::map_dfr(names(CMF_genes), function(motif_name) {
motif_genes <- unique(CMF_genes[[motif_name]])
# #print this result to compare
# print(motif_enrichment_dic)
# Count in motif
in_motif_dic <- sum(motif_genes %in% dic_set)
#number of genes inside the motif that are dic
in_motif_non_dic <- length(motif_genes) - in_motif_dic
#number of genes inside motif that are NOT dic
# Count outside motif
other_genes <- setdiff(all_tested_genes, motif_genes)
#checking the difference between all genes and motif genes
out_motif_dic <- sum(other_genes %in% dic_set)
#all genes outside motif that are dic
out_motif_non_dic <- length(other_genes) - out_motif_dic
#all genes outside the motif that are NOT dic
# Build contingency table
contingency_dic_cmf <- matrix(c(in_motif_dic, in_motif_non_dic,
out_motif_dic, out_motif_non_dic),
nrow = 2, byrow = TRUE)
#print my contingency table to check sample size
print(contingency_dic_cmf)
# Perform fisher's exact test (sample size too small for chi-square)
fisher_test_dic_cmf <- fisher.test(contingency_dic_cmf)
p_value <- fisher_test_dic_cmf$p.value
# Output summary
tibble(
Motif = motif_name,
In_Motif_dic = in_motif_dic,
In_Motif_Nondic = in_motif_non_dic,
Out_Motif_dic = out_motif_dic,
Out_Motif_Nondic = out_motif_non_dic,
Total_Genes = length(motif_genes),
DNA_Damage_Proportion = in_motif_dic / length(motif_genes) * 100,
P_Value = fisher_test_dic_cmf$p.value
)
}) %>%
mutate(Significance = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)) [,1] [,2]
[1,] 8 5630
[2,] 15 6434
[,1] [,2]
[1,] 14 6204
[2,] 9 5860
[,1] [,2]
[1,] 1 230
[2,] 22 11834# Recreate motif_long from motif_enrichment (with fixed column names)
motif_long_dic <- motif_enrichment_dic %>%
select(Motif, In_Motif_dic, In_Motif_Nondic, P_Value) %>% # include P_Value
pivot_longer(cols = c(In_Motif_dic, In_Motif_Nondic),
names_to = "Category", values_to = "Count") %>%
mutate(
Category = ifelse(Category == "In_Motif_dic", "Yes", "No"),
Fraction = Count / ave(Count, Motif, FUN = sum),
# Add significance stars inline
Significant = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)
)
#now I want to make sure that the yes is at the top so I have to reorder
motif_long_dic$Category <- factor(motif_long_dic$Category, levels = c("Yes", "No"))
#ensure that the stars are also on top for significance
label_positions_dic <- motif_long_dic %>%
filter(Category == "Yes") %>%
mutate(y_pos = Fraction + 1.01) # 101% above the bar for visual clarity
# Plot with corrected star positioning
ggplot(motif_long_dic, aes(x = Motif, y = Fraction, fill = Category)) +
geom_bar(stat = "identity", position = "stack") +
geom_text(
data = label_positions_dic %>% filter(Significant != ""),
aes(x = Motif, y = y_pos, label = Significant),
size = 6,
color = "black",
fontface = "bold"
) +
scale_y_continuous(labels = scales::percent_format(scale = 1), limits = c(0, 1.08)) +
scale_fill_manual(values = c("Yes" = "#e41a1c", "No" = "#377eb8")) +
labs(
title = "Proportion of DOX Cardiotox Genes per Cormotif Motif",
x = "Cormotif Motif",
y = "Percentage",
fill = "DIC Gene"
) +
theme_minimal() +
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
plot.title = element_text(size = 16, hjust = 0.5)
)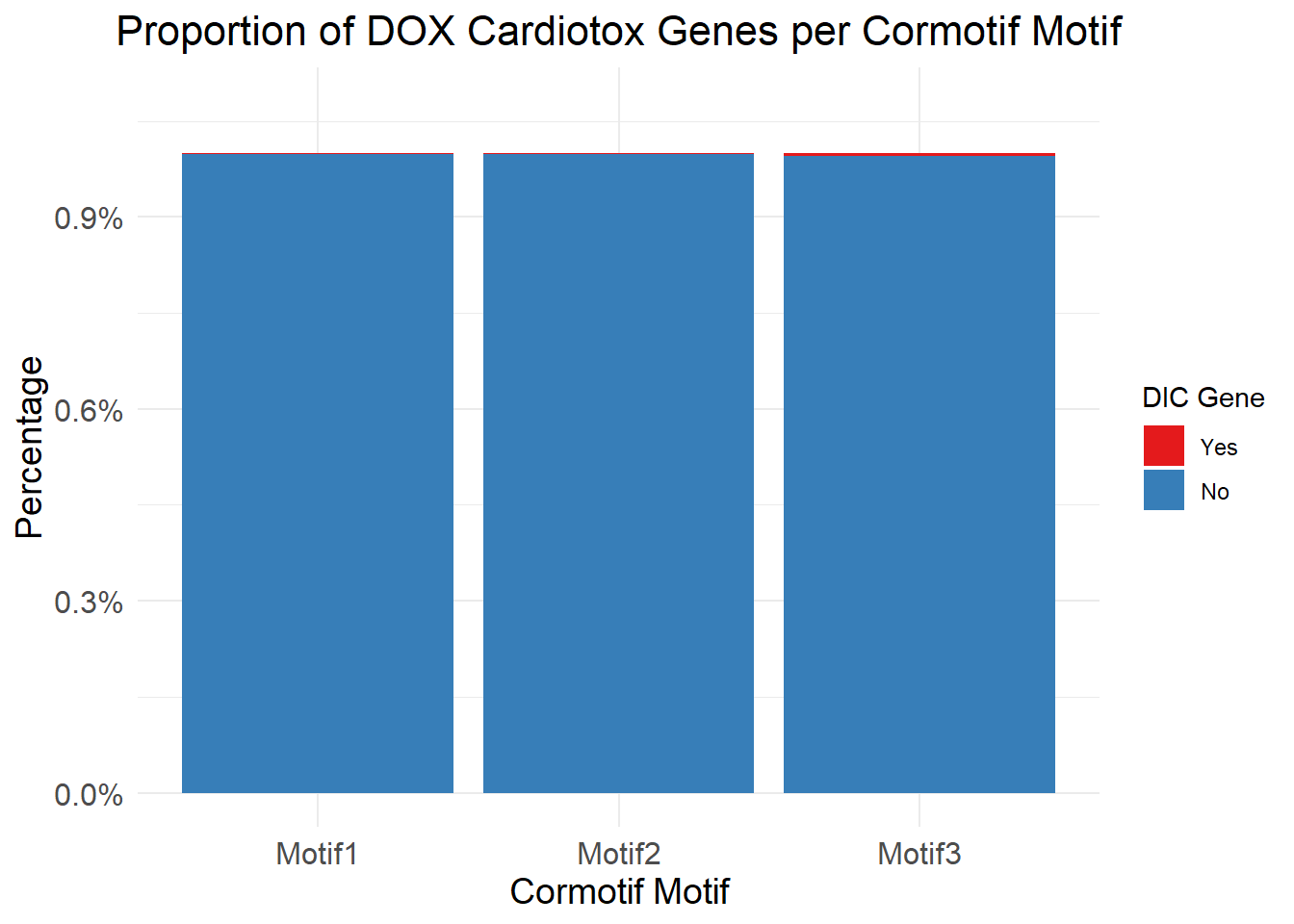
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#print this so I know the p-value and percentages
print(motif_long_dic)# A tibble: 6 × 6
Motif P_Value Category Count Fraction Significant
<chr> <dbl> <fct> <int> <dbl> <chr>
1 Motif1 0.299 Yes 8 0.00142 ""
2 Motif1 0.299 No 5630 0.999 ""
3 Motif2 0.409 Yes 14 0.00225 ""
4 Motif2 0.409 No 6204 0.998 ""
5 Motif3 0.359 Yes 1 0.00433 ""
6 Motif3 0.359 No 230 0.996 "" p53_target_genes <- readRDS("data/new/RUV/p53_targetgenes_entrezid.RDS")
total_p53_genes <- length(p53_target_genes) #Total number of p53 target genes
#should be 300
#define my Cormotif gene lists
CMF_genes <- list(
"Motif1" = motif1_genes_matrix, "Motif2" = motif2_genes_matrix, "Motif3" = motif3_genes_matrix
)
all_tested_genes <- unique(unlist(CMF_genes))
#this has the 12087 used for clustlike+ p.post together
#usual gene set is 14319
#compute my sets
p53_set <- unique(na.omit(p53_target_genes))
motif_enrichment_p53 <- purrr::map_dfr(names(CMF_genes), function(motif_name) {
motif_genes <- unique(CMF_genes[[motif_name]])
# #print this dataset
# print(motif_enrichment_p53)
# Count in motif
in_motif_p53 <- sum(motif_genes %in% p53_set)
#number of genes inside the motif that are p53
in_motif_non_p53 <- length(motif_genes) - in_motif_p53
#number of genes inside motif that are NOT p53
# Count outside motif
other_genes <- setdiff(all_tested_genes, motif_genes)
#checking the difference between all genes and motif genes
out_motif_p53 <- sum(other_genes %in% p53_set)
#all genes outside motif that are p53
out_motif_non_p53 <- length(other_genes) - out_motif_p53
#all genes outside the motif that are NOT p53
# Build contingency table
contingency_p53_cmf <- matrix(c(in_motif_p53, in_motif_non_p53,
out_motif_p53, out_motif_non_p53),
nrow = 2, byrow = TRUE)
#print the results so I can visualize the contingency tables for my motifs
print(contingency_p53_cmf)
# Perform fishers exact test (sample size too small for chi-square)
fisher_test_p53_cmf <- fisher.test(contingency_p53_cmf)
p_value <- fisher_test_p53_cmf$p.value
# Output summary
tibble(
Motif = motif_name,
In_Motif_p53 = in_motif_p53,
In_Motif_Nonp53 = in_motif_non_p53,
Out_Motif_p53 = out_motif_p53,
Out_Motif_Nonp53 = out_motif_non_p53,
Total_Genes = length(motif_genes),
DNA_Damage_Proportion = in_motif_p53 / length(motif_genes) * 100,
P_Value = fisher_test_p53_cmf$p.value
)
}) %>%
mutate(Significance = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)) [,1] [,2]
[1,] 122 5516
[2,] 102 6347
[,1] [,2]
[1,] 84 6134
[2,] 140 5729
[,1] [,2]
[1,] 18 213
[2,] 206 11650# Recreate motif_long from motif_enrichment (with fixed column names)
motif_long_p53 <- motif_enrichment_p53 %>%
select(Motif, In_Motif_p53, In_Motif_Nonp53, P_Value) %>% # include P_Value
pivot_longer(cols = c(In_Motif_p53, In_Motif_Nonp53),
names_to = "Category", values_to = "Count") %>%
mutate(
Category = ifelse(Category == "In_Motif_p53", "Yes", "No"),
Fraction = Count / ave(Count, Motif, FUN = sum),
# Add significance stars inline
Significant = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)
)
#now I want to make sure that the yes is at the top so I have to reorder
motif_long_p53$Category <- factor(motif_long_p53$Category, levels = c("Yes", "No"))
#ensure that the stars are also on top for significance
label_positions_p53 <- motif_long_p53 %>%
filter(Category == "Yes") %>%
mutate(y_pos = Fraction + 1.01) # 101% above the bar for visual clarity
# Plot with corrected star positioning
ggplot(motif_long_p53, aes(x = Motif, y = Fraction, fill = Category)) +
geom_bar(stat = "identity", position = "stack") +
geom_text(
data = label_positions_p53 %>% filter(Significant != ""),
aes(x = Motif, y = y_pos, label = Significant),
size = 6,
color = "black",
fontface = "bold"
) +
scale_y_continuous(labels = scales::percent_format(scale = 1), limits = c(0, 1.08)) +
scale_fill_manual(values = c("Yes" = "#e41a1c", "No" = "#377eb8")) +
labs(
title = "Proportion of p53 Target Genes per Cormotif Motif",
x = "Cormotif Motif",
y = "Percentage",
fill = "p53 Gene"
) +
theme_minimal() +
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
plot.title = element_text(size = 16, hjust = 0.5)
)Warning: Removed 1 row containing missing values or values outside the scale range
(`geom_text()`).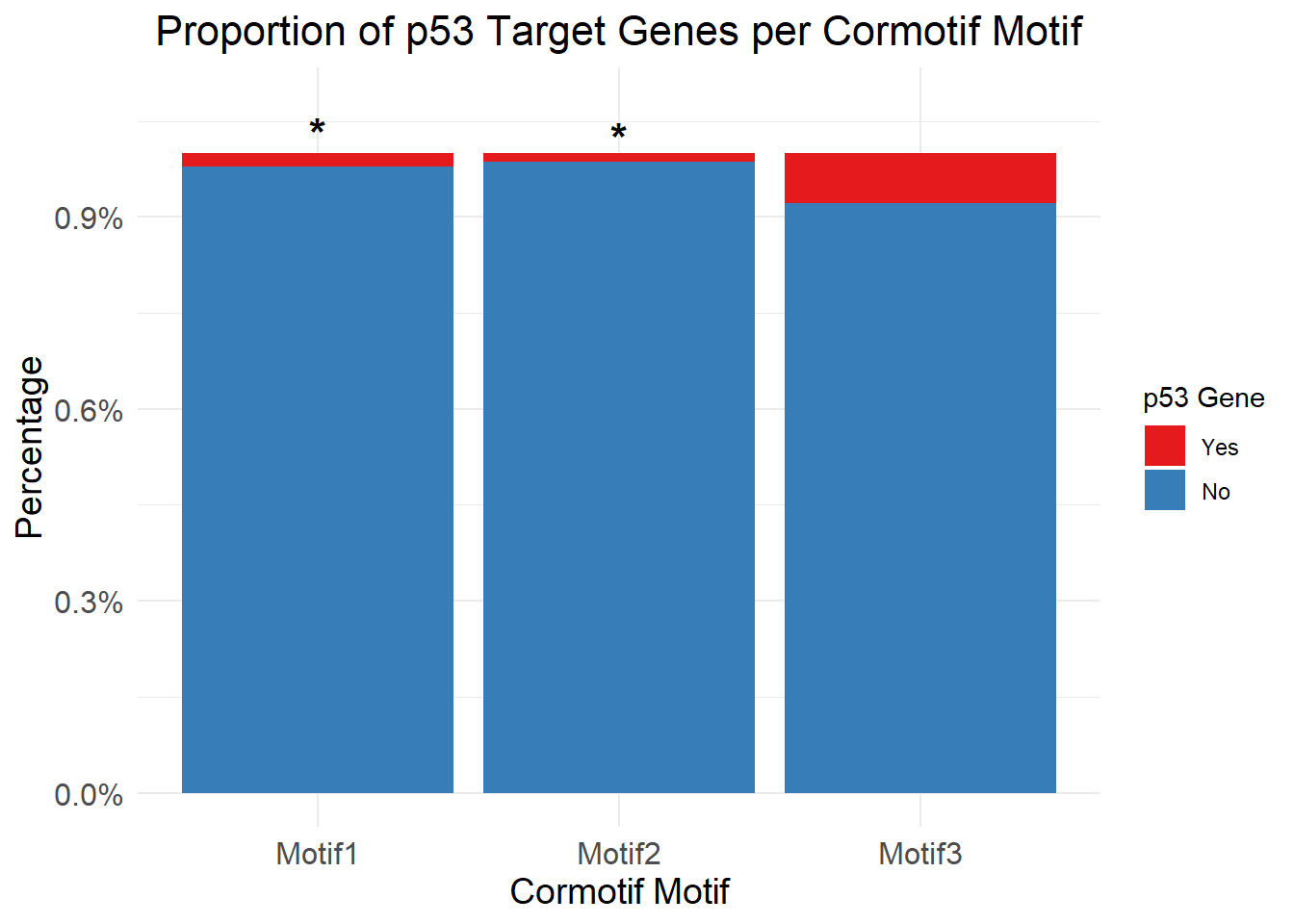
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
#print these data to check percentages and p-value
print(motif_long_p53)# A tibble: 6 × 6
Motif P_Value Category Count Fraction Significant
<chr> <dbl> <fct> <int> <dbl> <chr>
1 Motif1 0.0213 Yes 122 0.0216 *
2 Motif1 0.0213 No 5516 0.978 *
3 Motif2 0.0000262 Yes 84 0.0135 *
4 Motif2 0.0000262 No 6134 0.986 *
5 Motif3 0.000000285 Yes 18 0.0779 *
6 Motif3 0.000000285 No 213 0.922 * ##Cormotif Proportion Barplots New Motifs RUVs Corrected ###DDR Genes Proportion Barplot RUVs Cormotif
# DNA_damage_genes <- readRDS("data/new/RUV/DNA_damage_genes_symbolentrez.RDS")
DNA_damage_genes <- readRDS("data/new/RUV/DDR_genes_entrezid.RDS")
total_DNA_damage_genes <- length(DNA_damage_genes) #Total number of DNA damage genes
#should be 65 genes
#define my Cormotif gene lists
CMF_genes_RUV <- list(
"Motif1" = final_genes_1_RUV,
"Motif2" = final_genes_2_RUV
)
all_tested_genes_RUV <- unique(unlist(CMF_genes_RUV))
#this has the 12087 used for clustlike+ p.post together
#usual gene set is 14319
#compute my sets
ddr_set <- unique(na.omit(DNA_damage_genes))
motif_enrichment_RUV <- purrr::map_dfr(names(CMF_genes_RUV), function(motif_name) {
motif_genes_RUV <- unique(CMF_genes_RUV[[motif_name]])
#print these results
# print(motif_enrichment_RUV)
# Count in motif
in_motif_ddr_RUV <- sum(motif_genes_RUV %in% ddr_set)
#number of genes inside the motif that are DDR
in_motif_non_ddr_RUV <- length(motif_genes_RUV) - in_motif_ddr_RUV
#number of genes inside motif that are NOT DDR
# Count outside motif
other_genes_RUV <- setdiff(all_tested_genes_RUV, motif_genes_RUV)
#checking the difference between all genes and motif genes
out_motif_ddr_RUV <- sum(other_genes_RUV %in% ddr_set)
#all genes outside motif that are DDR
out_motif_non_ddr_RUV <- length(other_genes_RUV) - out_motif_ddr_RUV
#all genes outside the motif that are NOT DDR
# Build contingency table
contingency_ddr_cmf_RUV <- matrix(c(in_motif_ddr_RUV, in_motif_non_ddr_RUV,
out_motif_ddr_RUV, out_motif_non_ddr_RUV),
nrow = 2, byrow = TRUE)
#print the contingency table to check sample size
print(contingency_ddr_cmf_RUV)
# Perform fisher's exact test - sample size too small and unreliable for chi-square
fisher_test_ddr_cmf_RUV <- fisher.test(contingency_ddr_cmf_RUV)
p_value_RUV <- fisher_test_ddr_cmf_RUV$p.value
# Output summary
tibble(
Motif = motif_name,
In_Motif_DDR = in_motif_ddr_RUV,
In_Motif_NonDDR = in_motif_non_ddr_RUV,
Out_Motif_DDR = out_motif_ddr_RUV,
Out_Motif_NonDDR = out_motif_non_ddr_RUV,
Total_Genes = length(motif_genes_RUV),
DNA_Damage_Proportion = in_motif_ddr_RUV / length(motif_genes_RUV) * 100,
P_Value = fisher_test_ddr_cmf_RUV$p.value
)
}) %>%
mutate(Significance = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)) [,1] [,2]
[1,] 24 7578
[2,] 16 485
[,1] [,2]
[1,] 16 485
[2,] 24 7578# Recreate motif_long from motif_enrichment (with fixed column names)
motif_long_RUV <- motif_enrichment_RUV %>%
select(Motif, In_Motif_DDR, In_Motif_NonDDR, P_Value) %>% # include P_Value
pivot_longer(cols = c(In_Motif_DDR, In_Motif_NonDDR),
names_to = "Category", values_to = "Count") %>%
mutate(
Category = ifelse(Category == "In_Motif_DDR", "Yes", "No"),
Fraction = Count / ave(Count, Motif, FUN = sum),
# Add significance stars inline
Significant = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)
)
#now I want to make sure that the yes is at the top so I have to reorder
motif_long_RUV$Category <- factor(motif_long_RUV$Category, levels = c("Yes", "No"))
#ensure that the stars are also on top for significance
label_positions_RUV <- motif_long_RUV %>%
filter(Category == "Yes") %>%
mutate(y_pos = Fraction + 1.01) # 101% above the bar for visual clarity
# Plot with corrected star positioning
ggplot(motif_long_RUV, aes(x = Motif, y = Fraction, fill = Category)) +
geom_bar(stat = "identity", position = "stack") +
geom_text(
data = label_positions_RUV %>% filter(Significant != ""),
aes(x = Motif, y = y_pos, label = Significant),
size = 6,
color = "black",
fontface = "bold"
) +
scale_y_continuous(labels = scales::percent_format(scale = 1), limits = c(0, 1.08)) +
scale_fill_manual(values = c("Yes" = "#e41a1c", "No" = "#377eb8")) +
labs(
title = "Proportion DDR Genes per Motif RUVs clustlike + p.post",
x = "Cormotif Motif",
y = "Percentage",
fill = "DDR Gene"
) +
theme_minimal() +
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
plot.title = element_text(size = 16, hjust = 0.5)
)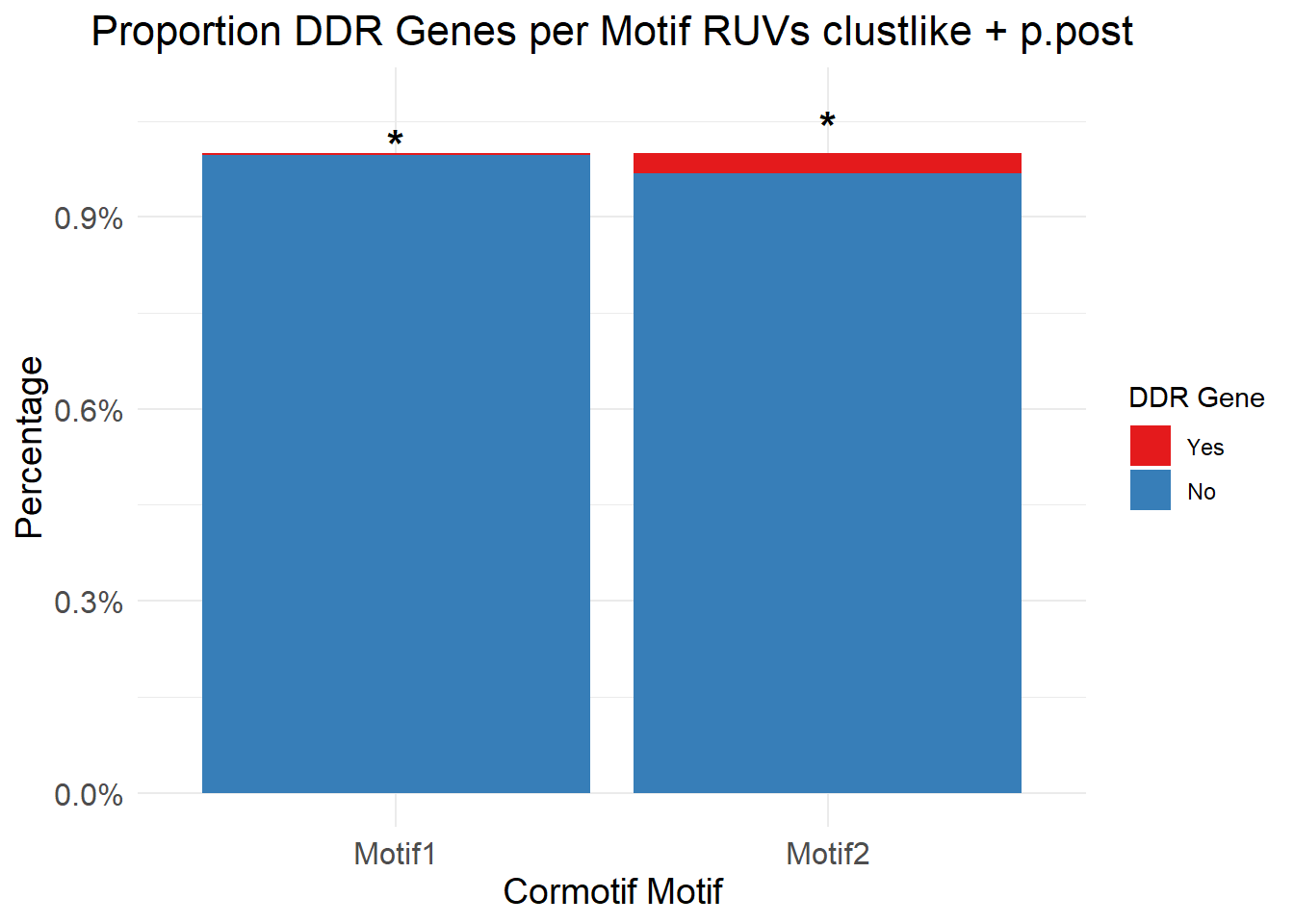
| Version | Author | Date |
|---|---|---|
| 0ae67cb | emmapfort | 2025-07-10 |
#print this to see p-value and percentages
print(motif_long_RUV)# A tibble: 4 × 6
Motif P_Value Category Count Fraction Significant
<chr> <dbl> <fct> <int> <dbl> <chr>
1 Motif1 5.67e-10 Yes 24 0.00316 *
2 Motif1 5.67e-10 No 7578 0.997 *
3 Motif2 5.67e-10 Yes 16 0.0319 *
4 Motif2 5.67e-10 No 485 0.968 * ###DIC Genes Cormotif RUVs
DOX_cardiotox_genes <- readRDS("data/new/RUV/DIC_genes_entrezid.RDS")
total_DIC_genes <- length(DOX_cardiotox_genes) #Total number of DNA damage genes
#should be 29
#define my Cormotif gene lists
CMF_genes_RUV_clust <- list(
"Motif1" = clust1_genes_RUV, "Motif2" = clust2_genes_RUV
)
all_tested_genes <- unique(unlist(CMF_genes_RUV_clust))
#this has the 14117 genes used for clustlike only
#usual gene set is 14319
#compute my sets
dic_set <- unique(na.omit(DOX_cardiotox_genes))
motif_enrichment_dic_RUV_clust <- purrr::map_dfr(names(CMF_genes_RUV_clust), function(motif_name) {
motif_genes_dic_RUV_clust <- unique(CMF_genes_RUV_clust[[motif_name]])
#print this to get a summary of results
# print(motif_enrichment_dic_RUV_clust)
# Count in motif
in_motif_dic_RUV_clust <- sum(motif_genes_dic_RUV_clust %in% dic_set)
#number of genes inside the motif that are dic
in_motif_non_dic_RUV_clust <- length(motif_genes_dic_RUV_clust) - in_motif_dic_RUV_clust
#number of genes inside motif that are NOT dic
# Count outside motif
other_genes_dic_RUV_clust <- setdiff(all_tested_genes, motif_genes_dic_RUV_clust)
#checking the difference between all genes and motif genes
out_motif_dic_RUV_clust <- sum(other_genes_dic_RUV_clust %in% dic_set)
#all genes outside motif that are dic
out_motif_non_dic_RUV_clust <- length(other_genes_dic_RUV_clust) - out_motif_dic_RUV_clust
#all genes outside the motif that are NOT dic
# Build contingency table
contingency_dic_cmf_RUV_clust <- matrix(c(in_motif_dic_RUV_clust,
in_motif_non_dic_RUV_clust,
out_motif_dic_RUV_clust,
out_motif_non_dic_RUV_clust),
nrow = 2, byrow = TRUE)
#print
print(contingency_dic_cmf_RUV_clust)
# Perform fisher's exact test (sample size too small for chi-square)
fisher_test_dic_cmf_RUV_clust <- fisher.test(contingency_dic_cmf_RUV_clust)
p_value <- fisher_test_dic_cmf_RUV_clust$p.value
# Output summary
tibble(
Motif = motif_name,
In_Motif_dic = in_motif_dic_RUV_clust,
In_Motif_Nondic = in_motif_non_dic_RUV_clust,
Out_Motif_dic = out_motif_dic_RUV_clust,
Out_Motif_Nondic = out_motif_non_dic_RUV_clust,
Total_Genes = length(motif_genes_dic_RUV_clust),
DNA_Damage_Proportion = in_motif_dic_RUV_clust / length(motif_genes_dic_RUV_clust) * 100,
P_Value = fisher_test_dic_cmf_RUV_clust$p.value
)
}) %>%
mutate(Significance = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)) [,1] [,2]
[1,] 22 13578
[2,] 1 516
[,1] [,2]
[1,] 1 516
[2,] 22 13578# Recreate motif_long from motif_enrichment (with fixed column names)
motif_long_dic_RUV_clust <- motif_enrichment_dic_RUV_clust %>%
select(Motif, In_Motif_dic, In_Motif_Nondic, P_Value) %>% # include P_Value
pivot_longer(cols = c(In_Motif_dic, In_Motif_Nondic),
names_to = "Category", values_to = "Count") %>%
mutate(
Category = ifelse(Category == "In_Motif_dic", "Yes", "No"),
Fraction = Count / ave(Count, Motif, FUN = sum),
# Add significance stars inline
Significant = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)
)
#now I want to make sure that the yes is at the top so I have to reorder
motif_long_dic_RUV_clust$Category <- factor(motif_long_dic_RUV_clust$Category, levels = c("Yes", "No"))
#ensure that the stars are also on top for significance
label_positions_dic_RUV_clust <- motif_long_dic_RUV_clust %>%
filter(Category == "Yes") %>%
mutate(y_pos = Fraction + 1.01) # 101% above the bar for visual clarity
# Plot with corrected star positioning
ggplot(motif_long_dic_RUV_clust, aes(x = Motif, y = Fraction, fill = Category)) +
geom_bar(stat = "identity", position = "stack") +
geom_text(
data = label_positions_dic_RUV_clust %>% filter(Significant != ""),
aes(x = Motif, y = y_pos, label = Significant),
size = 6,
color = "black",
fontface = "bold"
) +
scale_y_continuous(labels = scales::percent_format(scale = 1), limits = c(0, 1.08)) +
scale_fill_manual(values = c("Yes" = "#e41a1c", "No" = "#377eb8")) +
labs(
title = "Proportion DOX Cardiotox Genes per Motif RUVs clustlike",
x = "Cormotif Motif",
y = "Percentage",
fill = "DIC Gene"
) +
theme_minimal() +
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
plot.title = element_text(size = 16, hjust = 0.5)
)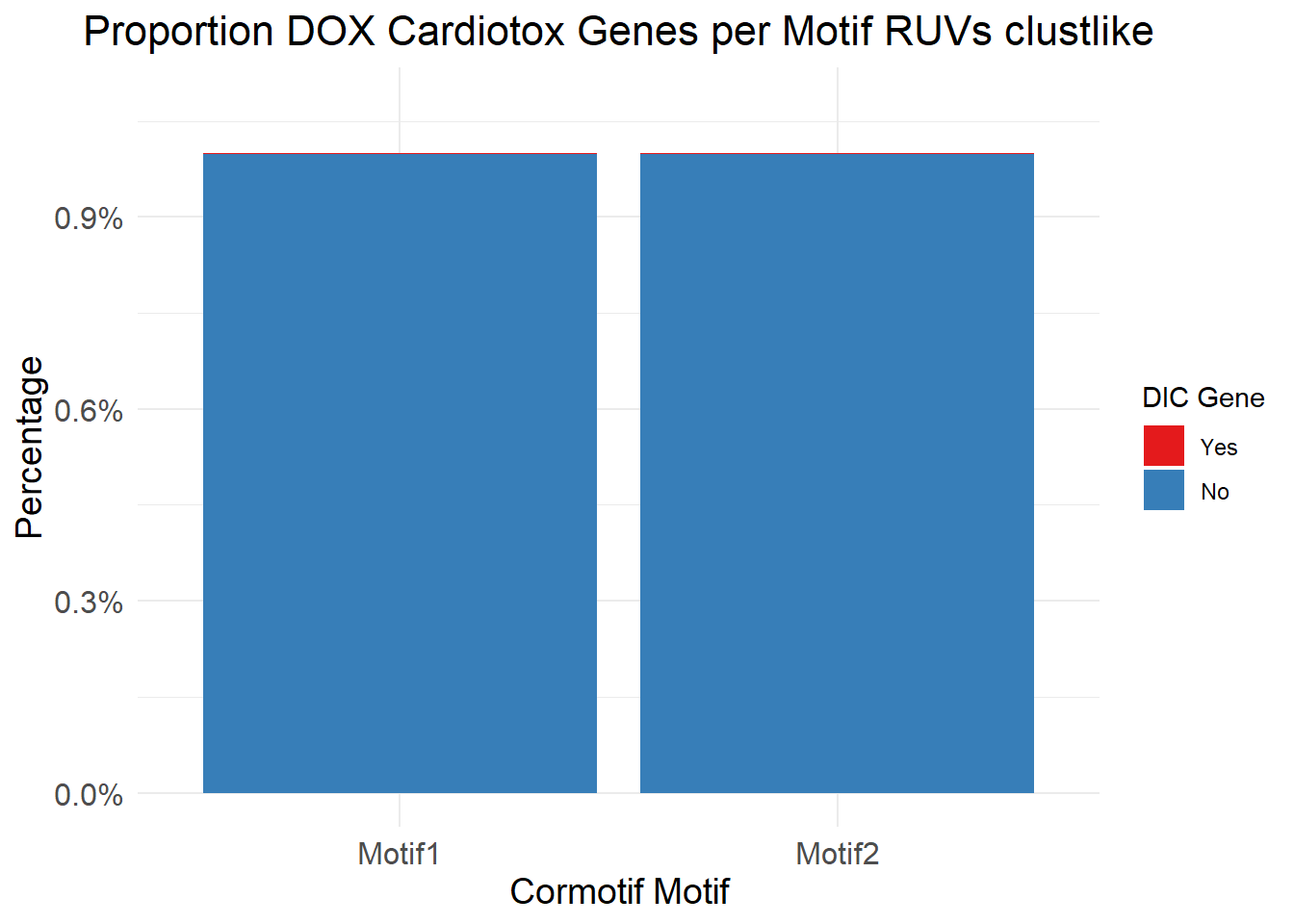
| Version | Author | Date |
|---|---|---|
| 0ae67cb | emmapfort | 2025-07-10 |
#print these data to check p-value and percentages
print(motif_long_dic_RUV_clust)# A tibble: 4 × 6
Motif P_Value Category Count Fraction Significant
<chr> <dbl> <fct> <int> <dbl> <chr>
1 Motif1 0.576 Yes 22 0.00162 ""
2 Motif1 0.576 No 13578 0.998 ""
3 Motif2 0.576 Yes 1 0.00193 ""
4 Motif2 0.576 No 516 0.998 "" ###DIC Genes RUVs Cormotif Proportion Barplot clustlike + p.post
DOX_cardiotox_genes <- readRDS("data/new/RUV/DIC_genes_entrezid.RDS")
total_DIC_genes <- length(DOX_cardiotox_genes) #Total number of DNA damage genes
#should be 29
#define my Cormotif gene lists
CMF_genes_RUV <- list(
"Motif1" = final_genes_1_RUV, "Motif2" = final_genes_2_RUV
)
all_tested_genes <- unique(unlist(CMF_genes_RUV))
#this has the 12087 used for clustlike+ p.post together
#usual gene set is 14319
#compute my sets
dic_set <- unique(na.omit(DOX_cardiotox_genes))
motif_enrichment_dic_RUV <- purrr::map_dfr(names(CMF_genes_RUV), function(motif_name) {
motif_genes_dic_RUV <- unique(CMF_genes_RUV[[motif_name]])
# print(motif_enrichment_dic_RUV)
# Count in motif
in_motif_dic_RUV <- sum(motif_genes_dic_RUV %in% dic_set)
#number of genes inside the motif that are dic
in_motif_non_dic_RUV <- length(motif_genes_dic_RUV) - in_motif_dic_RUV
#number of genes inside motif that are NOT dic
# Count outside motif
other_genes_dic_RUV <- setdiff(all_tested_genes, motif_genes_dic_RUV)
#checking the difference between all genes and motif genes
out_motif_dic_RUV <- sum(other_genes_dic_RUV %in% dic_set)
#all genes outside motif that are dic
out_motif_non_dic_RUV <- length(other_genes_dic_RUV) - out_motif_dic_RUV
#all genes outside the motif that are NOT dic
# Build contingency table
contingency_dic_cmf_RUV <- matrix(c(in_motif_dic_RUV, in_motif_non_dic_RUV,
out_motif_dic_RUV, out_motif_non_dic_RUV),
nrow = 2, byrow = TRUE)
#print contingency table
print(contingency_dic_cmf_RUV)
# Perform fisher's exact test
fisher_test_dic_cmf_RUV <- fisher.test(contingency_dic_cmf_RUV)
p_value <- fisher_test_dic_cmf_RUV$p.value
# Output summary
tibble(
Motif = motif_name,
In_Motif_dic = in_motif_dic_RUV,
In_Motif_Nondic = in_motif_non_dic_RUV,
Out_Motif_dic = out_motif_dic_RUV,
Out_Motif_Nondic = out_motif_non_dic_RUV,
Total_Genes = length(motif_genes_dic_RUV),
DNA_Damage_Proportion = in_motif_dic_RUV / length(motif_genes_dic_RUV) * 100,
P_Value = fisher_test_dic_cmf_RUV$p.value
)
}) %>%
mutate(Significance = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)) [,1] [,2]
[1,] 10 7592
[2,] 0 501
[,1] [,2]
[1,] 0 501
[2,] 10 7592# Recreate motif_long from motif_enrichment (with fixed column names)
motif_long_dic_RUV <- motif_enrichment_dic_RUV %>%
select(Motif, In_Motif_dic, In_Motif_Nondic, P_Value) %>% # include P_Value
pivot_longer(cols = c(In_Motif_dic, In_Motif_Nondic),
names_to = "Category", values_to = "Count") %>%
mutate(
Category = ifelse(Category == "In_Motif_dic", "Yes", "No"),
Fraction = Count / ave(Count, Motif, FUN = sum),
# Add significance stars inline
Significant = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)
)
#now I want to make sure that the yes is at the top so I have to reorder
motif_long_dic_RUV$Category <- factor(motif_long_dic_RUV$Category, levels = c("Yes", "No"))
#ensure that the stars are also on top for significance
label_positions_dic_RUV <- motif_long_dic_RUV %>%
filter(Category == "Yes") %>%
mutate(y_pos = Fraction + 1.01) # 101% above the bar for visual clarity
# Plot with corrected star positioning
ggplot(motif_long_dic_RUV, aes(x = Motif, y = Fraction, fill = Category)) +
geom_bar(stat = "identity", position = "stack") +
geom_text(
data = label_positions_dic_RUV %>% filter(Significant != ""),
aes(x = Motif, y = y_pos, label = Significant),
size = 6,
color = "black",
fontface = "bold"
) +
scale_y_continuous(labels = scales::percent_format(scale = 1), limits = c(0, 1.08)) +
scale_fill_manual(values = c("Yes" = "#e41a1c", "No" = "#377eb8")) +
labs(
title = "Proportion DOX Cardiotox Genes per Motif RUVs clustlike",
x = "Cormotif Motif",
y = "Percentage",
fill = "DIC Gene"
) +
theme_minimal() +
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
plot.title = element_text(size = 16, hjust = 0.5)
)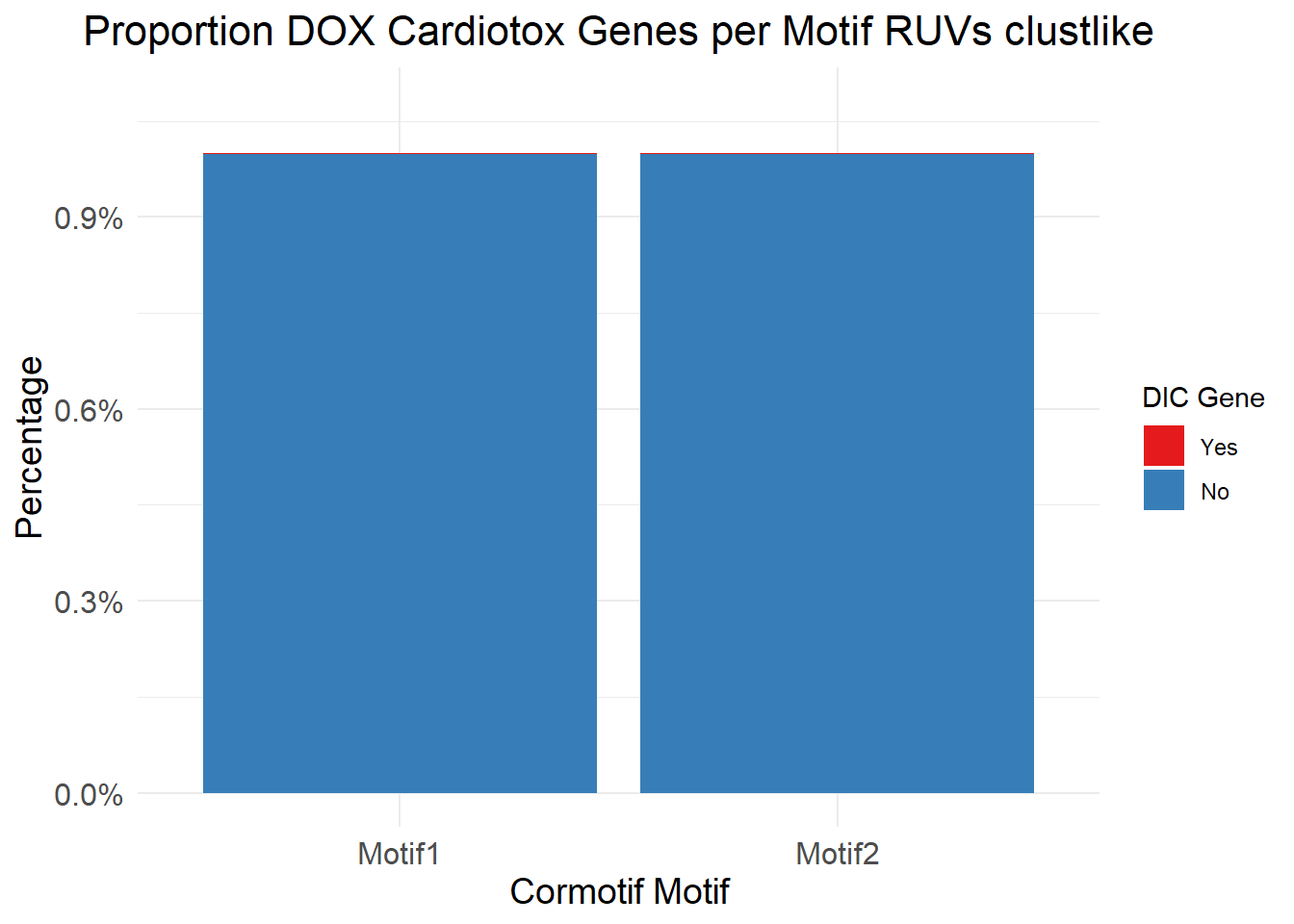
###p53 Target Genes Cormotif RUVs
p53_target_genes <- readRDS("data/new/RUV/p53_targetgenes_entrezid.RDS")
total_p53_genes <- length(p53_target_genes) #Total number of p53 target genes
#should be 300
#define my Cormotif gene lists
CMF_genes_RUV_clust <- list(
"Motif1" = clust1_genes_RUV, "Motif2" = clust2_genes_RUV
)
all_tested_genes_RUV_clust <- unique(unlist(CMF_genes_RUV_clust))
#this has the 14117 used for clustlike only
#usual gene set is 14319
#compute my sets
p53_set <- unique(na.omit(p53_target_genes))
motif_enrichment_p53_RUV_clust <- purrr::map_dfr(names(CMF_genes_RUV_clust), function(motif_name) {
motif_genes_p53_RUV_clust <- unique(CMF_genes_RUV_clust[[motif_name]])
# Count in motif
in_motif_p53_RUV_clust <- sum(motif_genes_p53_RUV_clust %in% p53_set)
#number of genes inside the motif that are p53
in_motif_non_p53_RUV_clust <- length(motif_genes_p53_RUV_clust) - in_motif_p53_RUV_clust
#number of genes inside motif that are NOT p53
# Count outside motif
other_genes_p53_RUV_clust <- setdiff(all_tested_genes_RUV_clust, motif_genes_p53_RUV_clust)
#checking the difference between all genes and motif genes
out_motif_p53_RUV_clust <- sum(other_genes_p53_RUV_clust %in% p53_set)
#all genes outside motif that are p53
out_motif_non_p53_RUV_clust <- length(other_genes_p53_RUV_clust) - out_motif_p53_RUV_clust
#all genes outside the motif that are NOT p53
# Build contingency table
contingency_p53_cmf_RUV_clust <- matrix(c(in_motif_p53_RUV_clust,
in_motif_non_p53_RUV_clust,
out_motif_p53_RUV_clust,
out_motif_non_p53_RUV_clust),
nrow = 2, byrow = TRUE)
#print my contingency table
print(contingency_p53_cmf_RUV_clust)
# Perform fisher's exact test
fisher_test_p53_cmf_RUV_clust <- fisher.test(contingency_p53_cmf_RUV_clust)
p_value_RUV_clust <- fisher_test_p53_cmf_RUV_clust$p.value
# Output summary
tibble(
Motif = motif_name,
In_Motif_p53 = in_motif_p53_RUV_clust,
In_Motif_Nonp53 = in_motif_non_p53_RUV_clust,
Out_Motif_p53 = out_motif_p53_RUV_clust,
Out_Motif_Nonp53 = out_motif_non_p53_RUV_clust,
Total_Genes = length(motif_genes_p53_RUV_clust),
DNA_Damage_Proportion = in_motif_p53_RUV_clust / length(motif_genes_p53_RUV_clust) * 100,
P_Value = fisher_test_p53_cmf_RUV_clust$p.value
)
}) %>%
mutate(Significance = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)) [,1] [,2]
[1,] 261 13339
[2,] 29 488
[,1] [,2]
[1,] 29 488
[2,] 261 13339# Recreate motif_long from motif_enrichment (with fixed column names)
motif_long_p53_RUV_clust <- motif_enrichment_p53_RUV_clust %>%
select(Motif, In_Motif_p53, In_Motif_Nonp53, P_Value) %>% # include P_Value
pivot_longer(cols = c(In_Motif_p53, In_Motif_Nonp53),
names_to = "Category", values_to = "Count") %>%
mutate(
Category = ifelse(Category == "In_Motif_p53", "Yes", "No"),
Fraction = Count / ave(Count, Motif, FUN = sum),
# Add significance stars inline
Significant = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)
)
#now I want to make sure that the yes is at the top so I have to reorder
motif_long_p53_RUV_clust$Category <- factor(motif_long_p53_RUV_clust$Category, levels = c("Yes", "No"))
#ensure that the stars are also on top for significance
label_positions_p53_RUV_clust <- motif_long_p53_RUV_clust %>%
filter(Category == "Yes") %>%
mutate(y_pos = Fraction + 1.01) # 101% above the bar for visual clarity
# Plot with corrected star positioning
ggplot(motif_long_p53_RUV_clust, aes(x = Motif, y = Fraction, fill = Category)) +
geom_bar(stat = "identity", position = "stack") +
geom_text(
data = label_positions_p53_RUV_clust %>% filter(Significant != ""),
aes(x = Motif, y = y_pos, label = Significant),
size = 6,
color = "black",
fontface = "bold"
) +
scale_y_continuous(labels = scales::percent_format(scale = 1), limits = c(0, 1.08)) +
scale_fill_manual(values = c("Yes" = "#e41a1c", "No" = "#377eb8")) +
labs(
title = "Proportion p53 Target Genes per Motif RUVs clustlike",
x = "Cormotif Motif",
y = "Percentage",
fill = "p53 Gene"
) +
theme_minimal() +
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
plot.title = element_text(size = 16, hjust = 0.5)
)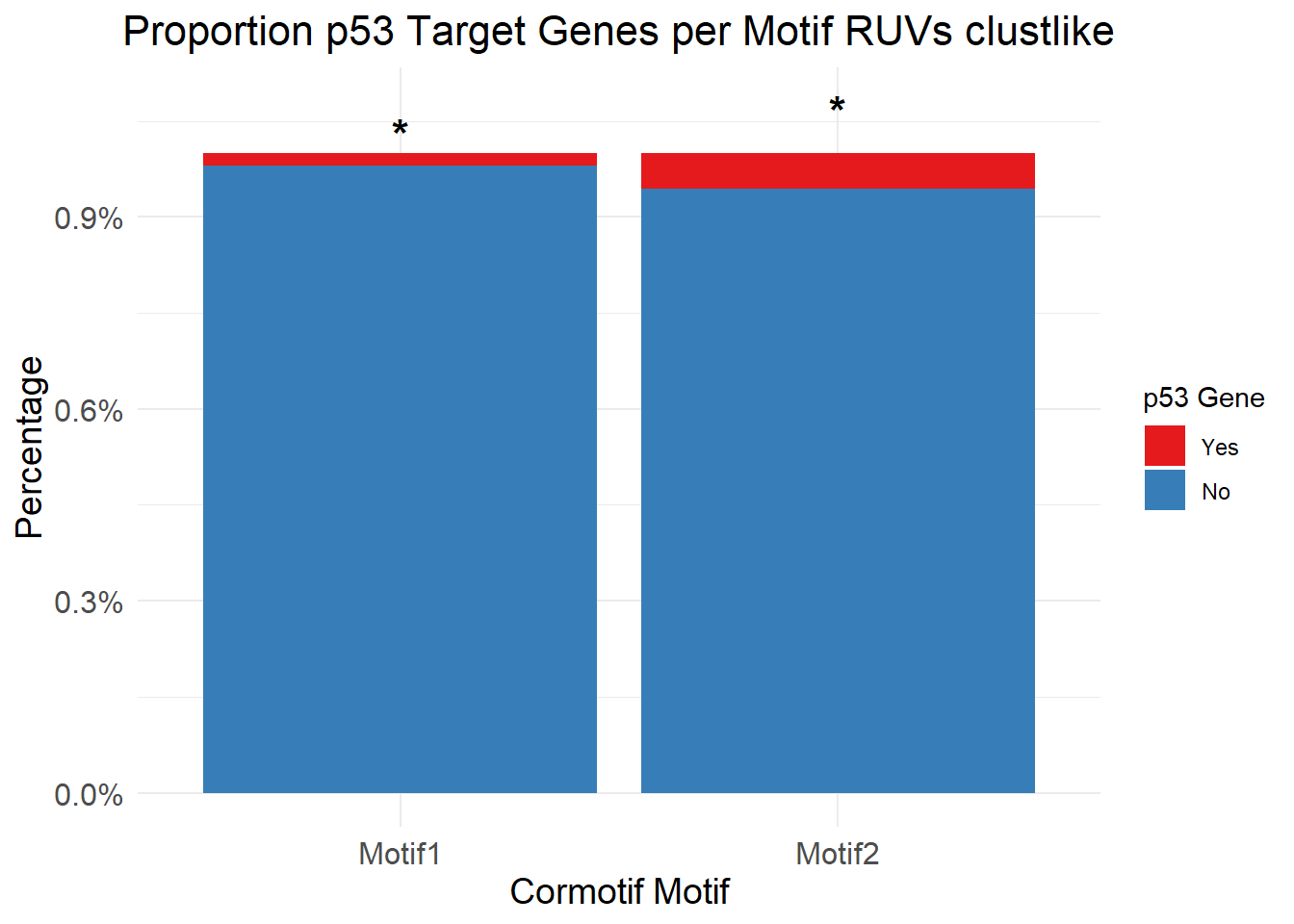
| Version | Author | Date |
|---|---|---|
| 0ae67cb | emmapfort | 2025-07-10 |
#print this dataframe to look at p-value and percentage
print(motif_long_p53_RUV_clust)# A tibble: 4 × 6
Motif P_Value Category Count Fraction Significant
<chr> <dbl> <fct> <int> <dbl> <chr>
1 Motif1 0.000000948 Yes 261 0.0192 *
2 Motif1 0.000000948 No 13339 0.981 *
3 Motif2 0.000000948 Yes 29 0.0561 *
4 Motif2 0.000000948 No 488 0.944 * ###p53 Target Genes Cormotif RUVs p.post + clustlike
p53_target_genes <- readRDS("data/new/RUV/p53_targetgenes_entrezid.RDS")
total_p53_genes <- length(p53_target_genes) #Total number of p53 target genes
#should be 300
#define my Cormotif gene lists
CMF_genes_RUV <- list(
"Motif1" = final_genes_1_RUV,
"Motif2" = final_genes_2_RUV
)
#7602 genes motif 1 + 501 genes motif 2
all_tested_genes_RUV <- unique(unlist(CMF_genes_RUV))
#this has the 8103 genes used for clustlike + p.post
#usual gene set is 14319
#compute my sets
p53_set <- unique(na.omit(p53_target_genes))
motif_enrichment_p53_RUV <- purrr::map_dfr(names(CMF_genes_RUV), function(motif_name) {
motif_genes_p53_RUV <- unique(CMF_genes_RUV[[motif_name]])
#print results
# print(motif_enrichment_p53_RUV)
# Count in motif
in_motif_p53_RUV <- sum(motif_genes_p53_RUV %in% p53_set)
#number of genes inside the motif that are p53
in_motif_non_p53_RUV <- length(motif_genes_p53_RUV) - in_motif_p53_RUV
#number of genes inside motif that are NOT p53
# Count outside motif
other_genes_p53_RUV <- setdiff(all_tested_genes_RUV, motif_genes_p53_RUV)
#checking the difference between all genes and motif genes
out_motif_p53_RUV <- sum(other_genes_p53_RUV %in% p53_set)
#all genes outside motif that are p53
out_motif_non_p53_RUV <- length(other_genes_p53_RUV) - out_motif_p53_RUV
#all genes outside the motif that are NOT p53
# Build contingency table
contingency_p53_cmf_RUV <- matrix(c(in_motif_p53_RUV, in_motif_non_p53_RUV,
out_motif_p53_RUV, out_motif_non_p53_RUV),
nrow = 2, byrow = TRUE)
#print my contingency tables
print(contingency_p53_cmf_RUV)
# Perform fisher's exact test
fisher_test_p53_cmf_RUV <- fisher.test(contingency_p53_cmf_RUV)
p_value_RUV <- fisher_test_p53_cmf_RUV$p.value
# Output summary
tibble(
Motif = motif_name,
In_Motif_p53 = in_motif_p53_RUV,
In_Motif_Nonp53 = in_motif_non_p53_RUV,
Out_Motif_p53 = out_motif_p53_RUV,
Out_Motif_Nonp53 = out_motif_non_p53_RUV,
Total_Genes = length(motif_genes_p53_RUV),
DNA_Damage_Proportion = in_motif_p53_RUV / length(motif_genes_p53_RUV) * 100,
P_Value = fisher_test_p53_cmf_RUV$p.value
)
}) %>%
mutate(Significance = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)) [,1] [,2]
[1,] 170 7432
[2,] 28 473
[,1] [,2]
[1,] 28 473
[2,] 170 7432# Recreate motif_long from motif_enrichment (with fixed column names)
motif_long_p53_RUV <- motif_enrichment_p53_RUV %>%
select(Motif, In_Motif_p53, In_Motif_Nonp53, P_Value) %>% # include P_Value
pivot_longer(cols = c(In_Motif_p53, In_Motif_Nonp53),
names_to = "Category", values_to = "Count") %>%
mutate(
Category = ifelse(Category == "In_Motif_p53", "Yes", "No"),
Fraction = Count / ave(Count, Motif, FUN = sum),
# Add significance stars inline
Significant = case_when(
P_Value < 0.05 ~ "*",
TRUE ~ ""
)
)
#now I want to make sure that the yes is at the top so I have to reorder
motif_long_p53_RUV$Category <- factor(motif_long_p53_RUV$Category, levels = c("Yes", "No"))
#ensure that the stars are also on top for significance
label_positions_p53_RUV <- motif_long_p53_RUV %>%
filter(Category == "Yes") %>%
mutate(y_pos = Fraction + 1.01) # 101% above the bar for visual clarity
# Plot with corrected star positioning
ggplot(motif_long_p53_RUV, aes(x = Motif, y = Fraction, fill = Category)) +
geom_bar(stat = "identity", position = "stack") +
geom_text(
data = label_positions_p53_RUV %>% filter(Significant != ""),
aes(x = Motif, y = y_pos, label = Significant),
size = 6,
color = "black",
fontface = "bold"
) +
scale_y_continuous(labels = scales::percent_format(scale = 1), limits = c(0, 1.08)) +
scale_fill_manual(values = c("Yes" = "#e41a1c", "No" = "#377eb8")) +
labs(
title = "Proportion p53 Target Genes per Motif RUVs clustlike + p.post",
x = "Cormotif Motif",
y = "Percentage",
fill = "p53 Gene"
) +
theme_minimal() +
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
plot.title = element_text(size = 16, hjust = 0.5)
)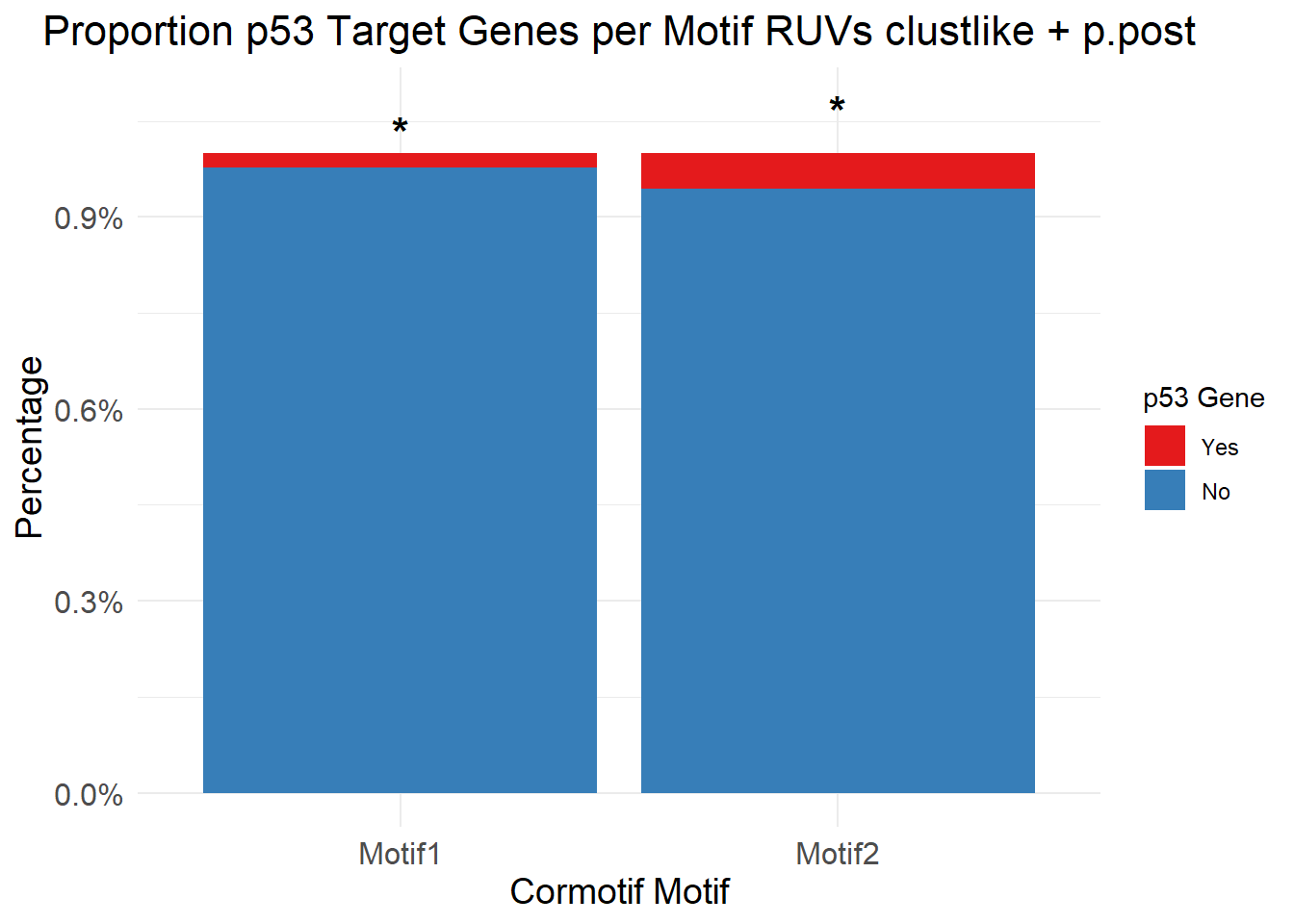
| Version | Author | Date |
|---|---|---|
| 54b2620 | emmapfort | 2025-07-08 |
print(motif_long_p53_RUV)# A tibble: 4 × 6
Motif P_Value Category Count Fraction Significant
<chr> <dbl> <fct> <int> <dbl> <chr>
1 Motif1 0.0000332 Yes 170 0.0224 *
2 Motif1 0.0000332 No 7432 0.978 *
3 Motif2 0.0000332 Yes 28 0.0559 *
4 Motif2 0.0000332 No 473 0.944 * #Final Cormotif RUVs Analysis We will continue forward with the RUVs corrected data based on log2cpm of normalized counts We will then use the Cormotif gene sets based on p.post and clustlike together ##DDR Proportion Bar plots Redo
#I am going to alter my strategy to identify the proportion of genes within one motif that are or are not DDR, do the same for the second motif, and then test between the two for differences in their proportions
#read in my DDR gene set
DNA_damage_genes <- readRDS("data/new/RUV/DDR_genes_entrezid.RDS")
ddr_set <- unique(na.omit(DNA_damage_genes))
#DDR genes inside each motif
in_motif1_DDR <- sum(final_genes_1_RUV %in% ddr_set)
in_motif2_DDR <- sum(final_genes_2_RUV %in% ddr_set)
#non-DDR genes inside each motif
in_motif1_nonDDR <- length(final_genes_1_RUV) - in_motif1_DDR
in_motif2_nonDDR <- length(final_genes_2_RUV) - in_motif2_DDR
#make contingency tables for fishers exact tests
contingency_motifs_DDR <- matrix(c(
in_motif1_DDR, in_motif1_nonDDR,
in_motif2_DDR, in_motif2_nonDDR),
nrow = 2, byrow = TRUE
)
#print the contingency tables
print(contingency_motifs_DDR) [,1] [,2]
[1,] 24 7578
[2,] 16 485#perform a fisher's exact test (small sample size)
fisher_test_motifs_DDR <- fisher.test(contingency_motifs_DDR)
#print fisher's exact test results
print(fisher_test_motifs_DDR)
Fisher's Exact Test for Count Data
data: contingency_motifs_DDR
p-value = 5.674e-10
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
0.04859885 0.19492860
sample estimates:
odds ratio
0.09606922 #make a summary table of all of these data to plot
motif_summary_DDR <- tibble(
Motif = c("Motif1", "Motif2"),
DDR = c(in_motif1_DDR, in_motif2_DDR),
non_DDR = c(in_motif1_nonDDR, in_motif2_nonDDR)
) %>%
pivot_longer(cols = c(DDR, non_DDR), names_to = "Category", values_to = "Count") %>%
mutate(
Category = ifelse(Category == "DDR", "Yes", "No"),
Fraction = Count / ave(Count, Motif, FUN = sum),
Significant = ifelse(Category == "Yes" & fisher_test_motifs_DDR$p.value < 0.05, "*", "")
)
#position significance stars just above bars
label_positions_DDR <- motif_summary_DDR %>%
filter(Category == "Yes") %>%
mutate(y_pos = Fraction + 1.01)
#change the ordering so that yes is on the top
motif_summary_DDR$Category <- factor(motif_summary_DDR$Category, levels = c("Yes", "No"))
#plot the proportions in a barplot
ggplot(motif_summary_DDR, aes(x = Motif, y = Fraction, fill = Category)) +
geom_bar(stat = "identity", position = "stack") +
geom_text(
data = label_positions_DDR %>% filter(Significant != ""),
aes(x = Motif, y = y_pos, label = Significant),
size = 6,
color = "black",
fontface = "bold"
) +
scale_y_continuous(labels = scales::percent_format(scale = 1), limits = c(0, 1.08)) +
scale_fill_manual(values = c("Yes" = "#e41a1c", "No" = "#377eb8")) +
labs(
title = "Proportion of DDR Genes in Motif 1 vs Motif 2 (RUVs clust + p.post)",
x = "Cormotif Motif",
y = "Percentage",
fill = "DDR Gene"
) +
theme_minimal() +
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
plot.title = element_text(size = 16, hjust = 0.5)
)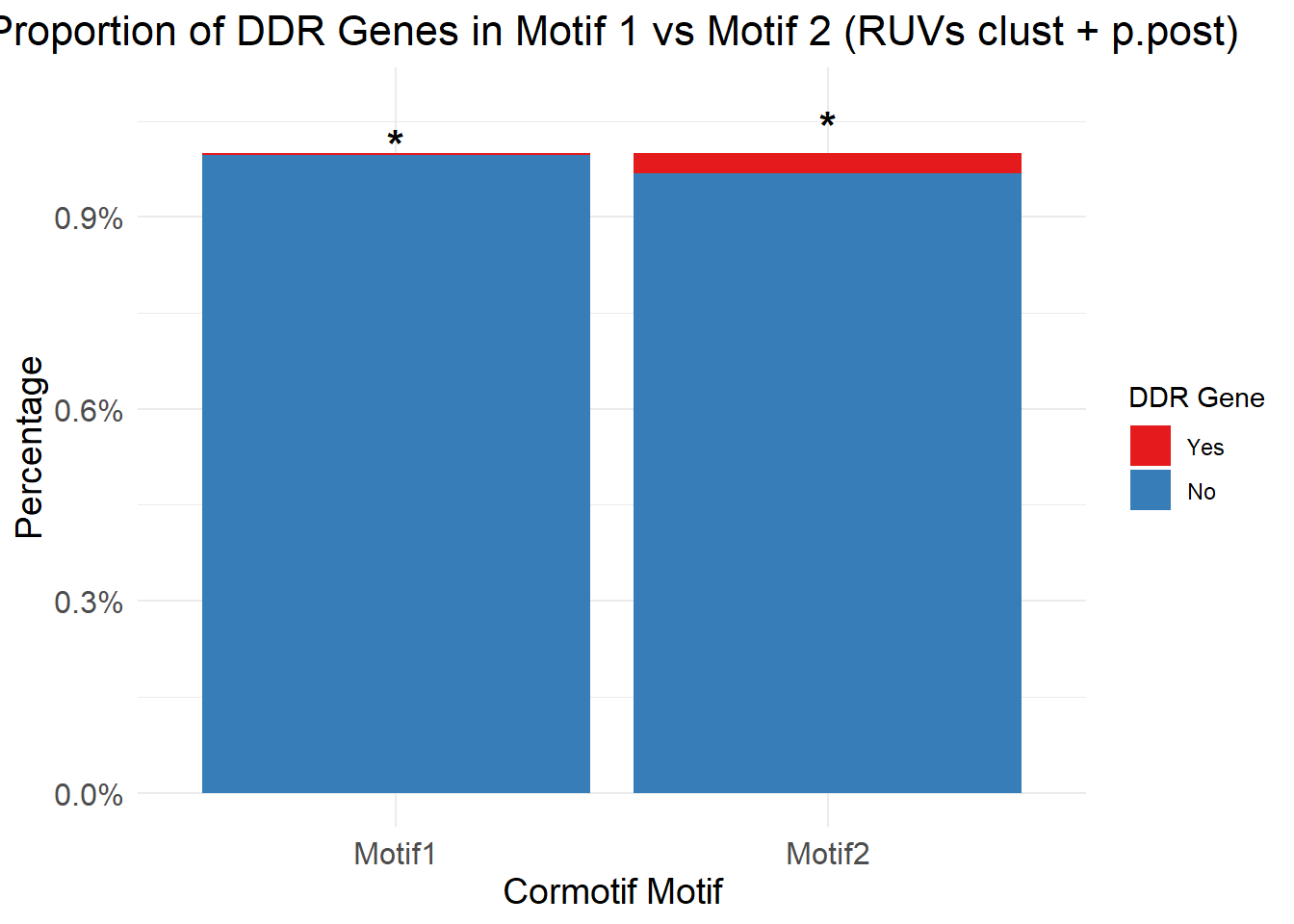
#print the proportions and p values
prop_motif1_DDR <- in_motif1_DDR / length(final_genes_1_RUV)
prop_motif2_DDR <- in_motif2_DDR / length(final_genes_2_RUV)
p_val_DDR <- fisher_test_motifs_DDR$p.value
cat(sprintf("Motif1: %.2f%% DDR genes (%d/%d)\n", 100*prop_motif1_DDR, in_motif1_DDR, length(final_genes_1_RUV)))Motif1: 0.32% DDR genes (24/7602)cat(sprintf("Motif2: %.2f%% DDR genes (%d/%d)\n", 100*prop_motif2_DDR, in_motif2_DDR, length(final_genes_2_RUV)))Motif2: 3.19% DDR genes (16/501)cat(sprintf("DDR Fisher's Exact Test p-value: %.5f\n", p_val_DDR))DDR Fisher's Exact Test p-value: 0.00000cat(sprintf("Odds ratio: %.3f\n", fisher_test_motifs_DDR$estimate))Odds ratio: 0.096##DIC Proportion Bar plots Redo
#read in my DIC gene set
DOX_cardiotox_genes <- readRDS("data/new/RUV/DIC_genes_entrezid.RDS")
dic_set <- unique(na.omit(DOX_cardiotox_genes))
#DIC genes inside each motif
in_motif1_DIC <- sum(final_genes_1_RUV %in% dic_set)
in_motif2_DIC <- sum(final_genes_2_RUV %in% dic_set)
#non-DIC genes inside each motif
in_motif1_nonDIC <- length(final_genes_1_RUV) - in_motif1_DIC
in_motif2_nonDIC <- length(final_genes_2_RUV) - in_motif2_DIC
#make contingency tables for fishers exact tests
contingency_motifs_DIC <- matrix(c(
in_motif1_DIC, in_motif1_nonDIC,
in_motif2_DIC, in_motif2_nonDIC),
nrow = 2, byrow = TRUE
)
#print the contingency tables
print(contingency_motifs_DIC) [,1] [,2]
[1,] 10 7592
[2,] 0 501#perform a fisher's exact test (small sample size)
fisher_test_motifs_DIC <- fisher.test(contingency_motifs_DIC)
#print fisher's exact test results
print(fisher_test_motifs_DIC)
Fisher's Exact Test for Count Data
data: contingency_motifs_DIC
p-value = 1
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
0.1474455 Inf
sample estimates:
odds ratio
Inf #make a summary table of all of these data to plot
motif_summary_DIC <- tibble(
Motif = c("Motif1", "Motif2"),
DIC = c(in_motif1_DIC, in_motif2_DIC),
non_DIC = c(in_motif1_nonDIC, in_motif2_nonDIC)
) %>%
pivot_longer(cols = c(DIC, non_DIC), names_to = "Category", values_to = "Count") %>%
mutate(
Category = ifelse(Category == "DIC", "Yes", "No"),
Fraction = Count / ave(Count, Motif, FUN = sum),
Significant = ifelse(Category == "Yes" & fisher_test_motifs_DIC$p.value < 0.05, "*", "")
)
#position significance stars just above bars
label_positions_DIC <- motif_summary_DIC %>%
filter(Category == "Yes") %>%
mutate(y_pos = Fraction + 1.01)
#change the ordering so that yes is on the top
motif_summary_DIC$Category <- factor(motif_summary_DIC$Category, levels = c("Yes", "No"))
#plot the proportions in a barplot
ggplot(motif_summary_DIC, aes(x = Motif, y = Fraction, fill = Category)) +
geom_bar(stat = "identity", position = "stack") +
geom_text(
data = label_positions_DIC %>% filter(Significant != ""),
aes(x = Motif, y = y_pos, label = Significant),
size = 6,
color = "black",
fontface = "bold"
) +
scale_y_continuous(labels = scales::percent_format(scale = 1), limits = c(0, 1.08)) +
scale_fill_manual(values = c("Yes" = "#e41a1c", "No" = "#377eb8")) +
labs(
title = "Proportion of DIC Genes in Motif 1 vs Motif 2 (RUVs clust + p.post)",
x = "Cormotif Motif",
y = "Percentage",
fill = "DIC Gene"
) +
theme_minimal() +
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
plot.title = element_text(size = 16, hjust = 0.5)
)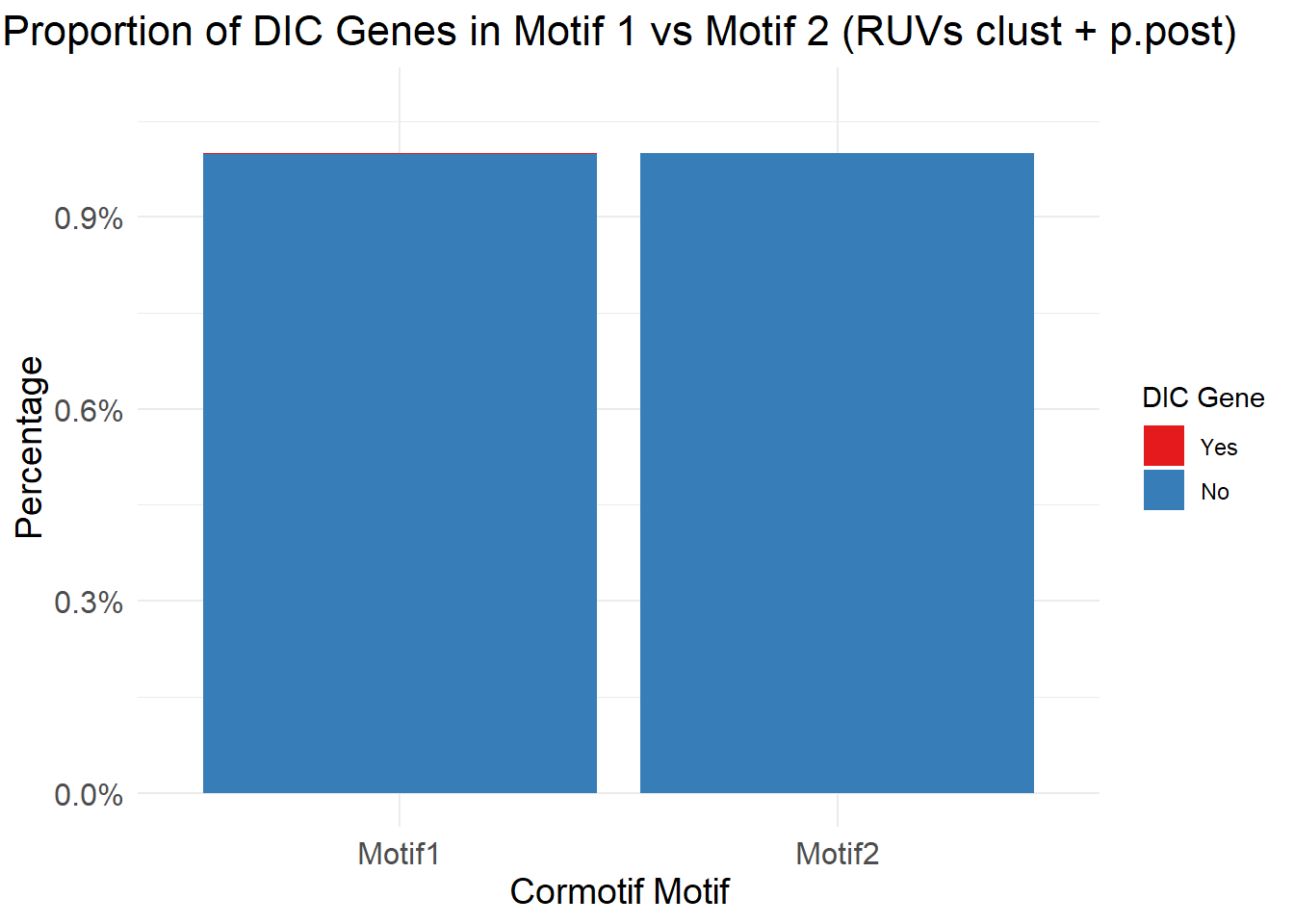
#print the proportions and p values
prop_motif1_DIC <- in_motif1_DIC / length(final_genes_1_RUV)
prop_motif2_DIC <- in_motif2_DIC / length(final_genes_2_RUV)
p_val_DIC <- fisher_test_motifs_DIC$p.value
cat(sprintf("Motif1: %.2f%% DIC genes (%d/%d)\n", 100*prop_motif1_DIC, in_motif1_DIC, length(final_genes_1_RUV)))Motif1: 0.13% DIC genes (10/7602)cat(sprintf("Motif2: %.2f%% DIC genes (%d/%d)\n", 100*prop_motif2_DIC, in_motif2_DIC, length(final_genes_2_RUV)))Motif2: 0.00% DIC genes (0/501)cat(sprintf("DIC Fisher's Exact Test p-value: %.5f\n", p_val_DIC))DIC Fisher's Exact Test p-value: 1.00000cat(sprintf("Odds ratio: %.3f\n", fisher_test_motifs_DIC$estimate))Odds ratio: Inf##p53 Targets Proportion Barplots Redo
#read in my p53 gene set
p53_target_genes <- readRDS("data/new/RUV/p53_targetgenes_entrezid.RDS")
p53_set <- unique(na.omit(p53_target_genes))
#p53 genes inside each motif
in_motif1_p53 <- sum(final_genes_1_RUV %in% p53_set)
in_motif2_p53 <- sum(final_genes_2_RUV %in% p53_set)
#non-p53 genes inside each motif
in_motif1_nonp53 <- length(final_genes_1_RUV) - in_motif1_p53
in_motif2_nonp53 <- length(final_genes_2_RUV) - in_motif2_p53
#make contingency tables for fishers exact tests
contingency_motifs_p53 <- matrix(c(
in_motif1_p53, in_motif1_nonp53,
in_motif2_p53, in_motif2_nonp53),
nrow = 2, byrow = TRUE
)
#print the contingency tables
print(contingency_motifs_p53) [,1] [,2]
[1,] 170 7432
[2,] 28 473#perform a fisher's exact test (small sample size)
fisher_test_motifs_p53 <- fisher.test(contingency_motifs_p53)
#print fisher's exact test results
print(fisher_test_motifs_p53)
Fisher's Exact Test for Count Data
data: contingency_motifs_p53
p-value = 3.322e-05
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
0.2548563 0.6054431
sample estimates:
odds ratio
0.3864804 #make a summary table of all of these data to plot
motif_summary_p53 <- tibble(
Motif = c("Motif1", "Motif2"),
p53 = c(in_motif1_p53, in_motif2_p53),
non_p53 = c(in_motif1_nonp53, in_motif2_nonp53)
) %>%
pivot_longer(cols = c(p53, non_p53), names_to = "Category", values_to = "Count") %>%
mutate(
Category = ifelse(Category == "p53", "Yes", "No"),
Fraction = Count / ave(Count, Motif, FUN = sum),
Significant = ifelse(Category == "Yes" & fisher_test_motifs_p53$p.value < 0.05, "*", "")
)
#position significance stars just above bars
label_positions_p53 <- motif_summary_p53 %>%
filter(Category == "Yes") %>%
mutate(y_pos = Fraction + 1.01)
#change the ordering so that yes is on the top
motif_summary_p53$Category <- factor(motif_summary_p53$Category, levels = c("Yes", "No"))
#plot the proportions in a barplot
ggplot(motif_summary_p53, aes(x = Motif, y = Fraction, fill = Category)) +
geom_bar(stat = "identity", position = "stack") +
geom_text(
data = label_positions_p53 %>% filter(Significant != ""),
aes(x = Motif, y = y_pos, label = Significant),
size = 6,
color = "black",
fontface = "bold"
) +
scale_y_continuous(labels = scales::percent_format(scale = 1), limits = c(0, 1.08)) +
scale_fill_manual(values = c("Yes" = "#e41a1c", "No" = "#377eb8")) +
labs(
title = "Proportion of p53 Genes in Motif 1 vs Motif 2 (RUVs clust + p.post)",
x = "Cormotif Motif",
y = "Percentage",
fill = "p53 Gene"
) +
theme_minimal() +
theme(
axis.title = element_text(size = 14),
axis.text = element_text(size = 12),
plot.title = element_text(size = 16, hjust = 0.5)
)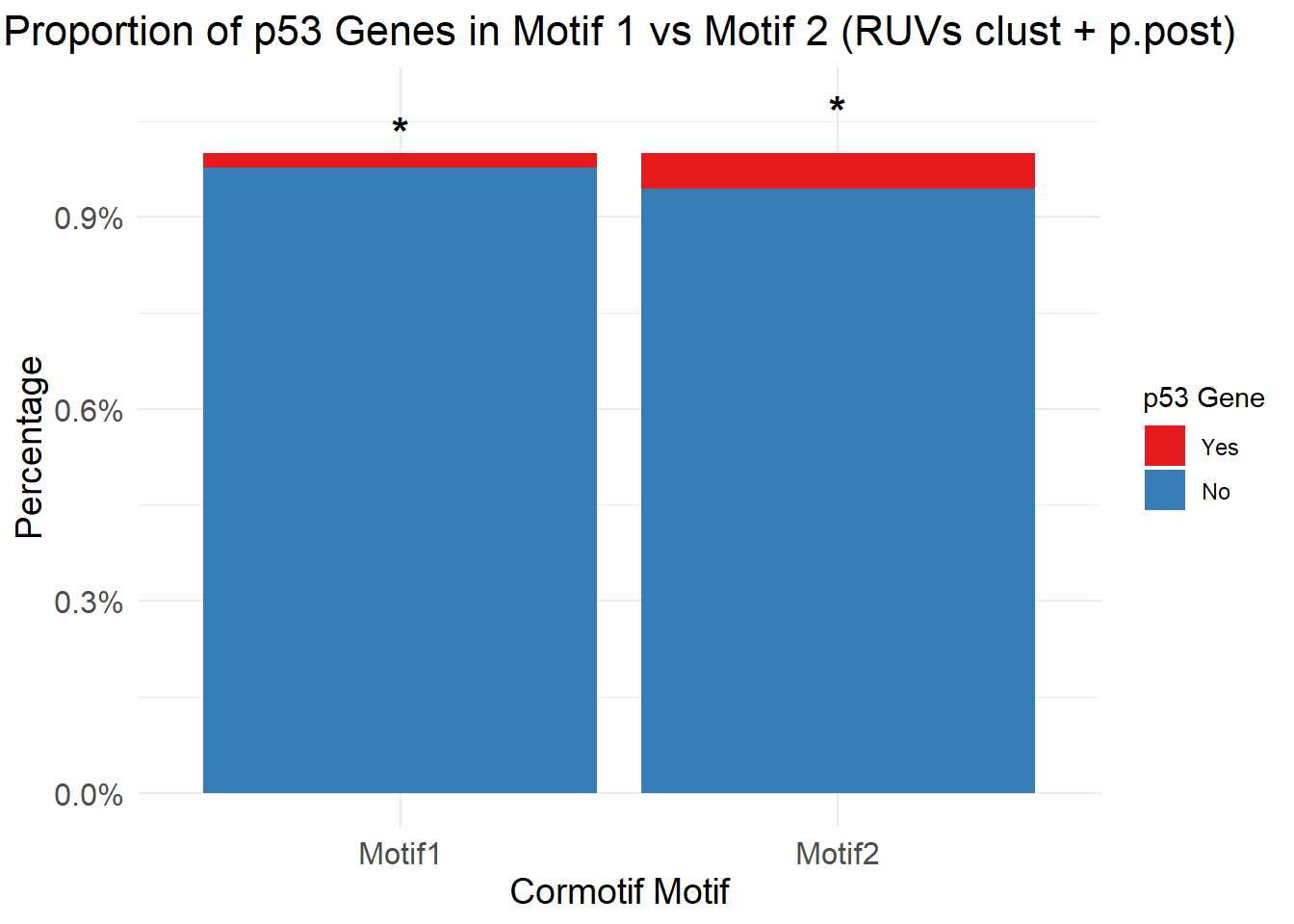
#print the proportions and p values
prop_motif1_p53 <- in_motif1_p53 / length(final_genes_1_RUV)
prop_motif2_p53 <- in_motif2_p53 / length(final_genes_2_RUV)
p_val_p53 <- fisher_test_motifs_p53$p.value
cat(sprintf("Motif1: %.2f%% p53 genes (%d/%d)\n", 100*prop_motif1_p53, in_motif1_p53, length(final_genes_1_RUV)))Motif1: 2.24% p53 genes (170/7602)cat(sprintf("Motif2: %.2f%% p53 genes (%d/%d)\n", 100*prop_motif2_p53, in_motif2_p53, length(final_genes_2_RUV)))Motif2: 5.59% p53 genes (28/501)cat(sprintf("p53 Fisher's Exact Test p-value: %.5f\n", p_val_p53))p53 Fisher's Exact Test p-value: 0.00003cat(sprintf("Odds ratio: %.3f\n", fisher_test_motifs_p53$estimate))Odds ratio: 0.386
sessionInfo()R version 4.4.2 (2024-10-31 ucrt)
Platform: x86_64-w64-mingw32/x64
Running under: Windows 11 x64 (build 22000)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8
[2] LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8
[4] LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] rstatix_0.7.2 car_3.1-3
[3] carData_3.0-5 gprofiler2_0.2.3
[5] RUVSeq_1.40.0 EDASeq_2.40.0
[7] ShortRead_1.64.0 GenomicAlignments_1.42.0
[9] SummarizedExperiment_1.36.0 MatrixGenerics_1.18.1
[11] matrixStats_1.5.0 Rsamtools_2.22.0
[13] GenomicRanges_1.58.0 Biostrings_2.74.0
[15] GenomeInfoDb_1.42.3 XVector_0.46.0
[17] BiocParallel_1.40.0 VennDetail_1.22.0
[19] VennDiagram_1.7.3 futile.logger_1.4.3
[21] ggpubr_0.6.0 UpSetR_1.4.0
[23] ggVennDiagram_1.5.2 reshape2_1.4.4
[25] circlize_0.4.16 ComplexHeatmap_2.22.0
[27] org.Hs.eg.db_3.20.0 AnnotationDbi_1.68.0
[29] IRanges_2.40.0 S4Vectors_0.44.0
[31] corrplot_0.95 ggfortify_0.4.17
[33] ggrepel_0.9.6 biomaRt_2.62.1
[35] scales_1.4.0 edgebundleR_0.1.4
[37] edgeR_4.4.0 limma_3.62.1
[39] Biobase_2.66.0 BiocGenerics_0.52.0
[41] lubridate_1.9.4 forcats_1.0.0
[43] stringr_1.5.1 dplyr_1.1.4
[45] purrr_1.0.4 readr_2.1.5
[47] tidyr_1.3.1 tibble_3.2.1
[49] ggplot2_3.5.2 tidyverse_2.0.0
[51] workflowr_1.7.1
loaded via a namespace (and not attached):
[1] later_1.4.2 BiocIO_1.16.0 bitops_1.0-9
[4] filelock_1.0.3 R.oo_1.27.1 XML_3.99-0.18
[7] lifecycle_1.0.4 httr2_1.1.2 pwalign_1.2.0
[10] doParallel_1.0.17 rprojroot_2.0.4 MASS_7.3-61
[13] processx_3.8.6 lattice_0.22-6 crosstalk_1.2.1
[16] backports_1.5.0 magrittr_2.0.3 plotly_4.10.4
[19] sass_0.4.10 rmarkdown_2.29 jquerylib_0.1.4
[22] yaml_2.3.10 httpuv_1.6.16 DBI_1.2.3
[25] RColorBrewer_1.1-3 abind_1.4-8 zlibbioc_1.52.0
[28] R.utils_2.13.0 RCurl_1.98-1.17 rappdirs_0.3.3
[31] git2r_0.36.2 GenomeInfoDbData_1.2.13 svglite_2.1.3
[34] codetools_0.2-20 DelayedArray_0.32.0 xml2_1.3.8
[37] tidyselect_1.2.1 shape_1.4.6.1 UCSC.utils_1.2.0
[40] farver_2.1.2 BiocFileCache_2.14.0 jsonlite_2.0.0
[43] GetoptLong_1.0.5 Formula_1.2-5 iterators_1.0.14
[46] systemfonts_1.2.2 foreach_1.5.2 tools_4.4.2
[49] progress_1.2.3 Rcpp_1.0.14 glue_1.8.0
[52] gridExtra_2.3 SparseArray_1.6.0 xfun_0.52
[55] withr_3.0.2 formatR_1.14 fastmap_1.2.0
[58] latticeExtra_0.6-30 callr_3.7.6 digest_0.6.37
[61] timechange_0.3.0 R6_2.6.1 mime_0.13
[64] colorspace_2.1-1 jpeg_0.1-11 RSQLite_2.3.9
[67] R.methodsS3_1.8.2 utf8_1.2.5 generics_0.1.4
[70] data.table_1.17.0 rtracklayer_1.66.0 prettyunits_1.2.0
[73] httr_1.4.7 htmlwidgets_1.6.4 S4Arrays_1.6.0
[76] whisker_0.4.1 pkgconfig_2.0.3 gtable_0.3.6
[79] blob_1.2.4 hwriter_1.3.2.1 htmltools_0.5.8.1
[82] clue_0.3-66 kableExtra_1.4.0 png_0.1-8
[85] knitr_1.50 lambda.r_1.2.4 rstudioapi_0.17.1
[88] tzdb_0.5.0 rjson_0.2.23 curl_6.0.1
[91] cachem_1.1.0 GlobalOptions_0.1.2 parallel_4.4.2
[94] restfulr_0.0.15 pillar_1.10.2 vctrs_0.6.5
[97] promises_1.3.2 dbplyr_2.5.0 xtable_1.8-4
[100] cluster_2.1.6 evaluate_1.0.3 GenomicFeatures_1.58.0
[103] cli_3.6.3 locfit_1.5-9.12 compiler_4.4.2
[106] futile.options_1.0.1 rlang_1.1.6 crayon_1.5.3
[109] ggsignif_0.6.4 labeling_0.4.3 aroma.light_3.36.0
[112] interp_1.1-6 ps_1.9.1 getPass_0.2-4
[115] plyr_1.8.9 fs_1.6.6 stringi_1.8.7
[118] viridisLite_0.4.2 deldir_2.0-4 lazyeval_0.2.2
[121] Matrix_1.7-1 hms_1.1.3 bit64_4.5.2
[124] KEGGREST_1.46.0 statmod_1.5.0 shiny_1.10.0
[127] igraph_2.1.4 broom_1.0.8 memoise_2.0.1
[130] bslib_0.9.0 bit_4.5.0